ABSTRACT
Resveratrol is among the best-known secondary plant metabolites because of its antioxidant, anti-inflammatory, and anticancer properties. It also is an important allelopathic chemical widely credited with the protection of plants from pathogens. The ecological role of resveratrol in natural habitats is difficult to establish rigorously, because it does not seem to accumulate outside plant tissue. It is likely that bacterial degradation plays a key role in determining the persistence, and thus the ecological role, of resveratrol in soil. Here, we report the isolation of an Acinetobacter species that can use resveratrol as a sole carbon source from the rhizosphere of peanut plants. Both molecular and biochemical techniques indicate that the pathway starts with the conversion of resveratrol to 3,5-dihydroxybenzaldehyde and 4-hydroxybenzaldehyde. The aldehydes are oxidized to substituted benzoates that subsequently enter central metabolism. The gene that encodes the enzyme responsible for the oxidative cleavage of resveratrol was cloned and expressed in Escherichia coli to establish its function. Its physiological role in the resveratrol catabolic pathway was established by knockouts and by the reverse transcription-quantitative PCR (RT-qPCR) demonstration of expression during growth on resveratrol. The results establish the presence and capabilities of resveratrol-degrading bacteria in the rhizosphere of the peanut plants and set the stage for studies to evaluate the role of the bacteria in plant allelopathy.
IMPORTANCE In addition to its antioxidant properties, resveratrol is representative of a broad array of allelopathic chemicals produced by plants to inhibit competitors, herbivores, and pathogens. The bacterial degradation of such chemicals in the rhizosphere would reduce the effects of the chemicals. Therefore, it is important to understand the activity and ecological role of bacteria that biodegrade resveratrol near the plants that produce it. This study describes the isolation from the peanut rhizosphere of bacteria that can grow on resveratrol. The characterization of the initial steps in the biodegradation process sets the stage for the investigation of the evolution of the catabolic pathways responsible for the biodegradation of resveratrol and its homologs.
KEYWORDS: biodegradation, environmental microbiology, resveratrol, stilbene
INTRODUCTION
Plants produce chemicals implicated in myriad ecological functions, including allelopathy and cross talk between plant roots and microbial members of the rhizosphere (1, 2). Such chemicals can be produced in any parts of plants, including roots, where they can affect target species in the surrounding soil (3, 4). It is well established that the exudation of specific compounds by plants selects for specific microbial partners and thus regulates the adjacent soil microbial community (5, 6). This chemical cross talk between plant roots and rhizosphere bacteria can lead to beneficial associations that foster plant growth or associations that negatively impact plant health (3, 7, 8).
Stilbenes, products of the phenylpropanoid pathway and well-known allelopathic chemicals, have been extensively investigated because of their roles in plant disease resistance (9, 10). Resveratrol also has well-established pharmaceutically relevant properties, including antioxidant, anti-inflammatory, and anticancer effects (11–14). Like other stilbene compounds, resveratrol is considered an allelochemical due to its antimicrobial activity and roles in plant survival and competition (15). It is produced by a number of plants, including grapes (Vitis vinifera) and peanuts (Arachis hypogaea) (11, 16, 17) and is thought to protect the plants from agriculturally significant infection by fungi (18–20), although its role has been questioned (21). Resveratrol synthesis in peanuts, especially in peanut roots, is induced by biotic and abiotic factors (22, 23), including injury and attack by fungi. It can accumulate to high concentrations in peanut tissues (22–24), but little information is available on its persistence in soil. It is difficult to impossible to measure resveratrol concentrations and flux in situ in the rhizosphere, perhaps because of biodegradation by bacteria such as those reported here. In axenic hairy root cultures where biodegradation is precluded, 300 to 500 μg of resveratrol/gram dry weight (equivalent to 2 mM) can be produced (16).
The biotransformation of most stilbenes has been well documented, but the growth of bacteria on resveratrol has not been reported (25). Carotenoid cleavage oxygenase (CCO)-like enzymes from bacteria (26, 27) and fungi (28, 29) can catalyze the cleavage of resveratrol to 4-hydroxybenzaldehyde and 3,5-dihydroxybenzaldehyde without further degradation. It seems likely that resveratrol is biodegradable in soil, but the degradation pathways, the bacteria involved in the biodegradation, and the ecological role of bacteria in attenuating the allelopathic effects of resveratrol are unknown. Understanding the biodegradation of resveratrol by bacteria near the plants that produce it is essential to evaluate its behavior and ecological roles in the soil and the plant. Therefore, we isolated an Acinetobacter strain capable of growing on resveratrol from the rhizosphere of peanut plants, characterized the initial steps in the catabolic pathway, and identified the enzyme responsible for the initial reaction.
RESULTS
Isolation and growth of Acinetobacter oleivorans strain JS678.
The six strains reported here were isolated from the rhizosphere of peanut plants (see Table S1 in the supplemental material) by enrichment with resveratrol. In related experiments (Riqing Yu and J. C. Spain, unpublished), the most probable number estimates of resveratrol-degrading bacteria in rhizosphere soil from the same plot ranged from 3.6 × 105 to 2.4 × 106/gram of soil, which suggests strongly that growth on resveratrol was an important process in the rhizosphere.
JS678 was chosen for its rapid growth on resveratrol as the sole carbon source. The 16S rRNA gene of JS678 has 99% nucleotide identity to that of Acinetobacter oleivorans DR1, so the isolate was named Acinetobacter oleivorans strain JS678. In our hands, strain DR1 (obtained from ATCC) did not grow on resveratrol. The isolate grew on resveratrol with a yield coefficient of 0.226 ± 0.008 g of protein/g of resveratrol but did not grow with the related stilbenes, pterostilbene and arachidin-3. During the growth of strain JS678 on resveratrol, 4-hydroxybenzaldehyde and 3,5-dihydroxybenzaldehyde accumulated transiently in the culture medium (Fig. 1). The accumulation indicated that the two compounds are early intermediates in the catabolic pathway. Both compounds serve as growth substrates for strain JS678 (Fig. 2) as well as for all other isolates (data not shown), which suggests that all the isolates employ a pathway similar to that of JS678. The growth yield was 0.235 ± 0.003 g of protein/g of 4-hydroxybenzaldehyde and 0.201 ± 0.026 g of protein/g of 3,5-dihydroxybenzaldehyde for JS678.
FIG 1.
