Abstract
The root cap functions in the perception of gravity, the protection of the root apical meristem, and facilitation of the passage of roots through the soil, but the genes involved in these functions are poorly understood. Here we report the isolation of a root-specific gene from the cap of maize (Zea mays L.) primary root by cDNA subtraction and differential screening. The gene zmGRP4 (Z. mays glycine rich protein 4) encodes a member of the glycine-rich proteins with a putative signal peptide at the amino terminus. The deduced molecular mass of mature zmGRP4 is 14.4 kD. In situ-hybridization analysis has shown zmGRP4 to be strongly expressed in the lateral root cap and weakly expressed in the root epidermis. A polyclonal antibody raised against recombinant zmGRP4 detected a protein of 36 kD in the insoluble protein fraction extracted from the root tip and the root proper, indicating posttranslational modification(s) of zmGRP4. Immunohistochemical analysis showed the accumulation of zmGRP4 in the mucilage that covers the root tip. These results indicate that lateral root-cap cells secrete modified zmGRP4 into the mucilage to which the protein may contribute to its characteristic physical properties.
The root cap covers the root meristem at the root apices of vascular plants but is absent in nonvascular plants such as liverworts and mosses. It is proposed that the root cap perceives gravity and protects the root apical meristem (Sievers and Braun, 1996). The root cap has a high regenerative capacity. When the root cap, which is sharply delineated from the root proper, is surgically removed from a maize (Zea mays L.) root, it regenerates completely within a few days (Barlow, 1975).
Anatomical studies suggest that the root cap consists of several distinct regions (Moore and McClelen, 1983; Dolan et al., 1993). The maize root cap, for example, can be divided into three regions: the calyptrogen, the columella root cap, and the lateral root cap. The calyptrogen faces the distal end of the quiescent center of the root apical meristem, is composed of approximately four cell layers, and serves as a root-cap meristem. The columella cells are generated by periclinal cell division from the central region of the calyptrogen. Sedimented large amyloplasts containing well-developed starch granules are characteristic of the columella cells. These amyloplasts function as statoliths in root gravitropism (Sievers and Braun, 1996). The lateral root cap surrounds the columella root cap. In maize roots with a closed-type construction, the lateral cap cells originate from the calyptrogen (Barlow, 1996). However, in Arabidopsis roots, which have an open-type construction, there is no discrete boundary between the root proper and the cap, and the lateral cap cells are derived from the same initials as the root epidermal cells (Dolan et al., 1993). The lateral root-cap cells are rich in the hypertrophied dictyosome cisternae that form large secretory vesicles (Mollenhauer et al., 1961). These cisternae may reflect the massive secretion of mucilage from the lateral cap cells, because the vesicle content was observed to be deposited between the plasma membrane and the outer tangential walls of the lateral cap cells (Morré et al., 1967).
In addition to these three tissues, sloughed-off cap cells and root mucilage may also be included as components, which together make up the cap region. Root-cap cells are continuously pushed toward the root-cap periphery and finally slough off into the external root environment. These detached cells are found at the root periphery, even at some distance from the root cap (Vermeer and McCully, 1982); they are metabolically active and have unique patterns of gene expression (Brigham et al., 1995). Several functions of the sloughed-off cap cells as a root-soil interface have been proposed (Hawes et al., 1998).
The root mucilage typically covers the root apex, is an amorphous and uneven gel, and ranges in thickness from 50 μm to 1 mm. The mucilage is secreted largely from the root cap, but the root epidermis is also covered by a thin film of mucilage, which is histochemically distinct from the cap-derived mucilage (Greaves and Darbyshire, 1972; Clarke et al., 1979; Foster, 1982; Vermeer and McCully, 1982). The matrix of maize mucilage consists of 95% polysaccharides and 5% protein (Harris and Northcote, 1970; Bacic et al., 1986).
To understand the functions of the root cap at the molecular level, we have identified genes that are specifically or predominantly expressed in maize root cap. One such gene is expressed strongly in the lateral root cap, and its gene product is secreted into and accumulates in the mucilage.
MATERIALS AND METHODS
Plant Material
Maize (Zea mays L. cv Merit) was supplied by the Asgrow Seed Company (Kalamazoo, MI). Seeds were soaked in tap water for 72 h in the dark at 30°C. After imbibition seeds were germinated on paper towels saturated with tap water for 1 to 2 d in the dark at 30°C. When the primary roots were 2 to 3 cm long, the cap and selected portions of the root were removed by a scalpel under a magnifying glass and immediately frozen in liquid N2. The tip region used in this study was the apical 5 mm of the root and included the root apical meristem and the whole root cap. The region of the root proper was between 1 and 3 cm from the distal end of the root.
Maize plantlets were grown under 18-h light/6-h dark or 24-h dark conditions at 30°C on layered wet paper towels in plastic pots.
RNA Isolation, cDNA Synthesis, and Subtractive Hybridization
Poly(A+) RNA was extracted directly from the root cap and the root proper of maize primary roots using Dynabeads oligo(dT25) (Dynal, Oslo, Norway). Several hundred nanograms of poly(A+) RNA were used to construct double-stranded cDNA using a cDNA synthesis kit (Pharmacia).
Subtractive hybridization was done essentially as described by Wang and Brown (1991) and Hashimoto et al. (1993). The double-stranded cDNAs were fragmented by AluI and RsaI and ligated to a PCR linker. cDNA fragments of 0.2 to 2.0 kb were amplified by PCR. The cDNA fragments from the root proper were then biotinylated with Photoprobe biotin (Vector Laboratories, Burlingame, CA) and used as the driver DNA.
One subtraction cycle consisted of five steps: hybridization of the excess driver DNA to the tracer DNA from the root cap for 20 h at 68°C; removal of nonhybridizing driver DNA by binding to streptavidin and extraction with organic solvent; another hybridization of the excess driver DNA to the remaining tracer DNA once again for 2 h at 68°C; removal of driver DNA as above; and PCR amplification of the tracer DNA. This subtraction cycle was repeated twice to produce subtracted root-cap cDNA fragments. Subtracted root-proper cDNA fragments were also generated in the same way, except that cDNA fragments from the root cap and the root proper were used, respectively, as the driver and the tracer DNAs.
