Abstract
Recently, FGFR1 was found to be overexpressed in osteosarcoma and represents an important target for precision medicine. However, because targeted cancer therapy based on FGFR inhibitors has so far been less efficient than expected, a detailed understanding of the target is important. We have here applied proximity-dependent biotin labeling combined with label-free quantitative mass spectrometry to identify determinants of FGFR1 activity in an osteosarcoma cell line. Many known FGFR interactors were identified (e.g. FRS2, PLCG1, RSK2, SRC), but the data also suggested novel determinants. A strong hit in our screen was the tyrosine phosphatase PTPRG. We show that PTPRG and FGFR1 interact and colocalize at the plasma membrane where PTPRG directly dephosphorylates activated FGFR1. We further show that osteosarcoma cell lines depleted for PTPRG display increased FGFR activity and are hypersensitive to stimulation by FGF1. In addition, PTPRG depletion elevated cell growth and negatively affected the efficacy of FGFR kinase inhibitors. Thus, PTPRG may have future clinical relevance by being a predictor of outcome after FGFR inhibitor treatment.
The fibroblast growth factor receptor (FGFR)1 family consists of four receptor tyrosine kinases (FGFR1–4), which are composed of an extracellular ligand binding part, a single transmembrane spanning stretch and an intracellular domain containing a tyrosine kinase (1, 2). Upon ligand (FGF) binding, dimerization causes the receptors to auto-transphosphorylate, leading to activation of downstream signaling cascades that regulate many key cellular responses such as proliferation, differentiation and cell migration. Importantly, aberrant FGF signaling is often involved in cancer development (1, 3). FGFR overexpression and activating mutations have recently been demonstrated to play an important role in several types of cancer, including sarcoma (e.g. osteosarcoma, rhabdomyosarcoma (RMS) and soft tissue sarcoma) (4–8). In addition, the FGFR-specific downstream signaling adaptor, the FGFR substrate 2 (FRS2), is overexpressed in liposarcoma and renders these cells sensitive to FGFR inhibitors (9, 10).
The incidence of sarcoma in adults is low (approx. 1% of all cancers), but more frequent in children and adolescents (approx. 10%) (8). There is little commercial interest in these small and heterogeneous patient groups, and for the same reasons, they are difficult to investigate and it is challenging to develop better treatments. There are, however, several initiatives to develop drugs specific for FGFRs that possibly could also be used to treat sarcomas with aberrant FGFR signaling (11). Most of these involve the development of specific small-molecular tyrosine kinase inhibitors and some have entered clinical trials for instance in patients with glioma, renal clear cell carcinoma, breast and lung cancer (ClinicalTrials.gov).
Unfortunately, in some cases such inhibitors fail even in the presence of the FGFR biomarker, for unknown reasons (12). There have also been reported effects of FGFR inhibitors in osteosarcoma cells without apparent FGFR aberrations, indicating that other mechanisms for FGFR vulnerability exist (9, 13). To increase the impact of FGFR inhibitors, it is crucial to understand in detail how their action on FGFR signaling and cell viability is determined.
As FGFR1 is overexpressed in 18.5% of osteosarcomas with poor response to chemotherapy and constitute a new and important therapeutic target for these patients (14, 15), we wanted to better understand how FGFR signaling is regulated. We, therefore, took advantage of the BioID proximity biotinylation system to identify determinants of FGFR1 signaling in osteosarcoma cells (16). Using this approach, we discovered that the tyrosine phosphatase receptor type G (PTPRG) negatively regulates FGFR1 activation in osteosarcoma. Cells depleted for PTPRG exhibit increased activation of FGFR and are more sensitive in mitogenic responses to FGF stimulation. Thus, PTPRG seems to be important for controlling excessive FGFR signaling, which corresponds well with previous reports that implicate PTPRG as a tumor suppressor (17, 18). Importantly, we found that PTPRG determines the sensitivity of cells to kinase inhibitors of FGFRs. We believe this may have clinical relevance as clinical cases with overexpressed FGFR1 combined with low expression of PTPRG have been reported.
EXPERIMENTAL PROCEDURES
Antibodies and Compounds
The following antibodies were used: rabbit anti-FGFR1 (ab76464), rabbit anti-Clathrin heavy chain (ab21679), mouse anti-COTL1 (ab187608), and rabbit anti-SLC20A1 (ab177147) from Abcam (Cambridge, UK); rabbit anti-FGFR1 (2144–1) from Epitomics (Burlingame, CA); rabbit anti-VAMP4 (136002) from SYSY (Goettingen, Germany); mouse anti-phospho-FGFR (Tyr653/654) (#3476), rabbit anti-FGFR1 (#9740), rabbit anti-DYKDDDDK (FLAG) tag (#2368), rabbit anti-phospho-PLCG1 (Tyr783) (#14008), mouse anti-phospho-ERK1/2 (Thr202/Tyr204) (#9106), rabbit anti-RSK2 (#5528), rabbit anti-OAS1 (#14498) and rabbit anti-PTPN1 (#5311) from Cell Signaling Technology (Leiden, The Netherlands); mouse anti-γ-tubulin (T6557), and mouse anti-FLAG M2 antibody (F-1804) from Sigma-Aldrich (St. Louis, MO); mouse anti-EEA1 (610456) from BD transduction laboratories (San Jose, CA); rabbit anti-phospho-PLCG1 (Tyr783) (sc-12943-R), rabbit anti-FRS2 (sc-8318), and mouse anti-PLCG1 (sc-7290) from Santa Cruz Biotechnology (Dallas, TX); rabbit anti-HA epitope tag (600–401-384) from Rockland (Limerick, PA); mouse anti-MYC Tag (05–724) from Merck Millipore (Burlington, MA), human anti-EEA1 antiserum was a gift from B. H. Toh (Monash University, Melbourne, Australia), HRP-Streptavidin (016–030-084), Alexa488-Streptavidin (016–540-084) and all secondary antibodies from Jackson Immuno-Research Laboratories (Cambridgeshire, UK). Rabbit anti-SHC4 antibody has been described previously (19).
Protease inhibitor mixture tablets (ethylenediaminetetraacetic acid (EDTA)-free, complete) were from Roche Diagnostics (Basel, Switzerland). DyLight 550 NHS Ester, Ez-link Sulfo-NHS-SS-biotin, PierceTM anti-HA magnetic beads and Dynabeads G protein were from Thermo Fisher Scientific (Waltham, MA). Hoechst 33342 was purchased from Life Technologies (Carlsbad, CA). Streptavidin Sepharose High Performance was from GE Healthcare Life Sciences (Chicago, IL). Mowiol, biotin, heparin, PD173074, active human PTPRG catalytic domain (SRP0223), active human PTPN12 catalytic domain (SRP5073), sodium orthovanadate, p-nitrophenyl phosphate (pNPP) and phosphatase inhibitors were from Sigma-Aldrich. AZD4547 was purchased from SelleckChem (Munich, Germany). FGF1 was prepared as previously described (20). FGF1 was labeled with DyLight 550 following the manufacturer's procedures. Recombinant GST, expressed in E. coli and purified using GSH Sepharose Fast Flow (GE Healthcare), was kindly provided by Dr. Coen Campsteijn from the Department of Molecular Cell Biology, Institute for Cancer Research, Oslo University Hospital.
Plasmids and siRNAs
pcDNA3.1-FGFR1-BirA* was made by cloning a PCR fragment containing the FGFR1 open reading frame and AgeI-HF and BamHI-HF flanking sites into pcDNA3.1 MCS-BirA*(R118G)-HA cut with AgeI-HF and BamHI-HF using pcDNA3-hFGFR1 as a template (21). pcDNA3.1 MCS-BirA(R118G)-HA was a gift from Kyle Roux (Addgene plasmid # 36047) (16). Construction of the pcDNA3.1/Zeo-BirA* was described previously (22). pEGFP-FGFR1 was made by cloning a PCR fragment containing the FGFR1 open reading frame and XhoI and ApaI flanking sites into pEGFP-N1 cut with XhoI and ApaI using pcDNA3-hFGFR1 as template. pcDNA3-SBP-FGFR1 was made by introducing a sequence coding for the streptavidin-binding peptide (SBP) after the signal peptide in FGFR1 using a GeneArt Strings DNA fragment (Thermo Fisher Scientific). The cleavage site of the signal peptide was predicted by SignalIP4.1 (23) to be between the amino acids CTA and RP in FGFR1. pCMV6-Entry vector containing PTPRG-MYC-FLAG was purchased from Origene (Rockville, MD) (RC_218964). PTPRG mutants were produced by site-directed mutagenesis using Pfu I HF (Agilent, Santa Clara, CA) with specific primers, followed by DpnI treatment. All restriction enzymes were from New England Biolabs (Ipswich, MA). PTPRG inactivating mutation (D1028A) was introduced with the primer 5′-TACACAGTGGCCTGCCATGGGAGTTCCCG-3′, whereas the primer 5′-CATTAGCCATGTCTCACCCGATAGTCTATATTTATTTCGGGTCCAGGCCGTGTGTCGGAACGAC-3′ was used to mutate 7 nucleotides and obtain siRNA-Resistant PTPRG (siRes#1 PTPRG) in both wild-type and D1028A mutant PTPRG. D1028A and siRes mutants were verified by sequencing. These plasmids are resistant to siRNA oligo s11549 (#1) PTPRG Silencer® Select. Silencer® Select siRNA oligos targeting PTPRG, s11549 (#1), s11550 (#2) and s11551 (#3); siRNA oligo targeting FGFR1 (s5177); siRNA oligos targeting PTPN1, s11506 (#1), s11507 (#2), s11508 (#3); siRNA oligos targeting PTPN13, s11527 (#1), s11528 (#2), s11529 (#3) and Silencer® select Negative Control No. 2 siRNA (scr) (4390846) were purchased from Life Technologies.
Cells and Transfection
To obtain U2OS cells stably expressing FGFR1-BirA* (U2OS-R1-BirA*), FGFR1-GFP (U2OS-R1-GFP), SBP-FGFR1 (U2OS-SBP-R1), FGFR2 (U2OS-R2) and U2OS-R1 stably expressing BirA* (U2OS-R1 + BirA*), Fugene liposomal transfection reagent was used according to the manufacturer's protocol. Clones were selected with 1 mg/ml geneticin (U2OS-R1-BirA*, U2OS-R1-GFP, and U2OS-SBP-R1) or 0.2 mg/ml Zeocin (U2OS-R1 + BirA*). Clones were chosen based on their receptor/BirA* expression levels analyzed by immunofluorescence and Western blotting. U2OS cells stably expressing FGFR1 (U2OS-R1), FGFR3 (U2OS-R3) and FGFR4 (U2OS-R4) have been described previously (24, 25). The G292 and RH30 cell lines were generous gifts from Prof. Ola Myklebost (Department of Tumor Biology, The Norwegian Radium Hospital). U2OS and G292 cells were propagated in DMEM or RPMI (respectively) supplemented with 10% fetal bovine serum, 100 U/ml penicillin, and 100 μg/ml streptomycin in a 5% CO2 atmosphere at 37 °C.
siRNA transfection was performed using Lipofectamine RNAiMAX Transfection Reagent (Invitrogen, Life Technologies) according to the manufacturer's protocol. 10 nm of siRNA was used and the experiments were performed 72 h after transfection. Transient expression of different plasmids was performed by transfecting cells with plasmid DNA using Fugene 6 Transfection Reagent (Roche Diagnostics) according to the manufacturer's protocol.
Affinity Capture of Biotinylated Proteins
Cells were incubated for 24 h in complete media supplemented with 50 μm biotin in the absence or presence of 100 ng/ml FGF1 and 10 U/ml heparin. After one PBS wash, cells (∼1 × 108 cells) were scraped in PBS supplemented with 100 mm Glycine. Cells were pelleted by centrifugation for 10 min at 4000 rpm and lysed in 1 ml lysis buffer (0.1 m NaCl, 10 mm Na2PO4, 1% triton X-100, and 1 mm EDTA, pH 7.4, supplemented with protease and phosphatase inhibitors). Lysates were then centrifuged at 14,000 rpm for 10 min at 4 °C. Supernatants were incubated with 300 μl Streptavidin-Sepharose High Performance for 2 h. Beads were collected and washed twice for 5 min at 4 °C in 1% Triton X-100, 500 mm NaCl, 1 mm EDTA, twice for 5 min at 4 °C in PBS with 0.1% Triton X-100, twice for 5 min at room temperature in 2% SDS, twice for 5 min at room temperature in 6 m Urea in PBS, and six times for 5 min at room temperature in 1 m NaCl, 25% Acetonitrile and twice for 5 min at room temperature in 20% Acteonitrile. Bound proteins were trypsin-digested on the beads.
Sample Preparation for Mass Spectrometry
Beads containing bound proteins were submitted to in solution trypsin digestion in 100 μl of 0.1% ProteaseMax (Promega, Madison, WI), containing 3.6 μg of trypsin (Modified, Promega). Trypsin reaction was performed overnight in a wet chamber at 37 °C. Reaction was quenched by adding 1% trifluoroacetic acid to the mixture (final concentration). Peptides were cleaned for mass spectrometry by the STAGE-TIP method (26) using a C18 resin disk (3 m Empore).
Mass Spectrometry
All experiments were performed on an Easy nLC1000 nano-LC system connected to a quadrupole - Orbitrap (QExactive) mass spectrometer (ThermoElectron, Bremen, Germany) equipped with a nanoelectrospray ion source (EasySpray/Thermo). For liquid chromatography separation we used an EasySpray column (C18, 2 μm beads, 100 Å, 75 μm inner diameter) (Thermo) capillary of 25 cm bed length. The flow rate used was 300 nL/min, and the solvent gradient was 2% B to 30% B in 120 min, then 90% B wash in 20 min. Solvent A was aqueous 0.1% formic acid, whereas solvent B was 100% acetonitrile in 0.1% formic acid. Column temperature was kept at 60 °C.