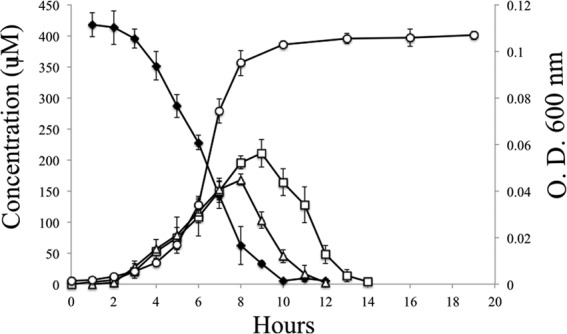
Growth of JS678 on resveratrol as the sole carbon source. ○, OD600; ◆, resveratrol; □, 3,5-dihydroxybenzaldehyde; △, 4-hydroxybenzaldehyde. Data represent the means and standard deviations from duplicate analyses.
FIG 2.
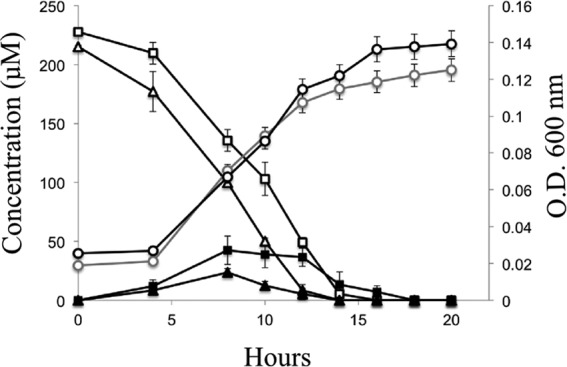
Transient accumulation of 4-hydroxybenzoate and 3,5-dihydroxybenzoate by JS678 growing on 4-hydroxybenzaldehyde or 3,5-dihydroxybenzaldehyde as the sole carbon source. □, 3,5-dihydroxybenzaldehyde; △, 4-hydroxybenzaldehyde; ■, 3,5-dihydroxybenzoate; ▲, 4-hydroxybenzoate; gray open circles, OD600 of cells grown with 3,5-dihydroxybenzaldehyde; ○, OD600 of cells grown with 4-hydroxybenzaldehyde. Data represent the means and standard deviations from duplicate analyses.
Identification of subsequent metabolites in the resveratrol degradation pathway.
When uninduced cells were provided with 4-hydroxybenzaldehyde or 3,5-dihydroxybenzaldehyde as their sole carbon source, 4-hydroxybenzoate or 3,5-dihydroxybenzoate accumulated transiently (Fig. 2). Thus, we hypothesized that the next step in the resveratrol degradation pathway is the conversion of the aldehydes to the corresponding acids, which also serve as growth substrates.
Respirometry with cells harvested during exponential growth on resveratrol or succinate (Table 1) indicated that resveratrol, 4-hydroxybenzaldehyde, 4-hydroxybenzoate, 3,4-dihydroxybenzoate, and 3,5-dihydroxybenzaldehyde stimulated the rapid uptake of oxygen with resveratrol-grown cells. The lack of stimulation in succinate-grown cells indicated that the enzymes involved in the resveratrol degradation pathway are inducible. The stoichiometry of oxygen utilization was consistent with the conversion of resveratrol to the corresponding aldehydes and acids in turn, since there is only one mole of oxygen difference in stoichiometry between the aldehydes and acids (two electrons made available for respiration) (Table 1). The low oxygen uptake with 3,5-dihydroxybenzoate suggested that there might be a transport barrier, since JS678 cells could grow on 3,5-dihydroxybenzoate. Additional experiments will be required to clarify the details of 3,5-dihydroxybenzaldehyde degradation.
TABLE 1.
Oxygen consumption by intact cells or dialyzed crude cell extracts of JS678
| Assay substrate(s)a | Oxygen consumption (nmol O2/min/mg protein [mol O2/mol of substrate])b |
|||
|---|---|---|---|---|
| Whole cells |
Dialyzed cell extract |
|||
| Succinate grown | Resveratrol grown | Succinate grown | Resveratrol grown | |
| Resveratrol | 13 ± 1.5 | 437 ± 4.5 (6.71 ± 0.22) | NDc | 620 ± 15 |
| 4-Hydroxybenzaldehyde | ND | 186 ± 1.5 (3.49 ± 0.09) | ND | ND |
| 3,5-Dihydroxybenzaldehyde | ND | 328 ± 0.2 (3.31 ± 0.27) | ND | ND |
| 4-Hydroxybenzoate | ND | 239 ± 3 (2.57 ± 0.34) | ND | ND |
| 3,5-Dihydroxybenzoate | ND | 6 ± 0.5 (2.51 ± 0.18) | 2 ± 0.1 | ND |
| 3,4-Dihydroxybenzoate | ND | 293 ± 12.1 (1.51 ± 0.06) | ND | 440 ± 15 |
| Gentisic acid | ND | ND | ND | ND |
| Hydroxyhydroquinone | ND | ND | ND | ND |
| Succinate | 161 ± 1 | 320 ± 1.5 | 140 ± 0.7 | ND |
| trans-Stilbene | ND | 3 ± 0.2 | ND | ND |
| Pterostilbene | ND | ND | ND | ND |
| Arachidin-3 | ND | ND | ND | ND |
| 4-Hydroxybenzaldehyde, NAD+ | NMd | NM | ND | ND |
| 3,5-Dihydroxybenzaldehyde, NAD+ | NM | NM | 1.8 ± 0.1 | 110 ± 10 |
| 4-Hydroxybenzoate, FAD, NADPH | NM | NM | ND | 110 ± 6 |
| 4-Hydroxybenzoate, NADPH | NM | NM | ND | ND |
| 3,5-Dihydroxybenzoate, NADPH | NM | NM | ND | ND |
| 3,5-Dihydroxybenzoate, NADH | NM | NM | ND | 127 ± 11 |
Substrates were added at a concentration of 100 μM, and two replicates were performed per experiment.
Data represent the means of duplicate analyses; the errors were calculated by taking the standard deviations between the data points.
ND, not detected.
NM, not measured.
Enzyme studies of resveratrol degradation in JS678.
