ABSTRACT
Ticks are important disease vectors, as they transmit a variety of human and animal pathogens worldwide. Symbionts that coevolved with ticks confer crucial benefits to their host in nutrition metabolism, fecundity, and vector competence. Although over 100 tick species have been identified in China, general information on tick symbiosis is limited. Here, we visualized the tissue distribution of Coxiella sp. and Rickettsia sp. in lab-reared Haemaphysalis longicornis and Rhipicephalus haemaphysaloides by fluorescent in situ hybridization. We found that Coxiella sp. colonized exclusively the Malpighian tubules and ovaries of H. longicornis, while Rickettsia sp. additionally colonized the midgut of R. haemaphysaloides. We also investigated the population structure of microbiota in Dermacentor silvarum ticks collected from Inner Mongolia, China, and found that Coxiella, Rickettsia, and Pseudomonas are the three dominant genera. No significant difference in microbiota composition was found between male and female D. silvarum ticks. We again analyzed the tissue localization of Coxiella sp. and Rickettsia sp. and found that they displayed tissue tropisms similar to those in R. haemaphysaloides, except that Rickettsia sp. colonized the nuclei of spermatids instead of ovaries in D. silvarum. Altogether, our results suggest that Coxiella sp. and Rickettsia sp. are the main symbionts in the three ticks and reside primarily in midgut, Malpighian tubules, and reproductive tissues, but their tissue distribution varies in association with species and sexes.
IMPORTANCE Tick-borne diseases constitute a major public health burden, as they are increasing in frequency and severity worldwide. The presence of symbionts helps ticks to metabolize nutrients, promotes fecundity, and influences pathogen infections. Increasing numbers of tick-borne pathogens have been identified in China; however, knowledge of native ticks, especially tick symbiosis, is limited. In this study, we analyze the distribution of Coxiella sp. and Rickettsia sp. in tissues of laboratory-reared Haemaphysalis longicornis and Rhipicephalus haemaphysaloides and field-collected Dermacentor silvarum. We found that the localization patterns of Coxiella sp. in three Chinese tick species were similar to those of other tick species. We also found a previously undefined intracellular localization of Rickettsia sp. in tick midgut and spermatids. In addition, we demonstrate that tissue tropisms of symbionts vary between species and sexes. Our findings provide new insights into the tissue localization of symbionts in native Chinese ticks and pave the way for further understanding of their functional capabilities and symbiotic interactions with ticks.
KEYWORDS: tissue localization, ticks, Coxiella, Rickettsia, microbiota
INTRODUCTION
Ticks are obligate bloodsucking invertebrates transmitting a variety of diseases all over the world. WHO estimates that 532,125 cases of Lyme disease in the United States and Western Europe and 10,000 to 12,000 cases of tick-borne encephalitis occurred globally in 2017 (1). In China alone, 33 tick-borne pathogens have been identified since 1982. This number, added to that of traditional tick-borne diseases, indicates that tick-borne diseases have emerged as a public health concern (2, 3). Human cases of Lyme borreliosis have been confirmed in almost all provinces except Tibet and Shanghai (2). Patients from 8 provinces have been diagnosed with babesiosis, with Babesia microti, Babesia divergens, and Babesia venatorum as the major disease-causing agents (2, 4). Severe fever with thrombocytopenia syndrome virus (SFTSV) is an emerging pathogen that has infected 2,543 humans, causing 154 deaths in China in 2013 (2). However, tick-borne diseases are still underestimated because of the complex distribution and the lack of effective diagnostic methods in China.
There are a total of 117 species of ticks in China, divided into 2 families, the Argasidae and the Ixodidae (5). Thirteen species belong to Argasidae, of which 4 species are able to transmit pathogens causing human illness (6). A total of 104 species in Ixodidae have been found in China, of which 32 species are confirmed to be disease-transmitting vectors (6). Of these 32 species, Haemaphysalis longicornis and Ixodes persulcatus are considered to be the most competent vectors, as they transmit at least 15 pathogens, including Borrelia burgdorferi, Theileria spp., Coxiella burnetii, Babesia spp., Anaplasma phagocytophilum, Ehrlichia, Bartonella, spotted-fever group rickettsiae (SFGR), Huaiyangshan virus, and the recently identified New bunyavirus, among others. The tick Dermacentor silvarum is another major vector associated with 13 emerging pathogens, including SFGR, Anaplasmataceae, Borrelia spp., and Babesia spp. (2).
In addition to pathogens, ticks also harbor microbiota that are critical for their physiology (7, 8). Coxiella, as an obligate symbiont, is present in at least two-thirds of tick species in the world (8, 9). This symbiont preferentially colonizes the ovaries and Malpighian tubules, suggesting its role in fecundity, metabolism, and osmoregulation (10–14). Its genome encodes almost complete pathways for the biosynthesis of major B vitamins and cofactors that are essential in tick physiology (15). Elimination of Coxiella by antibiotic treatment reduces the fecundity of Amblyomma americanum and H. longicornis and prevents Rhipicephalus microplus from developing into its adult form (16–18). Rickettsia is another obligate tick symbiont found in multiple tick species (8, 13, 14, 19). Its genome contains all the genes for folate de novo biosynthesis, suggesting its role in nutrition provision to ticks (20). Ticks also harbor other maternally transmitted symbionts, including Francisella, Arsenophonus, Rickettsiella, Cardinium, Spiroplasma, Lariskella, Midichloria, and Wolbachia, although their functions need to be further investigated (8). In addition to symbionts, other microbiota are present across several hard tick species, with 5 bacterial genera frequently observed. They are Pseudomonas, Sphingobacterium, Acinetobacter, Enterobacter, and Stenotrophomonas (7). The presence of these microbiota is essential in determining the infection outcome of different tick species (21). Increasing the quantities of the endosymbiont Rickettsia in the tick Dermacentor andersoni results in the reduction of Anaplasma marginale infection, while decreasing the quantities of Francisella leads to a reduction in infections by the pathogen Francisella novicida (22). Disturbing the homeostasis of gut microbiota in the tick Ixodes scapularis by antibiotic treatment reduces colonization by Borrelia burgdorferi (23). The presence of rickettsial endosymbionts protects I. scapularis from B. burgdorferi infection (24). Native ticks in China also harbor a variety of microbiota. Coxiella and Rickettsia are present in H. longicornis (Hebei), D. silvarum (Hebei), and R. microplus (13, 14, 25). Arsenophonus has been detected in H. longicornis (Hebei) and D. silvarum (Hebei) (13). In I. persulcatus, the genera Acinetobacter, Rickettsia, Pseudomonas, Chryseobacterium, and Sphingobacterium are the most abundant bacteria in unfed ticks (26). Studies of native Chinese ticks focus mainly on the detection of microbes using PCR and 16S rRNA sequencing techniques, while further information about their tissue distribution is limited.
In this study, we have investigated the localization of microbiota in 3 native Chinese ticks, H. longicornis, R. haemaphysaloides, and D. silvarum, using fluorescent in situ hybridization (FISH), and examined the microbial population structure of field-collected D. silvarum (from Inner Mongolia). We found that midgut, reproductive tissues, and Malpighian tubules are the three primary tissues in which microbes reside. Field-collected D. silvarum ticks have a complex microbiota, with Pseudomonas, Coxiella, and Rickettsia as the dominant genera.
RESULTS
Localization of microbiota in H. longicornis and R. haemaphysaloides.
