Abstract
Obesity is tightly linked to hepatic steatosis and insulin resistance. One feature of this association is the paradox of selective insulin resistance: insulin fails to suppress hepatic gluconeogenesis but activates lipid synthesis in the liver. How lipid accumulation interferes selectively with some branches of hepatic insulin signaling is not well understood. Here we provide a resource, based on unbiased approaches and established in a simple cell culture system, to enable investigations of the phenomenon of selective insulin resistance. We analyzed the phosphoproteome of insulin-treated human hepatoma cells and identified sites in which palmitate selectively impairs insulin signaling. As an example, we show that palmitate interferes with insulin signaling to FoxO1, a key transcription factor regulating gluconeogenesis, and identify altered FoxO1 cellular compartmentalization as a contributing mechanism for selective insulin resistance. This model system, together with our comprehensive characterization of the proteome, phosphoproteome, and lipidome changes in response to palmitate treatment, provides a novel and useful resource for unraveling the mechanisms underlying selective insulin resistance.
Obesity is often accompanied by liver steatosis and type 2 diabetes mellitus (T2D)1. Many aspects of the pathogenesis of T2D remain unclear, but a common feature is hepatic insulin resistance with impaired insulin signaling. These defects are selective and involve the failure of insulin to suppress of gluconeogenesis but not lipid synthesis (1). Evidence indicates that hepatic insulin resistance is caused by lipid species that accumulate in hepatocytes and selectively interfere with insulin signaling (2–4). Lipids implicated in causing hepatic insulin resistance include triglycerides (TGs), diacylglycerols (DGs), fatty-acyl CoAs, and ceramides. However, mouse models exhibit high hepatic levels of each of these lipids with or without insulin resistance (for examples, see (5–9). Thus, a consistent causal relationship for a specific lipid has not been found, and it remains unclear how lipid accumulation causes the paradox of selective insulin resistance in hepatocytes.
A major hurdle in tackling this question is the complexity of the systems that are studied. Although many studies of mice have revealed important insights into hepatic lipid metabolism, insulin signaling, and glucose metabolism, this model system also has limitations. Murine liver is a complex organ with multiple interactions of different cells (e.g. hepatocytes, stellate cells, reticuloendothelial cells, vascular cells) that are regulated by hormones and the neurological system. When layered with the complexity of many thousands of lipid species, varied over different cell types and cellular compartments, and the complexity of insulin signaling, it is not surprising that pathogenic mechanisms are not easy to discern.
Thus, we sought a complementary, reductionist approach to systematically study alterations in insulin signaling during lipid overload by establishing a simpler system that displays selective insulin resistance. We reasoned that cultured human hepatocytes treated with lipotoxic lipids might provide such a system. Among the various human hepatoma cell lines, HepG2 cells respond to insulin and have defects in insulin signaling on lipid accumulation (10). In addition, HepG2 cells are immortalized and, therefore, are fully compatible with stable isotope labeling (SILAC)-based phosphoproteomic analyses by mass spectrometry (11–13).
Using this model system, we show here that HepG2 cells treated with palmitate exhibit selective insulin resistance, and we characterize global changes by lipidomic, proteomic, and phosphoproteomic analyses. By further developing phosphoproteomic methodology with a protocol that omits peptide fractionation but includes multiple rounds of enrichment and mass spectrometry measurements for phosphopeptides, we quantified 18,000 sites in the phosphoproteome. Several thousand of these sites are responsive to insulin, and we show how palmitate treatment interferes with some of these changes. Our analyses provide a rich resource for generating and testing new hypotheses about the development of selective insulin resistance. As a proof-of-principle, we explored how palmitate interferes with a key node of insulin signaling regulating gluconeogenesis; we show that impaired FoxO1 signaling with palmitate treatment is associated with changes in its localization, suggesting a novel contributing mechanism for selective insulin resistance.
EXPERIMENTAL PROCEDURES
Experimental Design and Statistical Rationale
A total of three sample sets of HepG2 cells were analyzed and described in Results. Each sample set comprised of three biological replicates. Experimental controls from each sample set include nontreated wild-type HepG2 cells. Statistical analysis of three biological replicates using one sample t test with a Benjamini-Hochberg false discovery rate (FDR) of less than 5% were applied to provide statistical significance from insulin and palmitate treatments of HepG2 cells.
Protein Extraction and Phosphopeptide Enrichment for Mass Spectrometry
HepG2 cells were SILAC labeled and cultured in SILAC DMEM media with 10% (v/v) dialyzed FBS. For palmitate treatment, cells were seeded with 30% confluency, cultured for 18 h and treated with 0.25 mm palmitate for 24 h. Palmitate treated or untreated cells were stimulated with vehicle (50 μm HEPES, pH 8.0) or 100 nm insulin for 20 min and lysed in lysis buffer (6 m guanidinium hydrochloride (GdmCl), 10 mm Tris (2-carboxyethyl) phosphine (TCEP), 40 mm chloroacetamide, alkylating reagent (CAA), 100 mm Tris, pH 8.5). After heating at 95 °C for 5 min and sonicating, lysate was diluted 1:3 [lysate: dilution buffer (10% acetonitrile, 25 mm Tris, pH 9.0)], digested with LysC overnight, then further diluted to 1:5 (lysate/dilution buffer) and digested with trypsin. The resulting peptides were desalted with Sep-pak Waters C18 cartridge and enriched for phosphopeptides by incubating with 3 μl titanium dioxide (TiO2) beads suspended in DHB/buffer B (30 mg/1 ml) for 30 min. Beads were isolated by centrifugation and washed sequentially with 3% TFA and 30% acetonitrile, 0.3% TFA in 60% acetonitrile, and 0.3% TFA in 100% acetonitrile to remove peptides bound nonspecifically. The supernatant was used for the next round of phosphopeptide enrichment, and this cycle was repeated eight times. Phosphopeptides on beads were eluted by 16.5% ammonia in 20% acetonitrile, vacuum dried, resuspended in buffer A (0.1% formic acid) and loaded onto HPLC-MS/MS system for analysis on an Orbitrap Q-Exactive HF (Thermo Fisher Scientific) mass spectrometer coupled to an Easy nanoLC 1000 (Thermo Scientific) with a flow rate of 300 nl/min. The stationary phase buffer was 0.5% formic acid, and mobile phase buffer was 0.5% (v/v) formic acid in acetonitrile. Chromatography using increasing organic proportion was used for peptide separation (5–40% (v/v) acetonitrile over a 265 min gradient on a self-packed analytical column using PicoTipTM emitter (New Objective, Woburn, MA) using Reprosil Gold 120 C18 (Dr. Maisch, Ammerbuch-Entrigen, Germany) 1.9 μm particle size resin. The mass spectrometer operated in data dependent acquisition mode with a top ten method at a mass range of 300–2000 Da.
For proteome profiling, proteins of a SILAC labeled cell lysate were reduced using 5 mm dithiothreitol (Sigma-Aldrich) at 37 °C for 1 h, followed by alkylation of cysteine residues using 15 mm iodoacetamide (Sigma-Aldrich) in the dark at room temperature for 1 h. Excessive iodoacetamide was quenched using 10 mm dithiothreitol. Proteins were precipitated by the addition of nine volumes of ice-cold acetone and one volume of methanol and incubated at −80 °C for 2 h. Precipitated proteins were centrifuged for 1 h at 4500 × g at 4 °C. After washing with methanol, proteins were resolubilized in 100 mm NaOH aided by sonication at 4 °C and the solution was brought to pH 7.5 with 200 mm HEPES (free acid). Protein concentrations were determined using bicinchoninic acid assay kit (Pierce), followed by equal mixing of proteins at 1:1 ratio (light/heavy labels). Proteins were trypsinized using sequencing grade trypsin (Promega) at 37 °C for 16 h. Digested peptides were subsequently desalted using self-packed C18 STAGE tips (3 m EmporeTM) (14) for LC-MS/MS analysis.
Lipid Extraction
The lipid extraction protocol was modified from (15). Briefly, cell lysates (biological triplicates - 200 μg of protein each, containing the following standard lipid mix (Avanti Polar Lipids): 375 pmoles C17:1 lysophosphatidic acid; 225 pmoles C17:0/C20:4 phosphatidic acid; 170 pmoles C17:1 lysophosphatidylserine; 180 pmoles C17:0/C20:4 phosphatidylserine; 86 pmoles C17:1 lysophosphatidylethanolamine; 112 pmoles C17:0/C14:1 phosphatidylethanolamine; 95 pmoles C17:1 lysophosphatidylcholine; 112 pmoles C17:0/C20:4 phosphatidylcholine; 33.2 pmoles C17:1 lysophosphatidylinositol; 165 pmoles C17:0/C20:4 phosphatidylinositol; 105 pmoles C17:0/C14:1 phosphatidylglycerol; 180 pmoles C17:0/d18:1 ceramide; 140 pmoles C17:0/d18:1 sphingomyelin; 155 pmoles C12:0/d18:1 monohexosyl-ceramide; and 60 pmoles C17:1/C17:1/C17:1 triacylglycerol) were suspended in ice-cold HPLC-grade water, and transferred to 13 × 100-mm Pyrex culture tubes with polytetrafluoroethylene (PTFE)-lined screw caps. Then, HPLC-grade water, methanol, and chloroform were added to each vial, generating a final chloroform/methanol/water (C/M/W) ratio of 1:2:0.8 (v/v/v). Samples were vortexed vigorously for 5 min and centrifuged for 10 min at 1,800 × g at room temperature. After centrifugation, the supernatants were collected and transferred to fresh vials, and the remaining pellets were dried under nitrogen stream. The dry pellets were extracted with chloroform/methanol (2:1, v/v), centrifuged, and the resulting supernatants were combined with the corresponding supernatants from the first step of extraction (C/M/W 1:2:0.8 v/v/v). The combined supernatants were dried under nitrogen stream, and the resulting samples were submitted to Folch's partitioning by dissolving them in C/M/W (4:2:1.5, v/v/v), followed by vortexing and centrifuging, as described above. After centrifugation, the lower (organic) and upper (aqueous) phases were separated and transferred to fresh Pyrex glass test tubes. The Folch upper phase was then re-extracted with C/M (2/1; v/v), and the resulting organic phase was combined with the organic phase from the preceding step. The pooled organic phases were dried under a N2 stream and stored at −20 °C until use.