Screening of Differentially Expressed cDNAs
The subtracted root-cap cDNA fragments were digested with EcoRI, which cleaved the PCR linker, and inserted into the EcoRI site of pBluescript II SK(−) (Stratagene). These plasmids were introduced into the bacterial strain DH5α to construct a root-cap cDNA library, and 386 independent colonies were grown overnight in Luria-Bertani medium containing 50 μg mL−1 ampicillin at 37°C. From each culture a 50-μL aliquot was blotted in duplicate onto a membrane (Hybond N+, Amersham) using a filtration manifold system (GIBCO-BRL). After denaturation and neutralization, the duplicate filters were hybridized at 42°C for 16 h with either a 32P-labeled, subtracted root-cap cDNA probe or a 32P-labeled, nonsubtracted root-proper cDNA probe, in a hybridization buffer containing 50% formamide, 10% dextran sulfate, 1% SDS, 5× SSPE (1× SSPE: 180 mm NaCl, 1 mm EDTA, and 10 mm Na2HPO4, pH 7.5), 5× Denhardt's solution (1× Denhardt's solution: 0.02% [w/v] BSA, 0.02% [w/v] Ficoll, and 0.02% [w/v] PVP), and 100 μg mL−1 salmon testis DNA, and washed at 65°C in 0.1× SSPE and 0.1% SDS. Seventy-two positive cDNA clones, which hybridized only to the root-cap cDNA probe, were obtained.
To group the positive clones, the 72 recombinant bacterial cultures containing positive cDNA clones were blotted onto a Hybond N+ membrane and processed as described above. One cDNA clone, which had hybridized specifically and strongly to the root-cap cDNA probe, was chosen, labeled with 32P, and hybridized to the membrane. Positive clones were regarded as members of the same group. Next, another strongly and specifically hybridizing cDNA clone other than the members of this group was chosen and processed as above. Four hybridizations were done, resulting in four independent groups and 29 remaining cDNA clones. Representative clones from the four groups and the extra 29 cDNA clones were partially sequenced by a DNA sequencer (model 373A, Perkin-Elmer), using M13 reverse and universal primers. The sequence analysis classified the root-cap-positive cDNA clones into 23 groups.
Subtracted cDNA fragments from the root cap, nonsubtracted cDNA fragments from the root cap, and nonsubtracted cDNA fragments from the root proper were blotted in amounts of 0.05, 0.5, and 5 μg per slot onto a Hybond N+ membrane, as described above. Representative cDNA fragments from the 23 groups were used as the probes for hybridization. Ten cDNA fragments hybridized to the subtracted and nonsubtracted root-cap cDNA pools or to the subtracted root-cap cDNA pool, but not to the root-proper cDNA pool, and will be referred to as “root-cap abundant.” The other 13 clones either hybridized to the root-proper cDNA pool or did not hybridize to any cDNA pools.
A cDNA library of the maize primary root-tip region from within 1 mm of the distal-tip end was made in λZAPII (Stratagene) (Matsuyama et al., 1999). A total of 4 × 106 recombinants were independently screened with the 10 root-cap-abundant cDNA fragments as probes. Hybridization and other procedures were done as described above. Positive plaques were identified with 3 of the 10 root-cap-abundant probes. In this report, one cDNA representing six phage clones was analyzed. These positive recombinant phages were converted to pBluescript SK(−) plasmids by in vivo excision using the manufacturer's protocol (Stratagene). Both DNA strands of the longest insert of the six clones were sequenced. DNA and predicted amino acid sequences were analyzed with GeneWorks software (IntelliGenetics, Campbell, CA).
Genomic-DNA-Hybridization Analysis
Total genomic DNA was isolated from 3-d-old etiolated maize seedlings by cetyl-trimethyl-ammonium bromide extraction (Murray and Thompson, 1980). Genomic DNA (30 μg) was digested with restriction enzymes, electrophoresed on a 1% agarose gel, and blotted onto a Hybond N+ membrane. The membrane was hybridized to the full-length zmGRP4 (Z. mays GRP 4) cDNA probe and washed under the conditions described above.
Northern-Hybridization and RT-PCR Analysis
Total RNA was isolated from several tissues, including the root tip, root proper, young leaves from 2-week-old plants, and shoots from 3-d-old light-grown and etiolated plants using phenol:chloroform extraction and LiCl precipitation (Mohnen et al., 1985). Poly(A+) RNA was purified from total RNA using Oligotex-dT30 Super (Takara Shuzo, Tokyo, Japan). Poly(A+) RNA (1.5 μg per lane) was electrophoresed on a 1.2% formaldehyde agarose gel, blotted onto a Hybond N+ membrane, and hybridized to the full-length zmGRP4 cDNA probe under the conditions described above. After stripping the probe from the membrane by incubating at 67°C in a buffer containing 50% formamide, 10 mm Tris-HCl, and 10 mm EDTA, pH 8.0, the 32P-labeled PstI-SacI fragment of a ubiquitin cDNA (Christensen and Quail, 1989) was hybridized to the same membrane.
For RT-PCR analysis, total RNA was isolated from approximately 100 mg of root tip, root proper, shoot, and etiolated shoot using the RNeasy Plant Mini kit (Qiagen, Chatsworth, CA) and used to construct first-strand cDNA using the SUPERSCRIPT preamplification system (GIBCO-BRL). PCR primers for zmGRP4 were 5′-TTGTATCTCACAATGGCAGGC and 5′-GCGTTGGAATTCCAAGAACC (Fig. 1) and PCR primers for maize α-tubulin1 (Montoliu et al., 1989) were 5′-CTTGATCGCATCAGGAAGC and 5′-TCAGCAGAGATGACTGGAGC. PCR amplification was carried out for 18 cycles of denaturation at 94°C for 1 min, annealing at 60°C for 30 s, and elongation at 72°C for 2 min. Amplified zmGRP4 fragments were electrophoresed on a 1.2% agarose gel, blotted onto a Hybond N+ membrane, and hybridized to the full-length zmGRP4 cDNA probe as described above. Representative amplified DNA fragments were partially sequenced to confirm their identity.
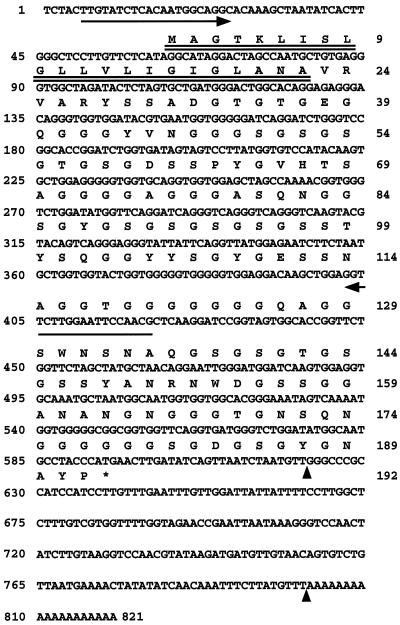
Figure 1.