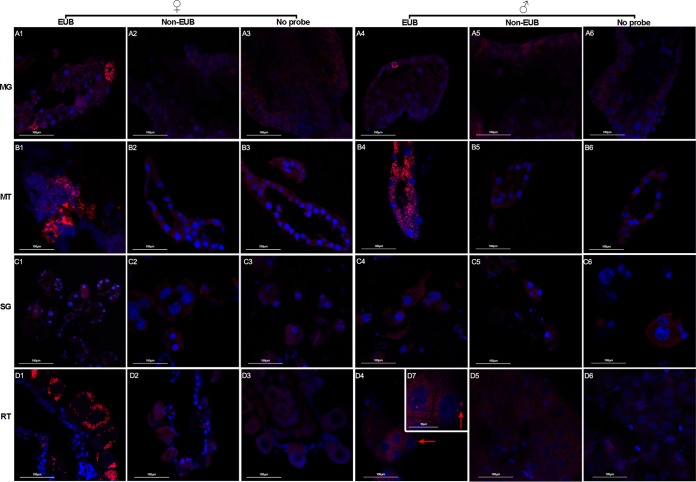
We first examined the tissue localization of microbiota in two tick species, H. longicornis and R. haemaphysaloides, originally from Yunnan Province, China, that had been maintained in the insectary for at least 2 years. To visualize the distribution of microbes in tissues of H. longicornis, whole bodies as well as dissected organs of female ticks were analyzed by FISH using a fluorescently labeled universal 16S rRNA probe (Fig. 1A to C). Intense staining was observed in Malpighian tubules and ovaries of whole-body sections (Fig. 1A to C). To further confirm the spatial distribution in the two organs, tissues, including midgut, salivary glands, Malpighian tubules, and ovaries were dissected 48 h following a blood meal and analyzed in the same manner as the whole-body sections. Similarly, strong fluorescent signals were detected in Malpighian tubules and ovaries (Fig. 1E1 and G1). No detectable signal was observed in midgut or salivary glands (Fig. 1D1 and F1). No signal was detected in controls with noneubacterial probe or without probe (Fig. 1D2 to G2 and D3 to G3). There was a high density of symbionts in the cytosol of Malpighian tubule cells but not in the tubule lumen (Fig. 1E1). Asynchronously developed oocytes were observed in the ovaries (Fig. 1G1). A large number of egg chambers at different developmental stages were attached to the ovarian wall. Symbionts were clustered around the nuclei in young oocytes but were concentrated on one side of the oocytes once matured (Fig. 1G1). This staining pattern indicates that Malpighian tubules and ovaries are the two major tissues harboring microbiota in female H. longicornis.
FIG 1.
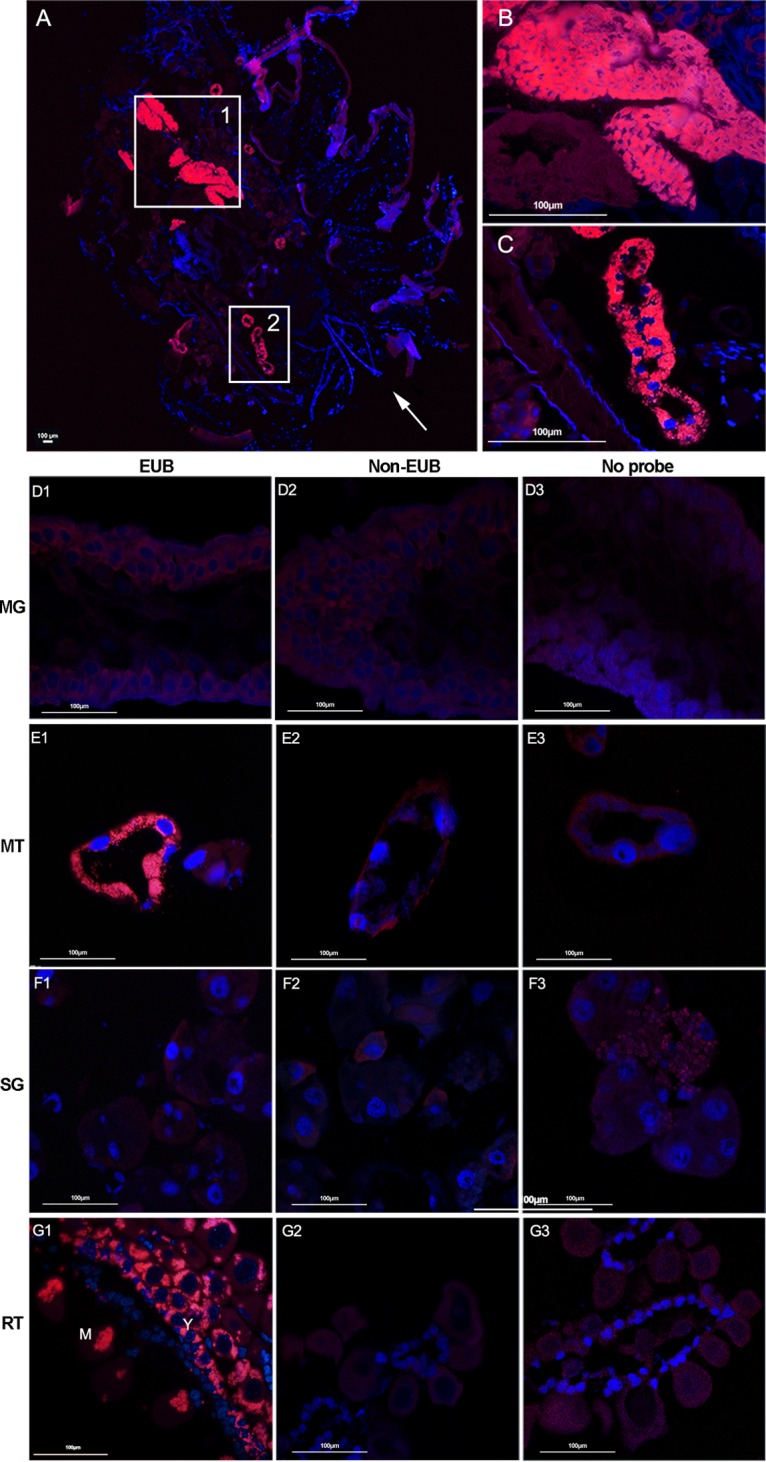
Localization of microbiota in whole body (A to C) and different tissues (D to G) of female H. longicornis by FISH analysis with universal 16S rRNA probe (red). Nuclei were stained with DAPI (blue). (A) Signals detected in whole body sections; the arrow denotes the direction from head to bottom; (B) close-up view of box 1 showing the ovary filled with microbiota; (C) close-up view of box 2 showing that the microbiota colonized the Malpighian tubules. Images are representative of at least 5 individual tick sections. Different tissues, including midgut (D), Malpighian tubules (E), salivary gland (F), and ovaries (G) were hybridized with universal 16S rRNA probe (EUB) (D1, E1, F1, and G1), noneubacterial probe (Non-EUB) (D2, E2, F2, and G2), and no probe (D3, E3, F3, and G3). MG, midgut; MT, Malpighian tubules; SG, salivary glands; RT, ovaries; Y, young oocytes; M, mature oocytes. Tissues from at least 5 individual ticks were pooled and used for FISH analysis. Images are representative of three independent experiments.
The tissue distribution of microbiota in female R. haemaphysaloides ticks was also investigated in the same way as that described above. We again detected intense fluorescent signals in Malpighian tubules and ovaries, with the same subcellular localization pattern as in H. longicornis (Fig. 2B1 and D1). In addition, we observed that midgut was the third organ that harbors microbiota (Fig. 2A1). Here the microbes clustered within a few midgut cells, leaving the midgut lumen free of bacteria. Again, no bacteria were detected in salivary glands (Fig. 2C1). Our results indicate that the R. haemaphysaloides microbiota shows a broader tissue tropism than does that of H. longicornis.
FIG 2.
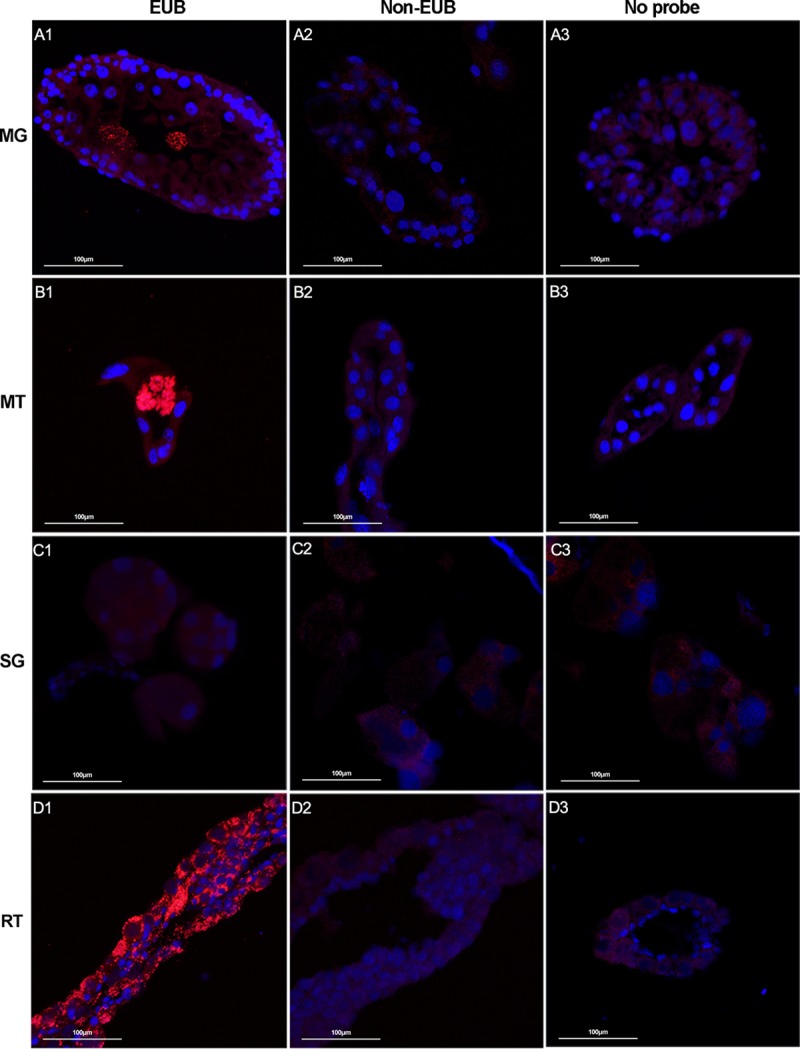
Localization of microbiota in different tissues of female R. haemaphysaloides by FISH analysis with universal 16S rRNA probe (red). Nuclei were stained with DAPI (blue). Fluorescent signals were examined in midgut (A), Malpighian tubules (B), salivary glands (C), and ovaries (D). Hybridizations with noneubacterial probe (Non-EUB) and without probe were used as negative controls. Tissues from at least 5 individual ticks were pooled and used for FISH analysis. Images are representative of three independent experiments.