UHPLC-ESI-MS/MS for Lipidomics Analyses
Lipid samples extracted as described above were diluted in 50 μl of C/M (2:1; v/v) and analyzed by UHPLC-ESI-MS using a modification of the method described by (16). UHPLC-ESI-MS/MS was conducted using a Dionex™ UltiMate 3000™ UHPLC system (Thermo Fisher Scientific) coupled to Q Exactive™ Hybrid Quadrupole-Orbitrap™ Mass Spectrometer (Thermo Fisher Scientific). Five microliter injections of samples were made onto a Accucore™ (Thermo Fisher Scientific) C18 LC column (2.6 μm solid-core particles, length of 30 mm). The eluent A consisted of acetonitrile/water (50:50 v/v) + 10 mm ammonium formate and 0.2% formic acid, and the eluent B consisted of methanol/isopropanol/water (10:88:20 v/v/v) + 2 mm ammonium formate and 0.01% formic acid. The following program was used for gradient elution of analytes (total run time of 46 min) at a constant flow rate of 500 μl per min. 35% eluent B - 45% eluent B over 5 min, 45% eluent B - 85% eluent B from min 5 - min 28, 85% eluent B - 100% eluent B from min 28 - min 38, followed by an immediate drop back to 35% eluent B, which is held constant up to min 46 of each run. The column temperature was set to 35 °C, and the autosampler tray temperature was set to 10 °C. The ion source was a HESI II (Thermo Fisher Scientific) set in the following conditions: sheath gas flow rate set to 60; auxiliary flow rate set to 20; sweep gas flow rate set to 1; spray voltage (KV) set to 3.00; capillary temperature set to 285 °C, S-Lens RF level set to 45, and auxiliary temperature set to 370 °C. The mass spectrometer acquisition settings were as follows: Samples were run in either positive or in negative ion modes, under otherwise identical conditions. The instrument was set for Full Scan - top 15 data-dependent MS/MS. Full scan was set for a range of 250–1800 m/z. The mass resolution was set to 70,000; AGC target was set to 1e6, the C-trap ion accumulation time was set to 120 ms, Data-dependent MS/MS was set to a mass resolution of 30,000, AGC target was set to 5e5, the C-trap ion accumulation time was set to 120 ms, select ion exclusion was set to 8 s, and the HCD (higher-energy collisional dissociation) fragmentation ramp was set to 15, 25, and 35 NCE (normalized collision energy).
Mass Spectrometry Data Analyses using MaxQuant
Mass spectrometry data from proteome and phosphosproteome analyses were processed by MaxQuant software version 1.5.2.8 (17) using the following setting: oxidized methionine residues, protein N-terminal acetylation and phosphorylation of Ser/Thr/Tyr (phosphoproteome only) as variable modification, cysteine carbamidomethylation as fixed modification, first search peptide tolerance 20 ppm, main search peptide tolerance 4.5 ppm. Protease specificity was set to trypsin with up to 2 missed cleavages were allowed. Only peptides longer than five amino acids were analyzed, and the minimal ratio count to quantify a protein is 2 (proteome only). The false discovery rate (FDR) was set to 1% for peptide and protein identifications. Database searches were performed using the Andromeda search engine integrated into the MaxQuant environment (18) against the UniProt-human database containing 71,579 entires (October 2017). “Matching between runs” algorithm with a time window of 0.7 min was employed to transfer identifications among samples processed using the same nanospray conditions. Protein tables were filtered to eliminate identifications from the reverse database and also common contaminants. Phosphosites were categorized based on the probability score: class I (> 0.75, more than 75% localization probability), class II (0.5–0.75, 50–75% localization probability), class III (0.25–0.5, 25–50% localization probability) and class IV (< 0.25, less than 25% localization probability). All mass spectrometry data generated for phosphoproteomics and proteomics are deposited to the ProteomeXchange Consortium via the PRIDE (19) partner repository (http://proteomecentral.proteomexchange.org/cgi/GetDataset) with the data set identifier PXD006395.
Identification and Quantitation of Lipid Species
Mass spectrometry generated by the LC-MS/MS runs for extracted lipids were analyzed using the LipidSearchTM Software (Thermo Fisher Scientific) for the identification of lipid species, combining accurate-mass information from parent ion with MS2 information. The following parameters were used for batch analysis: Database - Q Exactive; Search Type - Product; Experiment Type (Exp Type) - LC-MS; Parent Tolerance (Parent Tol) - 0.1 Da; NL/Prec Tol - 0.5 Da; Precursor Tolerance (Prec Tol) - 10.0 ppm; Product Tolerance (15.0) ppm; Merge Range (Min) - 0.0; Minimal Peak Width (min) - 0.0; Threshold Type - Relative; Product Ion - 1.0%; m-score threshold - 2.0; Recalculate Isotope - ON; R.T. Interval (min) - 0.01; Execute Quantitation - ON; m/z Tolerance (m/z tol) - −10.0/+ 10.0; Tolerance Type - ppm; R.T. range (min) - −0.05/+0.05; Toprank filter - ON; Main Node Filter - Main Isomer Peak; m-Score Threshold (Display) - 5.0; C-Score Threshold (Display) - 2.0; Fatty Acid Priority (FA Priority) - ON; ID Quality Filter (A, B, C). The following parameters were used for sample alignment: Search Type - Product; Experiment Type (ExpType) - LC - MS; Alignment Method - Max; R.T. Tolerance - 0.25; Calculate Unassigned Peak Area - ON; Filter Type - New Filter; Toprank Filter -ON; Main Node Filter - Main Isomer Peak; m-score threshold - 5.0; ID Quality - A, B, C. All lipid species identified using the LipidSearchTM software were manually curated after computational analysis, being either accepted, rejected, or reassigned.
Known quantities of each lipid standard were analyzed using the same UHPLC-ESI-MS/MS methods to generate a response factor (RF; peak area/pmol standard injected). The RF for each standard was divided by the CerG1 RF to calculate a molar relative response factor (MRRF) for each major lipid class. The MRRF for each class was normalized to the CerG1 peak area of each sample, and peak areas adjusted accordingly. Only those classes for which standards were detected in each run were considered for analysis. The in-built statistical tool for the quantitation of lipid species LipidSearchTM is student t test. For the calculation of lipid class fatty acid composition, the MainArea values output by LipidSearch™ were assigned to each FA moiety of a given species and summed using Microsoft Excel PivotTable. Area % is the summed FA value divided by the lipid class total, multiplied by 100 to reflect percentage within each lipid class. All lipidomics data have been deposited to the EMBL-EBI MetaboLights database (DOI: 10.1093/nar/gks1004. PubMed PMID: 23109552) with the identifier MTBLS582. The complete data set can be accessed at https://www.ebi.ac.uk/metabolights/MTBLS582 (20).
Bioinformatic Analyses
Microsoft Office Excel, R software and Perseus (Version 1.5.0.9, Max Planck Institute of Biochemistry, Munich) were used to perform bioinformatics analyses. Only proteins and phosphorylation sites passed one sample t test with a Benjamini-Hochberg false discovery rate of less than 5% were considered. The Gene ontology (GO) annotation enrichment analysis on phosphosites quantified by Fisher exact test was performed on Perseus and PANTHER (http://pantherdb.org/) was used to categorize proteins into different cell components, biological processes or signaling pathways. Motif analysis was performed on pLogo (21) using sequences of phosphopeptides in the phosphoproteome and human proteome as the background. To identify phosphosites impacted by insulin/palmitate, normalized heavy/light intensity ratios of quantified phosphosites were transformed to log2 data and a 2.5 median absolute deviations (MAD) threshold was used to reveal positively and negatively regulated sites on proteins and the rest sites were considered as unchanged.
Western Blotting and Immunostaining
For Western blotting, HepG2 cells were washed three times with 1× PBS and lysed in RIPA buffer. Protein concentrations of the lysates were quantified, and proteins were loaded onto 4–15% SDS-PAGE gels (Bio-Rad Laboratories) and transferred to nitrocellulose or PVDF membranes with an iBlot 2 gel transfer device (Thermo Fisher Scientific) for subsequent antibody labeling, following the manufacturer's instructions. For immunostaining, HepG2 cells were fixed by 4% paraformaldehyde for 20 min, blocked in 10% normal goat serum (Cell Signaling) at room temperature for 3 h and labeled with primary and secondary antibodies, according to manufacturer's manual. Images were taken with IN Cell Analyzer 6000 Cell Imaging System (GE Healthcare Life Sciences) and quantified with In Cell Developer Toolbox (Version 1.9.1) and ImageJ (Version 1.51i 17). Anti-phospho-eIF4B (Ser422), eIF4B, phosphoGSK-3 β, GSK-3β (27C10), FoxO1, phosphor-AKT (Ser 473) and AKT antibodies were from Cell Signaling Technology, anti-phospho-EIF4EBP1-S65 was from NeoScientific, anti-eIF4EBP1 antibody was from Abcam.