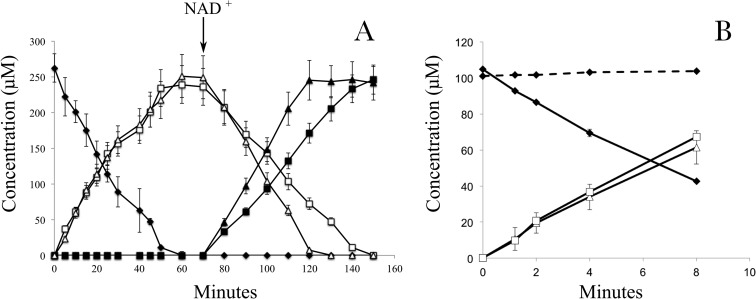
Enzyme assays were performed to determine the reactions and cofactors involved in the initial steps of resveratrol degradation. Dialyzed crude extracts prepared from resveratrol-grown cells of JS678 catalyzed the conversion of resveratrol to 4-hydroxybenzaldehyde and 3,5-dihydroxybenzaldehyde. 4-Hydroxybenzaldehyde was converted to 4-hydroxybenzoate and 3,5-dihydroxybenzaldehyde to 3,5-dihydroxybenzoate upon the addition of NAD+ (Fig. 3A). The rates of disappearance were 3.3 ± 0.4 μmol of resveratrol/min/mg of protein, 2.8 ± 0.2 μmol of 4-hydroxybenzaldehyde/min/mg of protein, and 1.6 ± 0.3 μmol of 3,5-dihydroxybenzaldehyde/min/mg of protein (Fig. 3A). Enzyme assays based on respirometry (Table 1) verified that oxygen is required for resveratrol cleavage. 4-Hydroxybenzoate catabolism required oxygen, flavin adenine dinucleotide (FAD; catalytic), and NADPH, which would be consistent with the properties of 4-hydroxybenzoate-3-monooxygenase (EC 1.14.13.33). No oxygen uptake was observed during the NAD+-dependent transformation of 4-hydroxybenzaldehyde, which is consistent with the characteristics of 4-hydroxybenzaldehyde dehydrogenases (EC 1.2.1.64). 3,5-Dihydroxybenzaldehyde and 3,5-dihydroxybenzoate stimulated oxygen uptake in the presence of NAD+ and NADH, respectively, but there was no oxygen utilization upon the addition of NADPH with 3,5-dihydroxybenzoate. 3,5-Dihydroxybenzoate concentrations did not decrease during the assays with NADH, which suggested that the oxygen utilization might be because of uncoupling (30); thus, the enzyme(s) that transforms 3,5-dihydroxybenzoate remains to be identified.
FIG 3.
Transformation of resveratrol by dialyzed crude extracts of JS678 1.8 ± 0.2 μg of protein/ml of reaction (A) or by extracts of an E. coli clone containing pJS701 (0.13 ± 0.05 μg of protein/ml of reaction) (B). The given compounds were analyzed by HPLC, and NAD+ (1 mM) was added at the indicated time. Dashed line represents results with dialyzed extracts of an E. coli clone containing only the pET15a vector. Data represent the means and standard deviations from duplicate analyses. ♦, resveratrol; □, 3,5-dihydroxybenzaldehyde; Δ, 4-hydroxybenzaldehyde; ■, 3,5-dihydroxybenzoate; ▲, 4-hydroxybenzoate.
Resveratrol cleavage oxygenase from JS678.
CCO homologs that can cleave stilbenes and stilbenoids are found in a variety of systems (26, 28, 31). The enzymes mediate the insertion of oxygen and the cleavage of stilbenes at the C-α=C-β double bond. Homologs that cleave metabolites from lignin degradation were previously named lignostilbene-α,β-dioxygenases (26). The above observation that an enzyme in cell extracts converts resveratrol to 4-hydroxybenzaldehyde and 3,5-dihydroxybenzaldehyde (Fig. 3A) is consistent with the activities of previously characterized homologs (26, 28, 31). Therefore, we screened the JS678 genome for putative CCO homologs. One gene encoded a protein with 43% amino acid identity to lignostilbene-α,β-dioxygenase from Sphingomonas paucimobilis TMY1009 (accession no. Q53353) (32). When expressed in Escherichia coli, the gene encoded an enzyme that catalyzed the cleavage of resveratrol, so it was designated resveratrol oxygenase (rzo, accession no. KY888940).
Resveratrol oxygenase was active in cells of resveratrol-grown JS678 (3.3 ± 0.4 μmol/min/mg of protein) and E. coli(pJS701) (61.7 ± 0.2 μmol/min/mg of protein), but activity was negligible in uninduced cells of JS678 or E. coli cells with pET15a containing no insert. The specific activity in JS678 was sufficient to account for the growth of the cells. The transformation of resveratrol by the heterologously expressed resveratrol oxygenase resulted in the stoichiometric accumulation of 4-hydroxybenzaldehyde and 3,5-dihydroxybenzaldehyde (Fig. 3B), indicating that the cloned enzyme catalyzed the same reaction as the inducible enzyme in extracts of JS678 (Fig. 3B). Knocking out the rzo gene resulted in a mutant, JS678-1 (Δrzo, Kmr), which was not capable of growth on resveratrol (Fig. 4). Additionally, reverse transcription-quantitative PCR (RT-qPCR) indicated an upregulation of rzo when JS678 was grown on resveratrol (Fig. 5). The above results established that resveratrol oxygenase is responsible for cleaving resveratrol in JS678.
FIG 4.
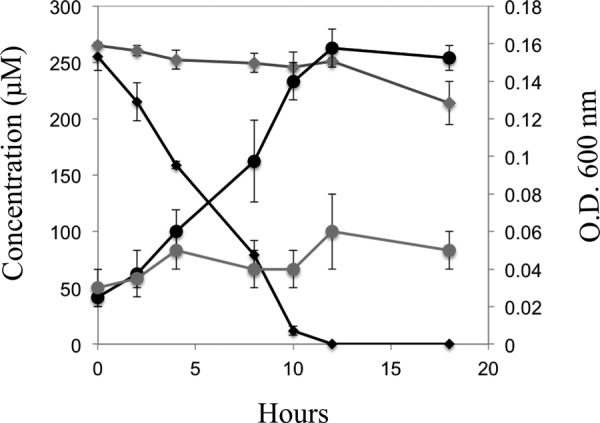
Transformation of resveratrol (diamonds) and cell growth (circles) by JS678 (black) and an rzo knockout clone, JS678-1 (Δrzo, Kmr) (gray). Data represent the means and standard deviations of duplicate analyses, and strains were grown with succinate prior to the analysis.
FIG 5.