The mass spectrometer was operated in the data-dependent mode to automatically switch between MS and MS/MS acquisition. Survey full scan MS spectra (from m/z 400 to 1,200) were acquired in the Orbitrap with resolution r = 70,000 at m/z 200 (after accumulation to a target of 3,000,000 ions in the quadruple). The method used allowed sequential isolation of the most intense multiply-charged ions, up to ten, depending on signal intensity, for fragmentation on the HCD cell using high-energy collision dissociation at a target value of 100,000 charges or maximum acquisition time of 100 ms. MS/MS scans were collected at 17,500 resolution at the Orbitrap cell. Target ions already selected for MS/MS were dynamically excluded for 30 s. General mass spectrometry conditions were: electrospray voltage, 2.1 kV; no sheath and auxiliary gas flow, heated capillary temperature of 250 °C, normalized HCD collision energy 25%. Ion selection threshold was set to 1e4 counts. Isolation width of 3.0 Da was used.
Protein Identification and Label-free Quantitation
MS raw files were submitted to MaxQuant software version 1.5.2.8 (27) for protein identification using its Andromeda engine. Parameters were set as follow: protein N-acetylation, methionine oxidation and phospho (Ser, Thr and Tyr) as variable modifications. First search error window of 20 ppm and both precursor and MS/MS main search error set to 6 ppm for precursor ions. Trypsin without proline restriction enzyme option was used, with two allowed miscleavages. Minimal unique peptides were set to 1, and FDR allowed was 0.01 (1%) for peptide and protein identification. Label-free quantitation was set with a retention time alignment window of 3 min. The Uniprot human database was used (download from October 2014, with 85,915 entries). Generation of reversed sequences was selected to assign FDR rates. Known contaminants as provided by MaxQuant and identified in the samples were excluded from the analysis. Proteins identified by a single-peptide were removed and not taken into consideration to any of the analysis that was performed. The complete datasets (raw files, and MaxQuant output files) have been uploaded to ProteomeXchange.
Experimental Design and Statistical Rationale
Six individual experiments were performed; three experiments consisting of samples C1 (U2OS-R1 cells), C2 (U2OS-R1 stably transfected with BirA*) and C3 (U2OS-R1 cells stably transfected with BirA* and stimulated with FGF1) and three experiments consisting of samples C1 (U2OS-R1 cells), S1 (U2OS-R1-BirA*) and S2 (U2OS-R1-BirA* stimulated with FGF1). All three samples in each of the six individual experiments were run three times (n = 3 for LC variability, n = 9 total number of replicates combined, in the case of C1: n = 6 for LC variability, n = 18 total number of replicates combined). In the case of one of the three experiments for C3 (U2OS-R1 cells stably transfected with BirA* and stimulated with FGF1) only one replicate was run (n = 3 for LC viability, n = 7 total number of replicates combined). The mean IBAQ values were calculated for each protein in each sample (C1, C2, C3, S1, and S2). Proteins identified in C1 were considered as background and the means of C3, S1, and S2 were compared with that of C1. Proteins were removed from the list if they were not significantly enriched at least ten times compared with C1 (p < 0.05, two-tailed t test). Proteins identified in C2 were considered as BirA* background and the means of C3, S1, and S2 were next compared with that of C2. Proteins were removed from the list if they were not significantly enriched at least ten times compared with C2 (p < 0.05, two-tailed t test). Proteins significantly enriched ten times or more in C3 compared with C1 and C2 were considered as proteins with potentially induced expression by FGF1 stimulation (p < 0.05, two-tailed t test). Proteins significantly enriched ten times or more in S1 compared with C1 and C2 were considered as proteins in proximity to FGFR1. S2 was in addition to being compared with C1 and C2 also compared with C3 and proteins were removed from the list if they were not significantly enriched at least ten times compared with C3 (p < 0.05, two-tailed t test). Proteins significantly enriched ten times or more in S2 compared with C1, C2, and C3 were considered as proteins in proximity to active FGFR1.
Western Blotting
After indicated treatment, cells were lysed in lysis buffer supplemented with protease and phosphatase inhibitors or directly in sample buffer and the lysates were then loaded for SDS-PAGE (4–20% gradient) and afterward transferred to a PVDF membrane (Bio-Rad, Hercules, CA) for Western blotting. Blots were developed with SuperSignal West Dura Extended Chemiluminescent Substrate (Thermo Scientific) and detected using ChemiDoc XRS+ (Bio-Rad). Western blots were quantified using the Gel analysis function in Image J (28).
RNA Isolation, cDNA Synthesis and Quantitative Real-time Polymerase Reaction (qRT-PCR)
Total RNA was isolated from cell lysate using RNeasy plus minikit and the QIAcube robot (Qiagen, Hilden, Germany) according to the manufacturer's protocol. Then 0.5–1 mg of RNA was used for cDNA synthesis using iScript cDNA synthesis kit. Quantitative real-time PCR was performed using QuantiTect SYBR Green PCR kit, cDNA template and the following QuantiTect primers: PTPRG (QT00060116), PTPN13 (QT00054446) and Succinate dehydrogenase (SDHA) (QT00059486). The qRT-PCR was run and analyzed using the Lightcycler 480 (Roche). Cycling conditions were 5 min at 95 °C followed by 45 cycles 10 s at 95 °C, 20 s at 60 °C and 10 s at 72 °C. Gene amplification was normalized to the expression of SDHA.
SBP Affinity Purification Assay
The cells (∼108) after indicated treatment were washed with ice-cold PBS, scrapped and centrifuged at 4000 rpm for 10 min. The pellet was lysed in lysis buffer with addition of protease and phosphatase inhibitors for 10 min on ice. The cleared lysates were incubated with streptavidin Sepharose for 2 h at 4 °C. followed by triple wash with lysis buffer. The protein complexes were released by SDS-PAGE loading buffer, separated in electrophoresis and analyzed by Western blotting.
Coimmunoprecipitation and Pulldown Assays
After indicated treatment, the cells were lysed in lysis buffer supplemented with protease and phosphatase inhibitors. Cell lysates were then subjected to immunprecipitation reactions with indicated antibody immobilized to Dyneabeads Protein G or with PierceTM anti-HA magnetic beads. In the case of affinity capture of biotinylated proteins for Western blotting, samples were treated as described above.
After washing, proteins were eluted in sample buffer, separated by SDS-PAGE and analyzed by Western blotting. Biotinylated proteins were eluted from streptavidin beads by boiling for 15 min in sample buffer containing 3 mm biotin.
In Vitro Phosphatase Assay
The enzymatic activity of recombinant PTPRG (catalytic domain, residues 801–1147) and PTPN12 (catalytic domain, residues 1–355) was probed by a standard colorimetric assay using p-nitrophenyl phosphate (pNPP) as substrate (29). The initial reaction rate was monitored colorimetrically (Abs. at 405 nm) within the first 10 min of reaction, where the data fell in the linear range. The reaction buffer and 300 nm GST in reaction buffer served as control to exclude substrate self-degradation and the effect of potential impurities related to the GST fusion protein purification system. One unit of phosphatase activity (1 U) was defined as the amount of enzyme that hydrolyzes 1 nmol of pNPP in 1 min at 30 °C in 50 μl reaction volume. Molar extinction coefficient of the reaction product (pNP) was assumed as 18,000 m−1cm−1.
After indicated treatment, U2OS-R1-BirA* cells were lysed in lysis buffer supplemented with protease and phosphatase inhibitors. Cell lysates were then subjected to immunprecipitation with PierceTM anti-HA magnetic beads (Thermo Scientific), which were subsequently washed with lysis buffer without phosphatase inhibitors and incubated at 37 °C with indicated recombinant phosphatases with addition of 2 mm DTT. The control samples were incubated with recombinant GST or in the presence of phosphatase inhibitor mixture, as indicated in the figure legend. The immunoprecipitates were then eluted in sample buffer, separated by SDS-PAGE and analyzed by Western blotting.
Light Microscopy
For confocal microscopy, cells grown on coverslips were treated as indicated and fixed in 4% formaldehyde. The cells were then permeabilized with 0.1% triton X-100, stained with indicated antibodies and mounted in mowiol. Confocal images were acquired with a 63× objective on Zeiss LSM 780 and Zeiss LSM 710 confocal microscopes (Jena, Germany). Images were prepared with Zeiss LSM Image Browser and CorelDRAW11 (Ottawa, Canada).
For wide-field (WF) microscopy and structured illumination microscopy (SIM), U2OS-R1 cells were grown on 1.5H glass coverslips and transiently transfected with plasmid encoding MYC/FLAG-tagged PTPRG or PTPRG-D1028A using Fugene 6 (according to the manufacturer's procedures), for approx. 20 h. The cells were serum starved for two hours (DMEM with penicillin and streptomycin but without serum), and then either fixed immediately or incubated with FGF1 (200 ng/ml) and heparin (10 U/ml) for 1 h and then fixed.
For total internal reflection fluorescence (TIRF) microscopy, U2OS-R1-GFP cells were grown in glass bottom culture dishes (MatTek, Ashland, MA). The cells were transfected with plasmid encoding MYC/FLAG-tagged PTPRG or PTPRG-D1028A or siRNA resistant versions of these (using Fugene 6) for 20 h. Next, the cells were serum-starved for 2 h and stimulated for 10 min with FGF1 and heparin, FGF1 and heparin in the presence of PD173074 (including 30 min pretreatment with PD173074), or no FGF1, and then the cells were fixed. In some cases, the cells were also transfected with scrambled siRNA or siRNA against PTPRG (siRNA #1) 2 days before plasmid transfection.
Next, cells were fixed in 4% formaldehyde (Sigma-Aldrich) in PBS (10 min at room temperature). The fixed cells were permeabilized with 0.05% saponin in PBS and stained with indicated combinations of primary antibodies diluted in PBS with 0.05% saponin, and anti-mouse/rabbit/human secondary antibodies labeled with Alexa Fluor 488, Alexa Fluor 568, or Alexa Fluor 647. Cells/coverslips for WF/SIM were also stained with Hoechst33342 and mounted on object slides with SlowFade Diamond Antifade Mountant (ThermoFisher). Stained cells for TIRF microscopy were maintained and imaged in PBS.
Wide-field, SIM, and TIRF imaging was performed on a Deltavision OMX V4 microscope (Applied Precision, Inc., Issaquah, WA) using an Olympus ×60 NA 1.42 Plan Apochromat objective for WF imaging and SIM, and an Olympus x60 NA 1.49 Plan Apo TIRF objective for TIRF imaging. The OMX is further equipped with an InSightSSI™ illumination module used for WF imaging, 405 nm, 488 nm, 568 nm, and 642 nm laserlines that were used for SIM and TIRF imaging, a Ring-TIRF module, and three cooled sCMOS cameras.
For WF imaging, z-stacks covering the whole cell were recorded with a z-spacing of 250 nm. For SIM, z-stacks were recorded with a z-spacing of 125 nm and for each focal plane, 15 raw images (five phases for three different angular orientations of the illumination pattern) were captured. WF images were deconvolved, SIM images were reconstructed, and all images were aligned using Softworx software (Applied Precision).
All TIRF images were captured using the same channel specific settings for Ring-TIRF diameter, laser intensity and exposure. The phospho-FGFR1 signal was quantified using Fiji/ImageJ software as follows; Cells were selected for quantification based on GFP intensity (indicating average/normal FGFR1 levels) and identified as untransfected or transfected with PTPRG/PTPRG-D1028A based on FLAG-staining. ROI's were defined by drawing the outline of selected cells, and the mean pixel value over an ROI in the phospho-FGFR1 specific channel was taken as the measure of the phospho-FGFR1 signal intensity of a cell. Images were subjected to background subtraction by a value set for each experiment. Data presented are the mean values of three or four independent experiments where 15–30 cells were measured for each condition in each experiment.
Further processing of images for presentations (projections, volume views, contrast adjustments, montages) were performed using Fiji/ImageJ software.
Cell Viability Assay
The cells were treated with indicated siRNAs and reseeded into 96-well plates the day before stimulation with FGF1 in serum-free medium, supplemented with 20 U/ml heparin. The cells were treated with FGF1 72 h after siRNA treatment. Cell viability was measured 48 h after stimulation using CellTiter-Glo assay (Promega). In the case of FGFR1 knockdown, cells were treated with FGFR1 siRNA or control siRNA (scr) for 72 h. During the last 48 h the cells were treated with 100 ng/ml FGF1 in the presence of 10 U/ml heparin. Cell viability was then measured using CellTiter-Glo assay.
Statistical Rationale
Data arised from series of three or more independent experiments as stated in figure legends. Results with p < 0.05 were considered statistically significant. Time-course and dose-response data series were analyzed using two-way ANOVA. Single end-point assay data were analyzed using one-way repeated measures (RM) ANOVA followed by Tukey post hoc test. For all experiments, the tests were performed on log transformed raw data. The tests were performed using GraphPad Prism 5 (GraphPad Software, La Jolla, CA) or Sigma plot (Systat Software, San Jose, CA).