RNA Extraction and Quantitative Real-time PCR
Total RNA from insulin- and palmitate-treated HepG2 cells were isolated with an RNeasy kit (Qiagen) according to manufacturer's instructions. cDNA was synthesized using iSCript cDNA synthesis kit (Bio-Rad), and quantitative real-time PCR was performed in triplicates using SYBR Green PCR Master Mix kit (Applied Biosystems). Primer sequences for quantitative real-time PCR can be found in supplementary data.
RESULTS
HepG2 Cells Pretreated with Palmitate Exhibit Selective Insulin Resistance
We sought to establish a reductionist model system to study insulin signaling in hepatocytes and selective insulin resistance. Criteria for our system included that it be an insulin-sensitive human hepatic cell line and, on treatment with lipotoxic lipids, exhibit insulin resistance with respect to insulin-mediated suppression of gluconeogenic genes, yet retain insulin sensitivity for other signaling events, such as genes regulating lipid synthesis. We also wanted an immortalized cell line to facilitate SILAC labeling for most accurate quantitative proteomic studies. Based on the large body of available literature (e.g. (10) and our own preliminary experiments (data not shown)), we chose to study insulin signaling in the human hepatoma cell line, HepG2, and treatment with palmitate as a lipotoxic lipid.
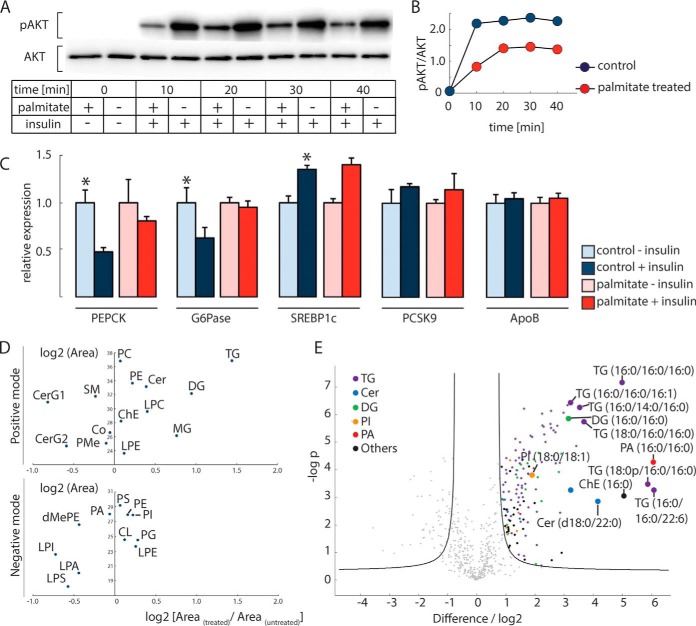
To test the insulin sensitivity of HepG2 cells, we treated them with 100 nm insulin for 20 min and detected robust phosphorylation of AKT at Ser473, a canonical marker for activated insulin signaling (22) (Fig. 1A and 1B). Pretreatment with 0.25 mm palmitate for 24 h reduced AKT phosphorylation at Ser473 (10) in these cells. In liver, insulin down-regulates the expression levels of PEPCK and G6Pase, which catalyze the rate-limiting and last step in gluconeogenesis, respectively, and up-regulates the levels of SREBP1c, a master regulator of lipid synthesis (23–25). In HepG2 cells, we found these gene expression responses were intact, and the down-regulation of PEPCK and G6Pase was blunted by palmitate treatment (Fig. 1C). However, palmitate did not affect the level of SREBP1c transcripts, therefore showing a selective effect of palmitate treatment on insulin signaling for gluconeogenesis, but not lipid synthesis. The expression of PCSK9, an SREBP2 target gene, trended higher in response to insulin but was not changed with palmitate treatment, and the expression of a control transcript, apolipoprotein B, was unaffected by insulin treatment.
Fig. 1.
HepG2 cells treated with palmitate exhibit selective insulin resistance accompanied by selected changes in the lipidome. A, Western blot analysis of Akt phosphorylation at S473. Insulin stimulation increased Akt phosphorylation, which is impaired by palmitate treatment. B, Quantification of data from (A). C, Relative mRNA expression of insulin response genes of gluconeogenesis and lipid synthesis in HepG2 cells. Measurements were made 2 h after 100 nm insulin stimulation. ApoB served as a control. Statistical significance was determined using student's t test; *, p < 0.05. D, Major lipid classes that are changed in palmitate-treated (versus untreated) cells. Results are shown separately for positive and negative ion modes. E, Specific lipid species that are changed in 24 h 0.25 mm palmitate-treated and untreated cells (monohexosylceramides, CerG1; dihexosylceramides, CerG2; sphingomyelins, SM; coenzyme, Co; phosphatidylmethanol, PMe; glycerophosphates, PA (lyso-PA, LPA); glycerophosphocholines, PC (lyso-PC, LPC); glycerophosphoethanolamines, PE (lyso-PE, LPE); glycerophosphoglycerols, PG; glycerophosphoinositols, PI (lyso-PI, LPI); glycerophosphoserines, PS (lyso-PS, LPS); cholesterol esters, ChE; ceramides, Cer; monoradylgerolipids, MG; diradylglycerolipids, DG; triradylglycerolipids, TG; dimethylphosphatidylethanolamine, dMePE; cardiolipins, CL).
To investigate lipid intermediates that might be responsible for the selective effects on insulin signaling, we profiled total cell lipid species by lipidomic analyses, using liquid chromatography coupled online to high-resolution mass spectrometry. We found that palmitate treatment of HepG2 cells for 24 h increased primarily levels of glycerolipids, especially DG and TG (Fig. 1D). Monoacylglycerols were also increased by ∼75%. Ceramides as a class were not greatly increased, and monohexosylceramides levels were decreased. Further quantification of specific lipid species (Fig. 1E) showed that the accumulated lipids were highly saturated: for example, TG (16:0/16:0/16:0) increased 32-fold, and DG (16:0/16:0) increased ∼9-fold. We also found increased levels of specific lipid species from lipid classes that were overall unchanged. These included 18-, 67-, and 3.7-fold increases of specific saturated ceramides (d18:0/22:0), phosphatidic acid (16:0/16:0), and PI (18:0/18:1), respectively. Thus, the most prominent changes induced by palmitate treatment were an increase in the levels of saturated species of glycerolipids. The broad changes in lipid species we measure are consistent with lipid changes found during the development of hepatic insulin resistance (9, 26–28), thus supporting the use of our cell model for studying selective insulin resistance.
Determination of a High-Content, Quantified Phosphoproteome in HepG2 Cells Treated with Insulin
Insulin signaling is characterized by a host of phosphorylation events that were systematically investigated in other cell types (29–31). We sought to comprehensively measure changes in the phosphoproteome in HepG2 cells after insulin treatment. Insulin stimulates many phosphorylation sites across a time frame of seconds to minutes (31). For this study, we examined the system after 20 min because we expected a wide range of both early and late signaling changes at this time (31) that would enable us to subsequently compare how palmitate treatment affects signaling.
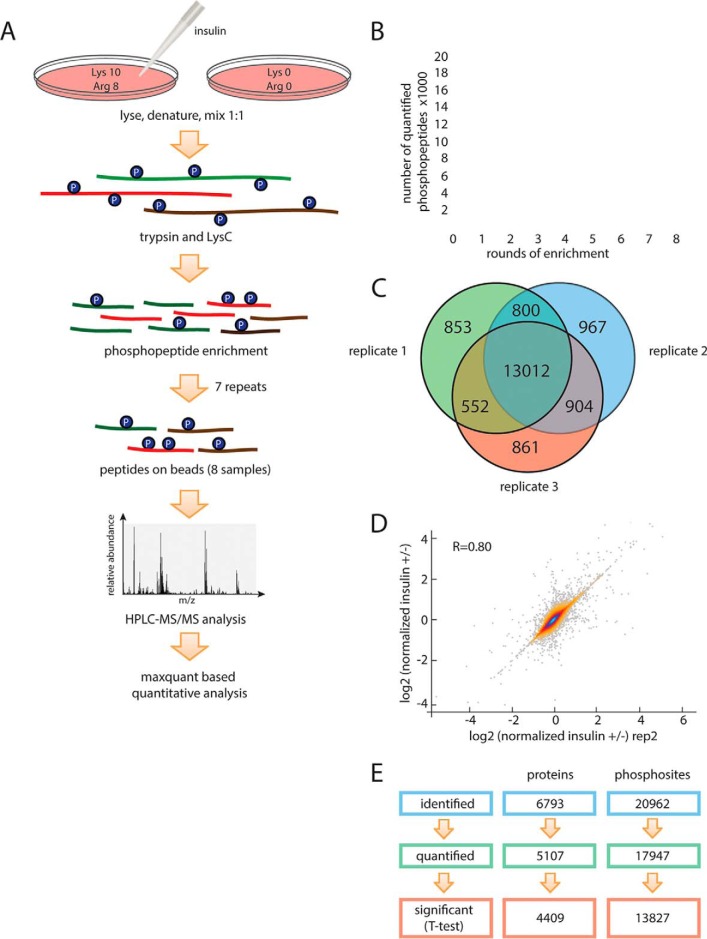
Most current proteomic approaches reply on sample fractionation by ion exchange chromatography to reduce sample complexity before HPLC-MS/MS analyses to obtain adequate coverage of the phosphoproteome (32, 33). These additional fractionation steps require substantial amounts of starting material, prolonged sample processing (increasing the risk of noninsulin mediated changes in phosphorylation), and can introduce material loss and variation among experiments. Taking advantage of the recent developments in mass spectrometry hardware that allow higher scan rates and an improved nanoflow HPLC/ESI strategy (34), we avoided fractionation of peptides by ion-exchange chromatography. Instead, we performed eight rounds of phospho-enrichment with TiO2 beads to increase peptide coverage (Figs. 2A and 2B). To allow for quantitation of phosphorylation changes, we SILAC-labeled HepG2 cells (35). After a 20-min insulin or mock treatment, cells were lysed and denatured, proteins were cleaved into small peptides by LysC and trypsin proteases. Phosphopeptides were enriched using a TiO2 resin in eight subsequent rounds of chromatography.
