Abstract
Coimmunoprecipitation (co-IP) is one of the most frequently used techniques to study protein-protein (PPIs) or protein-nucleic acid interactions (PNIs). However, the presence of coprecipitated contaminants is a well-recognized issue associated with single-step co-IPs. To overcome this limitation, we developed the two-step co-IP (TIP) strategy that enables sequential coimmunoprecipitations of endogenous protein complexes. TIP can be performed with a broad range of mono- and polyclonal antibodies targeting a single protein or different components of a given complex. TIP results in a highly selective enrichment of protein complexes and thus outperforms single-step co-IPs for downstream applications such as mass spectrometry for the identification of PPIs and quantitative PCR for the analysis of PNIs. We benchmarked TIP for the identification of CD95/FAS-interacting proteins in primary human CD4+ T cells, which recapitulated all major known interactors, but also enabled the proteomics discovery of PPM1G and IPO7 as new interaction partners. For its feasibility and high performance, we propose TIP as an advanced tool for the isolation of highly purified protein-protein and protein-nucleic acid complexes under native expression conditions.
Unraveling the complexity and dynamic behavior of protein-protein (PPIs)1 and protein-nucleic acid interactions (PNIs) that serve as central hubs for coordinated cellular events is one of the major objectives in cell biology and especially in proteome research (1). A powerful tool for the identification of protein networks is affinity purification coupled to mass spectrometry (AP-MS) applying highly sensitive, selective and fast scanning tandem MS systems (2). To elucidate correct complex stoichiometry, quantitative AP-MS is also used for relative and absolute protein quantification (3, 4).
Coimmunoprecipitation (co-IP) is a predominant method for affinity purification of PPIs and PNIs. This strategy employs antibodies directed against the protein of interest to enrich the protein and its interactors from a cellular lysate. Co-IP is typically coupled to several detection methods, such as Western blotting or mass spectrometry for identification of PPIs and nucleic acid amplification or sequencing for identification of PNIs (4). However, the gentle lysis and washing conditions essential for maintaining the integrity of the complexes result in copurification of nonspecific binders that affect all downstream applications. These false-positive interactors interfere with the identification of low abundant complex members, even though sophisticated mass spectrometry and data analysis methods help to discriminate between specific and nonspecific binders (5, 6).
Approaches that increase the purity of the samples largely overcome this issue, and a widespread strategy has exploited the fusion of two separate tags for affinity purification on a single protein. Using each tag sequentially, Tandem Affinity Purification (TAP) results in cleaner samples that can be used for identification by mass spectrometry, or for other downstream applications (7–15). Yet, the epitope tags need to be artificially added to the bait of interest, which in some cases may alter its functions and interactions. This also prevents the application of TAP to biological material (e.g. patient samples) that cannot be genetically manipulated.
To achieve high quality affinity isolations of protein-containing complexes without ectopic protein expression, we developed the two-step coimmunoprecipitation (TIP) method. TIP consists of two serial co-IPs using a biotinylated antibody (ab) directed against the endogenous protein. An anti-biotin resin enables the gentle elution of the protein complex prior to re-precipitation with protein A/G beads. Alternatively, the second co-IP can be performed with abs directed to the same bait or another component of the same complex. Here we exemplify the broad applicability of TIP by using several bait proteins and different readouts. We focused on the CD95/FAS “death inducing signaling complex” (DISC) and the IKK-complex, which have been both extensively studied and have essential functions in key immunological processes such as apoptosis signaling and NF-kB activation, respectively (16–19). In contrast to conventional single-step co-IP, TIP resulted in highly specific complex purifications as well as identification and quantification of all yet known DISC core components by MS. We also demonstrated that TIP could be streamlined by applying a biotinylated bridging (secondary) ab (bTIP), thus avoiding the need to modify the bait-specific antibodies. Finally, we underlined the functionality of TIP in primary human CD4+ T cells and purified CD95/FAS- and IKK-complex interacting proteins in this model system. We confirmed all known major binders with high confidence and identified protein phosphatase PP2Cgamma (PPM1G) and importin 7 (IPO7), which were so far not described to interact with CD95/FAS. Genetic ablation of PPM1G significantly sensitized cells to CD95/FAS-induced apoptosis, indicating that it represents a novel CD95/FAS-associated negative regulator for the induction of apoptosis by this prototypic cell death receptor.