RESULTS
Proteomic Screen Identifies Determinants of FGFR1 Activity
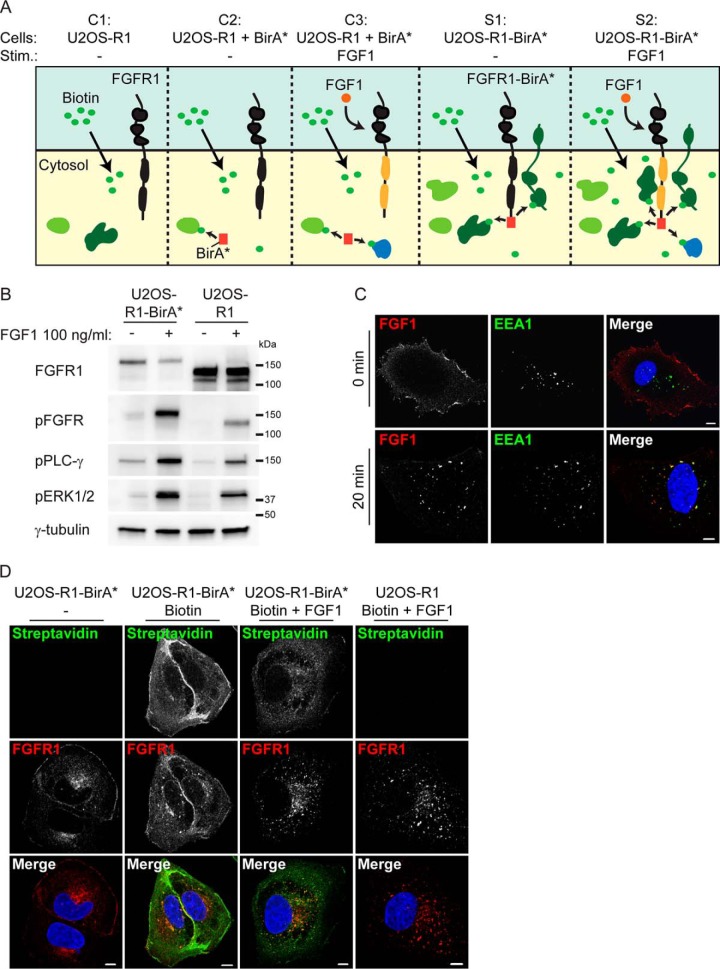
To identify proteins involved in the regulation of FGFR1 signaling in osteosarcoma cells, we performed a modified BioID screen by fusing a biotin ligase, BirA*, to the C-terminal tail of FGFR1. We recently validated and used this system to investigate signaling and trafficking of the related FGFR4 (22). When expressed in U2OS cells the biotin ligase biotinylates proteins in its proximity upon addition of biotin. The biotinylated proteins may be isolated by affinity to streptavidin and identified by quantitative proteomics using mass spectrometry (MS). Because FGFR1-regulating proteins could be proximal to the receptor both in its inactive or its active state, we included samples of unstimulated and FGF1-stimulated U2OS cells stably expressing FGFR1 fused to BirA* (U2OS-R1-BirA*) (see S1 and S2 in Fig. 1A).
Fig. 1.
A biotin ligase proteomic screen for determinants of FGFR1 activity in osteosarcoma cells. A, A schematic presentation of our BioID experiment. Upon addition of biotin to cells, proteins near the BirA* tag will be biotinylated. Biotinylated proteins are then isolated by Streptavidin pulldown and identified by quantitative LC MS/MS. The following five conditions are compared: C1 (U2OS-R1 cells), C2 (U2OS-R1 cells coexpressing BirA*), C3 (U2OS-R1 cells coexpressing BirA*, FGF1 stimulated), S1 (U2OS-R1-BirA* cells) and S2 (U2OS-R1-BirA* cells, FGF1 stimulated). Biotin is added in all conditions. Addition of FGF1 induces activation of the receptor and its downstream signaling (indicated in yellow). Proteins in proximity to the receptor are indicated by dark green and proteins further away from the receptor are indicated in light green. Proteins with increased expression upon FGF1 stimulation are indicated in blue. B, U2OS-R1 cells or U2OS-R1-BirA* cells were starved for 3 h in serum free media before stimulation for 20 min with 100 ng/ml FGF1 in the presence of heparin (20 U/ml). Cells were then lysed and the cellular material was analyzed by SDS-PAGE and Western blotting using the indicated antibodies. A p in front of the name of the antibody indicates that it recognizes the phosphorylated form of the protein. C, U2OS-R1-BirA* cells were allowed to bind DL550-FGF1 at 4 °C in the presence of heparin and then washed (to remove excess DL550-FGF1) and either fixed directly (0 min) or incubated for 20 min at 37 °C before fixation (20 min). Fixed cells were stained with anti-EEA1 antibody and Hoechst and examined by confocal microscopy. Scale bar 5 μm. D, U2OS-R1-BirA* and U2OS-R1 cells were either left untreated or treated with 50 mm biotin and/or 100 ng/ml FGF1 in the presence of 10 U/ml heparin as indicated for 24 h. The cells were then fixed and stained with anti FGFR1 antibody, Alexa 488 streptavidin and Hoechst. Merged images are shown in the bottom panel. Scale bar 5 μm.
Control samples included U2OS cells stably expressing FGFR1 wild-type (U2OS-R1, C1 in Fig. 1A) and U2OS cells stably coexpressing FGFR1 wild-type and control, nonfused BirA* (U2OS-R1 + BirA*, C2 in Fig. 1A). Proteins identified in these two conditions were considered as background. As FGFR signaling induces expression of certain proteins, increased expression of background proteins in FGF stimulated conditions could erroneously be considered as positive hits. We therefore also included samples of U2OS cells stably coexpressing FGFR1 wild-type and BirA* (U2OS-R1 + BirA*) and stimulated with FGF1 (C3 in Fig. 1A).
We first investigated whether the fusion of BirA* to FGFR1 could interfere with its functionality (Fig. 1B). After 20 min of stimulation with FGF1, we detected comparable phosphorylation of the receptors itself, as well as known downstream signaling molecules, such as PLCG1 and ERK1/2. Because, the total FGFR1 antibody used for these Western blots recognizes the wild-type receptor better than the tagged version, the staining underestimated the level of FGFR1-BirA* in these cells. Next, we tested whether the FGFR1-BirA* fusion protein can bind its ligand FGF1 at the cell surface and undergo endocytosis (Fig. 1C). Cells were kept on ice to facilitate binding of fluorophore-labeled FGF1 (DL550-FGF1) and heated to 37 °C to allow internalization. Then, the cells were stained and examined by confocal microscopy. DL550-FGF1 was clearly detected at the cell surface in cells kept on ice and next, after heating to 37 °C for 20 min, it was detected in intracellular structures colocalizing with EEA1. The results demonstrate that FGFR1-BirA* can bind FGF1 and internalize into early endosomes similarly to wild-type receptor (21).
Next, we tested the biotinylation efficiency of the fusion protein (Fig. 1D and supplemental Fig. S1). In the absence of biotin, little biotinylated proteins were detected in cells expressing FGFR1-BirA*. In the presence of biotin, a smear of bands representing biotinylated proteins was detected on Western blotting (supplemental Fig. S1) and a strong streptavidin staining was detected in cells by confocal microscopy (Fig. 1D). The streptavidin staining was stronger at the cell periphery close to the plasma membrane. When cells were treated with FGF1, the streptavidin staining was more dispersed in the cytoplasm, probably reflecting the transport of the receptor from the plasma membrane to endosomes upon FGF1 stimulation. Biotinylated proteins were barely visible in cells expressing FGFR1 wild-type and treated with biotin. Taken together these data indicate that the FGFR1-BirA* fusion protein is functional and active and efficiently biotinylates proteins in its proximity in the presence of biotin.
In the U2OS cells stably coexpressing nonfused BirA* and FGFR1 wild-type, a smear of bands representing biotinylated proteins were detected on Western blotting (supplemental Fig. S1) indicating that the biotin ligase is active in these cells. These cells express higher amounts of control BirA* than those expressing the FGFR1-BirA* fusion protein, which was an advantage to eliminate false positive hits in the screen.
Next, we performed the proximity labeling experiment and analyzed affinity-purified biotinylated proteins by label-free quantitative LC-MS/MS (Fig. 1A). Reassuringly, high reproducibility among the independent BirA*-MS experiments was confirmed by correlation analysis (supplemental Fig. S2).
Proteins significantly enriched more than 10× (p < 0.05) in S1 compared with C1 and C2 (Fig. 1A) were considered as proteins in proximity of nonstimulated FGFR1 (supplemental Table S1, supplemental Table S2), whereas proteins significantly enriched more than 10× (p < 0.05) in S2 compared with C1, C2, and C3 and, in addition enriched compared with S1, were considered as proteins with increased association or proximity to the active receptor (supplemental Table S1, supplemental Table S3). Proteins significantly enriched more than 10× (p < 0.05) in C3 compared with C1 and C2, were considered as proteins with increased expression upon FGF1 signaling (supplemental Table S1, (supplemental Table S4).
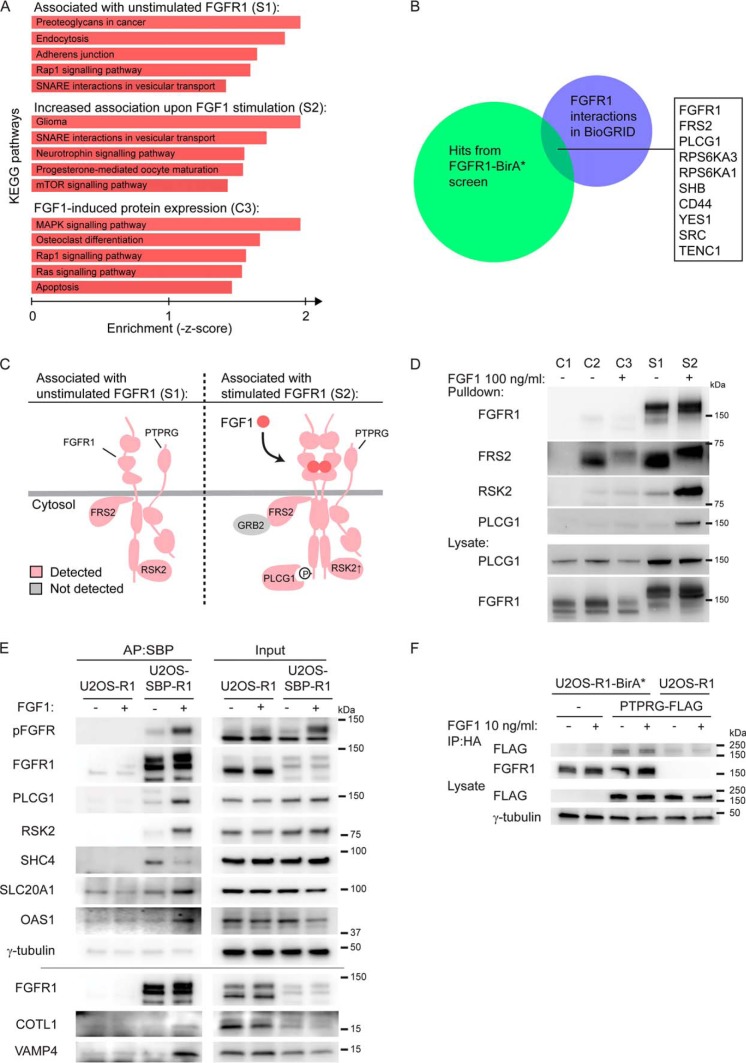
We then analyzed the data using Enrichr and KEGG Pathways applications (30), In the case of unstimulated receptors, the hits were enriched for plasma membrane functions (proteoglycans and adherens junctions) and membrane transport (endocytosis) (Fig. 2A), reflecting the known plasma membrane localization and the importance of trafficking of the receptors. Stimulated receptors also showed enrichment for membrane transport, but in addition several signaling pathways (Fig. 2A), as expected. In the case of FGF1-induced expression, we found enrichment of signaling pathways and, interestingly, induction of Osteoclast differentiation (Fig. 2A), which reflect the cell context of our analysis (osteosarcoma).
Fig. 2.
Analysis and validation of the proteomic screen A, KEGG pathways analyses were applied to the three datasets using Enrichr (http://amp. pharm.mssm.edu/Enrichr/) (30). B, A comparison of proteins identified in S1 and S2 with published interactions for FGFR1 denoted in BioGRID (35). C, Schematic illustration of selected proteins identified in the absence of FGF1 (S1) and in the presence of FGF1 stimulation (S2). Identified proteins are colored pink and some known proteins not detected are in gray. The enrichment of proteins in FGF1-stimulated cells versus unstimulated cells; PLCG1: 12.4X, RSK2: 4.0X, FGFR1: 1.0X, FRS2: 0.7X, PTPRG: 0.6X. D, U2OS-R1 control cells (C1), U2OS-R1 cells coexpressing BirA* (C2-C3), U2OS-R1-BirA* cells (S1-S2) were treated with biotin in the presence or absence of FGF1 for 24 h. The cells were then lysed and the biotinylated proteins were isolated by Streptavidin pulldown and analyzed by SDS-PAGE and Western blotting using the indicated antibodies. One representative of two independent experiment is shown. E, Validation of identified interactors by affinity purification (AP). U2OS-R1 and U2OS-SBP-R1 were starved for 2 h before addition of 10 ng/ml FGF1 in the presence of 20 U/ml heparin for 15 min, and then the cells were lysed. Proteins bound to SBP-FGFR1 were affinity-purified using streptavidin Sepharose and analyzed by Western blotting. One experiment of two is presented. F, U2OS-R1-BirA* and U2OS-R1 cells were transfected with PTPRG-MYC-FLAG plasmid for 24 h. U2OS-R1-BirA* cells not transfected with PTPRG were included as a control. Cells were then starved for 2 h and left untreated or treated with 10 ng/ml FGF1 in the presence of 20 U/ml heparin for 15 min. After that, the cells were lysed and the lysates were subjected to immunoprecipitation using anti-HA magnetic beads followed by SDS-PAGE and Western blotting with indicated antibodies. R1-BirA* is fused to an HA-tag in the C-terminal end.
Examining the hits form our proteomics analysis, we recognized several proteins previously found to interact with FGFR1 (Table I). Not surprisingly, FGFR1 itself was the top hit in our studies, but several other well known interaction partners of FGFR1 were identified (FRS2 (31), PLCG1 (32), RPS6KA3/RSK2 (33), and SRC (34)), thereby validating our approach (Table I, supplemental Tables S2 and S3). We compared our dataset with interactions to FGFR1 reported in the BioGrid database (35). Although there is some overlap between the two data sets (10 proteins, Fig. 2B, supplemental Table S5), there are considerable differences between the two. This underscore previously results showing that proximity biotinylation is a complementary approach compared with affinity purification (36).