Fig. 2.
Analysis of the phosphoproteome and proteome of HepG2 cells treated with insulin. A, Experimental workflow. Heavy (Arg 10, Lys 8) labeled and light (Arg 0, Lys 0) HepG2 cells were lysed, 1:1 mixed, and digested with trypsin and LysC. Digested peptides were enriched for phosphopeptides by incubating with TiO2 beads for eight rounds. Phosphopeptides obtained from each enrichment were desalted and analyzed by Q-Exactive HF mass spectrometry. B, Enrichment of phosphosites. Summed number of phosphosites quantified after round(s) of phosphoenrichment. C, Replicates of the analysis. Venn diagram of phosphosites quantified in individual replicates, respectively. More than 70% of phosphosites were quantified in all three replicates (n = 3). D, Reproducibility of the methodology. Quantitative reproducibility for duplicates are shown. E, Total phosphosites and proteins identified and quantified.
Our methodology proved efficient and robust. Starting with ∼3 mg of protein in a sample (cell lysate from one 15-cm culture dish), we accurately quantified more than 18,000 phosphosites after eight rounds of phosphopeptide enrichment. The reproducibility of our pipeline was high (Figs. 2C and 2D): ∼73% phosphopeptides were quantified in all three measured biological replicates and ∼77% quantified phosphopeptides passed a one-sample t test with a FDR of less than 5%.
In total, 1,103,068 high-resolution (Orbitrap higher-energy collision dissociation [HCD]) spectra were acquired, resulting in the identification of phosphopeptides corresponding to 20,961 phosphorylation sites on 17,241 phosphopeptides from 6781 proteins. The localization of phosphosites on the phosphopeptide was also highly accurate. MaxQuant categorizes phosphosites based on the probability score: class I (>0.75), class II (0.75–0.5), class III (0.5–0.25) and class IV (<0.25). 89.4% of the sites in our phosphoproteome were in class I, including 49.2% sites with scores higher than 0.99, and none of our phosphosites was in class IV. Our phosphoproteome data set was obtained using three biological replicates, and only the sites identified consistently (one sample t test, FDR<0.05) were used for further analyses. Distribution of singly, doubly, and multiple phosphorylated peptides in each round of TiO2 enrichment in HepG2 are illustrated in supplemental Fig. S1.
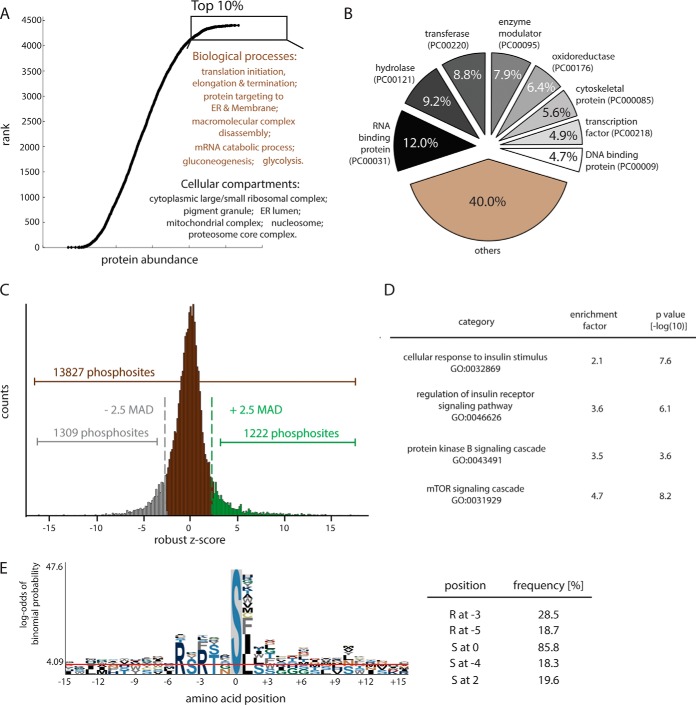
To better characterize the proteins expressed in HepG2 cells and to provide a reference for interpreting the phosphoproteome results, we also measured the proteome of HepG2 cells. We identified nearly 6800 expressed proteins, of which over 5000 were quantitated (Fig. 2E). In total, 13,827 phosphosites in 4409 proteins reported to meet minimum threshold for significance. Analysis of these proteins showed that those involved in cell growth and cell-cycle progression, such as translation initiation/elongation/termination and mRNA metabolism, were the most abundant proteins (see Fig. 3A) and RNA binding proteins (PC00031) were the largest protein class (see Fig. 3B). Additionally, we detected high levels of proteins involved in gluoconeogenesis and glycolysis, in accordance with the Warburg hypothesis that glycolytic metabolic pathways are hyperactive in cancer cells.
Fig. 3.
Analysis of the HepG2 phosphoproteome. A, Ranked abundance atlas of HepG2 hepatocyte proteins. GO enrichment analysis was performed by Fisher exact test with a threshold of Benjamini-Hochberg FDR 1%. B, Proteins in HepG2 proteome categorized by PANTHER classification system. C, Distribution of quantified phosphosites (Student's t test, p < 0.05). Applying a threshold of 2.5-median absolute deviations (MAD) to the log2-transformed data revealed 1222 positively regulated and 1309 negatively regulated phosphosites. D, Enrichment analysis of proteins with phosphosites up-regulated by insulin. E, Motif analysis for insulin up-regulation phosphosites reveals high representation of an Akt consensus site.
Measuring the Phosphoproteome of HepG2 Cells Treated with Insulin Reveals Novel Phosphosites
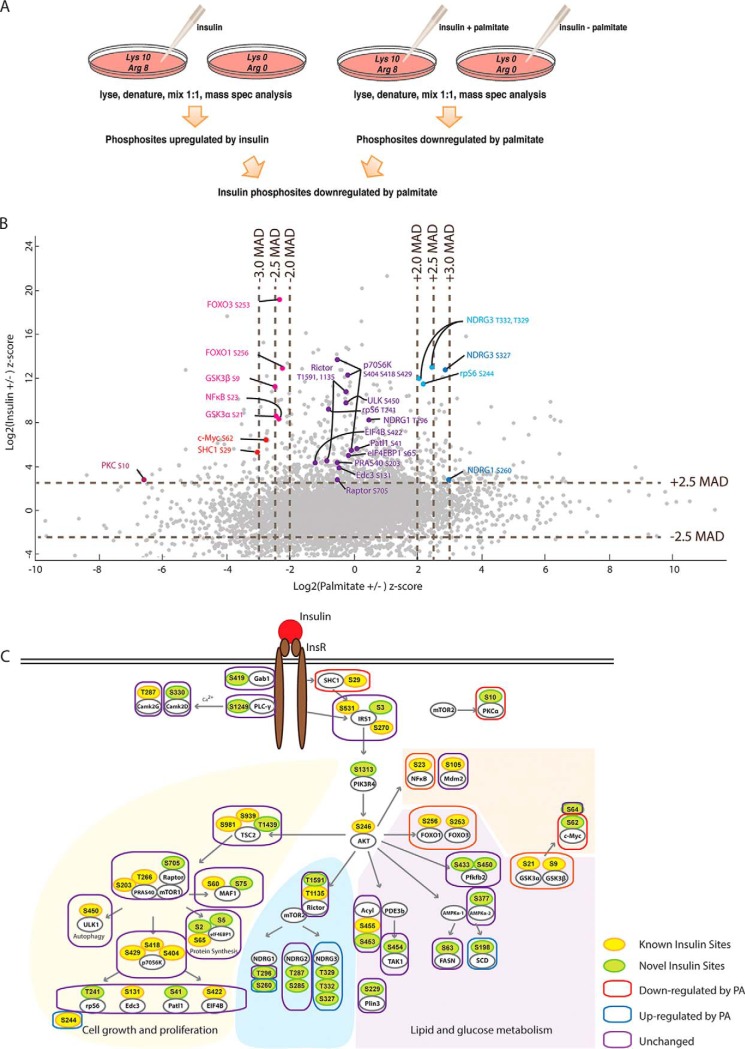
The results of our analysis of the insulin-responsive phosphoproteome are shown in Fig. 3C. By employing a significance threshold of 2.5 median absolute deviation (MAD), 1222 phosphosites were identified as up-regulated and 1309 were down-regulated on insulin stimulation. Using the Fisher exact test, we found that up-regulated phosphosites were specifically enriched for insulin signaling-relevant biological processes, including PKB/Akt and mTOR signaling cascades (Fig. 3D). A motif analysis showed that the most frequent sequence found in insulin-responsive phosphopeptides correspond to protein kinase AKT consensus motifs (RxRxxS/T; (36)), as is typical for Akt phosphorylation (Fig. 3E).