Localization of Coxiella sp. and Rickettsia sp. symbionts in H. longicornis and R. haemaphysaloides.
Coxiella sp. and Rickettsia sp. (species were not identified here; see Materials and Methods) are two of the main symbionts present in multiple tick species in China, including Haemaphysalis tibetensis, H. longicornis, D. silvarum, and R. microplus (13, 14, 25, 27, 28). To examine the presence and abundance of these species in lab-reared H. longicornis and R. haemaphysaloides, we quantified their relative abundance by real-time PCR using genus-specific primers (Fig. 3). Coxiella sp. was the dominant symbiont in both H. longicornis and R. haemaphysaloides. Rickettsia sp. was barely detected in H. longicornis, but it was present in both male and female R. haemaphysaloides ticks, although its relative abundance was significantly lower than that of Coxiella sp. (Fig. 3A and B).
FIG 3.
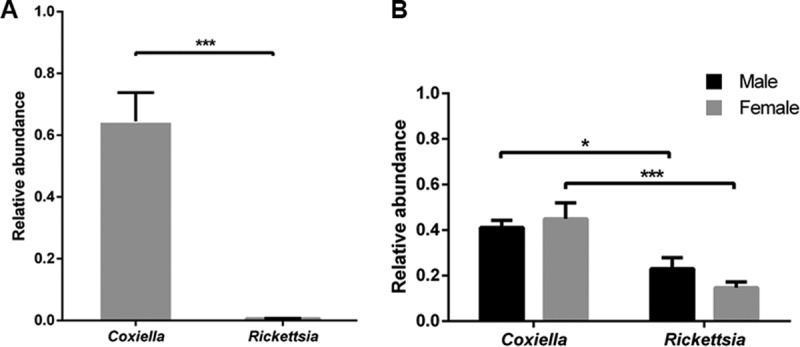
Relative abundance of Coxiella sp. and Rickettsia sp. in H. longicornis (A) and R. haemaphysaloides (B) determined by real-time quantitative PCR. Error bars indicate standard errors (n = 15). Significance was determined by Mann-Whitney test. *, P < 0.05; ***, P < 0.001.
We next examined the tissue distribution of these two symbionts in H. longicornis and R. haemaphysaloides colonies. As no Rickettsia sp. was detected in H. longicornis, sections of the same four tissues were hybridized only with a Coxiella-specific 23S rRNA probe (Fig. 4A1 to A4). We again found the same localization pattern as the one revealed by the universal 16S rRNA probe. Strong signals were detected exclusively in Malpighian tubules and ovaries (Fig. 4A2 and A4).
FIG 4.
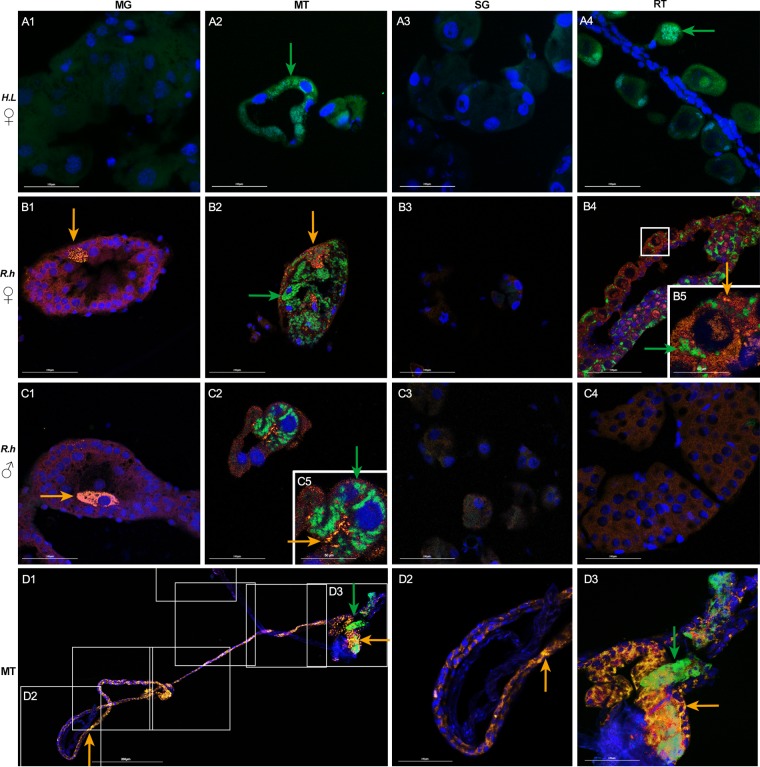
Localization of Coxiella sp. (green) and Rickettsia sp. (yellow) in H. longicornis (A) and R. haemaphysaloides (B to D) using species specific probes. Nuclei were stained with DAPI (blue). Merged images of DAPI and Coxiella sp. staining in female H. longicornis (A1 to A4). Merged images of DAPI, Coxiella sp., and Rickettsia sp. staining in female (B1 to B4) and male (C1 to C4) R. haemaphysaloides. Spliced view of whole-mount in situ hybridization in female Malpighian tubules of R. haemaphysaloides (D1 to D3). (A1, B1, C1), midgut (MG); (A2, B2, C2), Malpighian tubules (MT); (A3, B3, C3), salivary glands (SG); (A4), ovaries of H. longicornis (reproductive tissues [RT]); (B4), ovaries of R. haemaphysaloides; (C4), testes of R. haemaphysaloides; (B5) close-up view of boxed region, showing colocalization of Rickettsia sp. and Coxiella sp. in ovaries; (C5) close-up view of panel C2, colocalization of Rickettsia sp. and Coxiella sp. in Malpighian tubules. (D2, D3) Close-up view of boxed regions in D1. Green arrows denote Coxiella sp. and yellow arrows denote Rickettsia sp. Tissues from at least 5 individual ticks were pooled and used for FISH analysis. Each image shows a single focal plane. Images are representative of three independent experiments. Bars, 100 μm.
As the two symbionts were detected in R. haemaphysaloides, costaining of Coxiella sp. and Rickettsia sp. was performed in both female and males (Fig. 4B1 to C4). Again, intense signals of Coxiella sp. were observed in ovaries (Fig. 4B4 and B5) of female ticks and in Malpighian tubules of both male and female ticks (Fig. 4B2, C2, and C5). Rickettsia sp. was extensively distributed in midgut, Malpighian tubules, and ovaries (Fig. 4B1, C1, B2, C2, B4, and B5). The midgut was colonized by Rickettsia sp. exclusively. This bacterium clustered intracellularly in certain gut cells, leaving major parts of gut free of bacteria (Fig. 4B1 and C1). Rickettsia sp. in Malpighian tubule and ovary displayed a subcellular localization pattern similar to that of Coxiella sp., as the two bacteria reside intracellularly in Malpighian tubule cells, in interstitial cells between primary oocytes, and in cytoplasm of developing oocytes. As fluorescent signals of Coxiella sp. were detected only in a few sections of female Malpighian tubules while Rickettsia sp. was easily observed, we next analyzed the localization of the two symbionts in Malpighian tubules using whole-mount staining. We found that Coxiella sp. colonized only the distal region of Malpighian tubules, while Rickettsia sp. was widely spread all over the tissue (Fig. 4D1 to D3). Colocalization of these two symbionts is observed in ovaries and Malpighian tubules, indicating that a mutually beneficial relationship possibly exists between these two microbes (Fig. 4B2, B4, and C2). Species-specific labeling techniques confirm the different tissue tropism patterns of Coxiella sp. and Rickettsia sp. and the colocalization of these two symbionts in the same tissue.