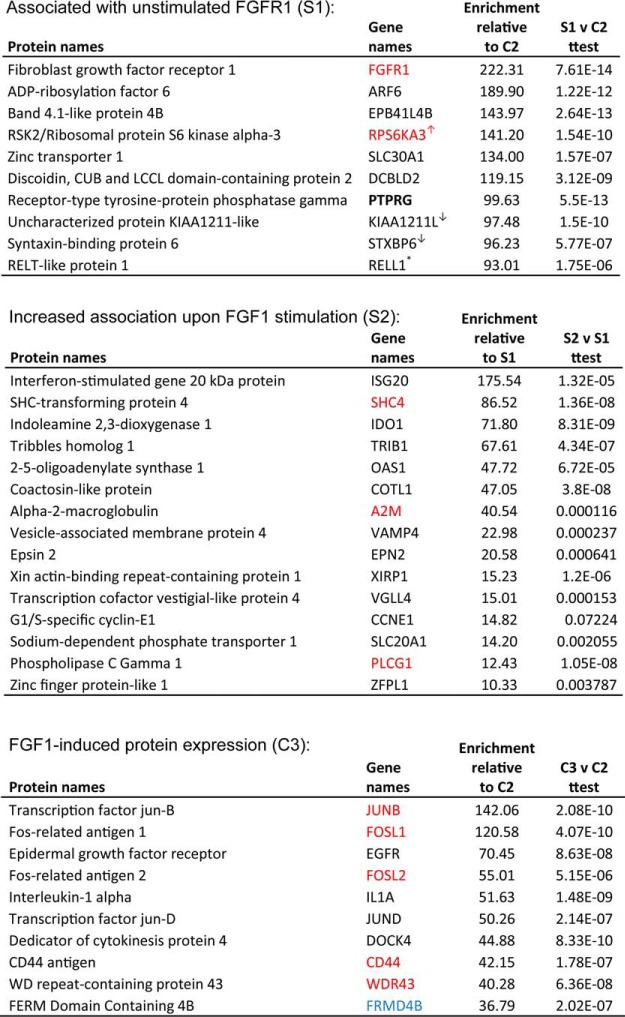
Table 1. Upper part: Top ten proteins of list 1; Associated with unstimulated FGFR1. Proteins significantly (p < 0.05) enriched in S1 (U2OS-R1-BirA* cells) compared to C1 (U2OS-R1 cells) and C2 (U2OS-R1 cells co-expressing BirA*). Proteins in red have previously been associated with FGFR1 as reported in BioGRID 3.4 (35). An arrow pointing upwards means that the protein was further enriched in samples stimulated with FGF1 whereas an arrow pointing down indicate that the protein was reduced in samples stimulated with FGF1. In bold, protein investigated further in this study. Middle part: Top 15 proteins of list 2; Increased association with FGF1 stimulation. Proteins significantly (p < 0.05) enriched in S2 (U2OS-R1-BirA* cells, FGF1 stimulated) compared to C1 (U2OS-R1 cells), C2 (U2OS-R1 cells coexpressing BirA*), C3 (U2OS-R1 cells coexpressing BirA*, FGF1 stimulated), and S1 (U2OS-R1-BirA* cells). Proteins in red have previously been associated with FGF signaling (35, 46, 53). Lower part: Top ten proteins of list 3; FGF induced gene expression. Proteins significantly (p < 0.05) enriched in C3 (U2OS-R1 + BirA* cells, FGF1 stimulated) compared to C1 (U2OS-R1 cells) and C2 (U2OS-R1 cells co-expressing BirA*). Proteins in red have previously been reported to be expressed upon FGF1 stimulation while proteins in blue were previously shown to be downregulated in response to FGF1 (54).
By comparing the data sets from samples before and after stimulation of FGF1, we could distinguish between proteins that are recruited to activated receptors and proteins that are constitutively associated with the receptor. Among the proteins enriched in samples of the activated receptor, we identified well-known FGFR downstream signaling proteins (PLCG1 and RSK2, Table I, Fig. 2C) (32, 33). FRS2 is known to be constitutively bound to FGFR1 (31) and was accordingly found to be in the proximity of both unstimulated and stimulated receptors (Fig. 2C). These data were confirmed by a small scale biotin ligase experiment where the biotinylated proteins that were pulled down by strepatavidin Sepharose were analyzed by Western blotting (Fig. 2D). PLCG1 and RSK2 were highly enriched in samples treated with FGF1, whereas FRS2 was found associated with both stimulated and unstimulated receptors. Thus, our screen can discriminate among interactions induced or increased by ligand activation to constitutively binding proteins.
As FGF1 treatment induces phosphorylation of intracellular proteins, we searched for phosphopeptides in our MS data. Although phosphopeptide enrichment was not performed in our experiment, we identified many phosphopeptides, which also clearly reflected the increased phosphorylation upon FGF1 treatment (supplemental Table S6). As expected, tyrosine phosphorylation of FGFR1 strongly increased upon addition of FGF1 (e.g. Tyr645 and Tyr655). Also, other phosphopeptides of relevant FGF signaling molecules were identified. For instance, phosphorylation of RPS6KA1 and RPS6KA3 (RSK1 and RSK2) was strongly increased upon FGF1 treatment (supplemental Table S6).
Because control BirA* is expressed uniformly in the cytosol and the nucleus, it biotinylates proteins in general and by comparing the control conditions with and without FGF1 stimulation, we could obtain an overview of which proteins are induced by FGF signaling in these cells. Proteins significantly enriched more than 10× (p < 0.05) in C3 compared with C1 and C2, were considered as proteins with increased expression upon FGF1 signaling (Table I, supplemental Table S4). Top hits among these were proteins known to be induced by FGF1 signaling, such as the transcription factors JUNB, FOSL1, and FOSL2 (Table I) (37).
To validate the obtained proximity labeling identifications we performed affinity purification experiments using U2OS cells stably expressing FGFR1 with an N-terminal streptavidin-binding peptide tag (U2OS-SBP-R1). As expected, we were able to copurify known FGFR1 interactors, PLCG1 and RSK2, in cells stimulated with FGF1 (Fig. 2E). Importantly, we were also able to detect several novel proteins that were found associated with activated FGF1 (S2) in our screen: sodium-dependent phosphate transporter 1 (SLC20A1), 2′-5′-oligoadenylate synthase 1 (OAS1), coactosin-like protein (COTL1) and vesicle-associated membrane protein 4 (VAMP4) (Fig. 2E). Moreover, we found SHC-transforming protein 4 (SHC4) copurified both with unstimulated and stimulated receptors. Intriguingly, a weaker band intensity for was observed for SHC4 in the activated FGFR1 in this assay (Fig. 2E), whereas the protein was found enriched upon receptor activation in the proximity screen (Table I). The discrepancy among the results for SHC4 may be explained by the different stimulation time: 24 h in the proximity screen and 15 min in case of the affinity assay. Possibly, the interaction dynamics is important in the case of SHC4.
A strong hit in our screen was the tyrosine phosphatases PTPRG (Table I) (38). Because PTPRG has previously been suggested to be a tumor suppressor (17), we chose to focus our attention to the possible regulatory role of PTPRG on FGFR1. Interestingly, PTPRG was found associated with both unstimulated and activated receptor (Fig. 2C, supplemental Table S2 and S3). To validate the interaction between PTPRG and FGFR1 that was suggested by the proximity labeling screen, we attempted coimmunoprecipitation of the two proteins. FGFR1-BirA* is fused to an HA-tag and U2OS-FGFR1-BirA* cells were transfected with MYC-FLAG-tagged PTPRG, lysed, and then immunoprecipitated with anti-HA antibodies and immunoblotted with anti-FLAG antibodies. The results demonstrated that PTPRG can efficiently be coimmunoprecipitated with FGFR1 indicating a physical interaction between PTPRG and FGFR1 (Fig. 2F).
FGFR1 is a Substrate for PTPRG
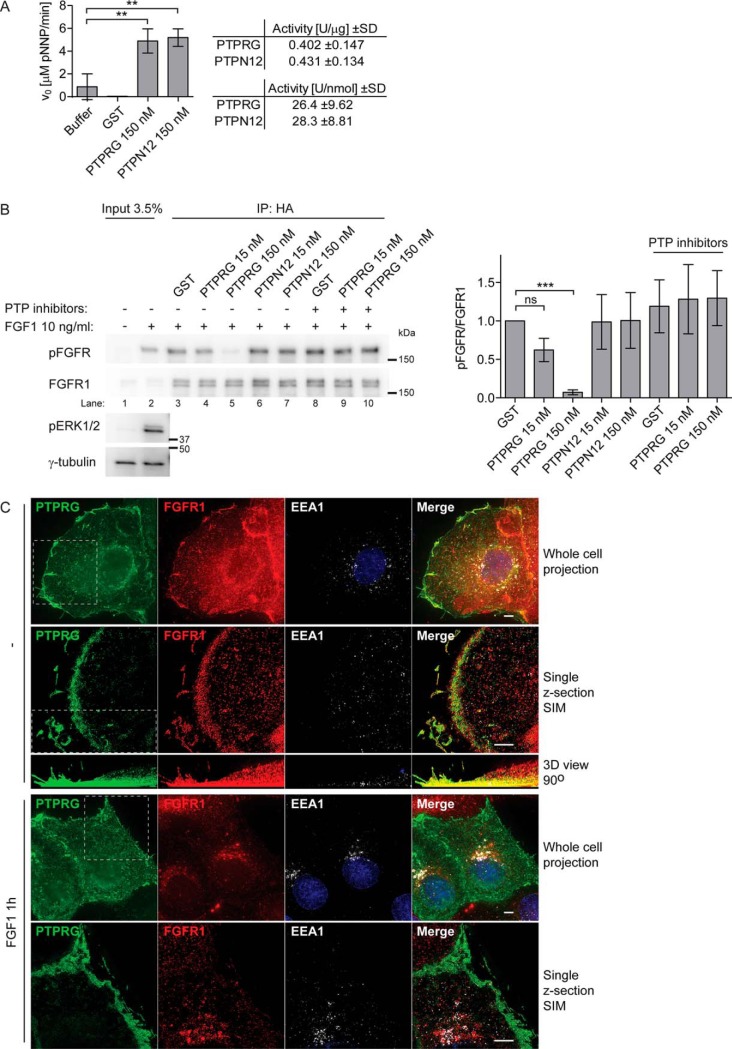
Because the PTPRG protein contains an active phosphatase domain, we considered FGFR1 as a potential substrate for PTPRG. To test this, we performed an in vitro phosphatase assay using activated FGFR1, which was immunoprecipitated from FGF1-treated U2OS-R1-BirA* cell lysates, and a recombinant PTPRG phosphatase domain in fusion with GST. As a control in the experiment, we also used the recombinant phosphatase domain of PTPN12, which has been shown to dephosphorylate other receptor tyrosine kinases (39), and exhibited phosphatase activity toward a nonspecific substrate (p-nitrophenyl phosphate, pNPP) comparable to PTPRG (Fig. 3A). After 45 min of incubation with 150 nm PTPRG we observed a significant decrease in the level of FGFR1 phosphorylation at Y653/Y654 residues, as detected by Western blotting, compared with incubation with 2-fold molar excess of GST (Fig. 3B, lane 5 compared with lane 3). Moreover, the dephosphorylation effect was less pronounced (and statistically insignificant) when we used 10-times lower concentration of the enzyme (Fig. 3B, lane 4), showing that the reduction in phospho-FGFR1 level is dependent on PTPRG concentration. We observed no changes in the phosphorylation of Y653/Y654 residues in FGFR1 using either 150 nm or 15 nm of PTPN12 (Fig. 3B, lane 6–7), suggesting a substrate specificity in the dephosphorylation of FGFR1 by PTPRG. Moreover, the dephosphorylation effect was not visible in the presence of a tyrosine-phosphatase inhibitor mixture (Fig. 3B, lane 9–10), confirming that the dephosphorylation directly relies on PTPRG enzymatic activity.
Fig. 3.
PTPRG binds and dephosphorylates FGFR1. A, Phosphatase activity of recombinant GST-PTPRG (catalytic domain) and GST-PTPN12 (catalytic domain) was estimated using pNPP assay. 2-fold molar excess of GST was used as a control. The initial rate of pNPP hydrolysis was measured colorimetrically (Abs. at 405 nm) during the first 10 min of reaction. The graph and table represent the mean ± S.D. of three independent experiments. B, U2OS-R1-BirA* cells were serum-starved for 2 h and then treated with 10 ng/ml FGF1 in the presence of 10 U/ml heparin for 15 min, lysed and the lysate was subjected to immunoprecipitation using anti-HA-tag antibodies. R1-BirA* is fused to an HA-tag in the C-terminal end. The beads containing immunoprecipitated FGFR1-BirA* were washed with lysis buffer without phosphatase inhibitor and subjected to on-beads dephosphorylation using indicated phosphatases or GST for 45 min, in the presence or absence of phosphatase inhibitors. After the incubation with phosphatases the immunoprecipiteted receptors were released from the beads and analyzed by SDS-PAGE followed by Western blotting using anti-pFGFR (Y653/Y654) antibodies. Western blots were quantified and bands corresponding to phosphorylated FGFR1 (pFGFR) were normalized to total FGFR1 immunoprecipitated and presented as fraction of GST without phosphatase inhibitors. The graph represents the mean ± S.D. of three independent experiments. The data were analyzed using one-way RM ANOVA followed by Tukey post hoc test. ***p ≤ 0.001, ns - not-significant. C, U2OS-R1 cells were transfected with PTPRG-MYC-FLAG, starved for 2 h and either left untreated or treated with 200 ng/ml FGF1 and 10 U/ml heparin for 1 h. The cells were fixed and stained with anti-FLAG, anti-FGFR1, anti-EEA1 antibodies and fluorophore (AF488, AF568, or AF647) labeled secondary antibodies and Hoechst. The cells were imaged in conventional wide-field mode and by SIM. Shown are Maximum Intensity Projections of whole cells (all z-sections) for deconvolved wide-field images, and a single selected optical section for SIM images, whereas all SIM z-sections were used for the 3D volume view, which was rotated 90° (side-view). Stippled lined squares indicate a region of the cell that is shown in a different mode in the panel below. Scale bars 4 μm.