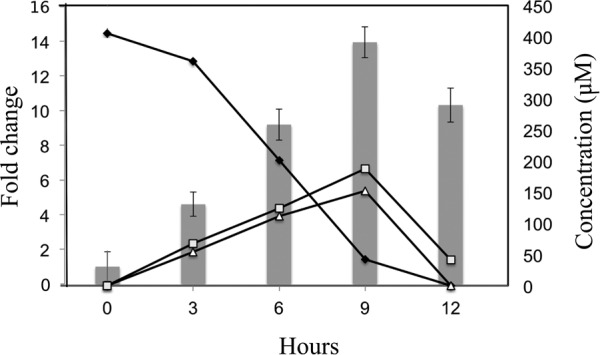
Expression of gene encoding resveratrol oxygenase determined by RT-qPCR. Cells of JS678 were grown on either resveratrol or succinate. The bars represent the fold upregulation in resveratrol-grown cells compared to succinate-grown cells. Data represent the means and standard deviations from triplicate analyses. ◆, resveratrol; □, 3,5-dihydroxybenzaldehyde; △, 4-hydroxybenzaldehyde.
A phylogenetic tree of the biochemically characterized carotenoid oxygenases indicated that the enzymes are phylogenetically diverse, and among the tested homologs, the ability to cleave resveratrol is widely distributed (see Fig. S1). A recent horizontal gene transfer between the fungi and the bacteria used for the analysis is not indicated by close relationships among the sequences. The carotenoid oxygenases that catalyze the cleavage of resveratrol do not form a distinct clade (Fig. S1), which indicates that identifying resveratrol oxygenases requires biochemical characterization. The enzyme from JS678 is the only one whose physiological role has been established rigorously. An investigation of additional resveratrol-degrading bacteria will be required to provide insight about the structure and function relationships among resveratrol oxygenase homologs.
DISCUSSION
Previous studies established the cleavage of stilbene derivatives by bacterial enzymes, e.g., lignostilbene by the lignostilbene-α,β-dioxygenase from Sphingomonas paucimobilis (31) and resveratrol by the enzymes Nov1 and Nov2 from Novosphingobium aromaticivorans DSM 12444 (26). In the latter case, the enzymes catalyze the cleavage of resveratrol at the C-α=C-β double bond, but the cells were not reported to grow on resveratrol. Peroxidase (33) and laccase (34) enzymes can also biotransform resveratrol in plants and fungi, respectively. The cleavage of resveratrol to 4-hydroxybenzaldehyde and 3,5-dihydroxybenzaldehyde by carotenoid oxygenase-like enzymes from fungi was also previously established (28). However, a recent study with Neurospora crassa showed that even though multiple carotenoid oxygenase homologs can be present in a genome, not all are involved in the cleavage of resveratrol (29).
The resveratrol oxygenase from JS678 seems to be relatively specific for resveratrol, because it is not active with resveratrol homologs produced by peanuts, including trans-stilbene, pterostilbene, and arachidin-3 (Table 1). The substrate specificity, inducibility of the activity, RT-PCR evidence for upregulation, heterologous expression, and knockout clone provide strong support for the physiological role of the enzyme in resveratrol degradation.
Resveratrol oxygenase belongs to a very large non-heme-iron-containing family of carotenoid cleavage oxygenases (35, 36). Previous alignments and crystal structure analyses of carotenoid oxygenases indicated that they have four conserved histidine residues consistent with a tetradentate iron coordination (35–38). All four histidine residues and three conserved glutamic acids responsible for iron binding are present in the sequence of Rzo (see Fig. S2 in the supplemental material). Further confirmation of this result was obtained using SWISS-MODEL (39) with the crystal structure of Nov1, a stilbene cleavage oxygenase from Novosphingobium aromaticivorans DSM 12444 (26, 38), as the template model. The predicted structure of Rzo is consistent with the requirement for one molecule of oxygen and one atom of iron (Fe3+) as ligands for the enzyme (see Fig. S3). In the present study with crude cell extracts, exogenously added iron (Fe3+) did not stimulate the enzyme activity, suggesting that iron was strongly bound to Rzo and was not lost during the handling or dialysis of the cell extracts.
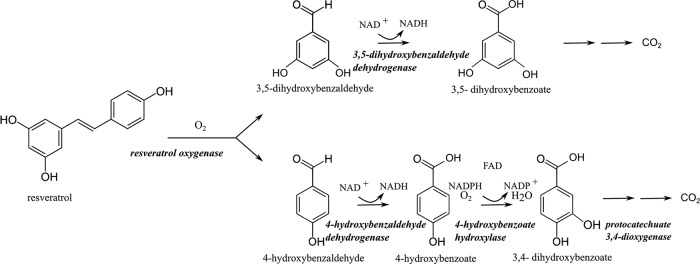
The above results indicate that resveratrol biodegradation in JS678 is initiated by resveratrol oxygenase (Fig. 6). The two products of the first reaction are 4-hydroxybenzaldehdye and 3,5-dihydroxybenzaldehyde, which is consistent with the previous understanding of CCO mechanisms (26, 27, 37). Both compounds accumulate but quickly degrade when the JS678 cells encounter resveratrol. The response should be similar to what would be expected in the rhizosphere, where the plants produce resveratrol in response to stress. 4-Hydroxybenzaldeyde is an intermediate of the p-cresol biodegradation pathway in bacteria, where it is oxidized to 4-hydroxybenzoate by an NAD+-dependent 4-hydroxybenzaldehyde dehydrogenase (encoded by pchA) (40–42). The subsequent reaction is catalyzed by 4-hydroxybenzoate hydroxylase (encoded by pobA), which uses NADPH and FAD as cofactors and produces 3,4-dihydroxybenzoate (protocatechuic acid) (43). Protocatechuate 3,4-dioxygenase (encoded by pcaG or pcaH) cleaves the ring of 3,4-dihydroxybenzoate to produce 3-carboxy-cis,cis-muconate (44, 45). In the draft genome of JS678, we were able to identify homologs of pchA, pobA, and pcaGH genes, but not genes encoding putative protocatechuate meta-cleavage enzymes such as protocatechuate 4,5- or 2,3-dioxygenases. The growth studies, respirometry, and enzyme assays performed in our study provided strong evidence for the operation of the 4-hydroxybenzaldeyde degradation pathway in JS678 (Fig. 6). Preliminary attempts to identify the genes encoding the enzymes of the downstream pathways in JS678 were not successful.
FIG 6.
Proposed resveratrol degradation pathway.