Population structure of microbiota in D. silvarum.
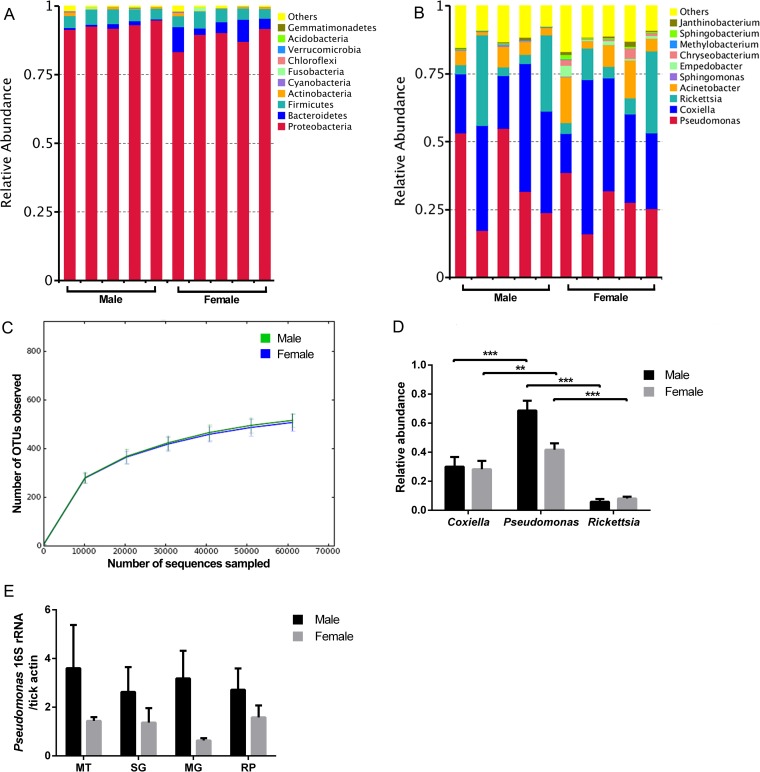
To understand the diversity of microbiota in field ticks, D. silvarum ticks, an important disease vector in the northern part of China, were collected from Inner Mongolia in March 2017. The population structure of microbiota in unfed male and female D. silvarum was analyzed using high-throughput sequencing of the 16S rRNA gene (V3-V4 region). The phylum Proteobacteria was dominant in this species, with relative abundance ranging from 83.3% to 97.5% (Fig. 5A). Ten bacterial genera, including Coxiella, Pseudomonas, Rickettsia, Acinetobacter, Sphingomonas, Empedobacter, Chryseobacterium, Methylobacterium, Sphingobacterium, and Janthinobacterium, were identified in all experimental groups but at different relative levels of abundance (Fig. 5B). Pseudomonas, Coxiella, and Rickettsia were the most abundant genera in D. silvarum. The rarefaction curves of operational taxonomic unit (OTU) numbers performed with male and female ticks confirmed that sequencing coverage was sufficiently extensive to fully evaluate the microbially diverse populations (Fig. 5C). Female and male ticks share most taxa. To examined if there was a difference in population structure of microbiota between male and female D. silvarum ticks, alpha diversity was calculated. There was no significant difference between male and female ticks as depicted by the Observed-species, Chao1, Shannon, and Simpson indices (Table 1). To further confirm the relative abundance of Pseudomonas, Coxiella, and Rickettsia estimated by 16S rRNA deep sequencing, real-time quantitative PCR (qPCR) was performed using universal 16S rRNA primers and genus-specific primers. In agreement with sequencing data, Pseudomonas sp., Coxiella sp., and Rickettsia sp. abundantly colonized D. silvarum, with relative abundances similar to those determined by 16S rRNA deep sequencing (Fig. 5D). To further confirm that Pseudomonas sp. resided within tick organs instead of being merely a cuticle contaminant, different tissues, including Malpighian tubules, salivary glands, midgut, and reproductive organs (ovaries/testes) were used to determine the presence and abundance of this bacterium (Fig. 5E). It was detected in all four tissues, with higher abundance in males than females. Our data show that Coxiella sp. and Rickettsia sp. are the main symbionts in D. silvarum and that Pseudomonas sp. is another dominant commensal with broad tissue distribution. There is no significant difference of microbiota structure between males and females.
FIG 5.
Population community of D. silvarum by 16S rRNA pyrosequencing. Phylum level (A) and genus level (B) abundance profiles for individual D. silvarum males and females prior to a blood meal. Each column represents one tick. (C) Rarefaction curve of the 16S rRNA gene sequences in male and female D. silvarum ticks based on OTUs determined at 97% similarity. (D) Relative abundance of Coxiella sp., Pseudomonas sp., and Rickettsia sp. in D. silvarum analyzed by qPCR. (E) Quantification of Pseudomonas sp. in different tick tissues by qPCR. Tissues from five individual tick were pooled for one biological replicate. Five biological replicates were used for qPCR analysis. Error bars indicate standard errors (n = 5). Statistical significance was determined using the Mann-Whitney test. **, P < 0.01; ***, P < 0.001.
TABLE 1.
Alpha diversity analysis of bacterial population structure in D. silvarum
| Tick sex or P value | Value (mean ± SD) |
|||
|---|---|---|---|---|
| Observed-species index | Chao1 index | Shannon index | Simpson index | |
| Male ticks | 515.4 ± 40.7 | 585.1 ± 35.6 | 2.95 ± 0.33 | 0.71 ± 0.02 |
| Female ticks | 506.6 ± 48.8 | 563.7 ± 55.4 | 3.42 ± 0.66 | 0.76 ± 0.09 |
| P valuea | 1.0000 | 0.6586 | 0.1061 | 0.1136 |
No significant difference was found between male and female ticks for any index of diversity.
Localization of symbionts in D. silvarum.
We next analyzed the tissue distribution of microbiota in D. silvarum. As in R. haemaphysaloides, where they are widely spread all over the tissues, high densities of microbes were detected intracellularly in the midgut, Malpighian tubules, and ovaries (Fig. 6A1 and A4, B1 and B4, and D1). Interestingly, fluorescent signals were also observed in the nuclei of spermatids (Fig. 6D4 and D7), suggesting the presence of symbionts in male testes.
FIG 6.
Localization of microbiota in different tissues of female and male D. silvarum ticks by FISH analysis with universal 16S rRNA probe (red). Nuclei were stained with DAPI (blue). Fluorescent signals were examined in midgut (A), Malpighian tubules (B), salivary glands (C), ovaries (D1 to D3), and testes (D4 to D7). Hybridizations with noneubacterial probe (Non-EUB) and without probe were used as negative controls. Red arrows denote residential bacteria. MG, midgut; MT, Malpighian tubules; SG, salivary glands; RT, reproductive tissue. Tissues from at least 5 individual ticks were pooled and used for FISH analysis. Images are representative of three independent experiments.