EXPERIMENTAL PROCEDURES
Cell Culture
HEK293T, HeLa and HCT116 cell lines were purchased from ATCC (CRL-11268, CCL-2 and CCL-247) and BJAB cell line from DSMZ (ACC 757). H9 and SKW6.4 cell lines were a gift from P.H. Krammer. The cells were grown in RPMI (Euroclone, Milan, Italy, cat. no. ECM2001L), DMEM (Euroclone cat. no. ECB2072L) and McCoy's (Thermo Fisher Scientific, Bremen, Germany, cat. no. 36600–021) supplemented with 10% FBS (Euroclone cat. no. ECS0180L) and antibiotics (Euroclone cat. no. ECB3001D) at 37 °C and 5% CO2.
Reagents for Single-step co-IP, TIP and bTIP
To perform the coimmunoprecipitation reactions we used IP-lysis buffer (30 mm Tris/HCl (pH 7.4), 120 mm NaCl, 10% Glycerol, 2 mm EDTA, 2 mm KCl and 1% NP40) supplemented with Complete EDTA-free protease inhibitor mixture tablet (Roche Diagnostics, Mannheim, Germany). For the elution we prepared 10 mm biotin stock solution (10 mm d-biotin, 10 mm bicine, 120 mm NaCl and 2 mm KCl) diluted in IP-lysis buffer reaching a final d-biotin concentration of 3.3 mm (pH 7.5). Unless otherwise indicated, standard salts and buffers were purchased from Sigma-Aldrich (Sigma-Aldrich, St. Louis, Missouri).
The resins used were: protein G beads (GE Healthcare Bio-Sciences, Uppsala, Sweden, cat. no. 17–6002-35), protein A/G beads (Santa Cruz Biotechnology, Santa Cruz, CA, cat. no. sc-2003) and anti-biotin beads (Sigma-Aldrich cat. no. A1559). The antibodies used for coimmunoprecipitations were: anti-IKKα (Santa Cruz Biotechnology cat. no. sc-7606), anti-CD95/FAS (APO-1–3 (20)) and anti-Caspase-8 (Santa Cruz Biotechnology cat. no. sc-6136). These were biotinylated as described below. For the control samples we used biotinylated isotype matched antibodies (Southern Biotech, Birmingham, AL, cat. no. 0104–08, cat. no. 0105–08 and from Santa Cruz Biotechnology cat. no. sc-2755). For bTIP experiments we used anti-IKKγ (Santa Cruz Biotechnology cat. no. sc-8330) and anti-CD95/FAS (APO-1–3 (20)) antibodies. In these experiments rabbit and IgG3 isotype matched antibodies were used (ProSci, Poway, CA, cat. no. 3703 and Southern Biotech cat. no. 0105–01). The secondary bridging antibodies were biotin conjugated goat anti-rabbit and goat anti-mouse antibodies (Thermo Fisher Scientific cat. no. A27035 and Invitrogen, Carlsbad, CA, cat. no. A28176).
For the application of TIP with two different antibodies, the following reagents were used: biotinylated or nonbiotinylated anti-CD95/FAS (APO-1–3 (20)), anti-FAS (Santa Cruz Biotechnology cat. no. sc-715), anti-IKKα (Santa Cruz Biotechnology cat. no. sc-7606), anti-IKKγ (Santa Cruz Biotechnology cat. no. sc-8330) and the isotype matched antibodies (Southern Biotech cat. no. 0105–08, 0105–01, ProSci cat. no. 3703 and Abcam, Cambridge, United Kingdom, cat. no. ab18469). Biotinylated bridging antibodies were the same as indicated above. The beads specifically recognizing the second antibodies were anti-rabbit beads (Rockland Bioscience, Limerick, Pottstown, PA, cat. no. 00–8800-25) and anti-mouse beads (Sigma-Aldrich cat. no. A6531). For single-step co-IP experiments we used anti-CD95/FAS (Biolegend, San Diego, CA, clone DX2, cat. no. 305602) and anti-IgG1 isotype control antibodies (Abcam cat. no. ab18443).
Coimmunoprecipitation Assays
Mass spectrometry samples were prepared using 1 × 108 BJAB or 2–3 × 107 CD4+ T cells, respectively. The cells were washed with cold DPBS (Euroclone cat. no. ECB4004L) and lysed in IP-lysis buffer for 20 min on a rotator at a density of 2 × 107 cells/ml (BJAB) and 4 × 107 cells/ml (CD4+ T cells). The lysates were cleared by centrifugation (14800 RPM for 20 min at 4 °C) and subjected to three pre-clearing steps. Two pre-clearing steps were performed using protein G beads precoupled with isotype control antibody (3 μg) and the last with empty beads (on a rotator for 30 min at 4 °C). Identical amounts of lysates were used for single-step co-IP, TIP and bTIP. Although the mock samples received no antibody, the stimulated and nonstimulated lysates for single-step co-IP and TIP were incubated with biotinylated antibodies (3 μg) (bait-specific and isotype control). To avoid problems of antibody aggregation, the lysates for the bTIP samples were first treated with nonbiotinylated primary antibodies (3 μg) on a rotator for 30 min at 4 °C. Afterward the biotin conjugated secondary antibodies (6 μg) were added and incubated for additional 30 min on a rotator at 4 °C. All other steps were performed under identical conditions. Finally, the anti-biotin beads were added.