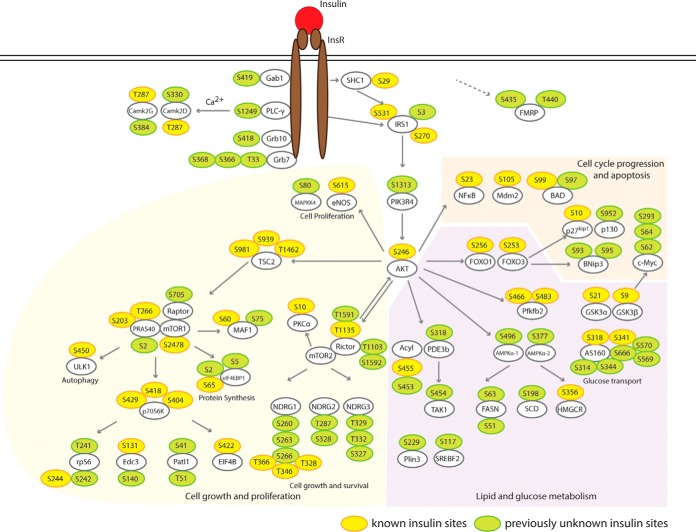
We constructed a cell signaling network based on quantitative information of phosphosites in our database that we mapped onto known pathways of insulin signaling. The results are shown in Fig. 4. This analysis revealed many known phosphosites for insulin signaling (shown in yellow), confirming these as sites in HepG2 cells, and additionally, identified many previously unrecognized sites (shown in green). The identification of these sites highlights the depth of coverage provided by our methodology.
Fig. 4.
Insulin-Signaling Cascade in HepG2 Hepatocytes. From our data set and previously characterized pathways of insulin signaling, we constructed a diagram of hepatocyte insulin signaling, including branches that control cell growth, glucose metabolism, lipid metabolism and apoptosis. Yellow: previously identified sites; Green: previously unknown sites.
Several of the newly identified insulin-induced phosphorylation sites are noteworthy. For example, our data set showed that insulin up-regulates phosphorylation of two sites (S435 and T440) on FMRP (fragile X mental retardation) protein by more than 10-fold (14.2- and 13.1-fold, respectively). FMRP plays a central role in neuronal development and loss of FMRP leads to fragile X syndrome (37), which includes mild to moderate intellectual disability. Recently, up-regulated insulin signaling has been found in a Drosophila FMRP mutant, and inhibited insulin signaling ameliorates fragile X syndrome (38). Moreover, FMRP function has been linked to circadian regulation of metabolic genes in murine liver (39, 40). The most common FMRP isoforms have a potential nuclear export sequence (NES) 425-EVDQLRLERLQIDEQ-441 and do not localize in the nucleus. However, isoforms 10 and 11 lack the NES but have 425-LQQRKRGRASCAEET-441 instead (41). The S435 and T441 phosphosites localize in this unique sequence of isoforms 10 and 11. Because insulin specifically phosphorylates FMRP isoforms 10 and 11, we hypothesize this modification might alter their subcellular localization.
We also detected strong changes in the phosphorylation of multiple sites on AS160 (Akt substrate of 160 kDa) protein: Ser314, Ser318, Ser341, Ser344, Ser569, Ser570, and Ser666. AS160 is crucial for translocating Glut4 glucose transporter to the plasma membrane (42). Two sites (Ser318 and Ser341) within the protein kinase AKT consensus motif (RXRXXS/T) are required for Glut4 translocation (43). The other phosphosites have not been previously characterized, probably because they are not fitting a canonical protein kinase AKT motif. Our data set shows that phosphorylation of AS160 by insulin is not limited to AKT-preferring consensus motif and previously unknown regulatory mechanisms may exist.
Our data set also uncovered a dramatic (11-fold) increase in phosphorylation of phosphoinositide 3-kinase regulatory subunit 4 (also named p150) at Ser1313. Very little is known about p150 except that it promotes PI3 kinase activity (44), thus suggesting a previously unknown layer of insulin-mediated phosphoinositide regulation.
Palmitate Pretreatment Disrupts Selected Nodes of Hepatic Insulin Signaling
We next investigated the effects of palmitate pretreatment on insulin signaling by treating “heavy” Lys/Arg SILAC-labeled HepG2 cells with 0.25 mm palmitate for 24 h and comparing them with “light” cells as a control. Both heavy and light cells were stimulated with 100 nm insulin for 20 min and then lysed for phosphoproteomic analyses (Fig. 5A). We combined this phosphoproteome with the insulin-treatment phosphoproteome and found that 9482 phosphosites were quantified in both phosphoproteomes, including 724 insulin-responsive phosphosites. By scatter plot analysis and applying a threshold of 2 MAD, we identified 107 insulin phosphosites that were impaired by palmitate treatment and 132 phosphosites that were enhanced (Fig. 5B). The remainder of the phosphosites were unaffected by insulin treatment.
Fig. 5.
Palmitate selectively affects insulin signaling cascade. A, Experimental workflow for the analyses of insulin responsive sites compromised by palmitate treatment. Heavy (Arg 10, Lys 8) labeled and light (Arg 0, Lys 0) HepG2 cells grown in medium containing palmitate and either control treated or incubated with insulin as shown. Mass spectrometry analysis was performed as outlined in Fig. 2A. B, Scatter plot of insulin (±) phosphoproteome and palmitate (±) phosphoproteome. Pink and Red: phosphosites downregulated by palmitate; Purple: phosphosites unaffected; Light blue and blue: phosphosites up-regulated by palmitate. C, Palmitate selectively impairs phosphosites in signaling branches controlling glucose metabolism but not cell proliferation and lipid metabolism.
Mapping of the affected sites onto the insulin signaling network revealed that palmitate pretreatment affected insulin signaling at specific nodes (Fig. 5C). For example, palmitate treatment impaired insulin-mediated phosphorylation of sites on proteins responsible for glucose metabolism, such as FoxO1 and GSKα/β, whereas sites on proteins involved in cell growth/proliferation and lipid metabolism were not affected or up-regulated. We also performed immunoblots to confirm protein phosphorylation findings of several targets for which antibodies were available (pS256-FoxO1, pS9-GSK3β, pS422-EIF4B, and pS65-EIF4eBP1) in response to insulin and palmitate treatments (supplemental Fig. S2). In each case, the phosphoproteomic findings were confirmed. Notably, although palmitate interfered with FoxO1 phosphorylation, which regulates gluconeogenesis, this treatment did not interfere broadly with insulin signaling, thereby validating HepG2 cells as exhibiting selective insulin resistance in this model.
Palmitate Pretreatment Results in Localization of FoxO1 to the Nucleus
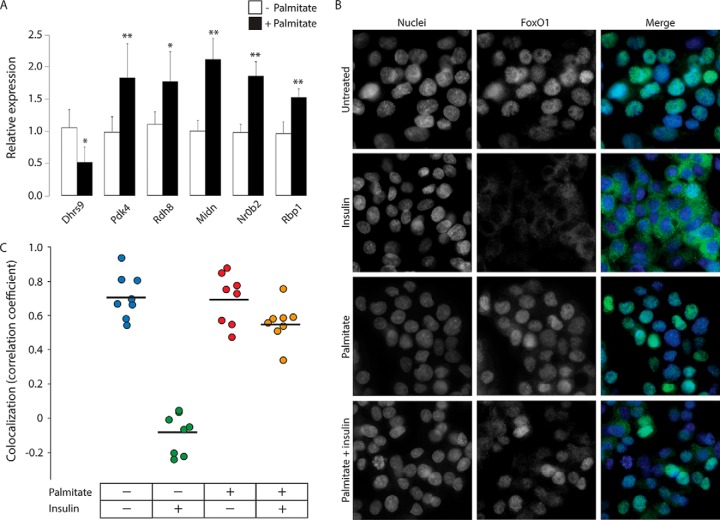
The finding that palmitate treatment impaired insulin signaling in a selective manner suggested the hypothesis that palmitate might interfere with specific signaling events rather than a cascade of events. FoxO1 protein localizes to the nucleus or cytoplasm, depending on its phosphorylation status (45). Insulin treatment normally causes phosphorylation of FoxO1, which in turn causes this transcription factor to localize to the cytoplasm, preventing it from stimulating gluconeogenic gene expression (46). Because we found that palmitate pretreatment causes impairment of insulin's effects on phosphorylation of FoxO1, we hypothesized that this impairment may be because of physical isolation of FoxO1 from kinases (i.e. that FoxO1 might be localized to the nucleus on palmitate treatment), rendering it insensitive to insulin signaling in the cytoplasm. We first examined how palmitate treatment affects FoxO1 regulation in HepG2 cells by examining mRNA levels of the FoxO1 target genes [Dhrs9, Pdk4, Rdh8, Midn, Nr0b2, and Rbp1 (47–52)] 2 h following palmitate treatment. We found that these genes undergo alterations in expression levels (in the expected directions) when cells were treated with palmitate (Fig. 6A). Despite the differential transcriptional activities of its target genes, immmunostaining of FoxO1 showed that the transcription factor is localized predominantly in nuclei in both untreated and palmitate-treated cells (Fig. 6B and 6C). Consistent with previous findings (46), we found that insulin treatment causes FoxO1 to be exported from the nucleus into the cytoplasm. Most significantly, palmitate pretreatment of HepG2 cells resulted in FoxO1 to be retained in the nuclei, even with insulin stimulation (Fig. 6B and 6C).