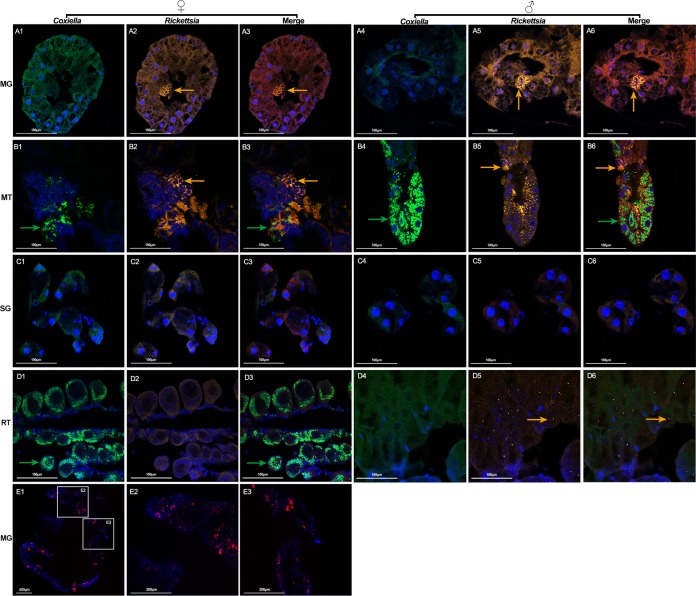
We next localized Coxiella sp. and Rickettsia sp. in the same four tissues, including midgut, Malpighian tubules, salivary glands, and ovaries/testes. In females, Coxiella sp. was detected in Malpighian tubules and ovaries (Fig. 7B1 and D1), while Rickettsia sp. was detected in midgut and Malpighian tubules (Fig. 7A2 and B2). In males, Malpighian tubules were the only tissues that were heavily infected with Coxiella sp. (Fig. 7B4). Rickettsia sp. was distributed in midgut, Malpighian tubules, and testes (Fig. 7A5, B5, and D5). It was interesting that each spermatid contained multiple Rickettsia sp. cells and that this bacterium preferentially localized in the nucleus of spermatids (Fig. 7D5). This is in agreement with results obtained by using a universal 16S rRNA probe, showing that bacteria colonized nuclei of spermatids. To further confirm the presence of Pseudomonas sp. in D. silvarum, we performed whole-mount in situ hybridization in midgut, which contained a relatively high abundance of Pseudomonas sp. (Fig. 7E1 to E3). Strong signals were observed in the midgut. Our results reveal that colocalization of Coxiella sp. and Rickettsia sp. is observed in Malpighian tubules in a manner similar to that seen in R. haemaphysaloides. Although female and male D. silvarum ticks share the same bacterial taxa, their localization in reproductive tissues is different.
FIG 7.
Costaining of Coxiella sp. (green), Rickettsia sp. (yellow), and Pseudomonas sp. (red) in D. silvarum. Nuclei were stained with DAPI (blue). Sections of midgut (A1 to A6), Malpighian tubules (B1 to B6), salivary glands (C1 to C6), ovaries (D1 to D3), and testes (D4 to D6) were hybridized with Coxiella sp.-specific 23S rRNA probe (A1 and A4, B1 and B4, C1 and C4, and D1 and D4) and Rickettsia sp.-specific 16S rRNA probe (A2 and A5, B2 and B5, C2 and C5, and D2 and D5) simultaneously. (A3 and A6, B3 and B6, C3 and C6, and D3 and D6) Merged images of DAPI, Coxiella sp., and Rickettsia sp. staining. Green arrows denote Coxiella sp. and yellow arrows denote Rickettsia sp. MG, midgut; MT, Malpighian tubules; SG, salivary glands; RT, reproductive tissue. (E1 to E3) Whole-mount in situ hybridization of D. silvarum midgut using Pseudomonas sp.-specific 16S rRNA probe. (E2, E3) Close-up view of boxed regions in panel E1. Tissues from at least 5 individual ticks were pooled and used for FISH analysis. Each image shows a single focal plane. Images are representative of three independent experiments.
In summary, our results from H. longicornis, R. haemaphysaloides, and D. silvarum suggest that Malpighian tubules and ovaries are two major organs harboring Coxiella sp. and Rickettsia sp. and that the midgut is colonized with Rickettsia sp. The tissue localization pattern of the same symbionts in these three tick species varies. Additionally, the two symbionts in the same tick species also display different tissue tropisms between sexes.
DISCUSSION
Emerging evidence shows that ticks harbor multiple symbionts that are important for their fecundity and vector competence (7, 8). However, information regarding tissue distribution and population diversity of the microbiota in most native ticks in China is far from complete. Here we visualized and compared the localization patterns of symbionts in three Chinese ticks, H. longicornis, R. haemaphysaloides, and D. silvarum, and analyzed the bacterial population structure in D. silvarum ticks collected from Inner Mongolia, China.
The presence of microorganisms in ticks was first described by Cowdry in 1925 (29). Multiple symbionts have been detected in different hard tick organs, with Coxiella, Rickettsia, Rickettsiella, Francisella, and Wolbachia as the most common ones that can be maternally transmitted (8, 21). In addition to symbionts, other microbiota belonging to the phyla Proteobacteria, Actinobacteria, Firmicutes, and Bacteroidetes have been identified in various tick species (7). In our study, Coxiella sp. and Rickettsia sp. are the two major symbionts in H. longicornis, R. haemaphysaloides, and D. silvarum. Malpighian tubules and ovaries are the two tissues that were massively colonized with symbionts in all three tick species. Malpighian tubules are responsible for the excretion of insoluble nitrogenous waste, the detoxification of metabolic wastes and xenobiotics in the hemolymph, and osmoregulation (30, 31). They are retained in both hemimetabolous and holometabolous arthropods throughout metamorphosis (32, 33). It is highly possible that Coxiella sp. and Rickettsia sp., by hiding in relatively stable Malpighian tubule cells, facilitate their transstadial transmission. Ovaries are the organs that are frequently colonized with symbionts, as the cytoplasm of the eggs provides ample space for symbionts to survive (34). The residence of Coxiella sp. or Rickettsia sp. in ovaries may facilitate its vertical transmission and ensure the fitness of the next generation. Interestingly, we also observed the intranuclear localization of Rickettsia sp. in spermatid of D. silvarum, suggesting that this bacterium may be paternally transmitted to the offspring during mating. Symbiont-like microorganisms are associated with the sperm cells and transmitted from male to female through copulation in Hyalomma marginatum, Hyalomma dromedarii, and Amblyomma hebraeum (35). In addition to ticks, Rickettsia sp. also infects sperm cells of multiple arthropods (36). It colonizes the spherical coenospermia of a kind of large bird spider (Pamphobeteus sp., Araneae) (36). As a facultative symbiont of a leafhopper, Nephotettix cincticeps, it also resides inside the nuclei of sperms without influencing male fecundity (37). Paternal transmission of beneficial symbionts is also observed in aphids (38). Regiella insecticola and Hamiltonella defensa are two male-borne symbionts that localize in testes and accessory glands in aphids. Both of them can be transferred to females during mating (38). Although it is still unclear how Rickettsia sp. infects the nucleus of a tick spermatid and what its influence on male fecundity is, the preferential colonization of this bacterium in D. silvarum spermatid nucleus indicates the distinct strategies that symbionts use to facilitate their vertical transmission. Such paternal transmission results in coinfections in the offspring that may have broad implications for symbiont-symbiont and symbiont-host interactions. The midgut is the third organ that is colonized with Rickettsia sp. in R. haemaphysaloides and D. silvarum. It is still unclear what type of midgut cells are colonized with this bacterium, and its influence on the hosts is not known. As the midgut is the principal digestive organ and the first tissue that encounters invading pathogens, this close association between Rickettsia sp. and midgut might be important for a tick's metabolism and vector competence.
The tissue localization pattern of symbionts differs in the three tick species. Coxiella sp. specifically colonizes Malpighian tubules and ovaries of all three tick species. This symbiont is widely distributed in multiple hard tick species collected from different countries, including Brazil, Colombia, Kenya, and China (8, 9, 39). In addition to colonizing Malpighian tubules and ovaries, it also infects the salivary glands of Amblyomma americanum and Amblyomma cajennense (11, 12). However, it is still unclear if and how the colonization of salivary gland in Coxiella sp. influences ticks. Rickettsia sp. is present in R. haemaphysaloides and D. silvarum but absent in H. longicornis. Rickettsia sp. resides preferentially in ovaries of R. haemaphysaloides but is absent in D. silvarum ovaries. Instead, it localizes in testes. The differential spatial localization patterns of these symbionts may result from various levels of interactions that these different ticks have with symbionts and from interactions between different symbionts during their long coevolution (8).
The population structure of native microbes also differs in the three tick species. H. longicornis in this study is colonized primarily with Coxiella sp., while the same tick species collected from Xiaowutai National Natural Reserve Area, in the northeast of China, harbors both Coxiella sp. and Rickettsia sp. (14). One possible reason is that H. longicornis in this study is parthenogenetic, with offspring developing from unfertilized eggs. The unique reproductive strategy that prevents the symbionts from paternally transmitting to offspring possibly leads to the absence of Rickettsia sp. in this tick strain (40). Both Coxiella sp. and Rickettsia sp. are detected in R. haemaphysaloides just as in Rhipicephalus microplus ticks collected in multiple places in China (25). Field-collected D. silvarum harbors a diverse microbiota, with Pseudomonas, Rickettsia, and Coxiella as the three dominant bacterial genera. The diversity and composition of tick microbiota are influenced by multiple factors, including environmental factors and host factors. The former factors include geographical location, habitat type, seasonal weather, and the vertebrates that they feed on. The latter factors include tick species, development stage, sex, and reproduction strategy (39). It is highly possible that the natural D. silvarum population is exposed to a more-complex environment and a greater variety of vertebrates during their development in the field.
