ABSTRACT
Bile acids are important cholesterol-derived nutrient signaling hormones, synthesized in the liver, that act as detergents to solubilize dietary lipids. Bile acid 7α-dehydroxylating gut bacteria generate the toxic bile acids deoxycholic acid and lithocholic acid from host bile acids. The ability of these bacteria to remove the 7-hydroxyl group is partially dependent on 7α-hydroxysteroid dehydrogenase (HSDH) activity, which reduces 7-oxo-bile acids generated by other gut bacteria. 3α-HSDH has an important enzymatic activity in the bile acid 7α-dehydroxylation pathway. 12α-HSDH activity has been reported for the low-activity bile acid 7α-dehydroxylating bacterium Clostridium leptum; however, this activity has not been reported for high-activity bile acid 7α-dehydroxylating bacteria, such as Clostridium scindens, Clostridium hylemonae, and Clostridium hiranonis. Here, we demonstrate that these strains express bile acid 12α-HSDH. The recombinant enzymes were characterized from each species and shown to preferentially reduce 12-oxolithocholic acid to deoxycholic acid, with low activity against 12-oxochenodeoxycholic acid and reduced activity when bile acids were conjugated to taurine or glycine. Phylogenetic analysis suggests that 12α-HSDH is widespread among Firmicutes, Actinobacteria in the Coriobacteriaceae family, and human gut Archaea.
IMPORTANCE 12α-HSDH activity has been established in the medically important bile acid 7α-dehydroxylating bacteria C. scindens, C. hiranonis, and C. hylemonae. Experiments with recombinant 12α-HSDHs from these strains are consistent with culture-based experiments that show a robust preference for 12-oxolithocholic acid over 12-oxochenodeoxycholic acid. Phylogenetic analysis identified novel members of the gut microbiome encoding 12α-HSDH. Future reengineering of 12α-HSDH enzymes to preferentially oxidize cholic acid may provide a means to industrially produce the therapeutic bile acid ursodeoxycholic acid. In addition, a cholic acid-specific 12α-HSDH expressed in the gut may be useful for the reduction in deoxycholic acid concentration, a bile acid implicated in cancers of the gastrointestinal (GI) tract.
KEYWORDS: oxo-bile acids, cholic acid, deoxycholic acid, 12α-hydroxysteroid dehydrogenase, bile acid 7α-dehydroxylation, human gut bacteria, Clostridium scindens, Clostridium hylemonae, Clostridium hiranonis
INTRODUCTION
Bile acids are synthesized from cholesterol in the liver, conjugated to the amino acid taurine or glycine, and secreted and stored in the gallbladder during the interdigestive period. During a meal, the gallbladder is hormonally stimulated to contract, releasing bile into the duodenum. In the small bowel, conjugated bile acids measure in the low-millimolar range and function to solubilize cholesterol, dietary lipids, and lipid-soluble vitamins (1). Conjugated bile acids are also directly antimicrobial, reducing the microbial load at the site of host nutrient absorption. In the terminal ileum, ∼95% of bile acids are returned to the liver after high-affinity transport by apical bile salt transporters into enterocytes and entrance into portal circulation after basolateral transport (2). However, roughly 400 to 800 mg per day of bile salts enters the large intestine, where anaerobic bacteria perform several important biotransformations.
The “gateway reaction” is hydrolysis of the amino acid conjugate by bacterial bile salt hydrolases (BSH) which are found in all major bacterial phyla and in the two main species of human gut archaea (3). Gut microbes are also capable of removing the 7α-hydroxyl group from host cholic acid (CA; 5β-cholanic acid-3α, 7α, 12α-triol), forming deoxycholic acid (DCA; 5β-cholanic acid-3α, 12α-diol), and from chenodeoxycholic acid (CDCA; 5β-cholanic acid-3α, 7α-diol), forming lithocholic acid (LCA; 5β-cholanic acid-3α-ol) (4, 5). DCA and LCA are the main components of the bile acid profile of stool in healthy humans (6) and have been associated causally with diseases of the gastrointestinal (GI) tract, including cancers of the colon (7), liver (8), and esophagus (9), as well as cholesterol gallstone disease in a subset of patients (10). 7β-Hydroxy bile acids, such as ursodeoxycholic acid (UDCA; 5β-cholanic acid-3α,7β-diol), are important in the therapy of the aforementioned GI diseases and function mainly to dilute and counteract the influence of microbe-derived DCA and LCA (11–13). UDCA synthesis from CA requires enzymatic oxidation of the 12α-hydroxyl group, followed by Wolff-Kishner reduction to remove the C-12 carbonyl group (14). Therefore, understanding the role of bacterial 12α-dehydrogenation in microbial bile acid biotransformation and identifying genes encoding 12α-hydroxysteroid dehydrogenases (HSDH) in gut microbes are important in understanding host disease processes and potentially in aiding in the biotechnological generation of therapeutic agents in disease treatment.
Strains of bile acid 7α-dehydroxylating bacteria (BA7) that have been isolated to date from human feces, or inferred by sequence, are within Clostridium clusters XIVa, IV, and XI (5). Those strains in Clostridium cluster XIVa that express high BA7 activity and encode a polycistronic bile acid-inducible (bai) operon include strains of C. scindens, C. hiranonis, and C. hylemonae (5). HSDH enzymes play an important role in secondary bile acid formation by these gut bacterial species. Bile acids are pumped inside the cell by a proton-dependent transport protein (BaiG) (15), ligated to coenzyme A (CoA) by the ATP-dependent CoA-ligase (BaiB) (16) or ATP-independent CoA-transferases (BaiF and BaiK) (17, 18). Diverse gut microbiota express 7α-HSDHs, which convert host primary bile acids to 7-oxo-bile acids (4). An NADP-dependent 7α-HSDH gene has been identified and characterized in C. scindens VPI 12708 (19) and is inferred by sequence similarity in C. scindens ATCC 35704, C. hylemonae DSM 15053, and C. hiranonis DSM 13275 (5). Additionally, after CoA ligation, two oxidation steps precede the rate-limiting 7α-dehydration step (BaiE) (20, 21), one of which, the baiA gene, encodes a 3α-HSDH (22, 23). The other, encoded by the baiCD and baiH genes, introduces a C-4=C-5 bond in 7α-hydroxyl and 7β-hydroxyl bile acid substrates, respectively (24). After 7α/β-dehydration, a series of reductions (catalyzed by the baiN gene product) occur to convert 5β-chol-4,6-dienoic acid-12α-ol-3-one to DCA or LCA (24, 25).
Roughly half of the host primary bile acids synthesized in the liver contain a 12α-hydroxyl group (1). Members of the gastrointestinal microbiota have evolved 12α-HSDHs capable of oxidizing and epimerizing the 12α-hydroxyl group from host CA and its microbial metabolites, including DCA (26). C. leptum, reported to express low BA7 activity (27), has also been reported to express a 12α-HSDH (28); however, the gene(s) encoding 12α-HSDH have not been identified. A previous bioinformatics analysis of bacterial HSDH enzymes identified a gene encoding a putative 12α-HSDH (accession no. WP_006441568.1) in C. hylemonae (29) based on a single deduced amino acid sequence from the only known gene encoding 12α-HSDH identified from Clostridium sp. strain ATCC 29733 (30, 31). This phylogeny also predicted that BA7 strains C. scindens ATCC 35704, C. hylemonae DSM 15053, and C. hiranonis DSM 13275 harbor 12α-HSDHs; however, the genes predicted to encode this activity have yet to be reported. A major clue to the expression of 12α-HSDH in C. hiranonis (formerly strain HD-17) came from a study where resting cells of C. hiranonis produced significant levels of 12-oxoLCA when incubated aerobically with CA (32). We therefore sought to evaluate the potential of C. scindens ATCC 35704, C. hylemonae DSM 15053, and C. hiranonis DSM 13275 to metabolize oxo-bile acids, and specifically to detect 12α-HSDH activity. Further, we sought to characterize purified recombinant 12α-HSDHs from these medically important gut microbes.
RESULTS
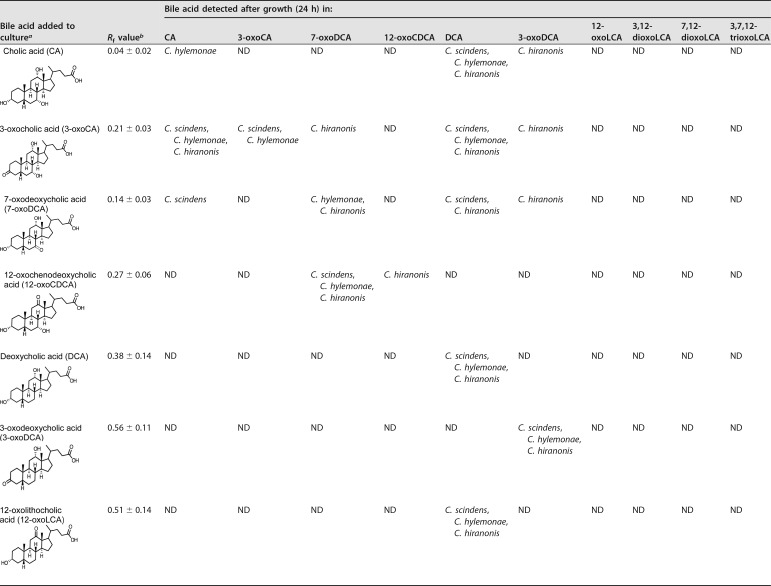
Transformation of 12-oxoLCA and other oxo-bile acids by C. scindens ATCC 35704, C. hylemonae DSM 15053, and C. hiranonis DSM 13275 in BHI cultures.
Studies to date have yet to examine the metabolism of 12-oxo-bile acid substrates in anaerobic cultures by BA7 strains. We therefore limited our screening of substrates to oxo-bile acids possessing a 12α-hydroxyl or 12-oxo group (i.e., CA derivatives) (Table 1). 12-oxoLCA was converted quantitatively to DCA in 24-h cultures of each strain (Table 1 and Fig. 1). In contrast, all three converted 12-oxoCDCA to 7-oxoDCA rather than to DCA. We did not detect the formation of 12-oxoLCA when CA or its C-3– or C-7–oxo derivative was the substrate, nor did we detect oxidation of the 12α-hydroxyl group when DCA was the substrate.