In the literature, the degradation of 3,5-dihydroxybenzaldehyde is less clear. In Streptomyces violaceoruber, the conversion of 3,5-dihydroxybenzaldehyde to the corresponding acid is catalyzed by a benzaldehyde dehydrogenase (encoded by ken6) as in the biosynthetic pathway of kendomycin (46). The JS678 genome contains a gene encoding an enzyme that has 39% amino acid identity to the enzyme encoded by ken6, but RT-qPCR experiments did not reveal an upregulation of the gene in resveratrol-grown cells (data not shown), and there was no indication that the activity was expressed constitutively. Thus, the gene that encodes 3,5-dihydroxybenzaldehyde dehydrogenase remains to be identified. It is clear that 3,5-dihydroxybenzoate is further degraded by intact cells (Fig. 2), but the mechanism is unknown. 3,5-Dihydroxybenzoate is metabolized to hydroxyhydroquinone under anoxic conditions in Thauera aromatica (47), but hydroxyhydroquinone seems not to stimulate oxygen uptake in JS678 cells (Table 1), suggesting that a novel pathway for 3,5-dihydroxybenzoate degradation is operating in JS678. The details of the 3,5-dihydroxybenzoate degradation pathway are under investigation.
The degradation of resveratrol in vivo could either have positive or negative effects on the plant fitness. Peanut plants produce many other stilbenes, including arachidin-1, arachidin-3, pterostilbene, and trans-stilbene (23, 48). The biological interactions among the plants, their fungal pathogens, the allelopathic chemicals, and the bacteria that degrade them are a mystery. It is also unclear why the peanut plants synthesize such a variety of chemicals. A plausible explanation is that the 3 entities are engaged in an evolutionary arms race, where plants are synthesizing novel derivatives of the allopathic chemicals, not only because fungi develop resistance to the chemicals, but also because bacteria evolve the ability to degrade them. Insight about the ecological relationships will require an investigation of in vivo interactions in the rhizosphere using molecular and bioinformatics approaches.
MATERIALS AND METHODS
Isolation and growth of bacteria.
Soil samples were collected from the rhizosphere of peanut plants (Arachis hypogea) from several locations around Dawson, GA. Samples were used as the inoculum in a selective enrichment prepared with Stanier's mineral salts basal medium (MSB) (49) containing resveratrol (200 μM) as the sole source of carbon. The concentrations of resveratrol were monitored by high-performance liquid chromatography (HPLC). After several additions of resveratrol, the cultures were diluted to extinction in microplates, and the strains were isolated on MSB agar plates with resveratrol (200 μM). The growth of JS678 was performed at the solubility limit of resveratrol in MSB (400 μM), and the yield experiments were performed with substrate concentrations of 400 μM.
Bacterial strains and plasmids.
Acinetobacter oleivorans strain JS678 was isolated in this study for its ability to use resveratrol as its sole carbon and energy source for growth. Escherichia coli DH5α (Gibco Life Technologies) was routinely used for plasmid propagation and cloning experiments. E. coli BL21(DE3) (Promega) was used as the host for the overexpression plasmid pET15a.
DNA techniques.
PCR, plasmid, and chromosomal DNA isolations, DNA fragment recovery, DNA ligations, transformations into E. coli, and restriction enzyme digestions were all carried out according to standard procedures (50) or by using specific recommendations by the suppliers of the molecular biology reagents (Qiagen GmbH, Promega, New England BioLabs, and Novagen). Genome sequencing of JS678 was performed by the Center for Genome Research and Biocomputing at Oregon State University. The genome was assembled using Velvet sequence assembler (51), and the assembled genes were curated by GeneMark (52) and annotated using UniProt. The cloned genes and 16S rRNA genes of isolates were sequenced by Genewiz, Inc. (Germantown, MD). Sequence databases were interrogated by using the Basic Local Alignment Search Tool (BLAST) program (53).
Enzyme assays.
JS678 was grown in MSB plus resveratrol (250 μM) with 2 to 4 repeated additions or with succinate (10 mM). Cells were harvested during exponential phase by centrifugation, washed twice in potassium phosphate buffer (pH 7.2, 20 mM), suspended in the same buffer, and passed twice through a French pressure cell (20,000 lb/in2). The cell lysates were clarified by centrifugation (20,000 × g at 4°C for 20 min) or by ultracentrifugation (160,000 × g at 4°C for 30 min). Protein concentrations were measured with a Pierce bicinchoninic acid (BCA) protein assay kit. The enzyme assays were carried out at 25°C in potassium phosphate buffer (pH 7.2, 20 mM) containing 1 to 2.5 mg of protein/ml and resveratrol, trans-stilbene, pterostilbene, arachidin-3, 3,5-dihydrohybenzaldehyde, or 4-hydroxybenzaldehyde (250 μM). Where indicated, NAD+ (250 μM), NADPH (250 μM), and FAD (50 μM) were added to the reaction mixtures. At appropriate intervals, samples were collected and trifluoroacetic acid (TFA) was added at a ratio of 1:100 to stop the reaction. The acidified reaction mixture was clarified by centrifugation, and the concentrations of substrates and products were determined by HPLC.
Cloning and expression of the rzo gene encoding resveratrol oxygenase.
The rzo gene was cloned and overexpressed by Molecular Cloning Laboratories (MCLAB) into a pET15a vector (Novagen, Gibbstown, NJ). The resulting construct was named pJS701 and transformed into E. coli BL21(DE3) [E. coli(pJS701)] for expression.
Cells of E. coli(pJS701) were grown in 250 ml of LB medium supplemented with ampicillin at 37°C. When the optical density at 600 nm (OD600) reached 0.5, isopropyl-β-d-1-thiogalactopyranoside (IPTG; 1.0 mM) was added, and the culture was incubated at room temperature for 4 h. The cells were harvested by centrifugation, washed twice with cold phosphate buffer (20 mM, pH 7.2), and stored on ice until used.
Construction of JS678 knockout mutants targeting rzo gene.
The rzo gene was interrupted by replacing nucleotide positions 69 to 1,144 with a kanamycin resistance gene. This was accomplished by first making a construct containing two portions of the rzo gene, corresponding to nucleotide positions 9 to 68 and 1,145 to 1,205, flanking the kanR gene (816 nucleotides) amplified from pET28a. Primers 5′-CTGGATCCATGAGTTTTACATTCCCCAATACTTCCGAGTTTACTGGACTTTACGAACCTTGCCGTATCATGAGCCATATTCAACGGGAAACG-3′ and 5′-CTGTCGACGTTTGGACATTAACACGAGCCAACTGATTAAAAAACTGGAAAGGATATTCTCCAAGATTT TTAGAAAAACTCATCGAGCATCAAATGAAACT-3′ were used to obtain the PCR product that was cut with BamHI and SalI and cloned into a pKNG101 suicide vector (54, 55) containing a streptomycin resistance marker. The resulting construct was transformed into E. coli SM10 λpir and then transferred to JS678 by mating. The mutants were grown with 5% sucrose to select for double crossovers and then screened via PCR using the primers 5′-CTGGATGCATGAGTTTTACATTCCCCAATACTTCCG-3′ and 5′-CTGTCGACTCATAATACTAGCAACCCAACCATCAC-3′. The mutation was confirmed by sequencing the PCR products, and the clone, designated Acinetobacter oleivorans JS678-1 (Δrzo, Kmr), was screened for its ability to grow on resveratrol under the conditions described above.