Nucleotide and deduced amino acid sequences of zmGRP4. A putative signal peptide is double underlined. The sequence between the vertical arrowheads was used as both antisense and sense probes for in situ-hybridization analysis. The two arrows indicate the positions of PCR primers used for RT-PCR analysis. The stop codon is shown by an asterisk.
In Situ-Hybridization Analysis
Maize primary root tips were fixed in 3% paraformaldehyde and 2% glutaraldehyde for 12 h at 4°C. After samples were dehydrated in a graded ethanol series and cleared in a graded xylene series, they were embedded in wax (Histoprep 580, Wako, Osaka, Japan) and sectioned at 10 μm by using a rotary microtome. Digoxigenin-labeled antisense and sense RNA probes were prepared from a 3′-untranslated region of zmGRP4 (see Fig. 1) using an RNA labeling kit (DIG, Boehringer Mannheim). Samples were incubated with the RNA probes at 50°C for 16 h, treated with RNase A (2.5 μg mL−1 in 0.5 m NaCl, 10 mm Tris-HCl, and 1 mm EDTA, pH 7.5) at 37°C for 30 min, and washed with several changes of 2× SSC (1× SSC: 150 mm NaCl and 15 mm Na3C6H5O7) and once with 0.1× SSC at 50°C. Signals were detected by a nucleic acid detection kit (DIG, Boehringer Mannheim). The color reaction was stopped with 10 mm Tris-HCl and 1 mm EDTA, pH 8.0. Sections were passed through an ethanol series and mounted for microscopic observation.
zmGRP4 Antiserum
A portion of the zmGRP4 cDNA encoding the carboxyl-terminal amino acid residues from 132 to 192 was subcloned into the EcoRI site of pET-32b(+) (Novagen), which would then express a fusion protein consisting of the N-terminal region of thioredoxin and the C-terminal region of zmGRP4. This plasmid was introduced into the BL21 (DE3) bacterial strain (Novagen), and the expression of the fusion protein was induced by 1 mm isopropyl β-d-thiogalactopyranoside at 37°C for 3 h in Luria-Bertani medium. Bacterial cells were harvested by centrifugation, suspended in 50 mm potassium phosphate buffer, pH 8.0, containing 1% Triton X-100 and 1 μg mL−1 lysozyme, incubated at 30°C for 15 min, and then ruptured by three cycles of freeze/thaw treatment and sonication for 1 min. After centrifugation of the homogenate, the fusion protein in the supernatant was separated by preparative SDS-PAGE using a PrepCell (model 491, Bio-Rad). The eluate fractions containing the fusion protein were concentrated with a YM-10 membrane filter (Amicon). The buffer of the concentrated protein solution was exchanged by using a PD-10 column (Pharmacia) equilibrated with buffer A (20 mm Tris-HCl and 10% glycerol, pH 7.0). The above solution was loaded onto a Mono-Q fast-protein liquid-chromatography column (Pharmacia) previously equilibrated with buffer A and eluted with a linear gradient of KCl from 0 to 0.5 m in buffer A. The fractions containing the fusion protein were concentrated with a YM-10 filter and desalted using a PD-10 column. The fusion protein had an approximate purity of 99.9%, as determined by staining with Coomassie Brilliant Blue R-250 after SDS-PAGE, and was used to raise antiserum in mice.
The reactivity of the antiserum against zmGRP4 was confirmed as follows. A BamHI-SmaI fragment of the zmGRP4 cDNA encoding the carboxyl-terminal amino acid residues from 137 to 192 was subcloned into pGEX-2T (Pharmacia). The resultant pGEX-zmGRP4 plasmid or pGEX-2T was introduced into the BL21 (DE3) bacterial strain, and the expression of either a chimeric protein consisting of zmGRP4 and GST, or GST alone, respectively, was induced at 37°C for 3 h with 1 mm isopropyl β-d-thiogalactopyranoside. Approximately 100 ng of total protein extracts from these bacterial cells was separated on a 15% SDS-polyacrylamide gel and transferred to an Immobilon PVDF membrane (Millipore). The membrane was blocked at room temperature in buffer B (100 mm Tris-HCl and 150 mm NaCl, pH 7.5) containing 5% skim milk powder for 1 h. The antiserum was diluted 1:5000 in buffer B and incubated with the membranes at room temperature for 1 h. After washing the antiserum several times with buffer B containing 0.2% Tween 20, the following procedures, including secondary antibody treatment and immunodetection, were performed by using an ECL Plus kit (Amersham) according to the manufacturer's instructions. The antiserum reacted strongly with the fusion protein consisting of zmGRP4 and GST, but not with GST alone.
Immunohistochemical Analysis
Fixed sections were prepared as described for in situ hybridization and blocked at room temperature in buffer B containing 0.2% Tween 20 and 3% BSA for 1 h. Anti-zmGRP4 serum or preimmune mouse serum was diluted 1:300 in blocking buffer and incubated with the sections at room temperature for 1 h. After washing the primary antibody several times in buffer B containing 0.2% Tween 20, anti-mouse IgG conjugated with alkaline phosphatase (Kirkegaard and Perry Laboratories, Gaithersburg, MD) was diluted 1:1000 in buffer B and incubated with the sections at room temperature for 1 h. After briefly washing the slides with buffer B, immunodetection was performed as described for in situ-hybridization analysis.
Immunoblot Analysis
Approximately 100 maize root tips were frozen with liquid N2 and ground to a fine powder with a pestle. For some sample preparations used in Figure 6B, after root mucilage including sloughed-off cap cells was gently wiped from the root tip with a paper towel, the root tips were immediately collected in liquid nitrogen. The pulverized root cells were extracted with buffer (50 mm Tris-HCl, 3 mm EDTA, 3 mm DTT, and 3 mm PMSF), and the suspension was centrifuged at 15,000g for 5 min. The supernatant was referred to as the soluble fraction. The pellet was resuspended in sample buffer (125 mm Tris-HCl, pH 6.8, containing 1% SDS) and used as the insoluble fraction. A total protein fraction was prepared by directly extracting the pulverized cells with sample buffer. Protein concentration was determined using the BCA protein assay reagent (Pierce).
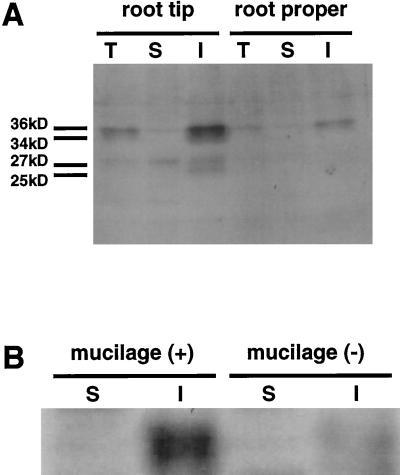
Figure 6.