TABLE 1.
Effect of structure on bile acid transformation by C. scindens ATCC 35704, C. hylemonae DSM 15053, and C. hiranonis DSM 13275
aThe final concentration of bile acid added to BHI culture was 0.1 mM.
bValues determined from TLC analysis represent the means ± standard deviation (SD) of three or more replications. ND, not detected. TLC solvent (cyclohexane-ethyl acetate-glacial acetic acid, 12:12:1 [vol/vol/vol]) was used for detection. The detection limits approximated ≤10 μM.
cThe presence or absence of the added bile acid following growth in BHI broth was determined with an additional TLC solvent (toluene–1-4 dioxane–glacial acetic acid, 70:20:2 [vol/vol/vol]). The detection limits approximated ≤10 μM.
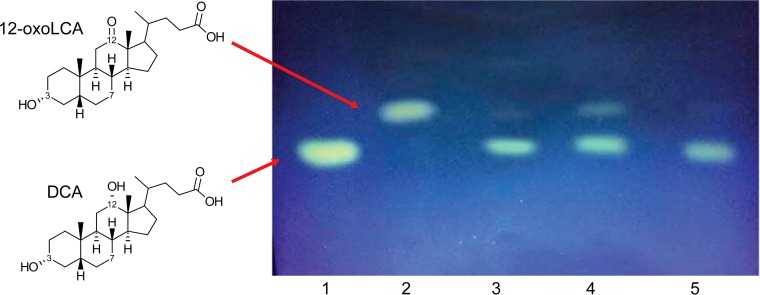
FIG 1.
Thin-layer chromatography (TLC) demonstrating the conversion of 12-oxolithocholic acid (12-oxoLCA) to deoxycholic acid (DCA) by anaerobic cultures of C. scindens ATCC 35704, C. hylemonae DSM 15053, and C. hiranonis DSM 13275. DCA and 12-oxoLCA standards were spotted in lanes 1 and 2, respectively. 12-oxoLCA was added to cultures of C. scindens ATCC 35704 (lane 3), C. hylemonae DSM 15053 (lane 4), and C. hiranonis DSM 13275 (lane 5).
All three strains reduced 3-oxoCA to CA, but only C. hiranonis DSM 13275 generated DCA (Table 1).
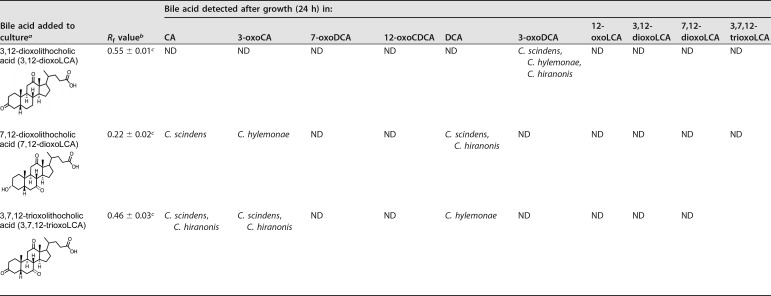
7-oxoDCA was converted to DCA by C. scindens ATCC 35704 and C. hiranonis DSM 13275 but not by C. hylemonae DSM 15053. DCA and 3-oxoDCA were not metabolized by these strains. Dioxo- and trioxo-bile acids were also evaluated for metabolism by BA7 bacteria. When 3,12-dioxoLCA was the substrate, all three strains reduced the 12-oxo-group and formed 3-oxoDCA. Substrates 7,12-dioxoLCA and 3,7,12-trioxoLCA were converted to a mixture of CA, DCA, and 3-oxoDCA. Collectively, these results indicated that C. scindens ATCC 35704, C. hylemonae DSM 15053, and C. hiranonis DSM 13275 reduce oxo-bile acids, and each strain possesses 12α-HSDH activity.
Cloning, overexpression, purification, and characterization of 12α-HSDH from C. scindens ATCC 35704, C. hylemonae DSM 15053, and C. hiranonis DSM 13275.
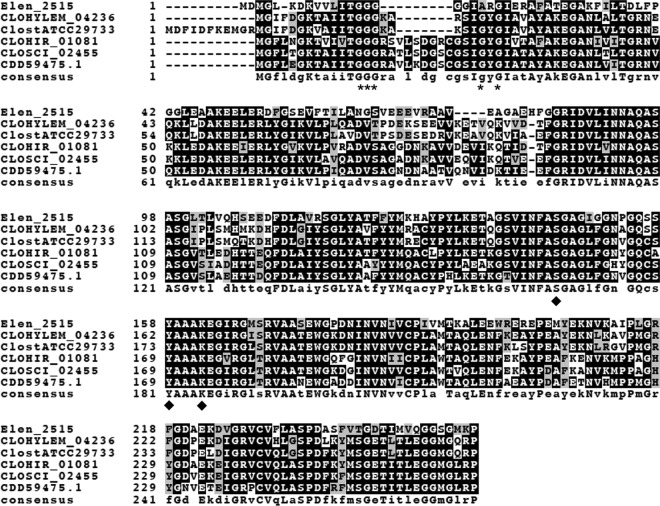
Recently, genes encoding 12α-HSDH were reported in Eggerthella lenta DSM 2243 (Elen_2515) (33) and Eggerthella CAG:298 (accession no. CDD59475) (34). We aligned the amino acid sequences of the 12α-HSDHs from Eggerthella strains as well as 12α-HSDH reported in Clostridium sp. ATCC 29733 (accession no. ERJ00208.1), with putative 12α-HSDH from C. hylemonae DSM 15053 (CLOHYLEM_04236), C. scindens ATCC 35704 (CLOSCI_02455), and C. hiranonis DSM 13275 (CLOHIR_01081). Multiple-sequence alignment is depicted in Fig. 2. Each protein in the alignment is a member of the 3-ketoacyl-(acyl-carrier-protein) short-chain dehydrogenase/reductase (SDR) family, defined by a conserved N-terminal Rossmann-fold responsible for NAD(P)-binding (GGGX5GXG) and catalytic triad (SYK) (Fig. 2). The genomic context of the genes encoding putative 12α-HSDH in each 7α-dehydroxylating bacterium is shown in Fig. 3.
FIG 2.
Boxshade representation of multiple-sequence alignment of 12α-hydroxysteroid dehydrogenases from Eggerthella lenta strains and Clostridium species. Depicted in the alignment are 12α-HSDH from Eggerthella lenta DSM 2243 (Elen_2515) (33), Eggerthella CAG:298 (accession no. CDD59475) (34), Clostridium sp. strain ATCC 29733 (ClostATCC29733), Clostridium scindens ATCC 35704 (CSCI), Clostridium hylemonae DSM 15053 (CHYL), and Clostridium hiranonis DSM 13275 (CHIR). Asterisks mark amino acids predicted to bind pyridine nucleotide cofactor, and diamonds indicate conserved active-site residues in the SDR family.
FIG 3.
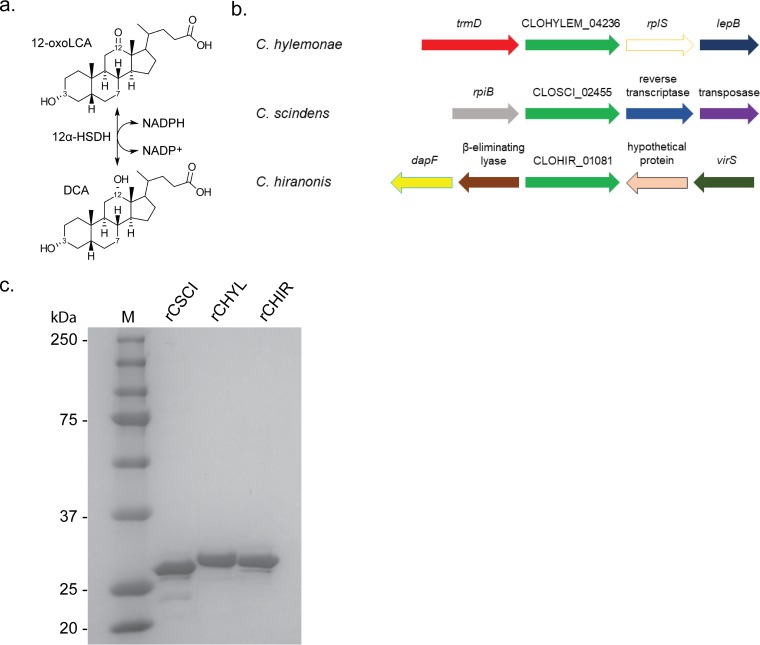
Overexpression and purification of recombinant 12α-hydroxysteroid dehydrogenases from bile acid 7α-dehydroxylating bacteria. (a) Reaction catalyzed by 12α-HSDH. (b) Genomic context of genes predicted to encode 12α-HSDH in BA7 bacteria. (c) SDS-PAGE of 2 μg purified recombinant 12α-HSDHs overexpressed in E. coli and affinity purified on Strep-Tactin resin. Lane M, molecular mass markers (values are in kilodaltons).
To determine whether the genes CLOHYLEM_04236 (259 amino acids), CLOSCI_02455 (266 amino acids), and CLOHIR_01081 (266 amino acids) encode novel 12α-HSDHs, we amplified the genes by PCR and cloned each gene into pET51(b)+ for overexpression in Escherichia coli. Each recombinant protein was overexpressed as an N-terminal streptavidin fusion protein, which was purified by affinity chromatography using Strep-Tactin resin and resolved by SDS-PAGE analysis (Fig. 3). Recombinant CLOHYLEM_04236 (rCHYL) had a theoretical subunit mass of 28.0 kDa and yielded an observed subunit mass of 28.5 ± 0.3 kDa on SDS-PAGE (three independent protein gels). rCLOSCI_02455 (rCSCI) from C. scindens ATCC 35704 had a deduced subunit molecular mass of 28.2 kDa, with an observed subunit mass of 27.3 ± 0.9 kDa, and rCLOHIR_01081 (rCHIR) (theoretical subunit mass, 28.5 kDa) yielded an observed subunit mass of 28.5 ± 0.1 kDa.