RT-qPCR.
RNA was extracted from resveratrol- and succinate-grown JS678 cells from triplicate biological samples using an EZNA total RNA kit II (Omega Bio-tek). RNA was converted to cDNA via an Applied Biosystems high-capacity RNA-to-cDNA kit. For qPCR, the manufacturer's protocol was used with an annealing temperature of 60°C, 2× Power SYBR green PCR master mix (Applied Biosystems, Carlsbad, CA), and an ABI 7500 fast real-time PCR system equipped with SDS v. 2.0.3 software. Primers were designed on the basis of the candidate genes obtained from the annotated genome in Table S2 in the supplemental material. The genes were not organized in a cluster in the genome. Succinate-grown cells were harvested and suspended in fresh succinate medium and in medium with resveratrol as the sole carbon source. At appropriate intervals, samples were analyzed for the expression levels of the candidate genes. The 16S rRNA expression levels were similar under all conditions and were used as an internal control. The upregulation of the gene encoding the putative resveratrol oxygenase was calculated by subtracting the expression levels in succinate-grown cells.
Analytical methods.
HPLC was performed on an Agilent 1100 system equipped with a diode array detector and a Merck C18 Chromolith column (100 mm by 4.6 mm). The mobile phase consisted of 98% part A (0.1% trifluoroacetic acid in water) and 2% part B (0.05% trifluoroacetic acid in acetonitrile) delivered at a flow rate of 1 ml/min for 1 min, and then the mobile phase composition was changed to 50% part A and 50% part B over 7 min at a flow rate of 1.5 ml/min. Compounds were identified and quantified by comparison with standards.
Respirometry.
Oxygen uptake was measured using a Clark-type electrode and a YSI model 3600 oxygen meter. JS678 cells were grown either with resveratrol or succinate, harvested by centrifugation, washed, and tested with the compounds of interest at a concentration of 100 μM. Measurements with dialyzed crude cell extracts were performed similarly.
Chemicals.
Resveratrol (≥99%) was from AK Scientific Inc., 4-hydroxybenzaldehyde (≥98%), 4-hydroxybenzoate (≥99%), trans-stilbene (96%), and pterostilbene (≥97%) were from Sigma-Aldrich, 3,5-dihydroxybenzaldehyde (≥98%) was from Acros Organics, and 3,5-dihydroxybenzoate (97%) was from Alfa Aesar. All chemicals were of the highest commercially available grade. Arachidin-3, extracted from peanut hairy root cultures (16), was provided by Fabricio Medina Bolivar from the University of Arkansas.
Supplementary Material
ACKNOWLEDGMENTS
We thank Renee Arias and Victor Sobolev from the National Peanut Research Laboratory in Dawson, Georgia, for providing soil samples and helpful discussions, Fabricio Medina Bolivar from the University of Arkansas for providing arachidin-3, Lars Jermiin at CSIRO Ecosystem Sciences (Canberra, Australia) for help with phylogeny, Sarah Craven who initiated the work, Shirley Nishino for technical assistance during the study, and Dan Shen at MCLAB and Janet Hatt and Lisa Waidner for helpful discussions.
This project was supported by Agriculture and Food Research Initiative competitive grant no. 2011-67019-30184 from the USDA National Institute of Food and Agriculture.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00104-18.
REFERENCES
- 1.Ehlers BK, Charpentier A, Grondahl E. 2014. An allelopathic plant facilitates species richness in the Mediterranean garrigue. J Ecol 102:176–185. doi: 10.1111/1365-2745.12171. [DOI] [Google Scholar]
- 2.Mishra S, Upadhyay RS, Nautiyal CS. 2013. Unravelling the beneficial role of microbial contributors in reducing the allelopathic effects of weeds. Appl Microbiol Biotechnol 97:5659–5668. doi: 10.1007/s00253-013-4885-y. [DOI] [PubMed] [Google Scholar]
- 3.Lorenzo P, Pereira CS, Rodriguez-Echeverria S. 2013. Differential impact on soil microbes of allelopathic compounds released by the invasive Acacia dealbata Link. Soil Biol Biochem 57:156–163. doi: 10.1016/j.soilbio.2012.08.018. [DOI] [Google Scholar]
- 4.Inderjit, Wardle DA, Karban R, Callaway RM. 2011. The ecosystem and evolutionary contexts of allelopathy. Trends Ecol Evol 26:655–662. doi: 10.1016/j.tree.2011.08.003. [DOI] [PubMed] [Google Scholar]
- 5.Lei N, Li J, Ni S, Chen J. 2014. Effects of clonal integration on microbial community composition and processes in the rhizosphere of the stoloniferous herb Glechoma longituba (Nakai) Kuprian. PLoS One 9:e108259. doi: 10.1371/journal.pone.0108259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang Q, Carrillo J, Jin H, Shang L, Hovick SM, Nijjer S, Gabler CA, Li B, Siemann E. 2013. Plant-soil biota interactions of an invasive species in its native and introduced ranges: implications for invasion success. Soil Biol Biochem 65:78–85. doi: 10.1016/j.soilbio.2013.05.004. [DOI] [Google Scholar]
- 7.Venturi V, Fuqua C. 2013. Chemical signaling between plants and plant pathogenic bacteria. Annu Rev Phytopathol 51:17–37. doi: 10.1146/annurev-phyto-082712-102239. [DOI] [PubMed] [Google Scholar]
- 8.Philippot L, Raaijmakers JM, Lemanceau P, van der Putten WH. 2013. Going back to the roots: the microbial ecology of the rhizosphere. Nat Rev Microbiol 11:789–799. doi: 10.1038/nrmicro3109. [DOI] [PubMed] [Google Scholar]
- 9.Lygin AV, Hill CB, Pawlowski M, Zernova OV, Widholm JM, Hartman GL, Lozovaya VV. 2014. Inhibitory effects of stilbenes on the growth of three soybean pathogens in culture. Phytopathology 104:843–850. doi: 10.1094/PHYTO-10-13-0287-R. [DOI] [PubMed] [Google Scholar]