Immunoblot analysis of zmGRP4. Each lane was loaded with 10 μg of protein extracted from the root tip or root proper. A, Total protein fraction (lanes T), Tris-buffer-soluble fraction (lanes S), and Tris-buffer-insoluble but SDS-buffer-soluble fraction (lanes I) were separated on a 15% SDS-polyacrylamide gel. B, The Tris-buffer-soluble fraction (S) and Tris-buffer-insoluble but SDS-buffer-soluble fraction (I) were extracted from intact root tips (+) or root tips from which mucilage had been removed (−).
Protein preparations (10 μg per lane) were separated on a 15% SDS-polyacrylamide gel. The remaining steps were performed as described above, except that 3% BSA was substituted for 5% skim milk powder in the blocking buffer.
RESULTS
Isolation of a Maize GRP cDNA That Is Highly Expressed in Root Cap
The root cap and the root proper are sharply delineated in the maize primary root, which has a closed-type construction. This anatomical feature is used to facilitate excision of maize root-cap tissues from the root proper using a scalpel (Barlow, 1975). We collected approximately 500 root caps and extracted poly(A+) RNA directly from the root cap and also from the root proper. The cDNAs specifically present in the root cap were enriched by subtracting the root-proper cDNA fragment pool from the root-cap cDNA fragment pool. Subsequently, the subtracted root-cap cDNA fragment library was duplicated and hybridized independently with the above root-proper cDNA fragment pool or the root-cap cDNA fragment pool as the probes. This differential screening recovered 72 cDNA fragments that hybridized specifically to the root-cap cDNA fragment pool, and these clones were classified into 23 groups by cross-hybridization and partial DNA sequencing. Further slot-blot hybridization with the above two probes confirmed that 10 cDNA groups were much more abundant in the root cap than in the root proper. Representative cDNA fragments from these 10 clones were used as the probes to screen a maize root-tip cDNA library, and three distinct cDNA clones were obtained. One of the three cDNA clones is reported here. The other two clones encoded a novel protein (Matsuyama et al., 1999) and a maize expansin.
Figure 1 shows the nucleotide and deduced amino acid sequences of a cDNA clone. The cDNA was 821 bp long and contained an open reading frame encoding a 16.9-kD polypeptide of 192 amino acids. The predicted protein had a hydrophobic putative signal peptide with a potential cleavage site between 22 and 23 amino acid residues (von Heijne, 1985) and was a member of the cell wall GRPs (Showalter, 1993). We will refer to this protein as zmGRP4. zmGRP4, excluding the putative signal peptide, was rich in Gly (40%), Ser (19%), Asn (7%), Ala (7%), and Tyr (6%). The high content of Gly, Ser, and Ala of zmGRP4 is consistent with the general characteristics of Gly-rich cell wall proteins.
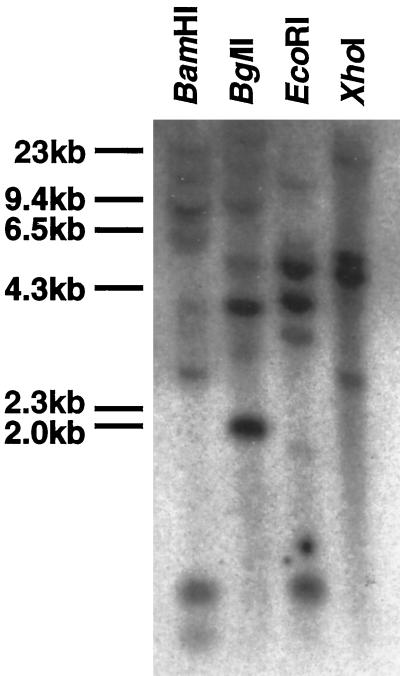
Genomic DNA blot-hybridization analysis was done with a full-length zmGRP4 cDNA probe at high-stringency conditions (Fig. 2). BamHI and EcoRI digested the zmGRP4 cDNA once, whereas BglII and XhoI did not digest the cDNA. Two to three strong bands and two to four weak bands were detected when the maize genome was digested with BamHI, BglII, EcoRI, or XhoI. Therefore, a small number of genes homologous to zmGRP4 are likely to exist in maize. In support of this, the amino acid sequence of another maize GRP cDNA (accession no. AF031083) and zmGRP4 share an 82% identical region of approximately 90 amino acid residues (data not shown). At the nucleotide sequence level, this region is 83% identical between these two GRPs.
Figure 2.
Genomic DNA-hybridization analysis of zmGRP4. Full-length zmGRP4 cDNA was used as a probe. Maize genomic DNA (20 μg) was digested with BamHI, BglII, EcoRI, or XhoI. The positions of the molecular markers are shown on the left.
zmGRP4 Is Expressed Strongly in the Lateral Root Cap and Weakly in the Epidermis of the Root Proper
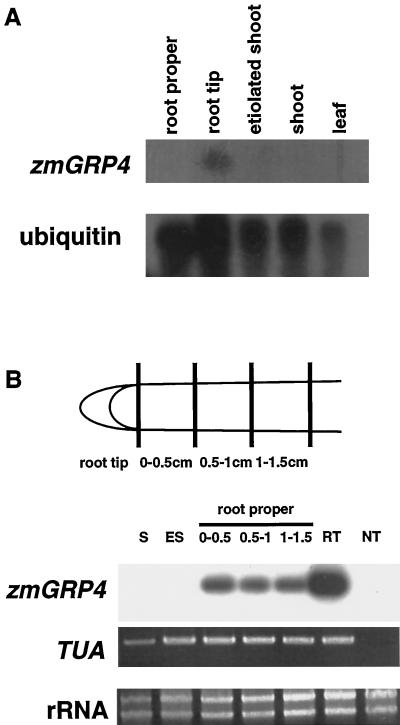
RNA blot-hybridization analysis with the full-length cDNA of zmGRP4 as the probe detected zmGRP4 expression in the root tip but not in the root proper, etiolated shoot, shoot, or mature leaf (Fig. 3A). Since RNA-blot hybridization may not detect low levels of zmGRP4 expression and may detect expression of zmGRP4-related gene(s) as well, RT-PCR was done to specifically amplify zmGRP4 RNA (Fig. 3B). Expression of zmGRP4 was detected in the root tip and root proper but not in the shoot and etiolated shoot. Although quantitative analysis is often difficult with RT-PCR, repeated RT-PCR analyses in which different amplification cycles were used (data not shown) confirmed that zmGRP4 is expressed more strongly in the root tip than in the root proper. Amplification of maize α-tubulin RNA indicated that approximately equal amounts of cDNA were used for each tissue sample.