TLC and mass spectrometry of enzyme reaction products.
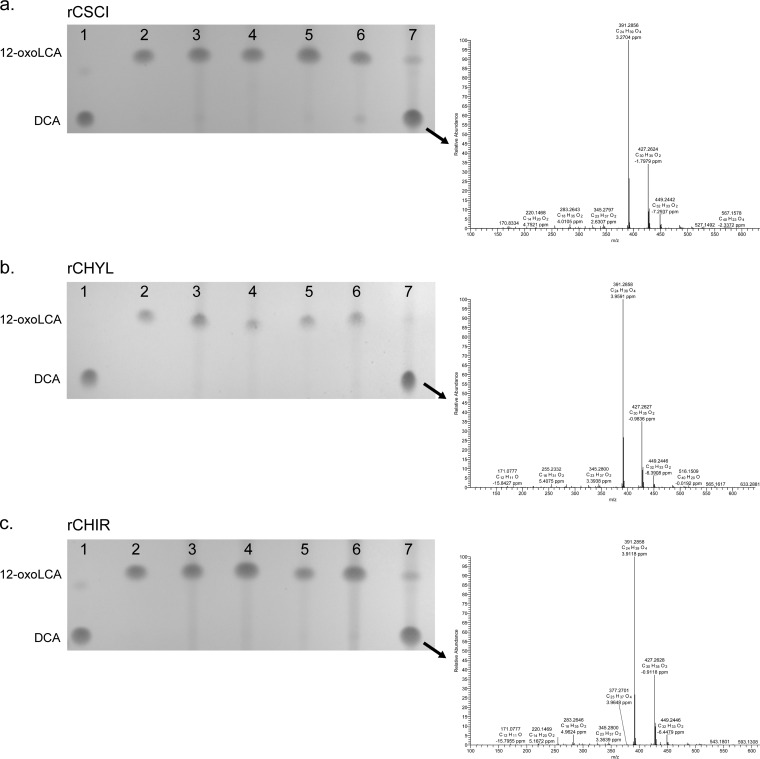
Next, we confirmed functional 12α-HSDH activity of purified rCSCI, rCHYL, and rCHIR in the presence of 12-oxoLCA substrate and reduced or oxidized pyridine nucleotide cofactors (Fig. 4). DCA standard yielded a major mass ion of 391.2858 m/z in negative-ion mode, consistent with the molecular mass of DCA at 392.57 atomic mass units (amu). rCSCI was NADPH dependent, according to thin-layer chromatography (TLC) separation of bile acid metabolites. The single reaction product was eluted from TLC silica and yielded a major mass ion of 391.2856 m/z. Similarly, rCHYL showed specificity for NADPH, with no significant formation of product with NADH after 24 h, as determined by TLC. A product, whose migration was identical to DCA, yielded a major mass ion in negative mode of 391.2856 m/z. When incubated with 12-oxoLCA in the presence of NADPH (but not NADH), purified rCHIR yielded a single product on TLC that comigrated with DCA, with a major mass ion of 391.2858 m/z.
FIG 4.
TLC and mass spectrometry of 12α-hydroxysteroid dehydrogenase overnight reaction products. In lanes 1 and 2, DCA and 12-oxoLCA were spotted as standards, respectively. Purified recombinant protein (12.5 nM rCHYL, 8 nM rCSCI, 8 nM rCHIR) was incubated overnight under the following conditions: lane 3, 12-oxoLCA plus NADPH, no enzyme; lane 4, 12-oxoLCA plus NAD+ plus enzyme; lane 5, 12-oxoLCA plus NADP+ plus enzyme; lane 6, 12-oxoLCA plus NADH plus enzyme; lane 7, 12-oxoLCA plus NADPH plus enzyme. Arrow depicts spot analyzed by electrospray ionization mass spectrometry in negative mode, and compared to authentic DCA standard. Formula weight for DCA is 392.58. (a to c) TLC of CSCI (a), CHYL (b), and CHIR (c) reaction products.
pH optimization of r12α-HSDHs.
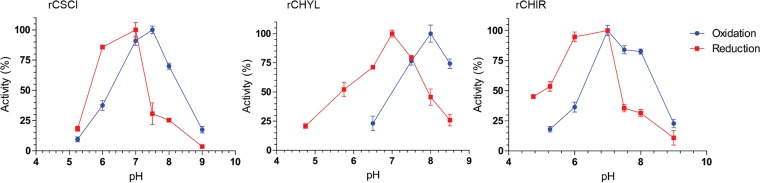
In order to optimize the enzyme-catalyzed C-12 oxidoreduction, we monitored the conversion of pyridine nucleotides at 340 nm in various buffer systems adjusted to pH increments from pH 4.5 to 10.0 (Fig. 5). The optimum pH for rCSCI in the oxidative direction with DCA as the substrate and NADP+ as the cosubstrate was 7.5, and the optimum in the reductive direction with 12-oxoLCA as the substrate and NADPH as the cosubstrate was 7.0. In the oxidative direction, the optimum pH for rCHYL with NADP+ as cofactor and DCA as the substrate was 8.0, while the optimum pH with NADPH and 12-oxoLCA was 7.0. The optimum pH for rCHIR in the oxidative direction was found to be 7.0 and was between 6.5 and 7.0 in the reductive direction.
FIG 5.
Optimal pHs for purified recombinant 12α-hydroxysteroid dehydrogenases. Substrates were 12-oxoLCA with NADPH as cofactor (reductive direction, red) or DCA with NADP+ as cofactor (oxidative direction, blue). See Materials and Methods for buffer compositions. Experiments were repeated three or more times; values represent means ± standard deviation (SD).
Kinetic analysis and substrate specificity of r12α-HSDHs.
Kinetic analysis was performed at the optimum pH for each enzyme and the particular reaction direction. In the oxidative direction, rCHYL, rCSCI, and rCHIR displayed comparable Km values for DCA at ∼150 μM, a concentration approximating levels of DCA in fecal water obtained from Western individuals (35) (Table 2; see also Fig. S1 in the supplemental material). The Km values for NADP+ were on the same order as those for DCA. The Km value for 12-oxoLCA in the reductive direction was an order of magnitude lower for rCHYL and ∼4.5- and 5-fold lower for rCSCI and rCHIR, respectively (Table 2). Vmax and kcat values were between 1.6- and 2.9-fold higher in the oxidative than the reductive direction. However, catalytic efficiencies (Km/kcat) were 2.8- to 3.5-fold greater in the reductive than oxidative direction.
TABLE 2.
Steady-state kinetic parameters of 12α-HSDH homologs
| Homolog | Kinetic parameter | Substrate or coenzymea |
|||
|---|---|---|---|---|---|
| 12-Oxolithocholic acid | NADPH | Deoxycholic acid | NADP+ | ||
| rCSCI | Km (μM) | 17.52 ± 0.32 | 34.01 ± 0.32 | 178.61 ± 0.66 | 79.88 ± 0.37 |
| kcat (s−1) | 31.13 ± 1.56 | 26.71 ± 1.22 | 90.93 ± 4.46 | 90.74 ± 4.35 | |
| Vmax (μmol · min−1 · mg−1) | 66.15 ± 3.31 | 56.74 ± 2.59 | 193.19 ± 9.47 | 192.79 ± 9.24 | |
| kcat/Km (μM−1 · s−1) | 1.77 ± 0.109 | 0.79 ± 0.042 | 0.51 ± 0.029 | 1.14 ± 0.063 | |
| rCHYL | Km (μM) | 32.70 ± 0.55 | 48.31 ± 0.37 | 147.80 ± 0.38 | 110.53 ± 0.38 |
| kcat (s−1) | 45.09 ± 2.19 | 44.52 ± 2.30 | 72.52 ± 3.55 | 66.72 ± 3.18 | |
| Vmax (μmol · min−1 · mg−1) | 96.76 ± 4.70 | 95.53 ± 4.94 | 155.63 ± 7.64 | 143.18 ± 6.82 | |
| kcat/Km (μM−1 · s−1) | 1.38 ± 0.08 | 0.92 ± 0.056 | 0.49 ± 0.028 | 0.60 ± 0.033 | |
| rCHIR | Km (μM) | 34.29 ± 0.34 | 40.37 ± 0.38 | 175.44 ± 0.50 | 127.94 ± 0.42 |
| kcat (s−1) | 72.09 ± 3.50 | 76.24 ± 3.88 | 130.62 ± 7.88 | 129.75 ± 6.68 | |
| Vmax (μmol · min−1 · mg−1) | 151.82 ± 7.37 | 160.57 ± 8.16 | 275.08 ± 16.60 | 273.26 ± 14.07 | |
| kcat/Km (μM−1 · s−1) | 2.10 ± 0.12 | 1.89 ± 0.11 | 0.74 ± 0.052 | 1.01 ± 0.060 | |
The concentrations used in the assays were based on Km values (refer to Materials and Methods). Values represent the means ± SD based on three or more replications.
In the reductive direction, recombinant NADPH-dependent 12α-HSDH had between 9.55 and 3.65% relative activity toward 12-oxoCDCA compared to 12-oxoLCA, suggesting hindrance by the 7α-hydroxyl group (Table 3). NADH was not a cosubstrate, as determined by spectrophotometric assay and overnight reaction mixtures separated by TLC (Fig. 3). In the oxidative direction, the relative activity toward CA was between 54.44 and 82.32% that of the activity toward DCA. rCSCI had 16.5-fold lower activity toward taurine-conjugated DCA (TDCA) than did DCA, while rCHIR and rCHYL showed 2.4-fold and 2.2-fold lower activity toward TDCA than DCA, respectively. Glycine conjugation resulted in a relative activity similar to taurine conjugation with rCSCI, but rCHYL and rCHIR showed 4.1-fold and 3.9-fold decreases in activity relative to DCA, respectively (Table 3).
TABLE 3.