For the samples only analyzed by Western blotting, we used 1 μg of primary and 2 μg of biotinylated secondary antibodies.
Single-step co-IP and the first step of TIP or bTIP samples were precipitated for 6–14 h at 4 °C on a rotator, followed by 4–6 washing steps in IP-lysis buffer. The single-step co-IP samples were then stored at −80 °C. The proteins bound to the anti-biotin beads were eluted with 3.3 mm biotin solution. We performed two consecutive elution steps in 500 μl volume for 10 min with rotation at 4 °C. After each elution, the samples were centrifuged (3500 RPM for 3 min at 4 °C) and the supernatant was carefully transferred into a new tube containing protein G beads. The final volume was adjusted to 1.5 ml with IP-lysis buffer and the second coimmunoprecipitation was performed for 6–14 h on a rotator at 4 °C. Afterward, the beads were washed as above and stored at −80 °C until analysis. To determine the optimal duration of the elution, the proteins from the supernatants after the biotin elution were precipitated using 10% end volume of TCA (Sigma-Aldrich cat. no. T0699) and compared with the bead-bound proteins.
Biotinylation of Antibodies
One to two micrograms of antibody in 100 μl PBS were biotinylated with a 50 fold molar excess of 10 mm EZ-Link Sulfo-NHS-LC-Biotin (Thermo Fisher Scientific cat. no. 21335). The antibody-biotin reaction was incubated for 2 h on ice and for additional 30 min at room temperature. To remove excess of unbound biotin, the mixture was dialyzed against PBS. To evaluate the antibody binding affinity, 2 × 105 BJAB cells were stained with a serial dilution of the abs in FACS buffer (PBS plus 0.5% BSA) for 30 min on ice. Afterward, the cells were washed 2 times with FACS buffer and stained with goat anti-mouse-PE antibody (Thermo Fisher Scientific cat. no. P852) for 20 min on ice. The cells were washed and analyzed by flow cytometry (FACS Canto, Becton Dickinson). The constant of dissociation (Kd) values were determined with Prism (GraphPad) software from the saturation binding curve analysis (21).
Sample Preparation for MS Analysis
The protein G beads were incubated in 25 μl reducing sample buffer at 70 °C for 10 min and a short (2 cm) gel electrophoresis run was performed using a 1.0 mm 12 or 15 wells, 4–12% BisTris Gel (Invitrogen) in MES buffer at 200 V, 300 mA, 50 W for 9 min 30 s. Gels were stained with Coomassie, the stained part of the gel lanes were excised in two pieces. Proteins in the individual gel slices were reduced, alkylated and in-gel digested with trypsin (Promega, Madison, WI). Before MS analysis, extracted peptides of the two gel pieces were combined, desiccated and dissolved in water, containing 2.5% hexafluoro-2-propanol (HFIP) (Sigma-Aldrich cat. no. 10522) and 0.1% formic acid (Biosolve, Valkenswaard, Netherlands, cat. no. 069141A8) prior to MS analysis.