Figure 3.
Expression of zmGRP4 in maize tissues. A, Northern-hybridization analysis. Poly(A+) RNA (1.5 μg) was isolated from 2- to 3-cm-long roots, 3-d-old etiolated shoots, and 3-d-old shoots, and leaves of 2-week-old plants. A maize ubiquitin probe served as a control to estimate the relative loading of RNA in each lane. B, RT-PCR analysis. S, Shoot; ES, etiolated shoot; root proper, 0- to 0.5-, 0.5- to 1.0-, and 1.0- to 1.5-cm regions distal from the excision site; RT, root tip; NT, negative control without reverse transcription. A maize α-tublin gene (TUA) served as a positive control.
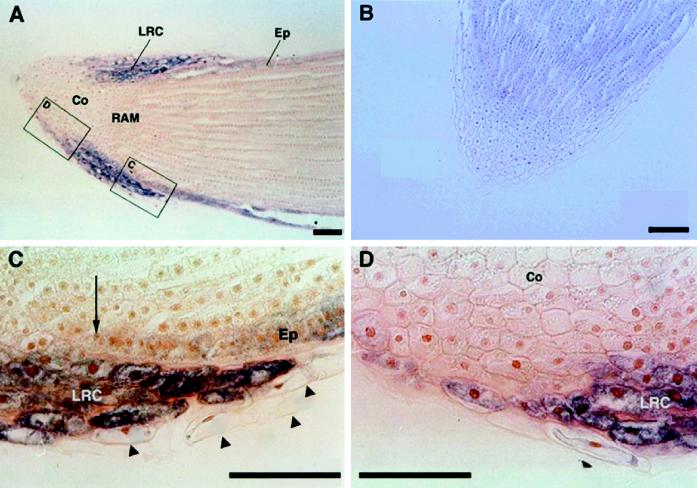
The 3′-untranslated region of the zmGRP4 cDNA was used as a probe for in situ-hybridization analysis to study the detailed expression pattern of zmGRP4 in maize primary root. The antisense probe detected strong zmGRP4 expression in the lateral root-cap cells and rather weak expression in epidermal cells of the root proper (Fig. 4A). The sense probe did not detect any hybridization signals (Fig. 4B). Peripheral cells that had been or were being detached from the lateral root cap showed little zmGRP4 expression (Fig. 4C, arrowheads), whereas weak zmGRP4 expression extended to several peripheral cells toward the central region of the root cap (Fig. 4D). zmGRP4 expression in epidermal cells of the root proper terminated in the region where zmGRP4 expression in the lateral root cap ended (Fig. 4C, arrow).
Figure 4.
In situ-hybridization analysis of zmGRP4. Longitudinal sections of maize primary root tip were hybridized with antisense (A, C, and D) or sense (B) digoxigenin-labeled zmGRP4-specific probes. The images in C and D were enlarged from the rectangles in A. An arrow shows the end of zmGRP4 expression in the epidermis, whereas arrowheads indicate sloughed-off cap cells. Co, Columella; LRC, lateral root cap; Ep, epidermis; RAM, root apical meristem. Scale bars = 100 μm.
zmGRP4 Accumulates in Root Mucilage
A polyclonal antibody was raised against a truncated zmGRP4 protein that contained amino acid residues 132 to 192. This carboxy-terminal region of zmGRP4 includes amino acid stretches of low Gly abundance and is expected to be specific for zmGRP4. The closest homolog of zmGRP, encoded by a maize expressed sequence tag (AF031083), is 58% identical in this region (data not shown).
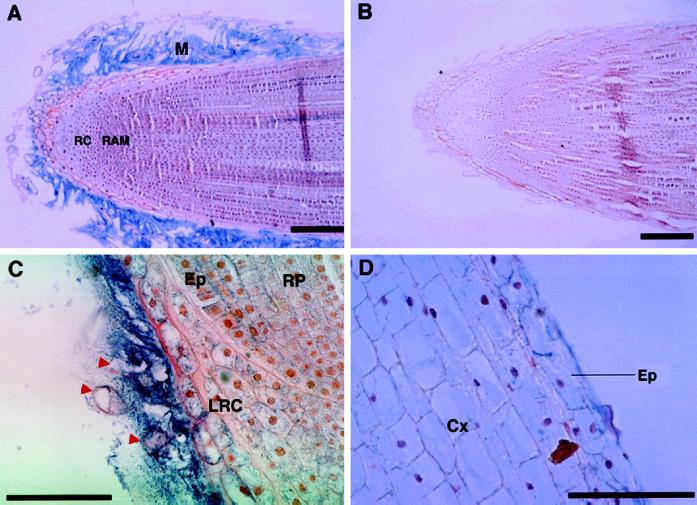
Immunohistochemical analysis using this antiserum showed that zmGRP4 is present specifically in the mucilage that covers the root tip (Fig. 5A). A preimmune mouse antiserum detected no signals (Fig. 5B). Longer exposure detected a relatively small amount of zmGRP4 in the lateral root-cap cells (Fig. 5C). Sloughed-off cap cells appeared to contain little zmGRP4 (Fig. 5C, red arrowheads). A weak signal was also observed in epidermal cells of the root proper in the distal 1 cm of the root tip (Fig. 5D). The presence of mucilage at the periphery of the root epidermis was not apparent because the layer of root epidermal mucilage is expected to be very thin (Foster, 1982).
Figure 5.
Immunolocalization of zmGRP4. Longitudinal sections of maize primary root tip were incubated with anti-zmGRP4 serum (A, C, and D) or preimmune serum (B). The lateral root-cap region adjacent to the root proper is shown with a higher magnification in C, whereas the root proper region 1 cm distal to the cap is shown in D. Alkaline-phosphatase reactions were done for 1 h in A and B, and for 3 h in C and D. Red arrowheads indicate sloughed-off cap cells. RC, Root cap; RAM, root apical meristem; M, mucilage; RP, root proper; LRC, lateral root cap; Cx, cortex; Ep, epidermis. Scale bars = 100 μm.
zmGRP4 May Be Posttranslationally Modified
Immunoblot analysis using the anti-zmGRP4 serum revealed that zmGRP4 exists in the maize root as a major band with an apparent molecular mass of 36 kD and a minor band with an apparent molecular mass of 34 kD in the insoluble fraction that was extracted with the SDS-containing buffer (Fig. 6A). Extraction with Tris buffer without SDS did not recover any zmGRP4 protein. The 36-kD form was much more abundant in the root tip than in the root proper. A few faint bands of 27 and 25 kD were also detected in the insoluble fraction from the root tip. Since the deduced molecular mass of the mature zmGRP4 is 14.4 kD, posttranslational modifications of zmGRP4 in maize root are suspected. Manual removal of root mucilage and detached cap cells from the root tip markedly reduced the abundance of the 36-kD zmGRP4 in the preparation (Fig. 6B). This strongly suggests that zmGRP4 accumulates in root mucilage mainly as a modified 36-kD protein.