Substrate specificity of purified 12α-HSDHs
| Substratea | Cofactor | rCSCIb |
rCHYLb |
rCHIRb |
|||
|---|---|---|---|---|---|---|---|
| Activity | Relative activity (%) | Activity | Relative activity (%) | Activity | Relative activity (%) | ||
| 12-oxoLCA | NADPH | 38.59 ± 2.38 | 100 | 79.42 ± 3.58 | 100 | 133.56 ± 4.91 | 100 |
| 12-oxoLCA | NADH | NA | NA | NA | NA | NA | NA |
| 12-oxoCDCA | NADPH | 3.69 ± 0.12 | 9.55 | 3.31 ± 0.11 | 4.16 | 4.88 ± 0.16 | 3.65 |
| DCA | NADP+ | 153.42 ± 6.2 | 100 | 127.68 ± 3.50 | 100 | 239.16 ± 2.89 | 100 |
| CA | NADP+ | 83.55 ± 1.55 | 54.44 | 102.79 ± 2.85 | 80.51 | 196.87 ± 4.36 | 82.32 |
| TDCA | NADP+ | 9.28 ± 0.57 | 6.05 | 57.56 ± 1.78 | 45.08 | 96.45 ± 6.48 | 40.33 |
| GDCA | NADP+ | 9.76 ± 1.56 | 6.36 | 30.86 ± 0.67 | 24.17 | 61.81 ± 3.61 | 25.85 |
| CDCA | NADP+ | NA | NA | NA | NA | NA | NA |
| DCA | NAD+ | NA | NA | NA | NA | NA | NA |
12-oxoLCA, 12-oxolithocholic acid; 12-oxoCDCA, 12-oxochenodeoxycholic acid; DCA, deoxycholic acid; CA, cholic acid; TDCA, taurodeoxycholic acid; GDCA, glycodeoxycholic acid; CDCA, chenodeoxycholic acid. For both the reductive and oxidative directions, the highest activity was set to 100%. The concentrations used in the assays were based on Km values (refer to Materials and Methods).
Values represent the means ± SD based on three or more replications. NA, no activity detected.
Phylogenetic analysis of bacterial 12α-HSDH.
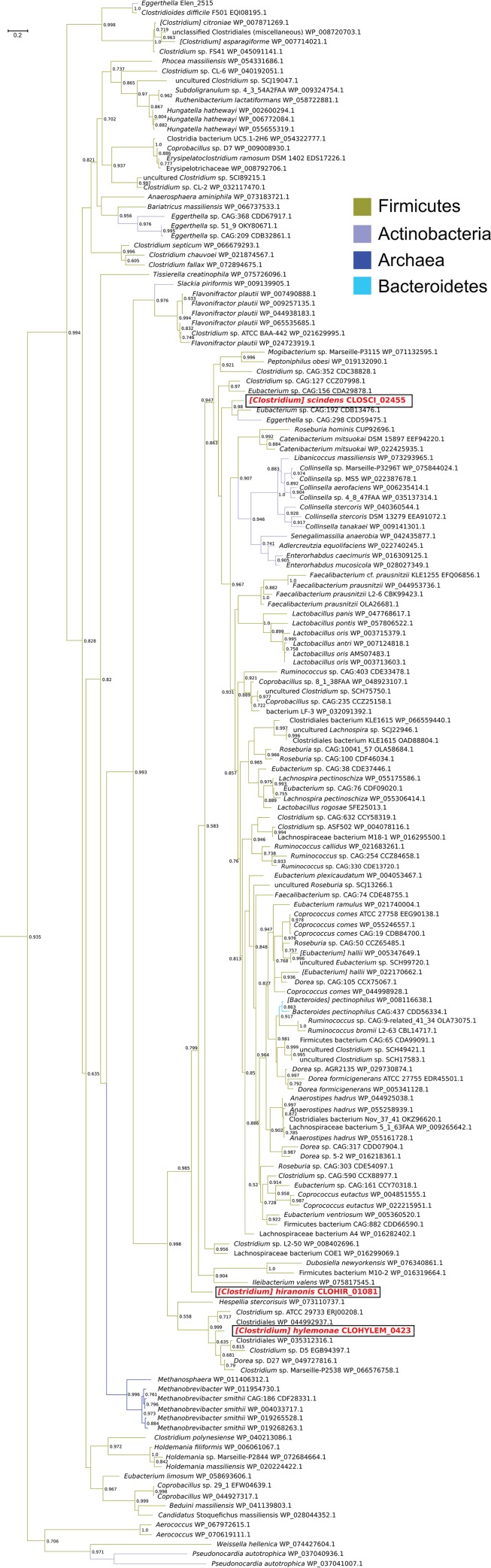
Next, we determined the phylogenetic positions of CSCI, CHIR, and CHYL relative to nearly 10,000 similar amino acid sequences in the NCBI NR database, by inferring a maximum likelihood tree, from which the subtree containing the sequences of interest was extracted for presentation (Fig. 6). CHYL clustered with other Clostridiales sequences, including the first characterized 12α-HSDH from Clostridium sp. ATCC 29733 (29). CHIR is a lone sequence which shares a common node with a larger set of clusters composed principally of Firmicutes and Actinobacteria. Within this larger cluster is CSCI, which is closely related to two metagenomic sequences, one sequence from Eubacterium sp. strain CAG:192 and one from Eggerthella sp. strain CAG:298 (accession no. CDD59475.1). We recently reported that the protein with accession number CDD59475.1 has 12α-HSDH activity (34).
FIG 6.
Subtree of phylogenetic analysis of 12α-hydroxysteroid dehydrogenases. Branch colors reflect bacterial phylum and the domain Archaea. Descriptors for 12α-HSDHs characterized in the present study are boxed and in red font. Accession numbers are shown to the right of the organism names.
The Actinobacteria represented in the phylogeny, from the family Coriobacteriaceae, are of particular interest due to previous reports of 12α-HSDH activity. Wegner et al. demonstrated that mouse fecal isolate Enterorhabdus mucosicola DSM 19490T expresses bile salt hydrolase, as well as 3α-HSDH and 12α-HSDH activities (36). Our phylogenetic analysis suggests that the NAD(P)-dependent oxidoreductase (accession no. WP_028027349.1) is the 12α-HSDH encoded by E. mucosicola, and that closely related Enterorhabdus caecimuris protein (accession no. WP_016309125.1) is also a 12α-HSDH. Within this cluster are proteins from other Coriobacteriaceae members, such as an equol-producing strain of Adlercreutzia equolifaciens (37) and an asaccharolytic strain of Senegalemassilia anaerobia. Sharing a node (support value of 0.946) with this cluster are additional Coriobacteriaceae family members in the genus Collinsella, including Collinsella aerofaciens (accession no. WP_006235414.1), which was recently reported to express 12α-HSDH activity (36). Another genus within the Coriobacteriaceae, Eggerthella, is represented at various points within the tree and deserves mention for two reasons. First, early reports establish E. lenta strains to be capable of oxidation and epimerization of bile acids, with clear 12α-HSDH activity (38–40). Second, in addition to the protein with accession number CDD59475.1 (34), we recently characterized Elen_2515 from E. lenta DSM 2243T, which was found to have bile acid 12α-HSDH activity (33).
While 12α-HSDH is well represented among Gram-positive members of the Actinobacteria and Firmicutes, the only taxa representing Gram-negative bacteria within this portion of the tree were strains of Bacteroides pectinophilus (Fig. 6). There is a large cluster of Bacteroides sp. proteins, including several from Bacteroides fragilis, which were further removed from the portion of the tree represented in Fig. 7 (see also Fig. S2). The B. fragilis proteins are members of the SDR family; however, they share only 31% amino acid identity with CSCI. Suspecting that the B. fragilis cluster represented in Fig. S2 might contain the 7α-HSDH reported for B. fragilis ATCC 25285, we compared the amino acid sequence of a 3-oxoacyl-[acyl-carrier-protein] reductase (accession no. WP_005799187.1) to that of 7α-HSDH (accession no. WP_005792012.1) (41). However, the B. fragilis 7α-HSDH shared only 34% amino acid identity with the protein with accession number WP_005799187.1, despite both proteins being members of the FabG family of proteins. The sequence identity between WP_005799187.1 and the 12α-HSDH from C. scindens (CSCI) was only 31%. Multiple forms of 7α-HSDH were reported among strains of B. fragilis (42), and further research will be needed to determine the functions of the proteins represented in this portion of the phylogeny.
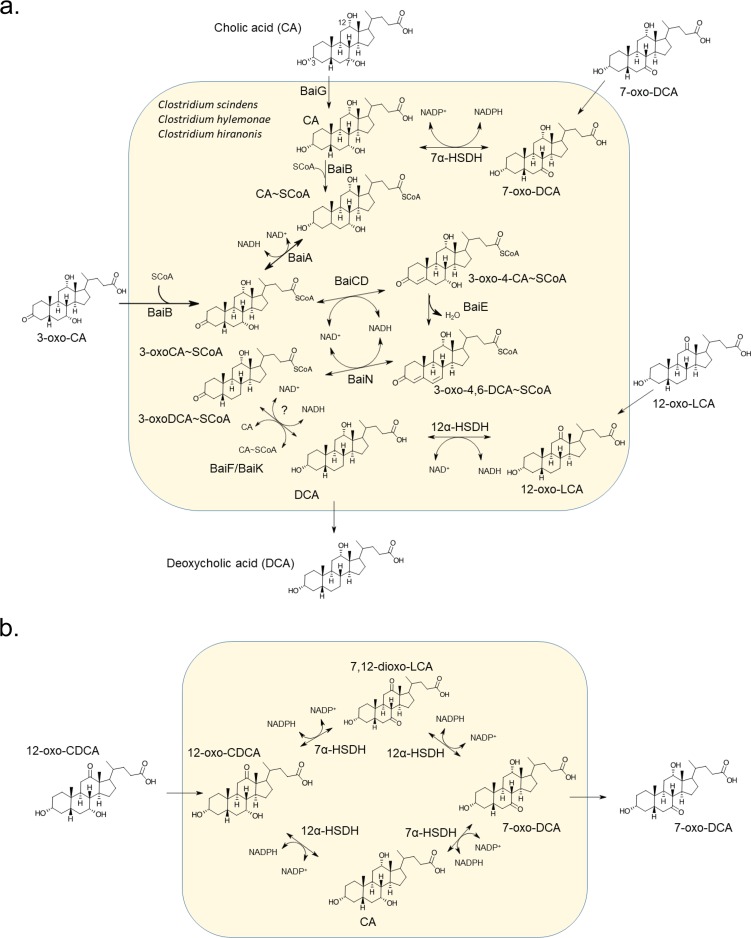
FIG 7.