- 10.Gindro K, Alonso-Villaverde V, Viret O, Spring J-L, Marti G, Wolfender J-L, Pezet R. 2012. Stilbenes: biomarkers of grapevine resistance to disease of high relevance for agronomy, oenology and human health, p 25–54. In Merillon JM, Ramawat KG (ed), Plant defence: biological control, vol 12 Springer, Geneva, Switzerland. [Google Scholar]
- 11.Sovak M. 2001. Grape extract, resveratrol, and its analogs: a review. J Med Food 4:93–105. doi: 10.1089/109662001300341752. [DOI] [PubMed] [Google Scholar]
- 12.Guerrero RF, Garcia-Parrilla MC, Puertas B, Cantos-Villar E. 2009. Wine, resveratrol and health: a review. Nat Prod Commun 4:635–658. [PubMed] [Google Scholar]
- 13.Halls C, Yu O. 2008. Potential for metabolic engineering of resveratrol biosynthesis. Trends Biotechnol 26:77–81. doi: 10.1016/j.tibtech.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 14.Wu JM, Wang ZR, Hsieh TC, Bruder JL, Zou JG, Huang YZ. 2001. Mechanism of cardioprotection by resveratrol, a phenolic antioxidant present in red wine (review). Int J Mol Med 8:3–17. [DOI] [PubMed] [Google Scholar]
- 15.Jeandet P, Clement C, Courot E, Cordelier S. 2013. Modulation of phytoalexin biosynthesis in engineered plants for disease resistance. Int J Mol Sci 14:14136–14170. doi: 10.3390/ijms140714136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abbott JA, Medina-Bolivar F, Martin EM, Engelberth AS, Villagarcia H, Clausen EC, Carrier DJ. 2010. Purification of resveratrol, arachidin-1, and arachidin-3 from hairy root cultures of peanut (Arachis hypogaea) and determination of their antioxidant activity and cytotoxicity. Biotechnol Prog 26:1344–1351. doi: 10.1002/btpr.454. [DOI] [PubMed] [Google Scholar]
- 17.Sanders TH, McMichael RW, Hendrix KW. 2000. Occurrence of resveratrol in edible peanuts. J Agric Food Chem 48:1243–1246. doi: 10.1021/jf990737b. [DOI] [PubMed] [Google Scholar]
- 18.Hasan MM, Cha M, Bajpai VK, Baek K-H. 2013. Production of a major stilbene phytoalexin, resveratrol in peanut (Arachis hypogaea) and peanut products: a mini review. Rev Environ Sci Biotechnol 12:209–221. doi: 10.1007/s11157-012-9294-7. [DOI] [Google Scholar]
- 19.Ribera AE, Zuniga G. 2012. Induced plant secondary metabolites for phytopatogenic fungi control: a review. J Soil Sci Plant Nutr 12:893–911. doi: 10.4067/S0718-95162012005000040. [DOI] [Google Scholar]
- 20.Hain R, Bieseler B, Kindl H, Schroder G, Stocker R. 1990. Expression of a stilbene synthase gene in Nicotiana tabacum results in synthesis of the phytoalexin resveratrol. Plant Mol Biol 15:325–335. doi: 10.1007/BF00036918. [DOI] [PubMed] [Google Scholar]
- 21.Sobolev VS, Khan SI, Tabanca N, Wedge DE, Manly SP, Cutler SJ, Coy MR, Becnel JJ, Neff SA, Gloer JB. 2011. Biological activity of peanut (Arachis hypogaea) phytoalexins and selected natural and synthetic stilbenoids. J Agric Food Chem 59:1673–1682. doi: 10.1021/jf104742n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chung IM, Park MR, Chun JC, Yun SJ. 2003. Resveratrol accumulation and resveratrol synthase gene expression in response to abiotic stresses and hormones in peanut plants. Plant Sci 164:103–109. doi: 10.1016/S0168-9452(02)00341-2. [DOI] [Google Scholar]
- 23.Sobolev VS, Neff SA, Gloer JB. 2010. New dimeric stilbenoids from fungal-challenged peanut (Arachis hypogaea) seeds. J Agric Food Chem 58:875–881. doi: 10.1021/jf903410e. [DOI] [PubMed] [Google Scholar]
- 24.Medina-Bolivar F, Condori J, Rimando AM, Hubstenberger J, Shelton K, O'Keefe SF, Bennett S, Dolan MC. 2007. Production and secretion of resveratrol in hairy root cultures of peanut. Phytochemistry 68:1992–2003. doi: 10.1016/j.phytochem.2007.04.039. [DOI] [PubMed] [Google Scholar]
- 25.Jilani G, Mahmood S, Chaudhry AN, Hassan I, Akram M. 2008. Allelochemicals: sources, toxicity and microbial transformation in soil—a review. Ann Microbiol 58:351–357. doi: 10.1007/BF03175528. [DOI] [Google Scholar]
- 26.Marasco EK, Schmidt-Dannert C. 2008. Identification of bacterial carotenoid cleavage dioxygenase homologues that cleave the interphenyl alpha,beta double bond of stilbene derivatives via a monooxygenase reaction. Chembiochem 9:1450–1461. doi: 10.1002/cbic.200700724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sui X, Golczak M, Zhang J, Kleinberg KA, Von Lintig J, Palczewski K, Kiser PD. 2015. Utilization of dioxygen by carotenoid cleavage oxygenase. J Biol Chem 290:30212–30223. doi: 10.1074/jbc.M115.696799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brefort T, Scherzinger D, Carmen Limon M, Estrada AF, Trautmann D, Mengel C, Avalos J, Al-Babili S. 2011. Cleavage of resveratrol in fungi: characterization of the enzyme Rco1 from Ustilago maydis. Fungal Genet Biol 48:132–143. doi: 10.1016/j.fgb.2010.10.009. [DOI] [PubMed] [Google Scholar]
- 29.Diaz-Sanchez V, Estrada AF, Carmen Limon M, Al-Babili S, Avalos J. 2013. The oxygenase CAO-1 of Neurospora crassa is a resveratrol cleavage enzyme. Eukaryot Cell 12:1305–1314. doi: 10.1128/EC.00084-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arunachalam U, Massey V, Vaidyanathan CS. 1992. p-Hydroxyphenylacetate-3-hydroxylase. A two protein component enzyme. J Biol Chem 267:25848–25855. [PubMed] [Google Scholar]
- 31.Kamoda S, Saburi Y. 1993. Structural and enzymatic comparison of lignostilbene-alpha, beta-dioxygenase isozymes, I, II, and III, from Pseudomonas paucimobilis TMY1009. Biosci Biotechnol Biochem 57:931–934. doi: 10.1271/bbb.57.931. [DOI] [PubMed] [Google Scholar]