Overall, our experiments demonstrate localization patterns of Coxiella sp. in three native tick species similar to those observed in other studies (8). We also show a specific intracellular localization pattern of Rickettsia sp. in midguts and spermatids, of which little is known in most of the tick species. Although emerging evidence shows that symbionts contribute to many aspects of host development, physiology, fecundity, and immunity, the molecular mechanisms through which the symbionts exert their beneficial influence are still largely undefined. Our work paves the way for future studies focusing on the interactions between coexisting symbionts and tripartite interaction between ticks, native microbes, and pathogens.
MATERIALS AND METHODS
Tick collecting and rearing.
Laboratory-reared H. longicornis (parthenogenesis strain) and R. haemaphysaloides ticks originally collected from Yunnan Province in 2015 and 2013, respectively, were maintained at the laboratory of National Institute of Parasitic Diseases in Shanghai as described previously (41). Adult D. silvarum ticks were collected from the Jiagedaqi forest, Inner Mongolia, in March 2017 and maintained in the same place as H. longicornis and R. haemaphysaloides. Tick species were identified by both morphological characterization and cytochrome c oxidase I (COI) genes as previously described (42). The work has been done according to guidelines for scientific ethical practices of the National Institute of Parasitic Diseases, Chinese Center for Disease Control and Prevention, based in Shanghai.
Tick dissection.
Ticks were dissected as previously described with some modifications (43). Briefly, ticks were surface sterilized by 75% ethanol twice, followed by 1× phosphate-buffered saline (PBS) twice. Forty-eight hours following blood feeding, individual ticks were placed dorsal side up on a drop of glue in the petri dish. Ticks were dissected in drops of 1× PBS. After removing upper cuticle, different tissues, including Malpighian tubules, midgut, ovaries/testes, and salivary glands, were collected and pooled for fixation.
Quantification of microbiota in ticks.
Ticks were surface sterilized by 75% ethanol twice, followed by 1× PBS twice. Total DNA from whole ticks or dissected tissues was extracted by the method of Holmes and Bonner as described previously (44). Fifteen individual unfed whole adults of each sample group were used for microbial quantification. Tissues from five individual adults 48 h following blood feeding were pooled for one biological replicate. Five biological replicates were used for qPCR analysis. Total bacterial density was quantified by universal 16S rRNA primers (45). The species of Coxiella and Rickettsia were unable to be identified using the primers in this study, so we refer to the two symbionts as Coxiella sp. and Rickettsia sp. Bacterial densities of Coxiella sp., Rickettsia sp., and Pseudomonas sp. were quantified using genus-specific primers (Table 2) (46–48). The copy number of each species and total microbiota was determined by standard curve. The relative abundance of each symbiont was normalized with total bacterial density. Quantitative real-time PCR was performed by the Roche LightCycler 96 real-time PCR detection system with the SYBR green qPCR master mix (Biomake, China) using the following conditions: 95°C for 5 min and 40 cycles of 30 s at 95°C, 30 s at 60°C, and 30 s at 72°C. Fluorescence readings were taken at 72°C after each cycle, followed by a melting curve (60°C to 95°C) to confirm the identity of the PCR product. Significance was determined using the Mann-Whitney test.
TABLE 2.
Primers used in this study
| Primer | Sequence | Gene | Organism(s) | Reference |
|---|---|---|---|---|
| RicCS-AF | 5′-GAGTGTAGTAGGGGATGATG-3′ | gltA | Rickettsia | 46 |
| RicCS-AR | 5′-CCACCATGTCAAGGGTTGGT-3′ | |||
| Cox sp434f | 5′-CCTTTTGAGCGTTGACGTTA-3′ | 16S rRNA | Coxiella | 47 |
| Cox sp1004r | 5′-CCAAAGGCACCAAGTCATTT-3′ | |||
| Pse435F | 5′-ACTTTAAGTTGGGAGGAAGGG-3′ | 16S rRNA | Pseudomonas | 48 |
| Pse686R | 5′-ACACAGGAAATTCCACCACCC-3′ | |||
| Eub27F | 5′-AGAGTTTGATCCTGGCTCAG-3′ | 16S rRNA | Eubacteria | 45 |
| Eub338R | 5′-CATGCTGCCTCCCGTAGGAGT-3′ | |||
| DsacF | 5′-TTCCAGCCCTCGTTCCTGGGTAT-3′ | actin | D. silvarum | 27 |
| DsacR | 5′-AATGATCTTGATCTTCATGGT-3′ |
Fluorescent in situ hybridization.
Samples, including whole tick bodies with their upper cuticles removed as well as dissected tissues, were fixed in 4% paraformaldehyde for 1 day. Sample preparation and FISH were performed as described previously (49). Five whole bodies of unfed H. longicornis and tissues from at least five partially engorged ticks of all three species were used for fluorescent in situ hybridization. Briefly, after fixation, samples were washed by 1× PBS 4 or 5 times for 2 to 3 h. After washing, samples were dehydrated in an ethanol series (70%, 95%, and 100%), followed by 100% butanol for 10 min, twice, and then stored in 100% butanol at 4°C for 1 day. Tissues were embedded in paraffin and sectioned at a thickness of 5 μm. Slides were hybridized with 10 ng/μl universal 16S rRNA probe (5′-GCTGCCTCCCGTAGGAGT-3′) labeled with Alexa Fluor 555 (Life Technology) (50). The probe for Coxiella sp. was Coxiella-specific 23S rRNA (5′-GACTTCCCACATCGTTT-3′) labeled with Alexa Fluor 488 (Life Technology) (10). The Rickettsia sp.-specific 16S rRNA probe, 5′-TCCACGTCACCGTCTTGC-3′, was labeled with Alexa Fluor 647 (Life Technology) (46). Costaining of Coxiella sp. and Rickettsia sp. was performed by hybridization with Coxiella-specific 23S rRNA and Rickettsia-specific 16S rRNA probes simultaneously. Noneubacterial probe (5′-CCGTCAATTCMTTTGAGTTT-3′) labeled with Alexa Fluor 555 (Life Technology) and no probe were used as negative controls (50). Whole tick bodies of H. longicornis hybridized with 10 ng/μl universal 16S rRNA probe were visualized using a Nikon Eclipse IVi microscope connected to a Nikon Digital Sight DS-U3 digital camera. Different tissues of all three species were visualized using a Zeiss LSM710 confocal microscope connected to a Nikon Digital Sight DS-U3 digital camera. One focal plane was taken of each image.
Whole-mount in situ hybridization was performed on dissected Malpighian tubules from female R. haemaphysaloides and midgut from male D. silvarum. Tissues were fixed in 4% paraformaldehyde for 1 day at 4°C. Sample preparation and hybridization were performed as described above with some modifications. Briefly, after fixation, samples were washed by 1× PBS 4 times for 20 min. Tissues were dehydrated in an ethanol series (70%, 95%, and 100%). Malpighian tubules were subjected to hybridization with 10 ng/μl Coxiella sp.- and Rickettsia sp.-specific probes simultaneously at 45°C for 6 h. Midgut was hybridized with 10 ng/μl Pseudomonas sp.-specific 16S rRNA probe, 5′-AATCCGACCTAGGCTCATC-3′, labeled with Alexa Fluor 555 (Life Technology) (51). Finally, tissues were mounted with 4,6-diamidino-2-phenylindole (DAPI; Invitrogen). Tissues were visualized using a Zeiss LSM710 confocal microscope connected to a Nikon Digital Sight DS-U3 digital camera. One focal plane was taken for each image.
Composition of microbiota in D. silvarum.
Field-collected D. silvarum ticks were surface sterilized with 70% ethanol twice and 1× PBS twice. Total DNA of 5 unfed individuals of each sex was extracted by the method of Holmes and Bonner as described previously (44). The composition of the microbiota of D. silvarum was analyzed using an Illumina HiSeq2500 platform in Novogene (Novogene, China) by primers targeting the V3-V4 region of bacterial 16S rRNA. No-template controls were included as controls. A total of 2,453,452 reads were detected. Raw sequence data were filtered according to the QIIME (V1.7.0,) quality-controlled process (52, 53). The clean tags were compared with the reference database (Gold database), and chimeric sequences were detected using the UCHIME algorithm and removed (54, 55). Operational taxonomic unit (OTU) analyses were performed by Uparse software (Uparse v7.0.1001) (56). Sequences with ≥97% similarity were assigned to the same OTUs. A total of 1,691 OTUs were obtained. For each representative sequence, the GreenGene Database 3 was used based on the RDP classifier (v2.2) algorithm to annotate taxonomic information (57, 58). Alpha diversity was applied in analyzing the complexity of species diversity for one sample through Observed-species, Chao1, Shannon, and Simpson indices. All these indices in our samples were calculated with QIIME (version 1.7.0) and displayed with R software (version 2.15.3). The Mann-Whitney test was used to assess significant differences.