Schematic model of bile acid metabolism by bile acid 7α-dehydroxylating bacteria. (a) See introduction for information regarding the core bile acid 7α-dehydroxylation pathway catalyzed by bile acid-inducible (Bai) enzymes. Transport of oxo-bile acid derivatives hypothesized to import via sodium dependent-bile acid transporter (BaiG). See Table 1 for anaerobic culture-based bile acid conversion studies. (b) Alternative routes for conversion of 12-oxochenodeoxycholic acid (12-oxoCDCA) to 7-oxodeoxycholic acid (7-oxoDCA), observed in bile acid conversion experiments (Table 1).
Archaea were also identified in an isolated cluster (Fig. 6) populated by human gut isolates and metagenomic sequences of Methanobrevibacter smithii and Methanosphaera stadtmanae DSM 3091, consistent with a previous report (29). Our phylogeny suggests that 12α-HSDH, like bile salt hydrolase (3), was horizontally transferred from the Firmicutes to these Archaea.
The low-activity BA7 bacteria C. leptum, Paraclostridium bifermentans, and Paeniclostridium sordellii were also represented in the larger phylogeny (Fig. S2). C. leptum was previously reported to express 12α-HSDH (28); however, the gene encoding this enzyme has not been identified. C. leptum did not appear in a previous phylogeny based on the 12α-HSDH from Clostridium sp. ATCC 29733 (30, 31). However, we located a C. leptum protein (accession no. WP_003532012.1) that shares 61% identity with a deduced protein from P. sordellii (accession no. WP_057547571.1), as well as C. perfringens (accession no. WP_096515955.1), an organism that has previously been shown to express 3α-, 7α-, and 12α-HSDH activities (43). When the protein with accession number WP_057547571.1 was aligned with the amino acid sequence of a 7α-HSDH previously characterized from P. sordellii, it was determined that they shared only 36.29% identity. Paraclostridium bifermentans encodes a gene with deduced amino acid sequence (accession no. WP_021433828.1) sharing 86% identity with WP_057547571.1 from P. sordellii. Further work will be required to determine if these proteins identified in the phylogeny have 12α-HSDH activity.
DISCUSSION
It was shown in previous studies that C. scindens VPI 12708 generates a complex mixture of metabolites from the bile acid CA during anaerobic growth in culture medium (Fig. 7) (4, 24). These studies identified 3α-HSDH and 7α-HSDH activities, and the genes encoding these enzymes have been reported in strains of C. scindens (19, 22), as well as C. hiranonis (44) and C. hylemonae (45). However, the formation of 12-oxo-intermediates has not been reported in anaerobic batch culture-based studies of these strains, but it was reported under aerobic conditions by resting cells of C. hiranonis (32). Oxidative conditions would be expected to funnel reductant toward detoxification of molecular oxygen, resulting in bile acid oxidation. C. leptum, which expresses BA7 activity but appears to lack several genes in the pathway harbored by C. scindens, C. hiranonis, and C. hylemonae (5), also expresses 12α-HSDH (28). Intriguingly, our extensive phylogenetic analysis may have identified the gene encoding 12α-HSDH in C. leptum; however, further work will be needed for confirmation.
The limitations of previous studies that cultured BA7 bacteria under low redox potential relate to their use of host-derived primary bile acids (CA, CDCA, and UDCA), rather than microbe-derived oxidized bile acid metabolites. Consistent with previous reports (24, 44, 45), 12α-HSDH activity was not observed when CA, 3-dehydro-CA, 7-oxoDCA, or DCA was added to anaerobic cultures of C. scindens, C. hylemonae, or C. hiranonis (Table 1). However, when we introduced 12-oxoLCA, 3,12-dioxoLCA, 7,12-dioxoLCA, and 3,7,12-trioxocholanic acid, we observed reduction of the 12-oxo-group, providing evidence for 12α-HSDH activity. Surprisingly, 12-oxoCDCA was not converted to DCA in batch cultures of C. scindens, C. hiranonis, and C. hylemonae. Instead, 12-oxoCDCA was converted quantitatively to 7-oxoDCA, whose formation could follow two metabolic routes, both depicted in Fig. 7. Both reduction of bile acid oxo-groups and 7α-dehydroxylation result in a net 2-electron reduction (4). We therefore speculate that a reduction of oxo-bile acids fulfills the redox balance needs of the organisms in batch culture. It could also be that these oxo-derivatives do not induce the bile acid 7α-dehydroxylation pathway. Future work will need to focus on understanding reductant flow and induction conditions in these gut bacteria when substrates are oxo-bile acids versus primary bile acids.
The purified recombinant 12α-HSDH enzymes from these strains displayed an order of magnitude lower activity toward 12-oxoCDCA than 12-oxoLCA (Table 2). This is consistent with kinetic analysis of 12α-HSDH enzymes from gut bacteria that lack BA7 activity and the bai operon (30, 33, 34). Unlike C. scindens, C. hiranonis, and C. hylemonae, non-BA7 gut bacteria appear to oxidize the C-12 of DCA (26, 33, 44, 45).
Our phylogenetic analysis updates and extends previous work on 12α-HSDH enzymes in gut bacteria (29). Indeed, we have recently verified that Eggerthella strains harbor distinct 12α-HSDH enzymes, both of which (Elen_2515 and the protein with accession no. CDD59475) are represented in our phylogeny (Fig. 6). Furthermore, newly reported members of the gut microbiota recently obtained through culturomics approaches are represented in our phylogeny, expanding the potential for exploring bile acid metabolism by diverse gut microbes. The widespread distribution of 12α-HSDH among Firmicutes, Coriobacteriaceae family members of the Actinobacteria, and gut Archaea may represent an important detoxification mechanism to protect against the antimicrobial nature of DCA. DCA is roughly 10-fold more toxic to many gut bacteria than CA (45). 12-oxoLCA is a commonly detected metabolite in human fecal samples; thus, the formation of this metabolite may have measurable effects on the microbial community and host physiology (46–48). Given their potential as redox-active centers, further research on oxo-bile acids is warranted.
The 12-oxo group of 12-oxoLCA would be expected to be an electron sink for bile acid 7α-dehydroxylating gut bacteria, and reduction of this group would be favorable in an anaerobic environment. However, this may be a proximate cause, and a different evolutionary pressure may have evolved to respond to C-12 oxidation by other members of the gut microbiota. Oxidation of bile acid hydroxyl groups reduces the detergent nature of the bile acid and its toxicity to gut bacteria (49–52). Therefore, 12α-HSDH activity in bile acid 7α-dehydroxylating gut bacteria may have arisen to maintain toxic levels of DCA in fecal water, perhaps to reduce competitors for key nutrients. Furthermore, the observations of Masuda and Oda (32) suggest that redox potential affects the direction of this reaction, and that oxidation of C-12 is more probable closer to the mucosa where oxygen partial pressures are increased (53).
At present, there are no structures for gut bacterial 12α-HSDHs. Such information would provide important insight into the key residues that might be altered to engineer an enzyme with increased activity toward CA, a step toward improving Wolff-Kishner reduction in the synthesis of the therapeutic bile acid UDCA. Further, our data suggest that shifting CA metabolism toward the formation of 12-oxoCDCA may reduce the rate of formation of DCA, a bile acid implicated in cancers of the liver (8) and colon (7), as well as cholesterol gallstone formation (10), and roles in cardiovascular disease are becoming apparent (54). Theoretical studies also suggest that oxo-bile acids, including 12-oxo-derivatives, are potential enzyme inhibitors, effectors of host nuclear receptors, and substrates for particular P450 monooxygenases (55). Further studies are needed to determine the role of microbial bile acid oxidation on host physiology.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
C. scindens ATCC 35704, C. hylemonae DSM 15053, and C. hiranonis DSM 13275 were obtained from −80°C glycerol stocks from culture collections at the University of Illinois at Urbana-Champaign (UIUC) and Eastern Illinois University (EIU). Each strain was grown anaerobically at 37°C in butyl rubber-stoppered crimp-sealed culture tubes (18 by 150 mm, series 2048; Bellco Glass, Inc., Vineland, NJ, USA; 27.2 ml approximate stoppered volume at 1 atm [101.29 kPa]) containing brain heart infusion (BHI) broth consisting of the following (grams per liter): BHI broth (BD Difco), 37; NaHCO3, 7.5; glucose, 2.0; yeast extract, 5.0; and resazurin, 0.001. The culture medium was prepared anaerobically by boiling and cooling the medium under 100% CO2, adding cysteine·HCI·H2O (0.5 g/liter) to the medium, and dispensing the medium under 100% CO2 into culture tubes (10 ml per tube). Tubes were subsequently crimp sealed and autoclaved. After autoclaving, the pH of the medium approximated 6.8. Stock solutions (10.6 mM) of bile acids were prepared in 100% methanol, added to tubes, sealed with stoppers, and degassed by sparging and then flushing the headspace gas with argon. Bile acid solutions were added (0.1 ml) via sterile needles and syringes to tubes of sterile BHI broth to achieve an initial concentration of 0.1 mM after inoculation. In all experiments, growth was initiated by injecting 0.5 ml of inoculum per culture. The final concentration of methanol in cultures approximated 0.9% and did not influence the growth of the strains being tested; growth was also not observed following the addition of methanolic bile acid solutions to tubes of sterile BHI broth in the absence of inoculum.
Escherichia coli DH5α (Turbo) competent cells were from New England BioLabs (NEB) (Ipswich, MA, USA) and E. coli BL21-CodonPlus(DE3)-RIPL cells were purchased from Stratagene (La Jolla, CA, USA) and grown in Luria-Bertani (LB) broth at 37°C.
Chemicals.