To gain insight into where in the cells PTPRG and FGFR1 interact, we used wide-field and structured illumination microscopy to investigate their colocalization. In U2OS-R1 cells expressing PTPRG, FGFR1 and PTPRG colocalized mainly at the plasma membrane in nonstimulated cells (Fig. 3C). Interestingly, at resting conditions FGFR1 and PTPRG strongly colocalized in protrusions of the cells, resembling filopodia and lamellopodia. When cells had been stimulated with FGF1 for one hour, FGFR1 was detected mainly in intracellular vesicular structures, including EEA1 positive endosomes. PTPRG, however, was predominantly observed at the cell surface also after stimulation, suggesting that PTPRG might mainly act on FGFR1 at the plasma membrane (Fig. 3C).
Regulation of FGFR1 Autophosphorylation by PTPRG Revealed by TIRF
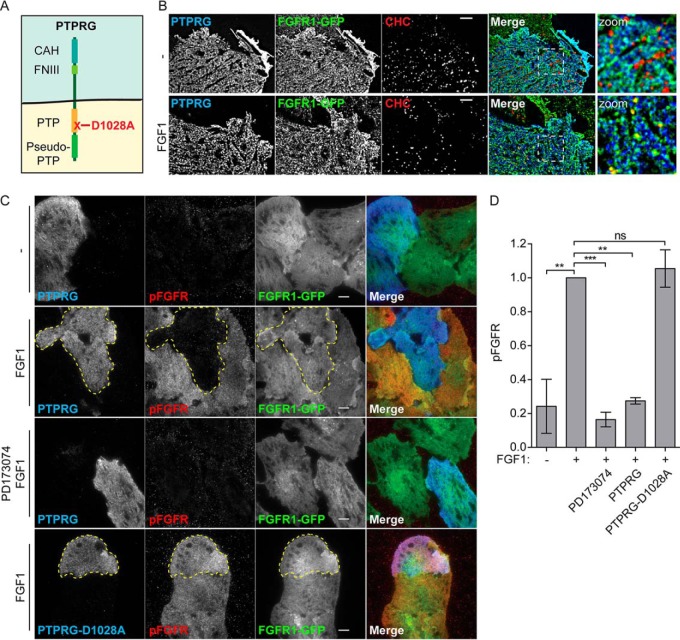
PTPRG is a large, transmembrane tyrosine phosphatase with an extracellular part containing a carbonic anhydrase-like (CAH) domain and an intracellular part consisting of one active and one inactive phosphatase domain (Fig. 4A) (40). Mutating the aspartate (D) at positon 1028 to alanine (A) inactivates the phosphatase activity of PTPRG (41).
Fig. 4.
PTPRG counters FGFR1 autophosphorylation A, Schematic presentation of PTPRG. PTPRG is a transmembrane protein with an extracellular carbonic anhydrase-like domain (CAH) and a fibronectin type III-like domain (FNIII). The intracellular part contains two protein tyrosine phosphatase domains (PTP) of which only one is active (indicated in orange). The other is called a pseudo-PTP. Mutation of aspartic acid 1028 to alanine inactivates the phosphatase activity. B, U2OS-R1-GFP cells transfected with PTPRG-myc-FLAG (for 20 h), serum starved for 2 h, and unstimulated (-) or stimulated with FGF1 for 15 min (FGF1), were fixed and stained with anti-myc and anti-Clathrin heavy chain, and imaged by TIRF. Merged images are overlays of PTPRG in blue, FGFR-GFP in green, and Clathrin in red. Blue and green overlay appears cyan. Green and red overlay appears yellow. Images were deconvolved, scale bar 4 μm. C, U2OS-R1-GFP cells were transfected with MYC-FLAG-tagged PTPRG or PTPRG-D1028A (for 20 h), starved for 2 h, and stimulated (or not) with FGF1 in the presence of heparin for 10 min (in one case in the presence of FGFR1 tyrosine kinase inhibitor PD173074) and then fixed and stained with anti-FLAG, anti-pFGFR1 (Y653/Y654), and fluorophore labeled secondary antibodies. The cells were imaged by TIRF. Merged images are overlays of PTPRG in blue, pFGFR1 in red, and FGFR1-GFP in green. Stippled lines indicate cells transfected with PTPRG or PTPRG-D1028A. Scale bars 8 μm. D, The signal intensities for pFGFR1 in PTPRG-transfected or -untransfected cells were measured for 15–30 cells for each condition in three independent experiments and is presented as the mean values ± S.D. where values had been normalized to the signal intensity of untransfected cells stimulated with FGF1. The data were analyzed using one-way RM ANOVA followed by Tukey post hoc test. ***p ≤ 0.001, **p ≤ 0.01, ns - not-significant.
Imaging by total internal reflection microscopy (TIRF), an imaging technique that reveals with high selectivity and clarity structures on, or close to, the cell surface, confirmed the localization of PTPRG at the plasma membrane. We observed a high degree of colocalization with FGFR1, but not with Clathrin Heavy Chain marking clathrin coated pits, which are entry sites for endocytosis (Fig. 4B, upper panel). Upon FGF1 stimulation, a partial shift of FGFR1 into clathrin coated pits was observed (yellow dots on the merged image, Fig. 4B, lower panel). This was not observed for PTPRG, suggesting that PTPRG is not coendocytosed with FGFR1.
Our biochemical analyses suggested that PTPRG acts directly to dephosphorylate FGFR1 and the plasma membrane localization of PTPRG suggests that it might do so mainly at the plasma membrane. To test this in cells, we used TIRF to monitor autophosphorylated FGFR1 at the plasma membrane. In this experiment, U2OS cells stably expressing FGFR1-GFP (U2OS-R1-GFP) were briefly stimulated with FGF1 before fixation and the activated FGFR1 was detected with anti-FGFR phospho-Tyr653/654 specific antibodies. The stimulation with FGF1 was sufficient to induce a robust activation of FGFR1 (pFGFR1) at the plasma membrane, which was fully inhibited by the FGFR kinase inhibitor PD173074, demonstrating that the observed immunofluorescent signal was specific (Fig. 4C). The cells were also transiently transfected with PTPRG, which had a dramatic effect, almost completely inhibiting the activity of FGFR1 at the plasma membrane. Overexpression of the inactive mutant PTPRG-D1028A however, had no effect on the FGFR1 activity (Fig. 4C). Quantification of the pFGFR1 levels detected by TIRF imaging, showed that overexpression of PTPRG reduced pFGFR1 levels by at least 70% (Fig. 4D). These experiments demonstrate that the enzymatic phosphatase activity of PTPRG counter the autophosphorylation of FGFR1.
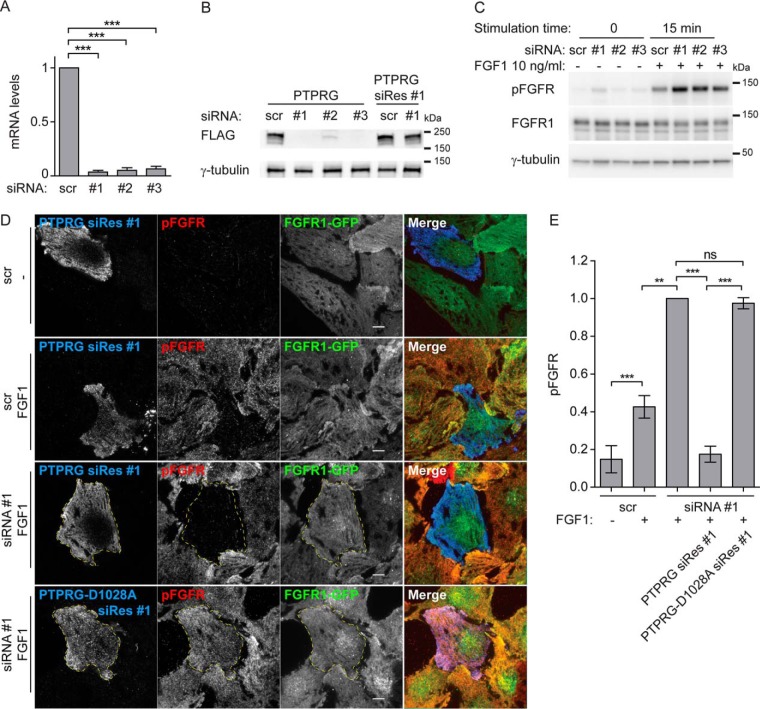
We then depleted PTPRG in U2OS-R1 cells using siRNA oligonucleotides, which were shown to efficiently deplete PTPRG (Fig. 5A and 5B). We also constructed PTPRG rescue mutants for siRNA oligo #1 that were resistant to siRNA depletion (PTPRG siRes #1 and PTPRG-D1028A siRes #1), (Fig. 5B). When PTPRG was knocked down, we observed a substantial increase in the levels of phosphorylated FGFR1 upon FGF1 stimulation (Fig. 5C). Moreover, using TIRF microscopy we could observe a similar effect at the plasma membrane (Fig. 5D and 5E). Depletion of PTPRG led to a strong increase in FGF-induced phosphorylation of FGFR1. This demonstrates that endogenous levels of PTPRG negatively regulate FGFR1 autophosphorylation. This effect could be totally reversed by transfecting the cells with the siRNA-resistant version of PTPRG, whereas the siRNA resistant version of the inactive mutant PTPRG-D1028A was not able to reverse the effect (Fig. 5D and 5E). Quantification of the pFGFR1 levels detected by TIRF imaging, showed that PTPRG knockdown increased pFGFR1 levels at least 2-fold (Fig. 5E), demonstrating that PTPRG is a highly efficient phosphatase for activated FGFR1.
Fig. 5.
PTPRG knockdown increases FGFR1 autophosphorylation. A, U2OS-R1 cells transfected with indicated siRNAs for 72 h were lysed and RNA isolation, cDNA synthesis and qRT-PCR were performed as described in materials and methods. The amount of mRNA was calculated relative to the housekeeping gene SDHA and is expressed as fraction of scr. The graphs represent the mean ± S.D. of three independent experiments. ***p ≤ 0.001. B, Cells transfected with indicated siRNAs for 18 h were transfected with MYC-FLAG-tagged PTPRG or siRNA-Resistant PTPRG (PTPRG siRes #1). 24 h later cells were lysed and the lysates were subjected to SDS-PAGE followed by Western blotting using denoted antibodies. C, U2OS-R1 cells were treated with PTPRG siRNAs (#1-#3) or control siRNA (scr) for 72 h. The cells were then serum-starved for 2 h before stimulation with 10 ng/ml FGF1 in the presence of 10 U/ml heparin for 15 min. Next, the cells were lysed and the lysates were subjected to SDS-PAGE followed by Western blotting using denoted antibodies. D, U2OS-R1-GFP cells were transfected with control (scr) or PTPRG-specific siRNA (siRNA #1) for a total of 72 h, transfected with MYC-FLAG-tagged siRNA resistant PTPRG (PTPRG siRes #1) or PTPRG-D1028A (PTPRG-D1028A siRes #1), for 20 h. The cells were then starved for 2 h, and stimulated (or not) with FGF1 in the presence of heparin for 10 min and then fixed and stained with anti-FLAG, and anti-phospho-FGFR (pFGFR) and fluorophore labeled secondary antibodies. The cells were imaged by TIRF. Merged images are overlays of PTPRG in blue, pFGFR in red, and FGFR1-GFP in green. Scale bars 8 μm. E, The signal intensities for pFGFR in PTPRG-transfected or -untransfected cells were measured for 15–30 cells for each condition in four independent experiments and is presented as the mean values ± S.D. where values had been normalized to the signal intensity of cells that were transfected with PTPRG-specific siRNA, but not expressing tagged PTPRG, and stimulated with FGF1. The data were analyzed using one-way RM ANOVA followed by Tukey post hoc test. ***p ≤ 0.001, **p ≤ 0.01, ns - not-significant.
In addition to PTPRG, our data set included two other phosphatases, PTPN1 and PTPN13 (supplemental Table S1 and S3). Depletion of cells for PTPN1 or PTPN13 did not significantly affect FGFR1 signaling (supplemental Fig. S3A). One siRNA against PTPN1 increased phosphorylation of FGFR1, but this was considered an off-target effect as it was only observed for one of three siRNAs with equal knock-down efficiencies. These phosphatases may therefore be in proximity to FGFR1, but do not act directly on FGFR1. Possibly, they could act on other tyrosine kinases associated with FGFR1 like SRC and YES1, which were hits in our proximity screen. SRC has previously been shown to be dephosphorylated by PTPN13 (42).