Availability of data.
The raw 16S rRNA gene sequences have been uploaded to the National Center for Biotechnology Information's Sequence Read Archive (accession numbers SAMN07607842 to SAMN07607860).
ACKNOWLEDGMENTS
We thank Jinlin Zhou from Shanghai Veterinary Research Institute for help with tick dissections and Ulrike Munderloh from the University of Minnesota for help with tissue identification.
This research was supported by National Natural Science Foundation of China (31472039 and 81601793), National Institutes of Health Grant (R01AI129819), National Research and Development Plan of China (No. 2016YFC1200500), and the Research Fund of the State Key Laboratory of Genetic Engineering, Fudan University.
We declare that we have no conflict of interest.
REFERENCES
- 1.WHO. 2017. Global vector control response 2017–2030. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 2.Fang LQ, Liu K, Li XL, Liang S, Yang Y, Yao HW, Sun RX, Sun Y, Chen WJ, Zuo SQ, Ma MJ, Li H, Jiang JF, Liu W, Yang XF, Gray GC, Krause PJ, Cao WC. 2015. Emerging tick-borne infections in mainland China: an increasing public health threat. Lancet Infect Dis 15:1467–1479. doi: 10.1016/S1473-3099(15)00177-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu XB, Na RH, Wei SS, Zhu JS, Peng HJ. 2013. Distribution of tick-borne diseases in China. Parasit Vectors 6:119. doi: 10.1186/1756-3305-6-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou X, Xia S, Huang JL, Tambo E, Zhuge HX, Zhou XN. 2014. Human babesiosis, an emerging tick-borne disease in the People's Republic of China. Parasit Vectors 7:509. doi: 10.1186/s13071-014-0509-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen Z, Yang X, Bu F, Yang X, Yang X, Liu J. 2010. Ticks (Acari: Ixodoidea: Argasidae, Ixodidae) of China. Exp Appl Acarol 51:393–404. doi: 10.1007/s10493-010-9335-2. [DOI] [PubMed] [Google Scholar]
- 6.Yu Z, Wang H, Wang T, Sun W, Yang X, Liu J. 2015. Tick-borne pathogens and the vector potential of ticks in China. Parasit Vectors 8:24. doi: 10.1186/s13071-014-0628-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Narasimhan S, Fikrig E. 2015. Tick microbiome: the force within. Trends Parasitol 31:315–323. doi: 10.1016/j.pt.2015.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bonnet SI, Binetruy F, Hernandez-Jarguin AM, Duron O. 2017. The tick microbiome: why non-pathogenic microorganisms matter in tick biology and pathogen transmission. Front Cell Infect Microbiol 7:236. doi: 10.3389/fcimb.2017.00236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Machado-Ferreira E, Vizzoni VF, Balsemao-Pires E, Moerbeck L, Gazeta GS, Piesman J, Voloch CM, Soares CA. 2016. Coxiella symbionts are widespread into hard ticks. Parasitol Res 115:4691–4699. doi: 10.1007/s00436-016-5230-z. [DOI] [PubMed] [Google Scholar]
- 10.Lalzar I, Friedmann Y, Gottlieb Y. 2014. Tissue tropism and vertical transmission of Coxiella in Rhipicephalus sanguineus and Rhipicephalus turanicus ticks. Environ Microbiol 16:3657–3668. doi: 10.1111/1462-2920.12455. [DOI] [PubMed] [Google Scholar]
- 11.Klyachko O, Stein BD, Grindle N, Clay K, Fuqua C. 2007. Localization and visualization of a Coxiella-type symbiont within the lone star tick, Amblyomma americanum. Appl Environ Microbiol 73:6584–6594. doi: 10.1128/AEM.00537-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Machado-Ferreira E, Dietrich G, Hojgaard A, Levin M, Piesman J, Zeidner NS, Soares CA. 2011. Coxiella symbionts in the Cayenne tick Amblyomma cajennense. Microb Ecol 62:134–142. doi: 10.1007/s00248-011-9868-x. [DOI] [PubMed] [Google Scholar]
- 13.Liu L, Li L, Liu J, Hu Y, Liu Z, Guo L, Liu J. 2013. Coinfection of Dermacentor silvarum Olenev (Acari: Ixodidae) by Coxiella-like, Arsenophonus-like, and Rickettsia-like symbionts. Appl Environ Microbiol 79:2450–2454. doi: 10.1128/AEM.03575-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu LM, Liu JN, Liu Z, Yu ZJ, Xu SQ, Yang XH, Li T, Li SS, Guo LD, Liu JZ. 2013. Microbial communities and symbionts in the hard tick Haemaphysalis longicornis (Acari: Ixodidae) from north China. Parasit Vectors 6:310. doi: 10.1186/1756-3305-6-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith TA, Driscoll T, Gillespie JJ, Raghavan R. 2015. A Coxiella-like endosymbiont is a potential vitamin source for the Lone Star tick. Genome Biol Evol 7:831–838. doi: 10.1093/gbe/evv016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhong J, Jasinskas A, Barbour AG. 2007. Antibiotic treatment of the tick vector Amblyomma americanum reduced reproductive fitness. PLoS One 2:e405. doi: 10.1371/journal.pone.0000405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang CM, Li NX, Zhang TT, Qiu ZX, Li Y, Li LW, Liu JZ. 2017. Endosymbiont CLS-HI plays a role in reproduction and development of Haemaphysalis longicornis. Exp Appl Acarol 73:429–438. doi: 10.1007/s10493-017-0194-y. [DOI] [PubMed] [Google Scholar]
- 18.Guizzo MG, Parizi LF, Nunes RD, Schama R, Albano RM, Tirloni L, Oldiges DP, Vieira RP, Oliveira WHC, Leite MS, Gonzales SA, Farber M, Martins O, Vaz IDS Jr, Oliveira PL. 2017. A Coxiella mutualist symbiont is essential to the development of Rhipicephalus microplus. Sci Rep 7:17554. doi: 10.1038/s41598-017-17309-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Treuren W, Ponnusamy L, Brinkerhoff RJ, Gonzalez A, Parobek CM, Juliano JJ, Andreadis TG, Falco RC, Ziegler LB, Hathaway N, Keeler C, Emch M, Bailey JA, Roe RM, Apperson CS, Knight R, Meshnick SR. 2015. Variation in the microbiota of Ixodes ticks with regard to geography, species, and sex. Appl Environ Microbiol 81:6200–6209. doi: 10.1128/AEM.01562-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hunter DJ, Torkelson JL, Bodnar J, Mortazavi B, Laurent T, Deason J, Thephavongsa K, Zhong J. 2015. The Rickettsia endosymbiont of Ixodes pacificus contains all the genes of de novo folate biosynthesis. PLoS One 10:e0144552. doi: 10.1371/journal.pone.0144552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de la Fuente J, Antunes S, Bonnet S, Cabezas-Cruz A, Domingos AG, Estrada-Pena A, Johnson N, Kocan KM, Mansfield KL, Nijhof AM, Papa A, Rudenko N, Villar M, Alberdi P, Torina A, Ayllon N, Vancova M, Golovchenko M, Grubhoffer L, Caracappa S, Fooks AR, Gortazar C, Rego ROM. 2017. Tick-pathogen interactions and vector competence: identification of molecular drivers for tick-borne diseases. Front Cell Infect Microbiol 7:114. doi: 10.3389/fcimb.2017.00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gall CA, Reif KE, Scoles GA, Mason KL, Mousel M, Noh SM, Brayton KA. 2016. The bacterial microbiome of Dermacentor andersoni ticks influences pathogen susceptibility. ISME J 10:1846–1855. doi: 10.1038/ismej.2015.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Narasimhan S, Rajeevan N, Liu L, Zhao YO, Heisig J, Pan J, Eppler-Epstein R, Deponte K, Fish D, Fikrig E. 2014. Gut microbiota of the tick vector Ixodes scapularis modulate colonization of the Lyme disease spirochete. Cell Host Microbe 15:58–71. doi: 10.1016/j.chom.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Steiner FE, Pinger RR, Vann CN, Grindle N, Civitello D, Clay K, Fuqua C. 2008. Infection and co-infection rates of Anaplasma phagocytophilum variants, Babesia spp., Borrelia burgdorferi, and the rickettsial endosymbiont in Ixodes scapularis (Acari: Ixodidae) from sites in Indiana, Maine, Pennsylvania, and Wisconsin. J Med Entomol 45:289–297. [DOI] [PubMed] [Google Scholar]