5β-Cholanic acid-3α, 7α, 12α-triol (CA), 5β-cholanic acid-3α, 12α-diol (DCA), 5β-cholanic acid-3α, 7α-diol (CDCA), 5β-cholanic acid-3α, 12α-diol N-(2-sulfoethyl)-amide (tauroDCA), glycoDCA, 5β-cholanic acid-3α, 7α, 12α-triol N-(2-sulfoethyl)-amide (tauroCA), 5β-cholanic acid-3α, 7α, 12α-triol N-(carboxymethyl)-amide (glycoCA), 5β-cholanic acid-3α, 7α-diol N-(2-sulfoethyl)-amide (tauroCDCA), and 5β-cholanic acid-3α, 7α-diol N-(carboxymethyl)-amide (glycoCDCA) were purchased from Sigma-Aldrich (St. Louis, MO, USA). 5β-Cholanic acid-3α, 7α-diol-12-one (12-oxoCDCA), 5β-cholanic acid-3α-ol-12-one (12-oxoLCA), 5β-cholanic acid-12α-ol-3-one (3-oxoDCA), 5β-cholanic acid-3α, 12α-diol-7-one (7-oxoDCA), 5β-cholanic acid-3,7,12-trione (3,7,12-trioxoLCA), 5β-cholanic acid-3α-ol-7,12-dione (7,12-dioxoLCA), and 5β-cholanic acid-3,12-dione (3,12-dioxoLCA) were purchased from Steraloids, Inc. (Newport, RI, USA) (56). Isopropyl β-d-1-thiogalactopyranoside (IPTG) was purchased from Gold Biotechnology (St. Louis, MO, USA). Strep-Tactin resin was purchased from IBA GmbH (Göttingen, Germany). All other reagents were of the highest possible purity and were purchased from Fisher Scientific (Pittsburgh, PA, USA).
Detection of bile acids in cultures.
TLC was used for the detection of bile acids in BHI cultures and in standards prepared from sterile BHI broth supplemented with a bile acid (24, 57, 58). Briefly, a 1-ml sample of culture or sterile broth standard was transferred to duplicate 2-ml microcentrifuge tubes (0.5 ml of sample per tube). To each sample, 100 μl of 3 N HCl and 500 μl of ethyl acetate were added. The microcentrifuge tubes were capped, mixed by vortexing, and spun in a microcentrifuge for 1 min at 14,000 rpm. The organic phase (top layer) from both samples was removed and transferred to a 20-ml glass scintillation vial. The extraction step was repeated for each sample, and the combined ethyl acetate extracts (∼2 ml) were dried at room temperature under a stream of nitrogen. Methanol (100 μl) was added to each vial, and the resuspended extracts were immediately spotted (50 μl) onto silica plates (Whatman AL SIL G, 250 μm thick, non-UV; Fisher Scientific). Individual and mixed methanolic standards (0.1 and 1.0 mM) were also prepared by dissolving pure bile acids directly in methanol and spotting (50 μl) onto silica plates. Unless indicated otherwise, TLC plates were developed in a tank containing 50 ml of solvent (cyclohexane, ethyl acetate, and glacial acetic acid, 12:12:1 [vol/vol/vol]). Once the solvent reached approximately 25 mm from the top of the plate, the plate was removed from the tank, sprayed with charring agent solution consisting of methanol (150 ml), water (150 ml), concentrated sulfuric acid (10 ml), and MnCl2·4H2O (1 g), heated to 115°C for 15 min, and visualized with long-UV light (365 nm). Bile acids were identified by comparing Rf values of bile acid standards to those of bile acids detected in cultures.
Isolation of genomic DNA.
Genomic DNA for PCR and molecular cloning applications was extracted from C. hiranonis DSM 13275, C. scindens ATCC 35704, and C. hylemonae DSM 15053 using the Fast DNA isolation kit from Mo Bio (Carlsbad, CA, USA), according to the manufacturer's protocol.
Heterologous expression of 12α-HSDH enzymes in E. coli and protein purification.
The pET-51b(+) vector was obtained from Novagen (San Diego, CA, USA). Restriction enzymes were purchased from NEB (Ipswich, MA). Inserts were generated by PCR amplification using cloning primers obtained from Integrated DNA Technologies, Inc. (Coralville, IA, USA). The cloning primers used in this study are listed in Table 4. Inserts for directional cloning were amplified using the Phusion high-fidelity polymerase (Stratagene, La Jolla, CA, USA) and cloned into pET-51b(+) after the insert and vector were doubly digested with the appropriate restriction endonuclease and treated with DNA ligase. Recombinant plasmid was transformed into chemically competent E. coli DH5α cells via a heat shock method, plated, and grown for 16 h at 37°C on lysogeny broth (LB) agar plates supplemented with ampicillin (100 μg/ml). A single colony from each transformation was inoculated into LB broth (5 ml) containing ampicillin (100 μg/ml) and grown to saturation. The cells were subsequently centrifuged (3,220 × g, 15 min, 4°C), and plasmids were extracted from the resulting cell pellet using the QIAprep Spin miniprep kit (Qiagen, Valencia, CA, USA). The sequence of the insert was determined by Sanger sequencing (W. M. Keck Center for Comparative and Functional Genomics at the University of Illinois at Urbana-Champaign).
TABLE 4.
Sequences of oligonucleotide cloning primers
| Gene IDa | Primer sequences (forward and reverse, 5′ to 3′) | Mol mass (kDa)b | Extinction coefficient (M−1 · cm−1) |
|---|---|---|---|
| CLOSCI_02455 | ATATAGGATCCTATGGGATTTTTAACAGGTAAGACAGCC | 28.24 | 32,110 |
| ATATAAAGCTTTTATGGCCTAAGCCCCATTCC | |||
| CLOHYLEM_04236 | ATATATGGTACCGATGGGTATATTTGACGGAAAAACAGCTA | 27.96 | 24,995 |
| ATATATAAGCTTTTATGGGCGCTGTCCCATGC | |||
| CLOHIR_01081 | ATATAGGATCCTATGGGATTTTTAAATGGAAAAACAG | 28.49 | 29,130 |
| ATATAGAGCTCTTAAGGTCTTAATCCCATTCCA |
ID, identification.
Deduced recombinant 12α-HSDH molecular mass.
For protein expression, the correct recombinant plasmids extracted from the E. coli DH5α cells were transformed into E. coli BL21 CodonPlus (DE3)-RIPL chemically competent cells by heat shock method and cultured for 24 h at 37°C on LB agar plates supplemented with ampicillin (100 μg/ml) and chloramphenicol (50 μg/ml). Selected colonies were inoculated into LB broth (10 ml) supplemented with antibiotics and grown at 37°C for 6 h with vigorous aeration. The precultures were then added to fresh LB broth (1 liter), supplemented with ampicillin (100 μg/ml), and aerated at 37°C until reaching an optical density at 600 nm (OD600) of 0.3. To induce recombinant protein expression, IPTG was added to each culture at a final concentration of 0.1 mM, and the temperature was decreased to 16°C. Following 16 h of incubation, cells were pelleted by centrifugation (4,000 × g, 30 min, 4°C) and resuspended in 30 ml of binding buffer (20 mM Tris-HCl, 150 mM NaCl, 10% glycerol, 10 mM 2-mercaptoethanol [pH 7.9]). The cell suspension was subjected to four passages through an EmulsiFlex C-3 cell homogenizer (Avestin, Ottawa, Canada), and the cell lysate was clarified by centrifugation at 20,000 × g for 30 min at 4°C.
The recombinant 12α-HSDHs were purified using Strep-Tactin resin (IBA GmbH), as per the manufacturing protocol. The protein was eluted with an elution buffer composed of 20 mM Tris-HCl, 150 mM NaCl, 10% glycerol, 10 mM 2-mercaptoethanol (pH 7.9), and 2.5 mM d-desthiobiotin. The purity of the proteins was assessed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and the protein bands were visualized by Coomassie brilliant blue G-250 staining. The concentrations of the proteins were calculated based on their extinction coefficients and molecular weights. The observed subunit weight for each was calculated by migration distance of purified protein to standard proteins in ImageJ (https://imagej.nih.gov/ij/index.html).
Enzyme assays.
The buffers used to investigate the optimal pHs of the recombinant 12α-HSDH enzymes (rCHYL, rCSCI, and rCHIR) contained 150 mM NaCl and varied contents of the following buffering agents: 50 mM sodium acetate (pH 4 to 6), 50 mM sodium phosphate (pH 6.5 to 7.5), and 50 mM Tris-Cl (pH 8 to 9). The linearity of the 12α-HSDH activity over time and the enzyme concentration were determined by monitoring the oxidation and reduction of NADP(H) aerobically at 340 nm (ε = 6,220 M−1 · cm−1) continuously for 1.5 min in a reaction mixture with 12α-HSDH and bile acid substrates. The reaction mixtures for rCHYL in the oxidative direction were composed of sodium phosphate buffer at pH 8.0, 18 nM enzyme with either 700 μM NADP+ and various concentrations of DCA or 900 μM DCA and differing NADP+ concentrations. The reductive direction consisted of pH 7.0 sodium phosphate buffer and 150 μM NADPH and different 12-oxoLCA concentrations or 100 μM 12-oxoLCA and various NADPH concentrations, with 12.5 nM enzyme. rCSCI oxidative reaction mixtures included 10 nM enzyme sodium phosphate buffer (pH 7.5), and 400 μM NADP+ with various concentrations of DCA or 1,500 μM DCA with different NADP+ concentrations. The reductive mixtures were sodium phosphate buffer at pH 7.0, 8 nM enzyme, and 150 μM NADPH with changing 12-oxoLCA concentrations or 50 μM 12-oxoLCA with differing NADPH concentrations. The oxidative reaction mixtures for rCHIR were sodium phosphate buffer (pH 7), and 10 nM enzyme with either 900 μM NADP+ and various DCA concentrations or 1,600 μM DCA and different NADP+ concentrations. The reductive mixtures were composed of sodium phosphate buffer (pH 7), 200 μM NADPH with various 12-oxoLCA concentrations or 150 μM 12-oxoLCA with differing NADPH concentrations, and 8 nM enzyme. The kinetic parameters were estimated by fitting the data to the Michaelis-Menten equation by the nonlinear regression method using the enzyme kinetics module in GraphPad Prism (GraphPad Software, La Jolla, CA, USA). The substrate specificity analysis was performed according to the previously stated reaction mixtures, using saturating concentrations of substrate and pyridine nucleotide.