Liquid Chromatography and Mass Spectrometry
Peptides from trypsin digestions were separated using a nanoAcquity ultra high-performance liquid chromatography (UPLC) system (Waters). Peptides were trapped with a constant flow of 5 μl/min on a 180 μm × 20 mm nanoAcquity UPLC 2G Trap Column filled with Symmetry C18, 5 μm particles. Sample separation was performed on a 100 μm × 100 mm nanoAcquity BEH C18, 1.7 μm analytical column applying a constant flow of 400 nL/min. Chromatography was carried out using either a 2h stepped linear gradient of solvent A (96.9% water, 3% DMSO, 0.1% formic acid) and solvent B (99.9% acetonitrile and 0.1% formic acid) in the following sequence: from 0 to 4% B in 1 min, from 4 to 30% B in 80 min, from 30 to 45% B in 10 min, from 45 to 90% B in 10 min, 10 min at 90% B, from 90 to 0% B in 0.1 min, and 9.9 min at 0% B or 3 h gradient from 0 to 4% B in 1 min, from 4 to 30% B in 140 min, from 30 to 45% B in 15 min, from 45 to 90% B in 5 min, 10 min at 90% B, from 90 to 0% B in 0.1 min, and 9.9 min at 0% B. The nanoUPLC was coupled online to a nano-electrospray source of a linear ion trap quadrupole (LTQ) Orbitrap XL mass spectrometer (Thermo Fisher Scientific). A Pico-Tip Emitter tip type 360 μm OD × 20 μm ID; 10 μm (New Objective) was used for sample ionization and introduction into the MS. The MS was operated in the sensitive mode with the following parameters: ESI voltage was set to 2400 V, the capillary temperature was 200 °C and the normalized collision energy was 35 V. The orbitrap filling time was set at maximum of 500 ms. Full scan MS spectra were acquired in a mass-to-charge ratio (m/z) range from 350–2000 in the profile mode with a mass resolution of 60,000 at m/z 400. Simultaneously, six most abundant precursor ions from the full-scan MS were selected for MS/MS fragmentation in the LTQ. MS/MS data were acquired in centroid mode. Only multiply charged (2+, 3+…) precursor ions were selected for MS/MS. The dynamic exclusion list was restricted to 500 entries, with a maximum retention period of 30 s and a relative mass window of 10 ppm.
Data Processing and Validation/Protein Identification and Quantification
Protein quantification was performed with the MaxQuant software 1.5.3.8 (22) wherein peptide identification was performed using the Andromeda (23) search engine integrated into the MaxQuant environment against the human SwissProt database (uniprot-organism_9606+reviewed_yes 03/2016, 20274 sequences). The peptide mass tolerance for database searches was set to 7 ppm and fragment mass tolerance to 0.4 Da. Cysteine carbamidomethylation was set as fixed modification. Variable modifications included oxidation of methionine, deamidation of asparagine and glutamine and protein N-terminal acetylation. Two missed cleavage sites in case of incomplete trypsin hydrolysis was allowed. Furthermore, proteins were considered as identified if at least two unique peptides were identified. Identification under the applied search parameters refers to false discovery rate (FDR) < 1% and a match probability of p < 0.01, where p is the probability that the observed match is a random event.
Data transformation and evaluation was performed with the Perseus software (version 1.5.2.4) (24). Contaminants as well as proteins identified by site modification only and proteins derived from decoy database containing reversed protein sequences were strictly excluded from further analysis. Protein ratios were calculated by label free quantification (LFQ) comparing experiment and control samples. Filtering for quantitative values was applied to have at least two valid LFQ values in three replicates of either the experiment or the control group. To avoid zero LFQ values for calculating protein expression levels, values equal zero were substituted by the lowest LFQ values of each data set regarded. For statistical analyses, 2-sample t-tests were performed to calculate differences in the protein abundance between these groups (p value <0.01). Proteins were only considered as significantly regulated and ranked as potential PPI complex partners if their abundance changed more than +4-fold.
Public Access to Mass Spectrometry Data
The raw data obtained by mass spectrometry were deposited in the PRIDE (PRoteomicsIDEntifications) data repository of the EMBL-EBI (25).
Software Tools
Protein quantification was performed with the MaxQuant software 1.5.3.8 (22).
Peptide identification was performed using the Andromeda software (23).
Data transformation and evaluation was performed with the Perseus software (version 1.5.2.4) (24).
Silver Staining
Silver staining was performed with the Silver Quest silver staining kit (Invitrogen cat. no. LC6070) according to the manufacturers' recommendation.
Chromatin IP Assays
HCT116 cells were plated in 10 cm dishes at a density of 5 × 106 in McCoy's medium plus 10% FBS at 37 °C and 5% CO2 overnight. Afterward, the cells were treated with 40 nm SN38 (Sigma-Aldrich cat. no. I1406) for 4 h at 37 °C. Chromatin was prepared using a standard protocol (Simple ChIP Plus Enzymatic Chromatin IP Kit). The isolated chromatin was coimmunoprecipitated using 2 μg of anti-p53 antibody (Santa Cruz Biotechnology cat. no. sc-126) or isotype control IgG2a antibody (Southern Biotech cat. no. 0103–01). ChIP was performed using reagents and the protocol of the Simple ChIP Plus Enzymatic Chromatin IP Kit (Cell Signaling Technology, Danvers, MA, cat. no. 9004). The bTChIP samples were incubated with 3 μg of biotinylated Ig-kappa-specific anti-mouse antibody (BD Pharmingen, San Diego, CA, cat. no. 559750) for 2 h and further incubated for other 4 h in the presence of anti-biotin beads (all steps performed on a rotator at 4 °C). After 3 washes in low salt ChIP buffer, the chromatin was eluted with biotin buffer as described above and again immunoprecipitated with Protein G beads on a rotator at 4 °C overnight. The single-step ChIP and bTChIP samples were processed according to the protocol supplied by Cell Signaling Technology and the DNA purification/precipitation step was performed with chloroform/phenol/isoamyl alcohol. The samples were analyzed by quantitative real time PCR with an Applied Biosystems Step One plus cycler. The signals obtained from each immunoprecipitation were expressed as a percent of the total input chromatin (2% × 2(C[T] 2%Input Sample - C[T] IP Sample), C[T] = CT = Threshold cycle of PCR reaction). The values shown represent average and S.E. of at least 3 independent biological replicates and p values were calculated by student t test.