DISCUSSION
zmGRP4 Is a New Member of Maize GRPs
Many structural cell wall proteins that have putative signal peptides and no catalytic domains have been reported in various plants (Showalter, 1993).These cell wall proteins are characterized by a high abundance of a single amino acid, repetitive sequence motifs, and a tendency to become insolubilized within the cell wall. The three major plant cell wall protein classes include HRGPs, PRPs, and GRPs. GRP cDNAs have been isolated from several plants, including three from maize. zmGRP is expressed in the epidermal cells of embryo, scutellar tissue, and young leaf, and induced by ABA, water stress, and wounding in leaves (Gómez et al., 1988). Since zmGRP does not have an amino-terminal signal peptide, it may be a cytosolic protein. Besides being rich in Gly, zmGRP has a putative RNA-binding motif (Gómez et al., 1988). Therefore, zmGRP and zmGRP4 belong to different subclasses of the GRP family. zmGRP3 (Goddemeier et al., 1998) has an N-terminal signal peptide but shows no significant homology to zmGRP4, except for abundant Gly residues. The expression of zmGRP3 was root specific, with the highest expression level in the meristematic and elongation regions (Goddemeier et al., 1998). RNA-blot analysis of zmGRP3 indicates that zmGRP3 and zmGRP4 are expressed in different regions of the maize root.
zmGRP4 Expression in the Root
The expression of cell wall proteins depends on cell type, developmental stage, and stress responses (Showalter, 1993). zmGRP4 is expressed strongly in the lateral root cap and weakly in root epidermis but scarcely in sloughed-off cap cells (Fig. 4). The immunohistochemical localization of zmGRP4 (Fig. 5) strongly indicates that zmGRP4 is synthesized in lateral root-cap and root-epidermal cells and then secreted into the mucilage. Lateral root-cap cells develop considerable hypertrophied Golgi cisternae and are the main site of mucilage secretion. Maize root epidermal cells are also reported to contain hypertrophied dictyosome cisternae and release mucilage (Clarke et al., 1979; Foster, 1982). Detached cap cells, however, have dictyosomes that are no longer hypertrophied (Clowes and Juniper, 1968). This close correlation between zmGRP4 expression and differentiation of secretion machinery suggests that zmGRP4 is secreted via hypertrophied Golgi cisternae into the mucilage. Likewise, bean GRP 1.8 was localized to dictyosomes of xylem parenchyma cells and was suggested to be exported into the walls of neighboring protoxylem vessels (Ryser and Keller, 1992).
Sloughed-off cells did not express zmGRP4, whereas the outermost cap periphery cells did express zmGRP4 (Fig. 4, C and D). A notable switch in gene expression was also reported to occur upon cap-border cell differentiation in pea (Brigham et al., 1995).
zmGRP4 Is Posttranslationally Modified
Many cell wall proteins are modified posttranslationally. For example, Pro residues of HRGP are enzymatically converted into Hyp residues, which are then glycosylated to various degrees (Cassab, 1998). zmGRP4 mainly existed as a 36-kD protein, whereas the deduced molecular mass of mature zmGRP4 is 14.4 kD. The high Gly content in GRPs may cause aberrant electrophoretic migration on SDS gels. When zmGRP4 was expressed in Escherichia coli as a GST-fusion protein, the recombinant fusion protein detected was approximately 2-kD larger than expected by SDS-PAGE analysis (T. Matsuyama and T. Hashimoto, unpublished results). However, this aberrant migration alone does not explain the more than 20-kD difference between the expected and observed size of zmGRP4 extracted from maize root tips.
Insolubilization of cell wall proteins has been observed in various developmental or stress-responsive processes (Cassab, 1998). Insolubilization of bean GRP 1.8 occurs during hypocotyl development (Keller et al., 1989). H2O2 generated by fungal elicitor or glutathione treatment of bean or soybean cells causes oxidative cross-linking and therefore the insolubilization of PRP (Bradley et al., 1992). Recovery of isodityrosine after hydrolysis of cross-linked HRGP indicates that the Tyr hydroxy groups in HRGP undergo intermolecular condensation via H2O2 (Fry, 1986). zmGRP4 contains a relatively high percentage of Tyr residues. Oxidative cross-linking between zmGRP4s themselves or between zmGRP4 and other proteins via Tyr residues might result in insolubilization and increased molecular mass of zmGRP4. It should also be noted that PRPs insolubilized by H2O2 were not extracted even in SDS-containing buffer (Brisson et al., 1994), and potential cross-linking of xylem GRPs with the aromatic residues of lignin has also been proposed (Showalter, 1993; Cassab, 1998). The absence of lignin and polyphenolics in root mucilage suggests that the cross-linking partners of zmGRP4 may be at least partly different from those of previously reported cell wall proteins.
Glycosylation is a common posttranslational modification found in secreted proteins. However, there are few reports of the potential glycosylation of GRPs (Showalter, 1993). Exceptions include a 30-kD GRP purified from soybean aleurone layers, which was reported to contain approximately 9% (w/w) sugars, including Man, Ara, Glc, Xyl, and Gal (Matsui et al., 1995). Purified soybean GRP showed a broad band after SDS-PAGE separation, indicating a microheterogeneity in the sugar component (Matsui et al., 1995). On the other hand, zmGRP4 extracted from maize root tips migrated as discrete bands on SDS-PAGE (Fig. 6). Since the deduced zmGRP4 amino acid sequence has no canonical N-glycosylation sites, the modification could be O-glycosylation with homogeneous sugar side chains, if zmGRP4 were to be glycosylated.
Possible Functions of zmGRP4 in Root Mucilage
Soil and sand sheaths usually cling tightly to the roots of field-grown grasses such as maize root. The sheath is thought to be formed by the binding of soil particles in mucilage originating from the root (Vermeer and McCully, 1982, and the refs. therein). Root hairs are probably not primarily responsible for the adhesion of soil aggregates. Mucilage, soil particles, sloughed-off root-cap cells, and some soil bacteria form the rhizosphere, and the chemical and physical properties of the mucilage should be very important in determining the nature of the rhizosphere.