TLC and mass spectrometry of bile acid metabolites.
Reaction mixtures were set up with 50 μM 12-oxoLCA substrate and NAD(P)(H) cofactor, either 12.5 nM rCHYL, 8 nM rCSCI, or 8 nM rCHIR, and the optimal pH 7.0 buffer up to 1 ml. The reaction mixtures were incubated overnight at room temperature and stopped with 150 μl of 1 N HCl. The mixtures were extracted through vortexing with two volumes of ethyl acetate for 1 min and then two more volumes and another 1 min of vortexing. The organic phase was recovered and evaporated under nitrogen gas. The residue was dissolved in 30 μl ethyl acetate and spotted on the TLC plate (silica gel IB2-F flexible TLC sheet, 20 by 20 cm, with 250-μm analytical layer; J.T. Baker, Avantor Performance Materials, LLC, PA, USA) with 8 μl of 10 mM DCA and 12-oxoLCA standards. The mobile phase consisted of 70:20:2 toluene–1,4-dioxane–acetic acid. The bile acid metabolites separated on the TLC were visualized using a 10% phosphomolybdic acid in ethanol spray and heating for 15 min at 100°C.
The bile acid metabolites were then extracted from TLC plates and sent for mass spectrometry (MS) analysis at the UIUC Mass Spectrometry Laboratory. The mass spectrometer (Thermo Scientific LTQ Orbitrap XL Hybrid Ion Trap-Orbitrap) was operated with an electrospray ionization (ESI) source in negative-ion mode. The sample was introduced through syringe pump injection at a flow rate of 10 μl · min−1. The capillary voltage was −28 V, with a source temperature of 235°C. The mass spectrogram data were processed with the Thermo Xcalibur software.
Phylogenetic analysis of bacterial 12α-HSDHs.
Sequences from the NCBI NR protein database were identified by BLASTP (limited to a maximum of 10,000 sequences, with an E value threshold of 1E−10) using as query protein with accession no. ERJ00208.1 (270 residues long) from Clostridium sp. ATCC 29733. Sequences shorter than 200 or longer than 350 amino acids were discarded to minimize fragments and other artifacts, as well as false positives (e.g., long sequences that have some similarity due to shared domains but are not likely to be homologues). The resulting 9,968 sequences were aligned using Muscle version 3.8.31 (59).
Maximum likelihood phylogenetic analysis was performed using FastTree version 2.1.8 (60), with gamma-distributed heterogeneity rates and employing the WAG substitution model. The resulting tree was drawn and processed in Dendroscope version 3.5.9 (61), and cosmetic adjustments were performed in Inkscape (http://inkscape.org).
Supplementary Material
ACKNOWLEDGMENTS
We gratefully acknowledge the financial support provided to J.M.R. for new faculty startup through the Department of Animal Sciences at the University of Illinois at Urbana-Champaign (grant Hatch ILLU-538-916). L.A.S. expresses her sincere gratitude to the Saudi Arabian Cultural Mission (SACM) for the financial support provided to her through the Department of Biological Sciences at Eastern Illinois University.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00235-18.
REFERENCES
- 1.Vlahcevic ZR, Heuman DM, Hylemon PB. 1996. Physiology and pathophysiology of enterohepatic circulation of bile acids, p 376–417. In Zakim D, Boyer T (ed), Hepatology: a textbook of liver disease, 3rd ed, vol 1 Saunders, Philadelphia, PA. [Google Scholar]
- 2.Dawson PA, Karpen SJ. 2015. Intestinal transport and metabolism of bile acids. J Lipid Res 56:1085–1099. doi: 10.1194/jlr.R054114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jones BV, Begley M, Hill C, Gahan CG, Marchesi JR. 2008. Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome. Proc Natl Acad Sci U S A 105:13580–13585. doi: 10.1073/pnas.0804437105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ridlon JM, Kang DJ, Hylemon PB. 2006. Bile salt biotransformations by human intestinal bacteria. J Lipid Res 47:241–259. doi: 10.1194/jlr.R500013-JLR200. [DOI] [PubMed] [Google Scholar]
- 5.Ridlon JM, Harris SC, Bhowmik S, Kang DJ, Hylemon PB. 2016. Consequences of bile salt biotransformations by intestinal bacteria. Gut Microbes 7:22–39. doi: 10.1080/19490976.2015.1127483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kakiyama G, Muto A, Takei H, Nittono H, Murai T, Kurosawa T, Hofmann AF, Pandak WM, Bajaj JS. 2014. A simple and accurate HPLC method for fecal bile acid profile in healthy and cirrhotic subjects: validation by GC-MS and LC-MS. J Lipid Res 55:978–990. doi: 10.1194/jlr.D047506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bernstein C, Holubec H, Bhattacharyya AK, Nguyen H, Payne CM, Zaitlin B, Bernstein H. 2011. Carcinogenicity of deoxycholic acid, a secondary bile acid. Arch Toxicol 85:863–871. doi: 10.1007/s00204-011-0648-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yoshimoto S, Loo TM, Atarashi K, Kanda H, Sato S, Oyadomari S, Iwakura Y, Oshima K, Morita H, Hattori M, Honda K, Ishikawa Y, Hara E, Ohtani N. 2013. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature 499:97–101. doi: 10.1038/nature12347. [DOI] [PubMed] [Google Scholar]
- 9.Wu J, Gong J, Geng J, Song Y. 2008. Deoxycholic acid induces the overexpression of intestinal mucin, MUC2, via NF-κB signaling pathway in human esophageal adenocarcinoma cells. BMC Cancer 8:333. doi: 10.1186/1471-2407-8-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berr F, Kullak-Ublick GA, Paumgartner G, Münzing W, Hylemon PB. 1996. 7α-Dehydroxylating bacteria enhance deoxycholic acid input and cholesterol saturation of bile in patients with gallstones. Gastroenterology 111:1611–1620. doi: 10.1016/S0016-5085(96)70024-0. [DOI] [PubMed] [Google Scholar]
- 11.Magouliotis DE, Tasiopoulou VS, Svokos AA, Chatedaki C, Sioka E, Zacharoulis D. 2017. Ursodeoxycholic acid in the prevention of gallstone formation after bariatric surgery: an updated systematic review and meta-analysis. Obes Surg 27:3021–3030. doi: 10.1007/s11695-017-2924-y. [DOI] [PubMed] [Google Scholar]
- 12.Peng S, Huo X, Rezaei D, Zhang Q, Zhang X, Yu C, Asanuma K, Cheng E, Pham TH, Wang DH, Chen M, Souza RF, Spechler SJ. 2014. In Barrett's esophagus patients and Barrett's cell lines, ursodeoxycholic acid increases antioxidant expression and prevents DNA damage by bile acids. Am J Physiol Gastrointest Liver Physiol 307:G129–G139. doi: 10.1152/ajpgi.00085.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim EK, Cho JH, Kim E, Kim YJ. 2017. Ursodeoxycholic acid inhibits the proliferation of colon cancer cells by regulating oxidative stress and cancer stem-like cell growth. PLoS One 12:e0181183. doi: 10.1371/journal.pone.0181183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sutherland JD, MacDonald IA, Forrest TP. 1982. The enzymatic and chemical synthesis of ursodeoxycholic and chenodeoxycholic acid from cholic acid. Prep Biochem 12:307–321. [DOI] [PubMed] [Google Scholar]
- 15.Mallonee DH, Hylemon PB. 1996. Sequencing and expression of a gene encoding a bile acid transporter from Eubacterium sp. strain VPI 12708. J Bacteriol 178:7053–7058. doi: 10.1128/jb.178.24.7053-7058.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mallonee DH, Adams JL, Hylemon PB. 1992. The bile acid-inducible baiB gene from Eubacterium sp. strain VPI 12708 encodes a bile acid-coenzyme A ligase. J Bacteriol 174:2065–2071. doi: 10.1128/jb.174.7.2065-2071.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ye HQ, Mallonee DH, Wells JE, Björkhem I, Hylemon PB. 1999. The bile acid-inducible baiF gene from Eubacterium sp. strain VPI 12708 encodes a bile acid-coenzyme A hydrolase. J Lipid Res 40:17–23. [PubMed] [Google Scholar]
- 18.Ridlon JM, Hylemon PB. 2012. Identification and characterization of two bile acid coenzyme A transferases from Clostridium scindens, a bile acid 7α-dehydroxylating intestinal bacterium. J Lipid Res 53:66–76. doi: 10.1194/jlr.M020313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baron SF, Franklund CV, Hylemon PB. 1991. Cloning, sequencing, and expression of the gene coding for bile acid 7α-hydroxysteroid dehydrogenase from Eubacterium sp. strain VPI 12708. J Bacteriol 173:4558–4569. doi: 10.1128/jb.173.15.4558-4569.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dawson JA, Mallonee DH, Björkhem I, Hylemon PB. 1996. Expression and characterization of a C24 bile acid 7α-dehydratase from Eubacterium sp. strain VPI 12708 in Escherichia coli. J Lipid Res 37:1258–1267. [PubMed] [Google Scholar]
- 21.Bhowmik S, Chiu HP, Jones DH, Chiu HJ, Miller MD, Xu Q, Farr CL, Ridlon JM, Wells JE, Elsliger MA, Wilson IA, Hylemon PB, Lesley SA. 2016. Structure and functional characterization of a bile acid 7α dehydratase BaiE in secondary bile acid synthesis. Proteins 84:316–331. doi: 10.1002/prot.24971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mallonee DH, Lijewski MA, Hylemon PB. 1995. Expression in Escherichia coli and characterization of a bile acid-inducible 3α-hydroxysteroid dehydrogenase from Eubacterium sp. strain VPI 12708. Curr Microbiol 30:259–263. doi: 10.1007/BF00295498. [DOI] [PubMed] [Google Scholar]
- 23.Bhowmik S, Jones DH, Chiu HP, Park IH, Chiu HJ, Axelrod HL, Farr CL, Tien HJ, Agarwalla S, Lesley SA. 2014. Structural and functional characterization of BaiA, an enzyme involved in secondary bile acid synthesis in human gut microbe. Proteins 82:216–229. doi: 10.1002/prot.24353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hylemon PB, Melone PD, Franklund CV, Lund E, Björkhem I. 1991. Mechanism of intestinal 7α-dehydroxylation of cholic acid: evidence that allo-deoxycholic acid is an inducible side-product. J Lipid Res 32:89–96. [PubMed] [Google Scholar]
- 25.Harris SC, Devendran S, Alves JMP, Mythen SM, Hylemon PB, Ridlon JM. 2017. Identification of a gene encoding a flavoprotein involved in bile acid metabolism by the human gut bacterium Clostridium scindens ATCC 35704. Biochim Biophys Acta 1863:276–283. doi: 10.1016/j.bbalip.2017.12.001. [DOI] [PubMed] [Google Scholar]
- 26.Edenharder R, Schneider J. 1985. 12β-Dehydrogenation of bile acids by Clostridium paraputrificum, C. tertium, and C. difficile and epimerization at carbon-12 of deoxycholic acid by co-cultivation with 12α-dehydrogenating Eubacterium lentum. Appl Environ Microbiol 49:964–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Doerner KC, Takamine F, LaVoie CP, Mallonee DH, Hylemon PB. 1997. Assessment of fecal bacteria with bile acid 7α-dehydroxylating activity for the presence of bai-like genes. Appl Environ Microbiol 63:1185–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harris JN, Hylemon PB. 1978. Partial purification and characterization of NADP-dependent 12α-hydroxysteroid dehydrogenase from Clostridium leptum. Biochim Biophys Acta 528:148–157. doi: 10.1016/0005-2760(78)90060-7. [DOI] [PubMed] [Google Scholar]
- 29.Kisiela M, Skarka A, Ebert B, Maser E. 2012. Hydroxysteroid dehydrogenases (HSDs) in bacteria: a bioinformatic perspective. J Steroid Biochem Mol Biol 129:31–46. doi: 10.1016/j.jsbmb.2011.08.002. [DOI] [PubMed] [Google Scholar]
- 30.MacDonald IA, Jellett JF, Mahony DE. 1979. 12α-Hydroxysteroid dehydrogenase from Clostridium group P strain C48-50 ATCC no. 29733: partial purification and characterization. J Lipid Res 20:234–239. [PubMed] [Google Scholar]
- 31.Aigner A, Gross R, Schmid R, Braun M, Mauer S. April 2011. Novel 12α-hydroxysteroid dehydrogenases, production and use thereof. US patent 20110091921A1.