- 25.Xu XL, Cheng TY, Yang H, Yan F. 2015. Identification of intestinal bacterial flora in Rhipicephalus microplus ticks by conventional methods and PCR-DGGE analysis. Exp Appl Acarol 66:257–268. doi: 10.1007/s10493-015-9896-1. [DOI] [PubMed] [Google Scholar]
- 26.Zhang XC, Yang ZN, Lu B, Ma XF, Zhang CX, Xu HJ. 2014. The composition and transmission of microbiome in hard tick, Ixodes persulcatus, during blood meal. Ticks Tick-Borne Dis 5:864–870. doi: 10.1016/j.ttbdis.2014.07.009. [DOI] [PubMed] [Google Scholar]
- 27.Liu L, Li L, Liu J, Yu Z, Yang X, Liu J. 2016. Population dynamics of multiple symbionts in the hard tick, Dermacentor silvarum Olenev (Acari: Ixodidae). Ticks Tick-Borne Dis 7:188–192. doi: 10.1016/j.ttbdis.2015.10.002. [DOI] [PubMed] [Google Scholar]
- 28.Wang R, Li N, Liu J, Li T, Liu M, Yu Z, Liu J. 2017. Symbiont dynamics of the Tibetan tick Haemaphysalis tibetensis (Acari: Ixodidae). Parasit Vectors 10:259. doi: 10.1186/s13071-017-2199-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cowdry EV. 1925. A group of microorganisms transmitted hereditarily in ticks and apparently unassociated with disease. J Exp Med 41:817–830. doi: 10.1084/jem.41.6.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hsu MH, Sauer JR. 1975. Ion and water-balance in feeding Lone Star tick. Comp Biochem Phys A 52:269–276. doi: 10.1016/S0300-9629(75)80085-5. [DOI] [PubMed] [Google Scholar]
- 31.Dow JA, Davies SA. 2006. The Malpighian tubule: rapid insights from post-genomic biology. J Insect Physiol 52:365–378. doi: 10.1016/j.jinsphys.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 32.Gautam NK, Verma P, Tapadia MG. 2017. Drosophila Malpighian tubules: a model for understanding kidney development, function, and disease. Results Probl Cell Differ 60:3–25. doi: 10.1007/978-3-319-51436-9_1. [DOI] [PubMed] [Google Scholar]
- 33.Faria VG, Sucena E. 2013. Wolbachia in the Malpighian tubules: evolutionary dead-end or adaptation? J Exp Zool B Mol Dev Evol 320:195–199. doi: 10.1002/jez.b.22498. [DOI] [PubMed] [Google Scholar]
- 34.Bright M, Bulgheresi S. 2010. A complex journey: transmission of microbial symbionts. Nat Rev Microbiol 8:218–230. doi: 10.1038/nrmicro2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.el Said A. 1992. Ultrastructure of symbiont-like microorganisms associated with the sperm of ixodid ticks. J Egypt Soc Parasitol 22:293–297. [PubMed] [Google Scholar]
- 36.Afzelius BA, Alberti G, Dallai R, Godula J, Witalinski W. 1989. Virus-infected and Rickettsia-infected sperm cells in arthropods. J Invertebrate Pathol 53:365–377. doi: 10.1016/0022-2011(89)90102-X. [DOI] [Google Scholar]
- 37.Watanabe K, Yukuhiro F, Matsuura Y, Fukatsu T, Noda H. 2014. Intrasperm vertical symbiont transmission. Proc Natl Acad Sci U S A 111:7433–7437. doi: 10.1073/pnas.1402476111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moran NA, Dunbar HE. 2006. Sexual acquisition of beneficial symbionts in aphids. Proc Natl Acad Sci U S A 103:12803–12806. doi: 10.1073/pnas.0605772103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Greay TL, Gofton AW, Paparini A, Ryan UM, Oskam CL, Irwin PJ. 2018. Recent insights into the tick microbiome gained through next-generation sequencing. Parasit Vectors 11:12. doi: 10.1186/s13071-017-2550-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Duron O, Binetruy F, Noel V, Cremaschi J, McCoy KD, Arnathau C, Plantard O, Goolsby J, Perez de Leon AA, Heylen DJA, Van Oosten AR, Gottlieb Y, Baneth G, Guglielmone AA, Estrada-Pena A, Opara MN, Zenner L, Vavre F, Chevillon C. 2017. Evolutionary changes in symbiont community structure in ticks. Mol Ecol 26:2905–2921. doi: 10.1111/mec.14094. [DOI] [PubMed] [Google Scholar]
- 41.Li LH, Zhu D, Zhang CC, Zhang Y, Zhou XN. 2016. Experimental transmission of Babesia microti by Rhipicephalus haemaphysaloides. Parasit Vectors 9:231. doi: 10.1186/s13071-016-1517-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chitimia L, Lin RQ, Cosoroaba I, Wu XY, Song HQ, Yuan ZG, Zhu XQ. 2010. Genetic characterization of ticks from southwestern Romania by sequences of mitochondrial cox1 and nad5 genes. Exp Appl Acarol 52:305–311. doi: 10.1007/s10493-010-9365-9. [DOI] [PubMed] [Google Scholar]
- 43.Edwards KGJ, Varela-Stokes A. 2009. Examination of the internal morphology of the Ixodid tick, Amblyomma maculatum Koch (Acari: Ixodidae); a “how-to” pictorial dissection guide. Midsouth Entomol 2:28–39. [Google Scholar]
- 44.Holmes DS, Bonner J. 1973. Preparation, molecular weight, base composition, and secondary structure of giant nuclear ribonucleic acid. Biochemistry 12:2330–2338. doi: 10.1021/bi00736a023. [DOI] [PubMed] [Google Scholar]
- 45.Armougom F, Raoult D. 2009. Exploring microbial diversity using 16S rRNA high-throughput methods. J Comput Sci Syst Biol 2:74–92. doi: 10.4172/jcsb.1000019. [DOI] [Google Scholar]
- 46.Sakurai M, Koga R, Tsuchida T, Meng XY, Fukatsu T. 2005. Rickettsia symbiont in the pea aphid Acyrthosiphon pisum: novel cellular tropism, effect on host fitness, and interaction with the essential symbiont Buchnera. Appl Environ Microbiol 71:4069–4075. doi: 10.1128/AEM.71.7.4069-4075.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lalzar I, Harrus S, Mumcuoglu KY, Gottlieb Y. 2012. Composition and seasonal variation of Rhipicephalus turanicus and Rhipicephalus sanguineus bacterial communities. Appl Environ Microbiol 78:4110–4116. doi: 10.1128/AEM.00323-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roosa S, Wauven CV, Billon G, Matthijs S, Wattiez R, Gillan DC. 2014. The Pseudomonas community in metal-contaminated sediments as revealed by quantitative PCR: a link with metal bioavailability. Res Microbiol 165:647–656. doi: 10.1016/j.resmic.2014.07.011. [DOI] [PubMed] [Google Scholar]
- 49.Attardo GM, Lohs C, Heddi A, Alam UH, Yildirim S, Aksoy S. 2008. Analysis of milk gland structure and function in Glossina morsitans: milk protein production, symbiont populations and fecundity. J Insect Physiol 54:1236–1242. doi: 10.1016/j.jinsphys.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Amann RI, Krumholz L, Stahl DA. 1990. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol 172:762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Watt M, Hugenholtz P, White R, Vinall K. 2006. Numbers and locations of native bacteria on field-grown wheat roots quantified by fluorescence in situ hybridization (FISH). Environ Microbiol 8:871–884. doi: 10.1111/j.1462-2920.2005.00973.x. [DOI] [PubMed] [Google Scholar]
- 52.Bokulich NA, Subramanian S, Faith JJ, Gevers D, Gordon JI, Knight R, Mills DA, Caporaso JG. 2013. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat Methods 10:57–59. doi: 10.1038/nmeth.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, Gormley N, Gilbert JA, Smith G, Knight R. 2012. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Haas BJ, Gevers D, Earl AM, Feldgarden M, Ward DV, Giannoukos G, Ciulla D, Tabbaa D, Highlander SK, Sodergren E, Methe B, DeSantis TZ, Human Microbiome C, Petrosino JF, Knight R, Birren BW. 2011. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res 21:494–504. doi: 10.1101/gr.112730.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Edgar RC. 2013. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 57.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]