PTPRG Down-regulates FGFR Activation in Osteosarcoma Cells
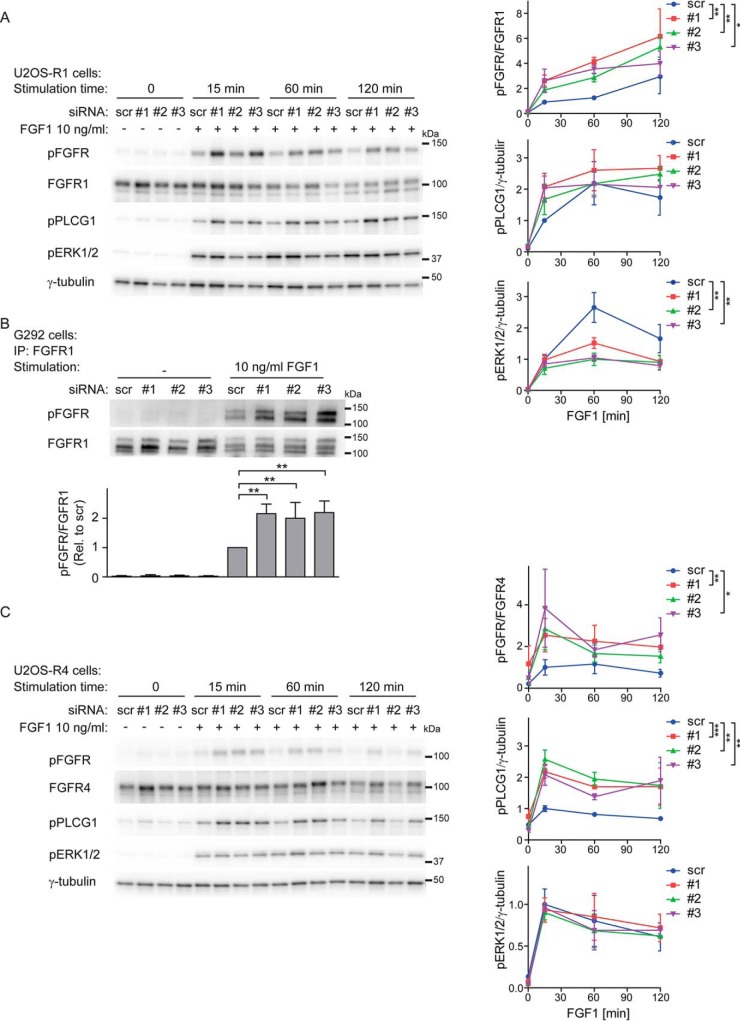
To further confirm the regulation of FGFR1 autophosphorylation by PTPRG and to analyze how this impinges on down-stream signaling pathways in osteosarcoma cells, we depleted cells for PTPRG using three different siRNAs and probed the activation of FGFR1 and its downstream signaling pathways by Western blotting using phospho-specific antibodies (Fig. 6).
Fig. 6.
Increased FGFR1 signaling upon PTPRG knockdown. A, U2OS-R1 cells were treated with PTPRG siRNAs (#1-#3) or control siRNA (scr) for 72 h. Then the cells were serum-starved for 2 h followed by stimulation with 10 ng/ml FGF1 in the presence of 10 U/ml heparin for various time points. The cells were then lysed and the lysates were subjected to SDS-PAGE followed by Western blotting using denoted antibodies. The Western blots were quantified and bands corresponding to phosphorylated proteins were normalized to total FGFR1 or loading control (γ-tubulin) (as indicated) and presented as fraction of scr, at 15 min stimulation time point. Means ± S.E. of three independent experiments are presented on the graphs. The time-course series were analyzed together using two-way ANOVA. **p ≤ 0.01, *p ≤ 0.05. B, G292 cells were left untreated or treated with 10 ng/ml FGF1 in the presence of 10 U/ml heparin for 20 min. The cells were then lysed and the cell lysates were subjected to immunoprecipitation using anti-FGFR1 antibody followed by SDS-PAGE and Western blotting with indicated antibodies. Quantification of Western blots are shown below. Bands corresponding to phosphorylated FGFR1 (pFGFR) were normalized to total FGFR1. The graph represents the mean ± S.D. of three independent experiments. **p ≤ 0.01. C, U2OS-R4 cells were treated as in (A), lysed and the lysates were subjected to SDS-PAGE followed by Western blotting using denoted antibodies. The Western blots were quantified and bands corresponding to phosphorylated proteins were normalized to total FGFR1 or loading control (γ-tubulin) (as indicated) and presented as fraction of scr, at 15 min stimulation time point. Means ± S.E. of three independent experiments are presented in the graphs. The time-course series were analyzed together using two-way ANOVA. ***p ≤ 0.001, **p ≤ 0.01, *p ≤ 0.05.
U2OS-R1 cells depleted of PTPRG displayed significantly increased levels of phosphorylated FGFR1 upon FGF1 stimulation for 15–120 min compared with control cells (Fig. 6A). Also the down-stream signaling molecule PLCG1 displayed somewhat (although insignificantly) increased activity when PTPRG was depleted. However, in the case of ERK1/2 activation, we observed no increase after 15 min of FGF1 treatment and even a decrease during the later time points.
We also estimated the significance of PTPRG in the context of other tyrosine phosphatases that possibly could dephosphorylate FGFR1 causing redundancy among phosphatases. We compared the effect of PTPRG depletion versus general inhibition of tyrosine phosphatases by high concentration of orthovanadate (2 mm). We found FGFR1 phosphorylation elevated ∼ 2.5× upon orthovanadate treatment after 15 min stimulation by FGF1 (10 ng/ml). PTPRG siRNA depletion led to slightly lower (∼ 2x) increase in FGFR1 activation (supplemental Fig. S4). Assuming that the high concentration of orthovanadate led to nearly complete inhibition of tyrosine phosphatases, we can conclude that PTPRG is a significant player in the dephosphorylation of activated FGFR1 accounting for ∼80% of tyrosine phosphatase activity on FGFR1 after 15 min stimulation.
We also evaluated the effect of PTPRG depletion in the osteosarcoma cell line G292, expressing endogenous FGFR1. Efficient knockdown was confirmed by real-time PCR (supplemental Fig. S3B). Because the expression level of FGFR1 is relatively low in this cell line, we immunoprecipitated the receptor using anti-FGFR1 antibodies and protein G coupled beads before analysis of phosphorylation levels by Western blotting. Upon 15 min of stimulation with FGF1 we observed increased activation of FGFR1 in PTPRG depleted cells (Fig. 6B), which shows that PTPRG can downregulate endogenous FGFR1 and confirms the previous findings.
To test if PTPRG also regulates other FGFR family members, we performed experiments using cell lines stably expressing FGFR4. We observed up-regulated autophosphorylation of FGFR4 during 15–120 min stimulation by FGF1 in U2OS-R4 cells depleted of PTPRG, and a parallel increase of PLCG1 phosphorylation (Fig. 6C), whereas no change was observed for ERK1/2 activation (Fig. 6C). PTPRG also regulated FGFR4 autophosphorylation in the rhabdomyosarcoma cell line RH30 expressing endogenous FGFR4 (supplemental Fig. S3C and S3D). In similar experiments, we used U2OS cells stably expressing FGFR2 and FGFR3 and we found elevated tyrosine phosphorylation levels of both receptors upon depletion of PTPRG (supplemental Fig. S3E and S3F). The results confirm that PTPRG down-regulates autophosphorylation of FGFRs. Moreover, an excessive activation of FGFRs, because of the loss of PTPRG, can lead to an elevation of downstream signaling pathways.
PTPRG Regulates the Biological Response to FGF1
Because our results demonstrate that PTPRG is responsible for dephosphorylation of activated FGFR, we hypothesized that the phosphatase could possibly alter the balance between phosphorylated and unphosphorylated forms of ligand-bound receptors. To confirm this hypothesis, we evaluated the levels of phospho-FGFR1 in PTPRG-depleted osteosarcoma cells stimulated with various concentrations of FGF1.
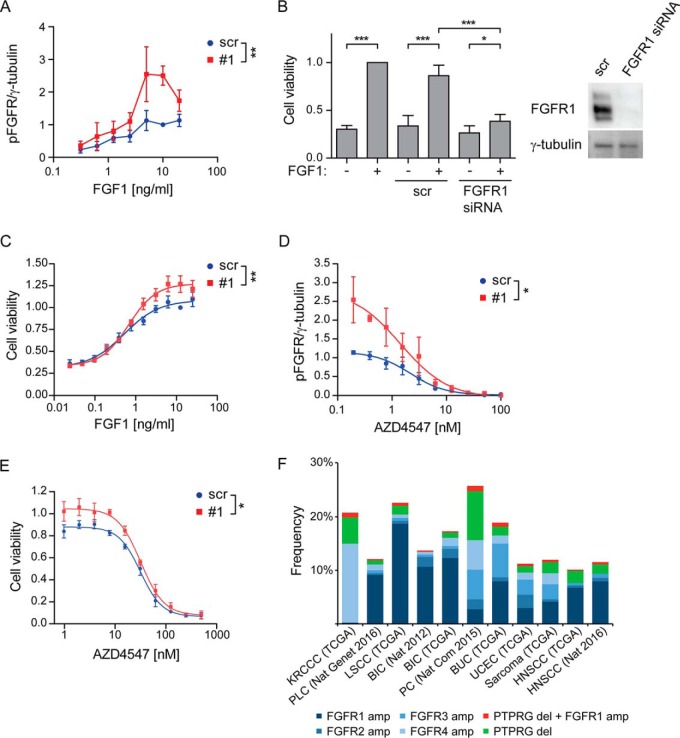
First, we evaluated whether depletion of PTPRG alters the sensitivity of FGFR1 toward FGF1 stimulation. After 15 min of treatment with 0–20 ng/ml FGF1, the levels of FGFR1 phosphorylation and activation of downstream signaling pathways were analyzed by Western blotting. We observed increased activation of FGFR1 and PLCG1 upon PTPRG knockdown under the applied range of FGF1 stimulation (supplemental Fig. S5A). No changes in ERK1/2 activation were observed. A detailed quantification of phospho-FGFR1 bands enabled us to detect a significant shift in the FGF1 dose-response curve toward higher pFGFR1 values when PTPRG was depleted (Fig. 7A). This demonstrates that PTPRG decreases the sensitivity of FGFR1 activation in response to FGF1. The effect of PTPRG depletion was more pronounced in parallel with increasing FGF1 concentrations. This agrees with the hypothesis that FGFR1 is a substrate for PTPRG.
Fig. 7.
PTPRG regulates cellular sensitivity to FGF1. A, U2OS-R1 cells were treated with siRNAs against PTPRG (#1) or control siRNA (scr) for 72 h. Then the cells were serum-starved for 2 h and stimulated with various concentrations of FGF1 in the presence of 10 U/ml heparin for 15 min and lysed. The lysates were subjected to SDS-PAGE followed by Western blotting using denoted antibodies (supplemental Fig. S5A). Western blots were quantified and bands corresponding to phosphorylated FGFR1 (pFGFR) were normalized to loading control (γ-tubulin) and presented as fraction of scr, 10 ng/ml stimulation. The graph represents the mean ± S.E. of three independent experiments. The concentration series were analyzed together using two-way ANOVA. **p ≤ 0.01. B, G292 cells were left untreated or treated with FGFR1 siRNA or control siRNA (scr) for 72 h. During the last 48 h the cells were treated with 100 ng/ml FGF1 in the presence of 10 U/ml heparin. Cell viability was then measured using CellTiter-Glo assay. The obtained data were normalized to nontransfected cells, stimulated with FGF1. The graph represents the mean ± S.D. of four independent experiments. ***p ≤ 0.001, *p ≤ 0.05. A Western blot showing the knockdown efficiency of FGFR1 after 72 h are presented to the right. C, G292 cells were treated with PTPRG siRNA (#1) or control siRNA (scr) for 72 h and then stimulated with different concentrations of FGF1 in the presence of 10 U/ml heparin for 48 h. Cell viability was then measured using CellTiter-Glo assay. The obtained data were normalized to scr, 12.5 ng/ml FGF1 and presented in the graph as means ± S.E. of 4 independent experiments. The fitted curve represents nonlinear regression analysis using Hill equation (dose-response with variable slope). The concentration series were analyzed together using two-way ANOVA. **p ≤ 0.01. D, U2OS-R1 cells were treated with siRNAs against PTPRG (#1) or control siRNA (scr) for 72 h, serum-starved for 2 h and stimulated with 10 ng/ml FGF1 in the presence of 10 U/ml heparin and various concentrations of AZD4547 for 15 min. The cells were then lysed and the lysates were subjected to SDS-PAGE followed by Western blotting using denoted antibodies (supplemental Fig. S5B). Western blots were quantified and bands corresponding to phosphorylated FGFR1 (pFGFR) were normalized to loading control (γ-tubulin) and presented as fraction of scr, 10 ng/ml stimulation. The graph represents the mean ± S.E. of three independent experiments. The fitted curve represents nonlinear regression analysis using Hill equation (dose-response with variable slope). The inhibitor concentration series were analyzed together using two-way ANOVA. *p ≤ 0.05. E, G292 cells were treated with PTPRG siRNA (#1) or control siRNA (scr) for 72 h and then stimulated with 10 ng/ml FGF1 in the presence of 10 U/ml heparin and various concentrations of AZD4547 for 48 h. Cell viability was then measured using CellTiter-Glo assay. The obtained data were normalized to scr without inhibitors and presented in the graph as means ± S.E. of three independent experiments. The fitted curve represents nonlinear regression analysis using Hill equation (dose-response with variable slope). The inhibitor concentration series were analyzed together using two-way ANOVA. *p ≤ 0.05. F, The graph shows the frequency of amplifications of the different FGFRs, deletions of PTPRG and the frequency of cases where at least one receptor is amplified and PTPRG is deleted. The figure is based on data generated by the TCGA Research Network (http://cancergenome.nih.gov/). The frequency is calculated according to the total number of cases for each study: KRCCC (Kidney Renal Clear Cell Carcinoma, 448 cases), PLC (Pan-Lung Cancer, 1144 cases), LSCC (Lung Squamous Cell Carcinoma), BIC (Breast Invasive Carcinoma, 482 cases and 1105 cases), PC (Pancreatic Cancer, 109 cases), BUC (Bladder Urothelial Carcinoma, 127 cases), UCEC (Uterine Corpus Endometrial Carcinoma, 242 cases), Sarcoma (243 cases), HNSCC (Head and Neck Squamous Cell Carcinoma, 504 and 279 cases).
We also tested whether the increased sensitivity of PTPRG-depleted cells toward FGF1 is biologically relevant. We chose the G292 cell line, expressing endogenous levels of FGFR1, and which growth in serum free media is dependent on FGF1 (Fig. 7B). We found that PTPRG-depleted cells displayed increased viability after treatment with various concentrations of FGF1 for 48 h (Fig. 7C). We also found that the difference was more pronounced with increasing concentrations of FGF1, in correspondence with the results obtained for analysis of FGFR1 phosphorylation by Western blotting (Fig. 7A and supplemental Fig. S5A). Our data indicate that PTPRG restricts the efficiency of the biological response of cells to FGF, and moreover, that down-regulation of PTPRG can serve as an advantage for cancer cells expressing FGFR1, allowing them to respond to lower FGF levels.