Root mucilage is composed of 99.9% water (Guinel and McGully, 1986). The dry mass of mucilage consists mainly of polysaccharides and polyuronic acids (Jones and Morré, 1967; Floyd and Ohlrogge, 1970; Paull et al., 1975). Although proteins have been detected in maize mucilage (Chaboud, 1983), their properties and possible roles have attracted little attention. Previous chemical analysis of maize mucilage, from which detached cap cells have been mostly removed, showed that the amino acid composition is rich in Gly (13.8% of total amino acids) (Bacic et al., 1986). We have shown here that zmGRP4 is a mucilage protein and possibly the major component of the protein fraction. Other well-characterized GRPs are localized in the vascular system, and in xylem in particular (Ryser and Keller, 1992, and the refs. therein). Ultrastructural localization, however, has demonstrated that bean GRP 1.8 is localized to unlignified primary walls of protoxylem cells, and a correlation between GRP 1.8 deposition and lignification was evidently lacking in bean hypocotyls (Ryser and Keller, 1992; Ryser et al., 1997). An apparent positive correlation of GRP deposition with expansive growth and an inverse correlation with lignification have been reported for petunia ptGRP1, which is deposited at the cell wall/membrane interface, rather than within the cell wall (Condit, 1993). Thus, these GRPs may provide elasticity to the stretching wall or some protective environment to cells under frictional stress. Some GRP sequences are predicted to adopt β-pleated sheets composed of varying numbers of antiparallel strands; such a structure could provide elasticity and tensile strength during vascular development (Showalter, 1993).
The soil sheath adhering along the entire length of field-grown maize roots is mostly permeated by mucilage, which is histochemically similar to that produced by the root cap (Vermeer and McCully, 1982). An experiment designed to measure the penetration resistance showed that maize roots receive much less frictional resistance than metal probes when growing into the soil (Bengough and McKenzie, 1997). One function of root mucilage, working together with sloughing root-cap cells, may be to decrease the frictional resistance during growth in the soil and to protect growing roots from abrasion by soil particles. If zmGRP4 has physical properties similar to other GRPs, it may provide elasticity to the root mucilage and may complement other mucilage components (e.g. polysaccharides and pectin) for a lubricant function.
Large amounts of fixed C are secreted into the rhizosphere from the surface of grass roots (Russell, 1977). The secreted C is mostly in the form of sugar, but a wide range of amino acids, organic acids, vitamins, and auxins are either released from the roots or synthesized by microorganisms in the root environment (Bar-Yosef, 1996). These organic compounds may support survival and growth of detached cap cells and soil bacteria. Some of the compounds even may be involved in interactions between particular plant genotypes and soil microorganisms. Secreted proteins in the rhizosphere may play similar roles. In this regard, distribution of GRPs in root mucilages of other maize genotypes and other plant species, and the stability of zmGRP4 in the rhizosphere should be interesting to examine in the future.
ACKNOWLEDGMENTS
We thank Dr. Katsumi Ueda for valuable advice on in situ-hybridization techniques and Dr. Robert Winz for critical reading of the manuscript. Dr. Peter Quail of the University of California, Berkeley, and Naoki Yasumura are acknowledged for the maize ubiquitin cDNA and the cDNA library from maize root tip, respectively.
Abbreviations:
- GRP
Gly-rich protein
- GST
glutathione S-transferase
- HRGP
Hyp-rich glycoprotein
- PRP
Pro-rich protein
- RT
reverse transcriptase
The accession number for the sequence described in this article is AB014475.
Footnotes
This work was supported in part by a Grant-in-Aid for Scientific Research on Priority Areas (“The Molecular Basis of Flexible Organ Plans in Plants,” no. 06278103) from the Ministry of Education, Science, Sports and Culture of Japan to T.H. T.M. was supported by a Japan Society for the Promotion of Science Research Fellowship for Young Scientists (no. 5487).
LITERATURE CITED
- Bacic A, Moody SF, Clarke AE. Structural analysis of secreted root slime from maize (Zea mays L.) Plant Physiol. 1986;80:771–777. doi: 10.1104/pp.80.3.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow PW. The root cap. In: Torrey JG, Clarkson DT, editors. The Development and Function of Roots. London: Academic Press; 1975. pp. 21–54. [Google Scholar]
- Barlow PW. Cellular patterning in root meristems: its origins and significance. In: Waisel Y, Eshel A, Kafkafi U, editors. Plant Roots: The Hidden Half, Ed 2. Madison, WI: Marcel Dekker; 1996. pp. 77–109. [Google Scholar]
- Bar-Yosef B. Root excretions and their environmental effects: influence on availability of phosphorus. In: Waisel Y, Eshel A, Kafkafi U, editors. Plant Roots: The Hidden Half, Ed 2. Madison, WI: Marcel Dekker; 1996. pp. 581–605. [Google Scholar]
- Bengough AG, McKenzie BM. Sloughing of root cap cells decreases the frictional resistance to maize (Zea mays L.) root growth. J Exp Bot. 1997;48:885–893. [Google Scholar]
- Bradley DJ, Kjellbom P, Lamb CJ. Elicitor- and wound-induced oxidative cross-linking of a proline-rich plant cell wall protein: a novel, rapid defense response. Cell. 1992;70:21–30. doi: 10.1016/0092-8674(92)90530-p. [DOI] [PubMed] [Google Scholar]
- Brigham LA, Woo H-H, Nicoll SM, Hawes MC. Differential expression of proteins and mRNAs from border cells and root tips of pea. Plant Physiol. 1995;109:457–463. doi: 10.1104/pp.109.2.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brisson LF, Tenhaken R, Lamb CJ. Function of oxidative cross-linking of cell wall structural proteins in plant disease resistance. Plant Cell. 1994;6:1703–1712. doi: 10.1105/tpc.6.12.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassab GI. Plant cell wall proteins. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:281–309. doi: 10.1146/annurev.arplant.49.1.281. [DOI] [PubMed] [Google Scholar]
- Chaboud A. Isolation, purification and chemical composition of maize root cap slime. Plant Soil. 1983;73:395–402. [Google Scholar]