Fig. 6.
FoxO1 localizes to nuclei of HepG2 cells during palmitate treatment. A, Relative mRNA expression levels of FoxO1 target genes in HepG2 cells following 24 h palmitate treatment. Statistical significance was determined by Mann-Whitney test (* p < 0.05 and ** p < 0.005, n = 6). B, Immunocytochemistry analyses of FoxO1 localization in 24-h palmitate- and/or 2-h insulin-treated HepG2 cells. Green: FoxO1; Blue: DAPI. C, Pearson's correlation coefficient of colocalization analyses between green-FoxO1 and blue-DAPI immunostainings (n = 8).
DISCUSSION
How lipid accumulation during hepatic steatosis leads to selectively impaired insulin signaling is an important, but complex problem. To reduce the complexity of this problem, we took a reductionist approach with a simple human hepatocyte cell model of selective insulin resistance induced by pretreatment with palmitate. We used this system to generate a resource providing the relative abundances of proteins, lipids, and many insulin signaling targets. We identified known and previously unknown phosphosites regulated by insulin and show that only some of these are altered by palmitate pretreatment. We also characterized the lipid changes in response to palmitate and found changes, for instance in DG and ceramides, consistent with previous studies of hepatic insulin resistance.
Among several experimentally amenable cell lines, we chose HepG2 hepatoma cells for our analyses of insulin signaling because we find these cells reproduce critical features of selective insulin resistance. Despite being a hepatoma cell line and having relatively low gluconeogenesis (53), the intact insulin signaling network and the resultant gene expression changes reflect the physiological situation during hepatic steatosis. Those changes lead to impairment of glucose metabolism regulation, but lipogenesis, as measured by a gene expression, is stimulated by insulin. Thus, palmitate treatment of HepG2 cells provides a highly simplified, but experimentally accessible model for studying selective insulin resistance observed in hepatic steatosis associated with obesity, where insulin signaling is similarly compromised (54). Because the key features of selective insulin resistance are reproduced, we believe that our data sets will provide a rich source for developing hypotheses for testing in primary hepatocytes or in vivo.
To analyze insulin signaling in this system in depth, we further developed sample preparation methods that involve state-of-the-art protocols for analyzing phosphoproteomes (55). By using multiple rounds of phosphopeptide enrichment without other peptide fractionation, we obtained ultra-deep phosphoproteome coverage with more than 18,000 phosphosites quantitated. Most likely, the gain in sensitivity is because of the increased retention of phosphopeptides with lower affinity for TiO2 resin (after higher affinity peptides were depleted in the early rounds of enrichment) and the repeated measurements. This methodology, which will be applicable to other signaling pathways, allowed us to analyze the response of the insulin signaling network after metabolic perturbation of cells. Surprisingly, after many important studies of insulin signaling (30, 31, 56, 57), our deep coverage of the phosphoproteome continued to reveal new targets of the insulin-signaling pathway. For example, we found a novel connection between insulin signaling and specific isoforms of FMRP proteins, which is corroborated by genetic interaction in the Drosophila model organism (38). Thus, our data set on insulin stimulation performed with adapted, new methodology provides a rich resource for novel biological discoveries.
Using the reductionist system, we found that palmitate impairs a surprisingly small number of this large set of insulin-responsive signaling nodes. Notably, we detected specific impairment of two key nodes of glucose metabolism by palmitate: the phosphorylation of the FoxO1 transcription factor and the kinase GSK3. Both factors regulate key steps in liver gluconeogenesis (58). Impairing their response to insulin thus can explain at least some aspects of selective insulin resistance. Our findings are consistent with previous reports that FoxO1 (Ser256) and GSK3β (Ser9) phosphorylation are impaired in hepatocytes from mice fed a high-fat diet (59). Moreover, FoxO1 phosphorylation at serine 256 is diminished in livers of steatosis patients (60).
In addition to FoxO1 and GSK3β, we also found decreased phosphorylation of several other sites during palmitate treatment, such as serine 23 on NF-κB, which might alter its stability. Similarly, SHC1 phosphorylation on serine 29, normally occurring directly after insulin receptor stimulation, was impaired after palmitate treatment. How these changes will affect signaling output is not yet clear, but our resource will enable future studies of these important signaling nodes.
Notably, although palmitate treatment caused a down-regulation of Akt S473 phosphorylation (found by immunoblot, as this site is in a peptide not accessible to mass spectrometry analyses), not all downstream Akt targets mirrored this down-regulation, and only a few proteins, such as FoxO1 and GSK3β, failed to respond to insulin-stimulated phosphorylation in palmitate-treated cells. At least two nonexclusive mechanisms may account for this resistance to insulin-mediated phosphorylation. One is the activation of a palmitate-activated phosphatase, which has only a subset of insulin-stimulated phosphosites as targets, and another is the sequestration of proteins in subcellular locations where they are not accessible to the upstream kinases involved in insulin signaling. Although there is ample evidence for phosphatase activation because of lipid changes (e.g. for ceramide- or palmitate-activated phosphatases) (61, 62), we also uncovered evidence for the control of substrate phosphorylation by subcellular localization. Specifically, we found that palmitate treatment prevents nuclear export of FoxO1 by insulin. At this site, the transcription factor will not be accessible to cytoplasmic Akt kinase, explaining why in the presence of palmitate, phosphorylation cannot occur. This model is consistent with previous studies in MIN6 cells (63). Furthermore, the palmitate-induced localization of FoxO1 into the nucleus resulted in the alteration of the transcription activities of its target genes. These data suggest the hypothesis that selective insulin resistance occurs, at least in part, because of changes in subcellular localization of key transcription factors, such as FoxO1, rendering them insensitive to insulin.
An important and unresolved question is precisely how lipids, such as palmitate, and other lipids accumulating during hepatic steatosis, mediate impairments in insulin signaling (2). Although different lipids might use different pathways to interfere with signaling, lipidomic analyses in our model system suggest that particularly the accumulation of saturated glycerophospholipids is at least one potent way to compromise the response to phospholipids. It is unclear how these changes lead to altered signaling. Changes in lipids of the plasma membrane, the endoplasmic reticulum (ER), or other intracellular organelles might activate signaling molecules (kinases or phosphatases) or stress responses, such as the unfolded protein response (UPR), that cause changes that interfere with insulin signaling in cells. Importantly, however, our data suggest that intervention at the step which leads to incorporation of palmitate into glycerophospholipids may be beneficial to avert the effects of saturated glycerophospholipids, such as DGs (64). Thus, either inhibiting acyl-CoA synthetase enzymes activating palmitate to palmitoyl-CoA, or inhibiting GPAT enzymes, esterifying palmitate to lysophosphatidic acid, might be a strategy to prevent palmitate interference with insulin signaling.
In summary, we provide a rich resource, based on a combination of unbiased approaches, for a reductionist model of selective insulin resistance. This resource is characterized by deep coverage of the phosphoproteome, proteome, and lipidome under various conditions. Our study reveals key nodes that are misregulated during selective insulin resistance and suggests novel hypotheses how this might lead to dysregulated glucose and lipid metabolism in physiology. Although by themselves these data cannot clarify the physiological importance of the nodes we find impaired, they afford new hypotheses and suggest a few, specific sites that are key to the development to selective, hepatic insulin resistance. Given the magnitude of this problem because of the raising epidemic of obesity linked hepatic steatosis, this likely will motivate further testing and developing strategies for treatment of selective insulin resistance, and ultimately type II diabetes.
DATA AVAILABILITY
The mass spectrometry proteomics data have been deposited to http://proteomecentral.proteomexchange.org/cgi/GetDataset (project ID: PXD006395) and (https://www.ebi.ac.uk/metabolights/MTBLS582 (project ID: MTBLS582).
Supplementary Material
Acknowledgments
We thank Florian Fröhlich, Natalie Krahmer, Mara Monetti and members of the Farese and Walther laboratory for comments on the manuscript, and Gary Howard for editorial assistance.
Footnotes
* This work was supported by R01 DK101579 (to R.V.F.) the Mathers foundation (to T.C.W). T.C.W is an investigator of the Howard Hughes Medical Institute.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- T2D
- type 2 diabetes mellitus
- CAA
- chloroacetamide alkylating reagent
- DG
- diacylglycerol
- DMEM
- Dulbecco's Modified Eagle's medium
- ER
- endoplasmic reticulum
- FDR
- false discovery rate
- FMRP
- fragile X mental retardation
- GdmCl
- guanidinium hydrochloride
- GO
- Gene ontology
- HCD
- high-energy collision dissociation
- MAD
- median absolute deviations
- NCE
- normalized collision energy
- NES
- nuclear export sequence
- PTFE
- polytetrafluoroethylene
- RIPA
- radioimmunoprecipitation assay
- SILAC
- stable isotope labeling with amino acids in cell culture
- TCEP
- Tris (2-carboxyethyl) phosphine
- TG
- triglyceride
- TiO2
- titanium dioxide
- UPR
- unfolded protein response.