- 32.Masuda N, Oda H. 1983. 7α-Dehydroxylation of bile acids by resting cells of an unidentified, Gram-positive, non-spore forming anaerobic bacterium. Appl Environ Microbiol 45:456–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harris SC, Devendran S, Méndez-García C, Mythen SM, Wright CL, Fields CJ, Hernandez AG, Cann I, Hylemon PB, Ridlon JM. 4 April 2018. Bile acid oxidation by Eggerthella lenta strains C592 and DSM 2243T. Gut Microbes doi: 10.1080/19490976.2018.1458180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mythen SM, Devendran S, Méndez-García C, Cann I, Ridlon JM. 2018. Targeted synthesis and characterization of a gene cluster encoding NAD(P)H-dependent 3α-, 3β-, and 12α-hydroxysteroid dehydrogenases from Eggerthella CAG:298, a gut metagenomic sequence. Appl Environ Microbiol 84:e02475-17. doi: 10.1128/AEM.02475-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ditscheid B, Keller S, Jahreis G. 2009. Faecal steroid excretion in humans is affected by calcium supplementation and shows gender-specific differences. Eur J Nutr 48:22–30. doi: 10.1007/s00394-008-0755-2. [DOI] [PubMed] [Google Scholar]
- 36.Wegner K, Just S, Gau L, Mueller H, Gérard P, Lepage P, Clavel T, Rohn S. 2017. Rapid analysis of bile acids in different biological matrices using LC-ESI-MS/MS for the investigation of bile acid transformation by mammalian gut bacteria. Anal Bioanal Chem 409:1231–1245. doi: 10.1007/s00216-016-0048-1. [DOI] [PubMed] [Google Scholar]
- 37.Maruo T, Sakamoto M, Ito C, Toda T, Benno Y. 2008. Adlercreutzia equolifaciens gen. nov., sp. nov., an equol-producing bacterium isolated from human feces, and emended description of the genus Eggerthella. Int J Syst Evol Microbiol 58:1221–1227. doi: 10.1099/ijs.0.65404-0. [DOI] [PubMed] [Google Scholar]
- 38.MacDonald IA, Jellett JF, Mahony DE, Holdeman LV. 1979. Bile salt 3α- and 12α-hydroxysteroid dehydrogenases from Eubacterium lentum and related organisms. Appl Environ Microbiol 37:992–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hirano S, Masuda N. 1981. Transformation of bile acids by. Eubacterium lentum. Appl Environ Microbiol 42:912–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Edenharder R, Mielek K. 1984. Epimerization, oxidation and reduction of bile acids by Eubacterium lentum. Syst Appl Microbiol 5:287–298. doi: 10.1016/S0723-2020(84)80031-4. [DOI] [Google Scholar]
- 41.Bennett MJ, McKnight SL, Coleman JP. 2003. Cloning and characterization of the NAD-dependent 7α-hydroxysteroid dehydrogenase from Bacteroides fragilis. Curr Microbiol 47:475–484. doi: 10.1007/s00284-003-4079-4. [DOI] [PubMed] [Google Scholar]
- 42.Hylemon PB, Sherrod JA. 1975. Multiple forms of 7α-hydroxysteroid dehydrogenase in selected strains of Bacteroides fragilis. J Bacteriol 122:418–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.MacDonald IA, Meier MC, Mahony DE, Costain GA. 1976. 3α-, 7α-, and 12α-hydroxysteroid dehydrogenase activities from Clostridium perfringens. Biochim Biophys Acta 450:142–153. doi: 10.1016/0005-2760(76)90086-2. [DOI] [PubMed] [Google Scholar]
- 44.Wells JE, Hylemon PB. 2000. Identification and characterization of a bile acid 7α-dehydroxylation operon in Clostridium sp. strain TO-931, a highly active 7α-dehydroxylating strain isolated from human feces. Appl Environ Microbiol 66:1107–1113. doi: 10.1128/AEM.66.3.1107-1113.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ridlon JM, Kang DJ, Hylemon PB. 2010. Isolation and characterization of a bile acid 7α-dehydroxylating operon in Clostridium hylemonae TN271. Anaerobe 16:137–146. doi: 10.1016/j.anaerobe.2009.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Setchell KDR, Lawson AM, Tanida N, Sjovall J. 1983. General methods for the analysis of metabolic profiles of bile acids and related compounds in feces. J Lipid Res 24:1085–1100. [PubMed] [Google Scholar]
- 47.Reddy BS, Sharma C, Simi B, Engle A, Laakso K, Puska P, Korpela R. 1987. Metabolic epidemiology of colon cancer: effect of dietary fiber on fecal mutagens and bile acids in healthy subjects. Cancer Res 47:644–648. [PubMed] [Google Scholar]
- 48.Mai V, Katki HA, Harmsen H, Gallaher D, Schatzkin A, Baer DJ, Clevidence B. 2004. Effects of a controlled diet and black tea drinking on the fecal microflora composition and the fecal bile acid profile of human volunteers in a double-blinded randomized feeding study. J Nutr 134:473–478. doi: 10.1093/jn/134.2.473. [DOI] [PubMed] [Google Scholar]
- 49.Begley M, Gahan CG, Hill C. 2005. The interaction between bacteria and bile. FEMS Microbiol Rev 29:625–651. doi: 10.1016/j.femsre.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 50.Hofmann AF, Roda A. 1984. Physicochemical properties of bile acids and their relationship to biological properties: an overview of the problem. J Lipid Res 25:1477–1489. [PubMed] [Google Scholar]
- 51.Devlin AS, Fischbach MA. 2015. A biosynthetic pathway for a prominent class of microbiota-derived bile acids. Nat Chem Biol 11:685–690. doi: 10.1038/nchembio.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Watanabe M, Fukiya S, Yokota A. 2017. Comprehensive evaluation of the bactericidal activities of free bile acids in the large intestine of humans and rodents. J Lipid Res 58:1143–1152. doi: 10.1194/jlr.M075143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Albenberg L, Esipova TV, Judge CP, Bittinger K, Chen J, Laughlin A, Grunberg S, Baldassano RN, Lewis JD, Li H, Thom SR, Bushman FD, Vinogradov SA, Wu GD. 2014. Correlation between intraluminal oxygen gradient and radial partitioning of intestinal microbiota. Gastroenterology 147:1055.e8–1063.e8. doi: 10.1053/j.gastro.2014.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brown JM, Hazen SL. 2018. Microbial modulation of cardiovascular disease. Nat Rev Microbiol 16:171–181. doi: 10.1038/nrmicro.2017.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Trifunović J, Borčić V, Mikov M. 2017. Bile acids and their oxo derivatives: potential inhibitors of carbonic anhydrase I and II, androgen receptor antagonists and CYP3A4 substrates. Biomed Chromatogr 31:e3870. doi: 10.1002/bmc.3870. [DOI] [PubMed] [Google Scholar]
- 56.Hofmann AF, Sjovall J, Kurz G, Radominska A, Schteingart CD, Tint GS, Vlahcevic ZR, Setchell KDR. 1992. A proposed nomenclature for bile acids. J Lipid Res 33:599–604. [PubMed] [Google Scholar]
- 57.Eneroth P. 1963. Thin-layer chromatography of bile acids. J Lipid Res 4:11–16. [PubMed] [Google Scholar]
- 58.Wall PE. 2007. Thin-layer chromatography: a modern practical approach. Royal Society of Chemistry, Cambridge, United Kingdom. [Google Scholar]
- 59.Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Price MN, Dehal PS, Arkin AP. 2010. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huson DH, Scornavacca C. 2012. Dendroscope 3: an interactive tool for rooted phylogenetic trees and networks. Syst Biol 61:1061–1067. doi: 10.1093/sysbio/sys062. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.