- Christensen AH, Quail PH. Sequence analysis and transcriptional regulation by heat shock of polyubiquitin transcripts from maize. Plant Mol Biol. 1989;12:619–632. doi: 10.1007/BF00044153. [DOI] [PubMed] [Google Scholar]
- Clarke KJ, McCully ME, Miki NK. A developmental study of the epidermis of young roots of Zea mays L. Protoplasma. 1979;98:283–309. [Google Scholar]
- Clowes FA, Juniper BE. Plant Cells. Oxford, UK: Blackwell Science; 1968. [Google Scholar]
- Condit CM. Developmental expression and localization of petunia glycine-rich protein 1. Plant Cell. 1993;5:277–288. doi: 10.1105/tpc.5.3.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan L, Janmaat K, Willemsen V, Linstead P, Poethig S, Roberts K, Scheres B. Cellular organisation of the Arabidopsis thaliana root. Development. 1993;119:71–84. doi: 10.1242/dev.119.1.71. [DOI] [PubMed] [Google Scholar]
- Floyd RA, Ohlrogge AJ. Gel formation on nodal root surfaces of Zea mays. I. Investigation of the gel's composition. Plant Soil. 1970;33:331–343. [Google Scholar]
- Foster RC. The fine structure of epidermal cell mucilages of roots. New Phytol. 1982;91:727–740. [Google Scholar]
- Fry SC. Cross-linking of matrix polymers in the growing cell walls of angiosperms. Annu Rev Plant Physiol. 1986;37:165–186. [Google Scholar]
- Goddemeier ML, Wulff D, Feix G. Root-specific expression of a Zea mays gene encoding a novel glycine-rich protein, zmGRP3. Plant Mol Biol. 1998;36:799–802. doi: 10.1023/a:1005998804622. [DOI] [PubMed] [Google Scholar]
- Gómez J, Sánchez-Martínez D, Stiefel V, Rigau J, Puigdomènech P, Pagès M. A gene plant hormone abscisic acid in response to water stress encodes a glycine-rich protein. Nature. 1988;334:262–264. doi: 10.1038/334262a0. [DOI] [PubMed] [Google Scholar]
- Greaves MP, Darbyshire JF. The ultrastructure of the mucilaginous layer on plant roots. Soil Biol Biochem. 1972;4:443–449. [Google Scholar]
- Guinel FC, McGully ME. Some water-related physical properties of maize root-cap mucilage. Plant Cell Environ. 1986;9:657–666. [Google Scholar]
- Harris PJ, Northcote DH. Patterns of polysaccharide biosynthesis in differentiating cells of maize root-tips. Biochem J. 1970;120:479–491. doi: 10.1042/bj1200479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto T, Hibi N, Yamada Y. Subtractive hybridization. Plant Tissue Cult Lett. 1993;10:307–313. [Google Scholar]
- Hawes MC, Brigham LA, Wen F, Woo H-H, Zhu Y (1998) Root border cells: phenomenology of signal exchange. In HE Flores, JP Lynch, D Eissenstat, eds, Radical Biology: Advances and Perspectives on the Function of Plant Roots. American Society of Plant Physiologists, Rockville, MD, pp 210–218
- Jones DD, Morré DJ. Golgi apparatus meditated polysaccharide secretion by outer cap cells of Zea mays. II. Isolation and characterization of the secretory product. Z Pflanzenphysiol. 1967;56:166–169. [Google Scholar]
- Keller B, Templeton MD, Lamb CJ. Specific localization of a plant cell wall glycine-rich protein in protoxylem cells of the vascular system. Proc Natl Acad Sci USA. 1989;86:1529–1533. doi: 10.1073/pnas.86.5.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui M, Toyosawa I, Fukuda M. Purification and characterization of a glycine-rich protein from the aleurone layer of soybean seeds. Biosci Biotechnol Biochem. 1995;59:2231–2234. doi: 10.1271/bbb.58.1920. [DOI] [PubMed] [Google Scholar]
- Matsuyama T, Yasumura N, Funakoshi M, Yamada Y, Hashimoto T (1999) Maize genes specifically expressed in the outermost cells of root cap. Plant Cell Physiol 40 (in press) [DOI] [PubMed]
- Mohnen D, Shinshi H, Felix G, Meins F. Hormonal regulation of β-1,3-glucanase messenger RNA levels in cultured tobacco tissues. EMBO J. 1985;4:1631–1635. doi: 10.1002/j.1460-2075.1985.tb03830.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollenhauer HH, Whaley WG, Leech JH. A function of the Golgi apparatus in outer rootcap cells. J Ultrastruct Res. 1961;5:193–200. doi: 10.1016/s0022-5320(61)90014-4. [DOI] [PubMed] [Google Scholar]
- Montoliu L, Rigau J, Puigdomènech P. A tandem of α-tubulin genes preferentially expressed in radicular tissues of Z. mays. Plant Mol Biol. 1989;14:1–15. doi: 10.1007/BF00015650. [DOI] [PubMed] [Google Scholar]
- Moore R, McClelen CE. Ultrastructural aspects of cellular differentiation in the root cap of Zea mays. Can J Bot. 1983;61:1566–1572. [Google Scholar]
- Morré DJ, Jones DD, Mollenhauer HH. Golgi apparatus meditated polysaccharide secretion by outer root cap cells of Zea mays. I. Kinetics and secretory pathway. Planta. 1967;74:286–301. doi: 10.1007/BF00384849. [DOI] [PubMed] [Google Scholar]
- Murray MG, Thompson WF. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980;8:4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paull RE, Johnson CM, Jones RL. Studies on the secretion of maize root cap slime. I. Some properties of the secreted polymer. Plant Physiol. 1975;56:300–306. doi: 10.1104/pp.56.2.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell RS. Plant Root Systems. London: McGraw-Hill; 1977. [Google Scholar]
- Ryser U, Keller B. Ultrastructural localization of a bean glycine-rich protein in unlignified primary walls of protoxylem cells. Plant Cell. 1992;4:773–783. doi: 10.1105/tpc.4.7.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryser U, Schorderet M, Zhao G, Studer D, Ruel K, Hauf G, Keller B. Structural cell-wall proteins in protoxylem development: evidence for a repair process mediated by a glycine-rich protein. Plant J. 1997;12:97–111. doi: 10.1046/j.1365-313x.1997.12010097.x. [DOI] [PubMed] [Google Scholar]
- Showalter AM. Structure and function of plant cell wall proteins. Plant Cell. 1993;5:9–23. doi: 10.1105/tpc.5.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers A, Braun M. The root cap: structure and function. In: Waisel Y, Eshel A, Kafkafi U, editors. Plant Roots: The Hidden Half, Ed 2. Madison, WI: Marcel Dekker; 1996. pp. 31–49. [Google Scholar]
- Vermeer J, McCully ME. The rhizosphere in Zea: new insight into its structure and development. Planta. 1982;156:45–61. doi: 10.1007/BF00393442. [DOI] [PubMed] [Google Scholar]
- von Heijne G. Signal sequences: the limits of variation. J Mol Biol. 1985;184:99–105. doi: 10.1016/0022-2836(85)90046-4. [DOI] [PubMed] [Google Scholar]
- Wang Z, Brown DD. A gene expression screen. Proc Natl Acad Sci USA. 1991;88:11505–11509. doi: 10.1073/pnas.88.24.11505. [DOI] [PMC free article] [PubMed] [Google Scholar]