REFERENCES
- 1. Brown M. S., and Goldstein J. L. (2008) Selective versus total insulin resistance: a pathogenic paradox. Cell Metab. 7, 95–96 [DOI] [PubMed] [Google Scholar]
- 2. Farese R. V. Jr, Zechner R., Newgard C. B., and Walther T. C. (2012) The problem of establishing relationships between hepatic steatosis and hepatic insulin resistance. Cell Metab. 15, 570–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Samuel V. T., and Shulman G. I. (2012) Mechanisms for insulin resistance: common threads and missing links. Cell 148, 852–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chaurasia B., and Summers S. A. (2015) Ceramides - Lipotoxic Inducers of Metabolic Disorders. Trends Endocrinol. Metab. 26, 538–550 [DOI] [PubMed] [Google Scholar]
- 5. Chakravarthy M. V., Pan Z., Zhu Y., Tordjman K., Schneider J. G., Coleman T., Turk J., and Semenkovich C. F. (2005) “New” hepatic fat activates PPARalpha to maintain glucose, lipid, and cholesterol homeostasis. Cell Metab. 1, 309–322 [DOI] [PubMed] [Google Scholar]
- 6. Monetti M., Levin M. C., Watt M. J., Sajan M. P., Marmor S., Hubbard B. K., Stevens R. D., Bain J. R., Newgard C. B., Farese R. V. Sr Hevener A. L., and Farese R. V. Jr. (2007) Dissociation of hepatic steatosis and insulin resistance in mice overexpressing DGAT in the liver. Cell Metab. 6, 69–78 [DOI] [PubMed] [Google Scholar]
- 7. Brown J. M., Betters J. L., Lord C., Ma Y., Han X., Yang K., Alger H. M., Melchior J., Sawyer J., Shah R., Wilson M. D., Liu X., Graham M. J., Lee R., Crooke R., Shulman G. I., Xue B., Shi H., and Yu L. (2010) CGI-58 knockdown in mice causes hepatic steatosis but prevents diet-induced obesity and glucose intolerance. J. Lipid Res. 51, 3306–3315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hoy A. J., Bruce C. R., Turpin S. M., Morris A. J., Febbraio M. A., and Watt M. J. (2011) Adipose triglyceride lipase-null mice are resistant to high-fat diet-induced insulin resistance despite reduced energy expenditure and ectopic lipid accumulation. Endocrinology 152, 48–58 [DOI] [PubMed] [Google Scholar]
- 9. Minehira K., Young S. G., Villanueva C. J., Yetukuri L., Oresic M., Hellerstein M. K., Farese R. V. Jr, Horton J. D., Preitner F., Thorens B., and Tappy L. (2008) Blocking VLDL secretion causes hepatic steatosis but does not affect peripheral lipid stores or insulin sensitivity in mice. J. Lipid Res. 49, 2038–2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gao D., Nong S., Huang X., Lu Y., Zhao H., Lin Y., Man Y., Wang S., Yang J., and Li J. (2010) The effects of palmitate on hepatic insulin resistance are mediated by NADPH Oxidase 3-derived reactive oxygen species through JNK and p38MAPK pathways. J. Biol. Chem. 285, 29965–29973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ong S. E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., and Mann M. (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1, 376–386 [DOI] [PubMed] [Google Scholar]
- 12. Ren F., Wu H., Lei Y., Zhang H., Liu R., Zhao Y., Chen X., Zeng D., Tong A., Chen L., Wei Y., and Huang C. (2010) Quantitative proteomics identification of phosphoglycerate mutase 1 as a novel therapeutic target in hepatocellular carcinoma. Mol. Cancer 9, 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sun Y., Mi W., Cai J., Ying W., Liu F., Lu H., Qiao Y., Jia W., Bi X., Lu N., Liu S., Qian X., and Zhao X. (2008) Quantitative proteomic signature of liver cancer cells: tissue transglutaminase 2 could be a novel protein candidate of human hepatocellular carcinoma. J. Proteome Res. 7, 3847–3859 [DOI] [PubMed] [Google Scholar]
- 14. Rappsilber J., Ishihama Y., and Mann M. (2003) Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 75, 663–670 [DOI] [PubMed] [Google Scholar]
- 15. Gazos-Lopes F., Oliveira M. M., Hoelz L. V., Vieira D. P., Marques A. F., Nakayasu E. S., Gomes M. T., Salloum N. G., Pascutti P. G., Souto-Padron T., Monteiro R. Q., Lopes A. H., and Almeida I. C. (2014) Structural and functional analysis of a platelet-activating lysophosphatidylcholine of Trypanosoma cruzi. PLoS Negl. Trop. Dis. 8, e3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Frohlich F., Petit C., Kory N., Christiano R., Hannibal-Bach H. K., Graham M., Liu X., Ejsing C. S., Farese R. V., and Walther T. C. (2015) The GARP complex is required for cellular sphingolipid homeostasis. Elife 4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cox J., and Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 18. Cox J., Neuhauser N., Michalski A., Scheltema R. A., Olsen J. V., and Mann M. (2011) Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805 [DOI] [PubMed] [Google Scholar]
- 19. Vizcaino J. A., Csordas A., Del-Toro N., Dianes J. A., Griss J., Lavidas I., Mayer G., Perez-Riverol Y., Reisinger F., Ternent T., Xu Q. W., Wang R., and Hermjakob H. (2016) 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 44, 11033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Haug K., Salek R. M., Conesa P., Hastings J., de Matos P., Rijnbeek M., Mahendraker T., Williams M., Neumann S., Rocca-Serra P., Maguire E., Gonzalez-Beltran A., Sansone S. A., Griffin J. L., and Steinbeck C. (2013) MetaboLights–an open-access general-purpose repository for metabolomics studies and associated meta-data. Nucleic Acids Res. 41, D781–D786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. O'Shea J. P., Chou M. F., Quader S. A., Ryan J. K., Church G. M., and Schwartz D. (2013) pLogo: a probabilistic approach to visualizing sequence motifs. Nat. Methods 10, 1211–1212 [DOI] [PubMed] [Google Scholar]
- 22. Alessi D. R., Andjelkovic M., Caudwell B., Cron P., Morrice N., Cohen P., and Hemmings B. A. (1996) Mechanism of activation of protein kinase B by insulin and IGF-1. EMBO J. 15, 6541–6551 [PMC free article] [PubMed] [Google Scholar]
- 23. Girard J. (2006) The inhibitory effects of insulin on hepatic glucose production are both direct and indirect. Diabetes 55, 5 [Google Scholar]
- 24. Yellaturu C. R., Deng X., Cagen L. M., Wilcox H. G., Mansbach C. M. 2nd, Siddiqi S. A., Park E. A., Raghow R., and Elam M. B. (2009) Insulin enhances post-translational processing of nascent SREBP-1c by promoting its phosphorylation and association with COPII vesicles. J. Biol. Chem. 284, 7518–7532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Repa J. J., Liang G., Ou J., Bashmakov Y., Lobaccaro J. M., Shimomura I., Shan B., Brown M. S., Goldstein J. L., and Mangelsdorf D. J. (2000) Regulation of mouse sterol regulatory element-binding protein-1c gene (SREBP-1c) by oxysterol receptors, LXRalpha and LXRbeta. Genes. Dev. 14, 2819–2830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Han M. S., Park S. Y., Shinzawa K., Kim S., Chung K. W., Lee J. H., Kwon C. H., Lee K. W., Lee J. H., Park C. K., Chung W. J., Hwang J. S., Yan J. J., Song D. K., Tsujimoto Y., and Lee M. S. (2008) Lysophosphatidylcholine as a death effector in the lipoapoptosis of hepatocytes. J. Lipid Res. 49, 84–97 [DOI] [PubMed] [Google Scholar]
- 27. van Herpen N. A., and Schrauwen-Hinderling V. B. (2008) Lipid accumulation in non-adipose tissue and lipotoxicity. Physiol. Behav. 94, 231–241 [DOI] [PubMed] [Google Scholar]
- 28. Yetukuri L., Katajamaa M., Medina-Gomez G., Seppanen-Laakso T., Vidal-Puig A., and Oresic M. (2007) Bioinformatics strategies for lipidomics analysis: characterization of obesity related hepatic steatosis. BMC Syst. Biol. 1, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kruger M., Kratchmarova I., Blagoev B., Tseng Y. H., Kahn C. R., and Mann M. (2008) Dissection of the insulin signaling pathway via quantitative phosphoproteomics. Proc. Natl. Acad. Sci. U.S.A. 105, 2451–2456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Humphrey S. J., Yang G., Yang P., Fazakerley D. J., Stockli J., Yang J. Y., and James D. E. (2013) Dynamic adipocyte phosphoproteome reveals that Akt directly regulates mTORC2. Cell Metab. 17, 1009–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Humphrey S. J., Azimifar S. B., and Mann M. (2015) High-throughput phosphoproteomics reveals in vivo insulin signaling dynamics. Nat. Biotechnol. 33, 990–995 [DOI] [PubMed] [Google Scholar]