- 32.Kamoda S, Terada T, Saburi Y. 2003. A common structure of substrate shared by lignostilbene dioxygenase isozymes from Sphingomonas paucimobilis TMY1009. Biosci Biotechnol Biochem 67:1394–1396. doi: 10.1271/bbb.67.1394. [DOI] [PubMed] [Google Scholar]
- 33.Calderon AA, Zapata JM, Barcelo AR. 1994. Peroxidase-mediated formation of resveratrol oxidation products during the hypersensitive-like reaction of grapevine cells to an elicitor from Trichoderma viride. Physiol Mol Plant Pathol 44:289–299. doi: 10.1016/S0885-5765(05)80031-1. [DOI] [Google Scholar]
- 34.Breuil AC, Jeandet P, Adrian M, Chopin F, Pirio N, Meunier P, Bessis R. 1999. Characterization of a pterostilbene dehydrodimer produced by laccase of Botrytis cinerea. Phytopathology 89:298–302. doi: 10.1094/PHYTO.1999.89.4.298. [DOI] [PubMed] [Google Scholar]
- 35.Sui X, Zhang J, Golczak M, Palczewski K, Kiser PD. 2016. Key residues for catalytic function and metal coordination in a carotenoid cleavage dioxygenase. J Biol Chem 291:19401–19412. doi: 10.1074/jbc.M116.744912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sui X, Kiser PD, Von Lintig J, Palczewski K. 2013. Structural basis of carotenoid cleavage: from bacteria to mammals. Arch Biochem Biophys 539:203–213. doi: 10.1016/j.abb.2013.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harrison PJ, Bugg TDH. 2014. Enzymology of the carotenoid cleavage dioxygenases: reaction mechanisms, inhibition and biochemical roles. Arch Biochem Biophys 544:105–111. doi: 10.1016/j.abb.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 38.McAndrew RP, Sathitsuksanoh N, Mbughuni MM, Heins RA, Pereira JH, George A, Sale KL, Fox BG, Simmons BA, Adams PD. 2016. Structure and mechanism of NOV1, a resveratrol-cleaving dioxygenase. Proc Natl Acad Sci U S A 113:14324–14329. doi: 10.1073/pnas.1608917113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guex N, Peitsch MC, Schwede T. 2009. Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: a historical perspective. Electrophoresis 30:S162–S173. doi: 10.1002/elps.200900140. [DOI] [PubMed] [Google Scholar]
- 40.Bossert ID, Whited G, Gibson DT, Young LY. 1989. Anaerobic oxidation of p-cresol mediated by a partially purified methylhydroxylase from a denitrifying bacterium. J Bacteriol 171:2956–2962. doi: 10.1128/jb.171.6.2956-2962.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cronin CN, Kim J, Fuller JH, Zhang XP, McIntire WS. 1999. Organization and sequences of p-hydroxybenzaldehyde dehydrogenase and other plasmid-encoded genes for early enzymes of the p-cresol degradative pathway in Pseudomonas putida NCIMB 9866 and 9869. DNA Seq 10:7–17. doi: 10.3109/10425179909033930. [DOI] [PubMed] [Google Scholar]
- 42.Hopper DJ. 1976. The hydroxylation of p-cresol and its conversion to p-hydroxybenzaldehyde in Pseudomonas putida. Biochem Biophys Res Commun 69:462–468. doi: 10.1016/0006-291X(76)90544-1. [DOI] [PubMed] [Google Scholar]
- 43.Entsch B, Vanberkel WJH. 1995. Structure and mechanism of para-hydroxybenzoate hydroxylase. FASEB J 9:476–483. doi: 10.1096/fasebj.9.7.7737455. [DOI] [PubMed] [Google Scholar]
- 44.Fujisawa H, Hayaishi O. 1968. Protocatechuate 3,4-dioxygenase I. Crystallization and characterization. J Biol Chem 243:2673–2679. [PubMed] [Google Scholar]
- 45.Hartnett C, Neidle EL, Ngai KL, Ornston LN. 1990. DNA sequences of genes encoding Acinetobacter calcoaceticus potocatechuate 3,4-dioxygenase: evidence indicating shuffling of genes and of DNA sequences within genes during their evolutionary divergence. J Bacteriol 172:956–966. doi: 10.1128/jb.172.2.956-966.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wenzel SC, Bode HB, Kochems I, Mueller R. 2008. A type I/type III polyketide synthase hybrid biosynthetic pathway for the structurally unique ansa compound kendomycin. Chembiochem 9:2711–2721. doi: 10.1002/cbic.200800456. [DOI] [PubMed] [Google Scholar]
- 47.Philipp B, Schink B. 2000. Two distinct pathways for anaerobic degradation of aromatic compounds in the denitrifying bacterium Thauera aromatica strain AR-1. Arch Microbiol 173:91–96. doi: 10.1007/s002039900112. [DOI] [PubMed] [Google Scholar]
- 48.Sobolev VS, Neff SA, Gloer JB. 2009. New stilbenoids from peanut (Arachis hypogaea) seeds challenged by an Aspergillus caelatus strain. J Agric Food Chem 57:62–68. doi: 10.1021/jf802891v. [DOI] [PubMed] [Google Scholar]
- 49.Stanier RY, Palleron NJ, Doudorof M. 1966. The aerobic pseudomonads: a taxonomic study. J Gen Microbiol 43:159–271. doi: 10.1099/00221287-43-2-159. [DOI] [PubMed] [Google Scholar]
- 50.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 51.Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu W, Lomsadze A, Borodovsky M. 2010. Ab initio gene identification in metagenomic sequences. Nucleic Acids Res 38:e132. doi: 10.1093/nar/gkp829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Altschul SF, Lipman DJ. 1990. Protein database searches for multiple alignments. Proc Natl Acad Sci U S A 87:5509–5513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kaniga K, Delor I, Cornelis GR. 1991. A wide-host-range suicide vector for improving reverse genetics in Gram-negative bacteria: inactivation of the blaA gene of Yersinia enterocolitica. Gene 109:137–141. doi: 10.1016/0378-1119(91)90599-7. [DOI] [PubMed] [Google Scholar]
- 55.Clemmer KM, Bonomo RA, Rather PN. 2011. Genetic analysis of surface motility in Acinetobacter baumannii. Microbiology 157:2534–2544. doi: 10.1099/mic.0.049791-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.