Altered Drug Sensitivity in Cells Depleted for PTPRG
Given that PTPRG counter the activity of FGFR by dephosphorylation, we wanted to test if PTPRG could influence the action of a small molecule tyrosine kinase inhibitor on FGFR1. Because our data suggest that PTPRG is involved in shifting the balance of receptor autophosphorylation to the inactive, nonphosphorylated state, the phosphatase could also affect kinase inhibition.
We first tested increasing concentrations of the FGFR kinase inhibitor AZD4547 on U2OS-R1 cells stimulated with a constant amount of FGF1 (10 ng/ml) and investigated the levels of activated FGFR1 and its downstream signaling molecules by Western blotting. We found that PTPRG-depleted cells displayed a higher level of phosphorylated FGFR1 and PLCG1 in the presence of the FGFR kinase inhibitor (supplemental Fig. S5B). Little effect was observed on ERK activation. Quantification of phospho-FGFR1 bands visualized a shift in the dose-response curve when PTPRG was knocked down (Fig. 7D). These data indicate that higher concentrations of the inhibitor are needed to prevent FGFR1 kinase activity when PTPRG is downregulated.
Next, we tested whether the disturbance in FGFR kinase inhibition, because of depleted PTPRG, translates into efficiency of the inhibitor to decrease cell growth. We knocked down PTPRG in G292 cells before stimulation with 10 ng/ml FGF1 in the presence of various concentrations of AZD4547. The experiment was performed using serum-free media to allow the cell growth to be dependent solely on FGF1. We found that PTPRG-depleted cells exhibited elevated viability after 48 h of FGF1 treatment in the presence of AZD4547 (Fig. 7E). The difference was dependent on the concentration of the inhibitor, being more pronounced at lower concentrations. Our findings suggest that higher concentrations of FGFR inhibitors are necessary to control FGFR activity in cells with low levels of PTPRG. Importantly, this effect would imply an advantage for cancer cells lacking PTPRG and serve as a possible resistance-mechanism to FGFR inhibitors.
To explore among different cancer types the frequency of cases where at least one FGFR is amplified and PTPRG is deleted, TCGA data generated by the TCGA Research Network (http://cancergenome.nih.gov/) was investigated. Specific gene information was extracted from 11 studies showing frequent FGFR amplifications by using the cBioportal (http://www.cbioportal.org/) (43, 44). The results show that the frequency of cases with amplified receptor and deleted PTPRG is between 0.2 and 0.9% and is found across many different cancer types (Fig. 7F). In total, 18 cases of the combination FGFR amplification/PTPRG deletion was identified in the 11 studies investigated, clearly demonstrating the relevance of our findings in human cancer.
DISCUSSION
FGFR inhibitors are now entering the clinic and it is crucial to understand how tumor cells respond to this treatment (11). We show here that PTPRG, a membrane bound tyrosine phosphatase, is an important modulator of FGFR tyrosine kinase activity. We demonstrate that PTPRG counter the activity of FGFR1 by direct dephosphorylation of the autoactivated, tyrosine phosphorylated FGFR1. The activity of PTPRG is also a determinant of the efficacy of FGFR inhibitors. We found that lowering the levels of PTPRG by specific siRNAs protected tumor cells against the clinically relevant FGFR kinase inhibitor AZD4547. It is therefore possible that PTPRG levels in cancer cells could be a predictor of outcome of FGFR kinase inhibition. Our data suggest that in clinical trials using FGFR inhibitors the level of PTPRG should be determined, to test the possibility that in tumors with low levels of PTPRG, kinase inhibitors may not be as efficient as in cells with normal levels of PTPRG. This may be particularly important when treating tumors with low doses of kinase inhibitors, which is normally the case because these inhibitors are associated with toxicity (e.g. hyperphosphatemia and tissue calcification) (11).
Sarcoma cells were here used to study the regulation of FGFR1, but it is likely that PTPRG also dephosphorylates FGFR1 in other cell types. For example, FGFR1 is overexpressed in breast cancers and is an attractive target with several clinical trials under way. Interestingly, TCGA data show that PTPRG is deleted and mutated in a subset of breast cancer patients (Fig 7E). Intriguingly, there are also reported cases where FGFR1 is overexpressed and PTPRG deleted, which could possibly be a bad combination for the patient. We also show that FGFR1 becomes hypersensitive to its ligand when PTPRG is down-regulated. It is therefore possible that FGFR1 can be aberrantly activated by low levels of ligand in the tumor microenvironment causing tumorigenic growth without overexpression or mutation of the receptor itself.
We also show that PTPRG regulates FGFR4, which is a potential drug target in rhabdomyosarcoma and hepatocellular carcinomas (3). The identification of PTPRG as a potent regulator of FGFR activity may therefore have implications for many cancer types.
Interestingly, we observed a difference in the response of two downstream signaling pathways to PTPRG depletion. Although the activity of PLCG1, similarly to that of FGFR, was up-regulated, ERK phosphorylation was mainly unchanged, and even reduced at later time points. The reason for this phenomenon could be that the MAPK pathway is subjected to several layers of both positive and negative regulation that may buffer for increased activity of the receptor. This may also imply that viability in osteosarcoma cells is modulated by other signaling pathways than the MAPK pathway.
Our studies suggest that PTPRG's main cellular localization is at the cell surface and that it is inefficiently endocytosed compared with the FGF1/FGFR1 activated complex. Concurrently, we find that PTPRG levels profoundly affect FGFR1 activity at the early timepoints (minutes) after FGF1 stimulation, which initiates at the plasma membrane. We also find that PTPRG levels affect FGFR1 and downstream signaling events even 2 h after the initial stimulation, and that this translates into biological effects such as cell viability several days after onset of FGF1 stimulation. It is not known in detail how the FGFR1 activity is affected by its subcellular localization, i.e. whether the rate of FGFR1 autophosphorylation is maintained or reduced after transfer from the plasma membrane to the endosomal membrane. Possibly, PTPRG levels exert a long-term effect on FGFR1 activity mainly by regulating its initial activation rate.
We used a modified BioID to investigate proximal proteins to FGFR1 that could potentially regulate FGFR signaling. The advantage of this method is that the biotinylation occurs in living cells and that the biotin tag makes it easier to pull down transient interactors and transmembrane proteins that may be difficult to detect in classic pull-down assays (45). Indeed, to our knowledge, PTPRG has not been found in any previous studies where FGFRs have been coprecipitated. Thus, as shown here, BioID may be used to identify important interactors that have not been found with the standard methods.
We have here concentrated our efforts on PTPRG, but we believe our proteomic data may be a resource for further studies of the regulation of FGFR signaling. For instance, in FGF1-stimulated cells, we identified known downstream effectors of activated FGFR (e.g. PLCG1 (32), RSK2 (33) and SHC4 (46)), but we also uncovered members of other signaling pathways (Table I, and supplemental Table S1). For instance, we found several members of the interferon-stimulated gene family, which may play a role in immunity. We also identified two cyclins (CCNE1, CCNB2) (Table I, supplemental Table S1 and S3) suggesting that FGFR1 may interact directly with these cell cycle regulators to stimulate proliferation. As we also have shown recently for FGFR4 (22), our BioID experiments revealed association between the FGFRs and many proteins involved in vesicular trafficking, reflecting the importance of intracellular transport for these receptors. We also identified A2M, which has previously been shown to bind the ligand FGF2 and thereby blocking its interaction with the receptors. Finally, we also identified a phosphate transporter (SLC20A1) that may be involved in the reabsorption of phosphate mediated by FGFR signaling (47). This may indicate a more direct activation of phosphate transporters than previously anticipated.
Analyzing proteins whose expression was induced by FGF1 signaling, we found several proteins that may confer negative feedback (Table I, supplemental Table S1, supplemental Table S4). One example includes the heparan sulfate proteoglycan CD44 that has been shown to regulate FGFR action (48). We also noticed a protein that has been shown to be a feedback inhibitor for EGFR family members (ERRFI1), which may possibly play a similar role for FGFRs.
There was some overlap between the proteins identified in proximity to FGF-stimulated FGFR1-BirA* and those identified in proximity to FGF-stimulated FGFR4-BirA* (22). At least 7 of the 33 proteins that were identified as 3 times enriched in FGF stimulated U2OS-R1-BirA* cells compared with unstimulated cells were also identified in the FGFR4 experiments. The somewhat low degree of overlap could reflect an actual biological difference among the receptors. In fact, FGFR1 and FGFR4 display clear differences in for instance intracellular trafficking and sorting and in signal intensity (21, 49). In addition, the criteria for sorting of the proteomic data were slightly different in the two experimental settings which could also contribute to differences in the lists. Especially, the cut-off for inclusion was lower in the case of FGFR4 experiments than in the more stringent FGFR1 experiments.
It is known that PTPRG has other targets than FGFR1 (50), but it remains an interesting question if additional tyrosine phosphatases are involved in directly regulating the activity of FGFR1. Indeed, two additional tyrosine phosphatases were discovered through our screen, whereas only PTPRG was shown to have an effect. The very strong effect of PTPRG knockdown on FGFR activity observed in our studies, indicates that PTPRG is a major regulator of FGFR, and indicates that there may be less redundancy among phosphatases than anticipated. This also implies that cells with low expression of PTPRG may be particularly vulnerable to excessive FGFR activity, which could lead to more aggressive cancer. We therefore believe that it will be important to study PTPRG as a predictor of outcome for disease caused by FGFRs.
DATA AVAILABILITY
The MS raw files were deposited to the public database ProteomeXchange (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository 51. The data can be found under project number: PXD006157. Annotated spectra were uploaded to MS-Viewer (http://msviewer.ucsf.edu/prospector/cgi-bin/msform.cgi?form=msviewer, search key: h5u2ahuncj) 52.
Supplementary Material
Acknowledgments
We thank Dr. Coen Campsteijn, Prof. Ola Myklebost and Prof. Claudio Sorio for sharing reagents and Prof. Knut Liestøl for guidance on the statistics.
Note Added in Proof
In the version of this article that was published as a Paper in Press on January 25, 2018, Dr. Nina Jones was inadvertently omitted as an author. This error has now been corrected.
Footnotes
* This work was supported by the Norwegian Cancer Society. J.W. held a Researcher fellowship (project 5756681), E.M.H a Career fellowship (project 6842225), M.K. a Postdoctoral fellowship (project 734183) and E.F. a Postdoctoral fellowship (project 5756681) from the Norwegian Cancer Society. This work was partially supported by The Research Council of Norway through its Centers of Excellence funding scheme, project number 262652. All MS data was collected at the Proteomics Core Facility, Rikshospitalet, which is funded by the Norwegian South-East Health Authority.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- FGFR
- fibroblast growth factor receptor
- PTPRG
- protein tyrosine phosphatase receptor type G
- TIRF
- total internal reflection fluorescence.