- 32. Hilger M., Bonaldi T., Gnad F., and Mann M. (2009) Systems-wide analysis of a phosphatase knock-down by quantitative proteomics and phosphoproteomics. Mol. Cell. Proteomics 8, 1908–1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pan C., Olsen J. V., Daub H., and Mann M. (2009) Global effects of kinase inhibitors on signaling networks revealed by quantitative phosphoproteomics. Mol. Cell. Proteomics 8, 2796–2808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Makarov A., Denisov E., and Lange O. (2009) Performance evaluation of a high-field Orbitrap mass analyzer. J. Am. Soc. Mass Spectrom. 20, 1391–1396 [DOI] [PubMed] [Google Scholar]
- 35. Ong S. E., and Mann M. (2005) Mass spectrometry-based proteomics turns quantitative. Nat. Chem. Biol. 1, 252–262 [DOI] [PubMed] [Google Scholar]
- 36. Pearce L. R., Komander D., and Alessi D. R. (2010) The nuts and bolts of AGC protein kinases. Nat. Rev. Mol. Cell Biol. 11, 9–22 [DOI] [PubMed] [Google Scholar]
- 37. Verkerk A. J., Pieretti M., Sutcliffe J. S., Fu Y. H., Kuhl D. P., Pizzuti A., Reiner O., Richards S., Victoria M. F., Zhang F. P., Eussen B. E., van Ommen G. B., Blonden L. A. J., Riggins G. J., Chastain J. L., Kunst C. B., Galjaard H., Caskey C. T., Nelson D. L., Oostra B. A., and Warren S. T. (1991) Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell 65, 905–914 [DOI] [PubMed] [Google Scholar]
- 38. Monyak R. E., Emerson D., Schoenfeld B. P., Zheng X., Chambers D. B., Rosenfelt C., Langer S., Hinchey P., Choi C. H., McDonald T. V., Bolduc F. V., Sehgal A., McBride S. M., and Jongens T. A. (2016) Insulin signaling misregulation underlies circadian and cognitive deficits in a Drosophila fragile X model. Mol. Psychiatry [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lumaban J. G., and Nelson D. L. (2015) The Fragile X proteins Fmrp and Fxr2p cooperate to regulate glucose metabolism in mice. Hum Mol. Genet. 24, 2175–2184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhang J., Fang Z., Jud C., Vansteensel M. J., Kaasik K., Lee C. C., Albrecht U., Tamanini F., Meijer J. H., Oostra B. A., and Nelson D. L. (2008) Fragile X-related proteins regulate mammalian circadian behavioral rhythms. Am. J. Hum. Genet. 83, 43–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dury A. Y., El Fatimy R., Tremblay S., Rose T. M., Cote J., De Koninck P., and Khandjian E. W. (2013) Nuclear Fragile X Mental Retardation Protein is localized to Cajal bodies. PLoS Genet. 9, e1003890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Garvey W. T., Maianu L., Zhu J. H., Brechtel-Hook G., Wallace P., and Baron A. D. (1998) Evidence for defects in the trafficking and translocation of GLUT4 glucose transporters in skeletal muscle as a cause of human insulin resistance. J. Clin. Invest. 101, 2377–2386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sano H., Kane S., Sano E., Miinea C. P., Asara J. M., Lane W. S., Garner C. W., and Lienhard G. E. (2003) Insulin-stimulated phosphorylation of a Rab GTPase-activating protein regulates GLUT4 translocation. J. Biol. Chem. 278, 14599–14602 [DOI] [PubMed] [Google Scholar]
- 44. Panaretou C., Domin J., Cockcroft S., and Waterfield M. D. (1997) Characterization of p150, an adaptor protein for the human phosphatidylinositol (PtdIns) 3-kinase. Substrate presentation by phosphatidylinositol transfer protein to the p150.Ptdins 3-kinase complex. J. Biol. Chem. 272, 2477–2485 [DOI] [PubMed] [Google Scholar]
- 45. Biggs W. H. 3rd, Meisenhelder J., Hunter T., Cavenee W. K., and Arden K. C. (1999) Protein kinase B/Akt-mediated phosphorylation promotes nuclear exclusion of the winged helix transcription factor FKHR1. Proc. Natl. Acad. Sci. U.S.A. 96, 7421–7426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nakae J., Kitamura T., Silver D. L., and Accili D. (2001) The forkhead transcription factor Foxo1 (Fkhr) confers insulin sensitivity onto glucose-6-phosphatase expression. J. Clin. Invest. 108, 1359–1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Shin D. J., Joshi P., Hong S. H., Mosure K., Shin D. G., and Osborne T. F. (2012) Genome-wide analysis of FoxO1 binding in hepatic chromatin: potential involvement of FoxO1 in linking retinoid signaling to hepatic gluconeogenesis. Nucleic Acids Res. 40, 11499–11509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Obrochta K. M., Krois C. R., Campos B., and Napoli J. L. (2015) Insulin regulates retinol dehydrogenase expression and all-trans-retinoic acid biosynthesis through FoxO1. J. Biol. Chem. 290, 7259–7268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wei D., Tao R., Zhang Y., White M. F., and Dong X. C. (2011) Feedback regulation of hepatic gluconeogenesis through modulation of SHP/Nr0b2 gene expression by Sirt1 and FoxO1. Am. J. Physiol. Endocrinol. Metab. 300, E312–E320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cheng Z., and White M. F. (2011) Targeting Forkhead box O1 from the concept to metabolic diseases: lessons from mouse models. Antioxid. Redox. Signal. 14, 649–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhao G., Jeoung N. H., Burgess S. C., Rosaaen-Stowe K. A., Inagaki T., Latif S., Shelton J. M., McAnally J., Bassel-Duby R., Harris R. A., Richardson J. A., and Kliewer S. A. (2008) Overexpression of pyruvate dehydrogenase kinase 4 in heart perturbs metabolism and exacerbates calcineurin-induced cardiomyopathy. Am. J. Physiol. Heart Circ. Physiol. 294, H936–H943 [DOI] [PubMed] [Google Scholar]
- 52. Long Y. C., Cheng Z., Copps K. D., and White M. F. (2011) Insulin receptor substrates Irs1 and Irs2 coordinate skeletal muscle growth and metabolism via the Akt and AMPK pathways. Mol. Cell Biol. 31, 430–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Okamoto T., Kanemoto N., Ban T., Sudo T., Nagano K., and Niki I. (2009) Establishment and characterization of a novel method for evaluating gluconeogenesis using hepatic cell lines, H4IIE and HepG2. Arch. Biochem. Biophys. 491, 46–52 [DOI] [PubMed] [Google Scholar]
- 54. Samuel V. T., Liu Z. X., Qu X., Elder B. D., Bilz S., Befroy D., Romanelli A. J., and Shulman G. I. (2004) Mechanism of hepatic insulin resistance in non-alcoholic fatty liver disease. J. Biol. Chem. 279, 32345–32353 [DOI] [PubMed] [Google Scholar]
- 55. Kulak N. A., Pichler G., Paron I., Nagaraj N., and Mann M. (2014) Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat. Methods 11, 319–324 [DOI] [PubMed] [Google Scholar]
- 56. Monetti M., Nagaraj N., Sharma K., and Mann M. (2011) Large-scale phosphosite quantification in tissues by a spike-in SILAC method. Nat. Methods 8, 655–658 [DOI] [PubMed] [Google Scholar]
- 57. Schmelzle K., Kane S., Gridley S., Lienhard G. E., and White F. M. (2006) Temporal dynamics of tyrosine phosphorylation in insulin signaling. Diabetes 55, 2171–2179 [DOI] [PubMed] [Google Scholar]
- 58. Puigserver P., Rhee J., Donovan J., Walkey C. J., Yoon J. C., Oriente F., Kitamura Y., Altomonte J., Dong H., Accili D., and Spiegelman B. M. (2003) Insulin-regulated hepatic gluconeogenesis through FOXO1-PGC-1alpha interaction. Nature 423, 550–555 [DOI] [PubMed] [Google Scholar]
- 59. Bae E. J., Xu J., Oh D. Y., Bandyopadhyay G., Lagakos W. S., Keshwani M., and Olefsky J. M. (2012) Liver-specific p70 S6 kinase depletion protects against hepatic steatosis and systemic insulin resistance. J. Biol. Chem. 287, 18769–18780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Valenti L., Rametta R., Dongiovanni P., Maggioni M., Fracanzani A. L., Zappa M., Lattuada E., Roviaro G., and Fargion S. (2008) Increased expression and activity of the transcription factor FOXO1 in nonalcoholic steatohepatitis. Diabetes 57, 1355–1362 [DOI] [PubMed] [Google Scholar]
- 61. Yuyama K., Mitsutake S., and Igarashi Y. (2014) Pathological roles of ceramide and its metabolites in metabolic syndrome and Alzheimer's disease. Biochim. Biophys. Acta 1841, 793–798 [DOI] [PubMed] [Google Scholar]
- 62. Wu Y., Song P., Xu J., Zhang M., and Zou M. H. (2007) Activation of protein phosphatase 2A by palmitate inhibits AMP-activated protein kinase. J. Biol. Chem. 282, 9777–9788 [DOI] [PubMed] [Google Scholar]
- 63. Martinez S. C., Tanabe K., Cras-Meneur C., Abumrad N. A., Bernal-Mizrachi E., and Permutt M. A. (2008) Inhibition of Foxo1 protects pancreatic islet beta-cells against fatty acid and endoplasmic reticulum stress-induced apoptosis. Diabetes 57, 846–859 [DOI] [PubMed] [Google Scholar]
- 64. Jornayvaz F. R., and Shulman G. I. (2012) Diacylglycerol activation of protein kinase Cepsilon and hepatic insulin resistance. Cell Metab. 15, 574–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to http://proteomecentral.proteomexchange.org/cgi/GetDataset (project ID: PXD006395) and (https://www.ebi.ac.uk/metabolights/MTBLS582 (project ID: MTBLS582).