REFERENCES
- 1. Turner N., and Grose R. (2010) Fibroblast growth factor signalling: from development to cancer. Nat. Rev. Cancer 10, 116–129 [DOI] [PubMed] [Google Scholar]
- 2. Haugsten E. M., Wiedlocha A., Olsnes S., and Wesche J. (2010) Roles of fibroblast growth factor receptors in carcinogenesis. Mol. Cancer Res. 8, 1439–1452 [DOI] [PubMed] [Google Scholar]
- 3. Wesche J., Haglund K., and Haugsten E. M. (2011) Fibroblast growth factors and their receptors in cancer. Biochem. J. 437, 199–213 [DOI] [PubMed] [Google Scholar]
- 4. Weekes D., Kashima T. G., Zandueta C., Perurena N., Thomas D. P., Sunters A., Vuillier C., Bozec A., El-Emir E., Miletich I., Patino-Garcia A., Lecanda F., and Grigoriadis A. E. (2016) Regulation of osteosarcoma cell lung metastasis by the c-Fos/AP-1 target FGFR1. Oncogene 35, 2852–2861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Guagnano V., Kauffmann A., Wohrle S., Stamm C., Ito M., Barys L., Pornon A., Yao Y., Li F., Zhang Y., Chen Z., Wilson C. J., Bordas V., Le D. M., Gaither L. A., Borawski J., Monahan J. E., Venkatesan K., Brummendorf T., Thomas D. M., Garcia-Echeverria C., Hofmann F., Sellers W. R., and Graus-Porta D. (2012) FGFR genetic alterations predict for sensitivity to NVP-BGJ398, a selective pan-FGFR inhibitor. Cancer Discov. 2, 1118–1133 [DOI] [PubMed] [Google Scholar]
- 6. Taylor J. G., Cheuk A. T., Tsang P. S., Chung J. Y., Song Y. K., Desai K., Yu Y., Chen Q. R., Shah K., Youngblood V., Fang J., Kim S. Y., Yeung C., Helman L. J., Mendoza A., Ngo V., Staudt L. M., Wei J. S., Khanna C., Catchpoole D., Qualman S. J., Hewitt S. M., Merlino G., Chanock S. J., and Khan J. (2009) Identification of FGFR4-activating mutations in human rhabdomyosarcomas that promote metastasis in xenotransplanted models. J. Clin. Invest. 119, 3395–3407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chudasama P., Renner M., Straub M., Mughal S. S., Hutter B., Kosaloglu Z., Schwessinger R., Scheffler M., Alldinger I., Schimmack S., Persigehl T., Kobe C., Jager D., von Kalle C., Schirmacher P., Beckhaus M. K., Wolf S., Heining C., Groschel S., Wolf J., Brors B., Weichert W., Glimm H., Scholl C., Mechtersheimer G., Specht K., and Frohling S. (2016) Targeting fibroblast growth factor receptor 1 for treatment of soft-tissue sarcoma. Clin. Cancer Res. 23, 962–973 [DOI] [PubMed] [Google Scholar]
- 8. Zhou W. Y., Zheng H., Du X. L., and Yang J. L. (2016) Characterization of FGFR signaling pathway as therapeutic targets for sarcoma patients. Cancer Biol. Med. 13, 260–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hanes R., Grad I., Lorenz S., Stratford E. W., Munthe E., Reddy C. C., Meza-Zepeda L. A., and Myklebost O. (2016) Preclinical evaluation of potential therapeutic targets in dedifferentiated liposarcoma. Oncotarget 7, 54583–54595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang K., Chu K., Wu X., Gao H., Wang J., Yuan Y. C., Loera S., Ho K., Wang Y., Chow W., Un F., Chu P., and Yen Y. (2013) Amplification of FRS2 and activation of FGFR/FRS2 signaling pathway in high-grade liposarcoma. Cancer Res. 73, 1298–1307 [DOI] [PubMed] [Google Scholar]
- 11. Dieci M. V., Arnedos M., Andre F., and Soria J. C. (2013) Fibroblast growth factor receptor inhibitors as a cancer treatment: from a biologic rationale to medical perspectives. Cancer Discov. 3, 264–279 [DOI] [PubMed] [Google Scholar]
- 12. Tabernero J., Bahleda R., Dienstmann R., Infante J. R., Mita A., Italiano A., Calvo E., Moreno V., Adamo B., Gazzah A., Zhong B., Platero S. J., Smit J. W., Stuyckens K., Chatterjee-Kishore M., Rodon J., Peddareddigari V., Luo F. R., and Soria J. C. (2015) Phase I Dose-Escalation Study of JNJ-42756493, an Oral Pan-Fibroblast Growth Factor Receptor Inhibitor, in Patients With Advanced Solid Tumors. J. Clinical Oncol. 33, 3401–3408 [DOI] [PubMed] [Google Scholar]
- 13. Campbell J., Ryan C. J., Brough R., Bajrami I., Pemberton H. N., Chong I. Y., Costa-Cabral S., Frankum J., Gulati A., Holme H., Miller R., Postel-Vinay S., Rafiq R., Wei W., Williamson C. T., Quigley D. A., Tym J., Al-Lazikani B., Fenton T., Natrajan R., Strauss S. J., Ashworth A., and Lord C. J. (2016) Large-Scale Profiling of Kinase Dependencies in Cancer Cell Lines. Cell Reports 14, 2490–2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fernanda Amary M., Ye H., Berisha F., Khatri B., Forbes G., Lehovsky K., Frezza A. M., Behjati S., Tarpey P., Pillay N., Campbell P. J., Tirabosco R., Presneau N., Strauss S. J., and Flanagan A. M. (2014) Fibroblastic growth factor receptor 1 amplification in osteosarcoma is associated with poor response to neo-adjuvant chemotherapy. Cancer Med. 3, 980–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Baroy T., Chilamakuri C. S. R., Lorenz S., Sun J. C., Bruland O. S., Myklebost O., and Meza-Zepeda L. A. (2016) Genome Analysis of Osteosarcoma Progression Samples Identifies FGFR1 Overexpression as a potential treatment target and CHM as a candidate tumor suppressor gene. Plos One 11, e0163859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Roux K. J., Kim D. I., Raida M., and Burke B. (2012) A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol. 196, 801–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. LaForgia S., Morse B., Levy J., Barnea G., Cannizzaro L. A., Li F., Nowell P. C., Boghosian-Sell L., Glick J., Weston A., and et al. (1991) Receptor protein-tyrosine phosphatase gamma is a candidate tumor suppressor gene at human chromosome region 3p21. Proc. Natl. Acad. Sci. U.S.A. 88, 5036–5040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shu S. T., Sugimoto Y., Liu S., Chang H. L., Ye W., Wang L. S., Huang Y. W., Yan P., and Lin Y. C. (2010) Function and regulatory mechanisms of the candidate tumor suppressor receptor protein tyrosine phosphatase gamma (PTPRG) in breast cancer cells. Anticancer Res. 30, 1937–1946 [PubMed] [Google Scholar]
- 19. Jones N., Hardy W. R., Friese M. B., Jorgensen C., Smith M. J., Woody N. M., Burden S. J., and Pawson T. (2007) Analysis of a Shc family adaptor protein, ShcD/Shc4, that associates with muscle-specific kinase. Mol. Cell. Biol. 27, 4759–4773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wesche J., Malecki J., Wiedlocha A., Ehsani M., Marcinkowska E., Nilsen T., and Olsnes S. (2005) Two nuclear localization signals required for transport from the cytosol to the nucleus of externally added FGF-1 translocated into cells. Biochemistry 44, 6071–6080 [DOI] [PubMed] [Google Scholar]
- 21. Haugsten E. M., Sorensen V., Brech A., Olsnes S., and Wesche J. (2005) Different intracellular trafficking of FGF1 endocytosed by the four homologous FGF receptors. J. Cell Sci. 118, 3869–3881 [DOI] [PubMed] [Google Scholar]
- 22. Haugsten E. M., Sorensen V., Kunova Bosakova M., de Souza G. A., Krejci P., Wiedlocha A., and Wesche J. (2016) Proximity labeling reveals molecular determinants of FGFR4 endosomal transport. J. Proteome Res. 15, 3841–3855 [DOI] [PubMed] [Google Scholar]
- 23. Petersen T. N., Brunak S., von Heijne G., and Nielsen H. (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786 [DOI] [PubMed] [Google Scholar]
- 24. Haugsten E. M., Malecki J., Bjorklund S. M., Olsnes S., and Wesche J. (2008) Ubiquitination of fibroblast growth factor receptor 1 is required for its intracellular sorting but not for its endocytosis. Mol. Biol. Cell 19, 3390–3403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Haugsten E. M., Zakrzewska M., Brech A., Pust S., Olsnes S., Sandvig K., and Wesche J. (2011) Clathrin- and dynamin-independent endocytosis of FGFR3–implications for signalling. PLoS ONE 6, e21708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rappsilber J., Ishihama Y., and Mann M. (2003) Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 75, 663–670 [DOI] [PubMed] [Google Scholar]
- 27. Cox J., and Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p. p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 28. Schneider C. A., Rasband W. S., and Eliceiri K. W. (2012) NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lorenz U. (2011) Protein tyrosine phosphatase assays. Current Protocols Immunol. Chapter 11, Unit 11 17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kuleshov M. V., Jones M. R., Rouillard A. D., Fernandez N. F., Duan Q., Wang Z., Koplev S., Jenkins S. L., Jagodnik K. M., Lachmann A., McDermott M. G., Monteiro C. D., Gundersen G. W., and Ma'ayan A. (2016) Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44, W90–W97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kouhara H., Hadari Y. R., Spivak-Kroizman T., Schilling J., Bar-Sagi D., Lax I., and Schlessinger J. (1997) A lipid-anchored Grb2-binding protein that links FGF-receptor activation to the Ras/MAPK signaling pathway. Cell 89, 693–702 [DOI] [PubMed] [Google Scholar]
- 32. Mohammadi M., Honegger A. M., Rotin D., Fischer R., Bellot F., Li W., Dionne C. A., Jaye M., Rubinstein M., and Schlessinger J. (1991) A tyrosine-phosphorylated carboxy-terminal peptide of the fibroblast growth factor receptor (Flg) is a binding site for the SH2 domain of phospholipase C-gamma 1. Mol. Cell. Biol. 11, 5068–5078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nadratowska-Wesolowska B., Haugsten E. M., Zakrzewska M., Jakimowicz P., Zhen Y., Pajdzik D., Wesche J., and Wiedlocha A. (2014) RSK2 regulates endocytosis of FGF receptor 1 by phosphorylation on serine 789. Oncogene 33, 4823–4836 [DOI] [PubMed] [Google Scholar]
- 34. Sandilands E., Akbarzadeh S., Vecchione A., McEwan D. G., Frame M. C., and Heath J. K. (2007) Src kinase modulates the activation, transport and signalling dynamics of fibroblast growth factor receptors. EMBO Rep. 8, 1162–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chatr-Aryamontri A., Oughtred R., Boucher L., Rust J., Chang C., Kolas N. K., O'Donnell L., Oster S., Theesfeld C., Sellam A., Stark C., Breitkreutz B. J., Dolinski K., and Tyers M. (2017) The BioGRID interaction database: 2017 update. Nucleic Acids Res. 45, D369–D379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lambert J. P., Tucholska M., Go C., Knight J. D., and Gingras A. C. (2015) Proximity biotinylation and affinity purification are complementary approaches for the interactome mapping of chromatin-associated protein complexes. J. Proteomics 118, 81–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Szklarczyk D., Franceschini A., Wyder S., Forslund K., Heller D., Huerta-Cepas J., Simonovic M., Roth A., Santos A., Tsafou K. P., Kuhn M., Bork P., Jensen L. J., and von Mering C. (2015) STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 43, D447–D452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sorio C., Mendrola J., Lou Z., LaForgia S., Croce C. M., and Huebner K. (1995) Characterization of the receptor protein tyrosine phosphatase gene product PTP gamma: binding and activation by triphosphorylated nucleosides. Cancer Res. 55, 4855–4864 [PubMed] [Google Scholar]
- 39. Sun T., Aceto N., Meerbrey K. L., Kessler J. D., Zhou C., Migliaccio I., Nguyen D. X., Pavlova N. N., Botero M., Huang J., Bernardi R. J., Schmitt E., Hu G., Li M. Z., Dephoure N., Gygi S. P., Rao M., Creighton C. J., Hilsenbeck S. G., Shaw C. A., Muzny D., Gibbs R. A., Wheeler D. A., Osborne C. K., Schiff R., Bentires-Alj M., Elledge S. J., and Westbrook T. F. (2011) Activation of multiple proto-oncogenic tyrosine kinases in breast cancer via loss of the PTPN12 phosphatase. Cell 144, 703–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Barnea G., Silvennoinen O., Shaanan B., Honegger A. M., Canoll P. D., D'Eustachio P., Morse B., Levy J. B., Laforgia S., Huebner K., and et al. (1993) Identification of a carbonic anhydrase-like domain in the extracellular region of RPTP gamma defines a new subfamily of receptor tyrosine phosphatases. Mol. Cell. Biol. 13, 1497–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhang W. D., Savelieva K. V., Tran D. T., Pogorelov V. M., Cullinan E. B., Baker K. B., Platt K. A., Hu S., Rajan I., Xu N. H., and Lanthorn T. H. (2012) Characterization of PTPRG in knockdown and phosphatase-inactive mutant mice and substrate trapping analysis of PTPRG in mammalian cells. Plos One 7, e45500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Glondu-Lassis M., Dromard M., Lacroix-Triki M., Nirde P., Puech C., Knani D., Chalbos D., and Freiss G. (2010) PTPL1/PTPN13 regulates breast cancer cell aggressiveness through direct inactivation of Src kinase. Cancer Res. 70, 5116–5126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gao J. J., Aksoy B. A., Dogrusoz U., Dresdner G., Gross B., Sumer S. O., Sun Y. C., Jacobsen A., Sinha R., Larsson E., Cerami E., Sander C., and Schultz N. (2013) Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signaling 6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cerami E., Gao J. J., Dogrusoz U., Gross B. E., Sumer S. O., Aksoy B. A., Jacobsen A., Byrne C. J., Heuer M. L., Larsson E., Antipin Y., Reva B., Goldberg A. P., Sander C., and Schultz N. (2012) The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discovery 2, 401–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Roux K. J., Kim D. I., and Burke B. (2013) BioID: A screen for protein-protein interactions. Curr. Protoc. Protein Sci. 74, 19–19 [DOI] [PubMed] [Google Scholar]
- 46. Smith M. J., Hardy W. R., Murphy J. M., Jones N., and Pawson T. (2006) Screening for PTB domain binding partners and ligand specificity using proteome-derived NPXY peptide arrays. Mol. Cell. Biol. 26, 8461–8474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Prie D., and Friedlander G. (2010) Genetic disorders of renal phosphate transport. New Engl. J. Med. 362, 2399–2409 [DOI] [PubMed] [Google Scholar]
- 48. Jones M., Tussey L., Athanasou N., and Jackson D. G. (2000) Heparan sulfate proteoglycan isoforms of the CD44 hyaluronan receptor induced in human inflammatory macrophages can function as paracrine regulators of fibroblast growth factor action. J. Biol. Chem. 275, 7964–7974 [DOI] [PubMed] [Google Scholar]
- 49. Wang J. K., Gao G., and Goldfarb M. (1994) Fibroblast growth factor receptors have different signaling and mitogenic potentials. Mol. Cell. Biol. 14, 181–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cheung A. K. L., Ip J. C. Y., Chu A. C. H., Cheng Y., Leong M. M. L., Ko J. M. Y., Shuen W. H., Lung H. L., and Lung M. L. (2015) PTPRG suppresses tumor growth and invasion via inhibition of Akt signaling in nasopharyngeal carcinoma. Oncotarget 6, 13434–13447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Vizcaino J. A., Csordas A., del-Toro N., Dianes J. A., Griss J., Lavidas I., Mayer G., Perez-Riverol Y., Reisinger F., Ternent T., Xu Q. W., Wang R., and Hermjakob H. (2016) 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 44, D447–D456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Baker P. R., and Chalkley R. J. (2014) MS-viewer: a web-based spectral viewer for proteomics results. Mol. Cell. Proteomics 13, 1392–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Dennis P. A., Saksela O., Harpel P., and Rifkin D. B. (1989) Alpha 2-macroglobulin is a binding protein for basic fibroblast growth factor. J. Biol. Chem. 264, 7210–7216 [PubMed] [Google Scholar]
- 54. Vasudevan H. N., Mazot P., He F., and Soriano P. (2015) Receptor tyrosine kinases modulate distinct transcriptional programs by differential usage of intracellular pathways. Elife 4, doi: 10.7554/eLife.07186 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The MS raw files were deposited to the public database ProteomeXchange (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository 51. The data can be found under project number: PXD006157. Annotated spectra were uploaded to MS-Viewer (http://msviewer.ucsf.edu/prospector/cgi-bin/msform.cgi?form=msviewer, search key: h5u2ahuncj) 52.