The primers used were: Human CDKN1A (Cell Signaling Technology cat. no. 6449), Puma (forward primer GCGAGACTGTGGCCTTGTGT reverse primer CGTTCCAGGGTCCACAAAGT) (26), alpha satellite (Cell Signaling Technology cat. no. 4486S) and RLP30 (Cell Signaling Technology cat. no.7014S).
Ethical Statement
The blood samples used in this research were obtained from healthy blood donors and anonymously provided by the Center for Immunohematology and Blood Bank of the Policlinico Umberto I (University “La Sapienza”) in Rome. The donors were informed and signed a written agreement allowing the use of the buffy coats for research purposes (M(DON)-Q AID Rev.3 del 17/02/2016). The consent form and the medical procedures were approved by the local Ethics Committee of the Policlinico Umberto I, Rome.
Primary Human CD4+ T Cell Isolation
Peripheral blood mononuclear cells (PBMCs) from healthy human donors were isolated by Ficoll-Histopaque (Sigma-Aldrich cat. no. 1077) density gradient centrifugation at 1500 RPMI for 20 min at room temperature. CD4+ T cells were purified from PBMCs using CD4 MACS beads (Miltenyi Biotec, Bergisch Gladbach, Germany, cat. no. 130–045-101). To perform TIP for the IKK-complex, 2 × 107 freshly isolated CD4+ T cells were either left untreated or treated with 500 ng/ml Ionomycin (Sigma-Aldrich cat. no. I3909) and 50 ng/ml PMA (Sigma-Aldrich cat. no. P8139) for 10 min. Then the cells were lysed in IP-lysis buffer, the lysates were cleared and TIP was performed.
For the isolation of the CD95/FAS-complex, CD4+ T cells were first activated and expanded. Freshly isolated CD4+ T cells were kept at a density of 2 × 106 cells/ml and incubated with 1 μg/ml PHA (Sigma-Aldrich cat. no. L9017) for 16h at 37 °C. Afterward the cells were cultivated in RPMI 1640 supplemented with 10% FBS and 25 U/ml IL-2 (Roche Diagnostics cat. no. 11147528001) for 6 days. For the CD95 isolation 3 × 107 cells were left untreated or were treated with ILZ-CD95L for 20 min at 37 °C, lysed and TIP was performed. The purity of CD4+ T cells (> 95%) was analyzed with surface staining using anti-CD4 antibody (Bio-Rad, Serotec, Hercules, CA, cat. no. MCA1267) and isotype antibody control (BD Biosciences, San Diego, CA, cat. no. 345815), data not shown.
CD95 Ligand Production
For the expression of the CD95L, a synthetic FLAG-peptide (DYKDDDDK) and a synthetic ILZ-sequence (IEKKIEAIEKKIEAIEKKIEAIEKKIE) were inserted into a eukaryotic expression vector (pcDNA3.1(+); Invitrogen) by oligonucleotide-cloning. Supernatant containing highly active recombinant FLAG-tagged ILZ-CD95L was produced in HEK293T cells upon transient transfection.
Knockdown of PPM1G
For the production of shRNAs, HEK293T were cotransfected with the packaging and pseudotyping DNAs (psPAX2 and pMD2.G) and pLKO.1 lentiviral shRNA vectors encoding for control and PPM1G targeting shRNAs. The packaging constructs were obtained from the ADDGENE consortium, the PPM1G targeting sequences used were TRCN0000001214 and TRCN0000001215 (Sigma-Aldrich). For transfection we used a standard calcium phosphate transfection method according to the manufacturer's recommendation (CalPhos, Clontech).
Cell Viability Assay
Control and PPM1G knockdown cells were seeded in equal numbers in a 96-well plate and treated with CD95L, Staurosporine (Enzo Life Sciences, Farmingdale, NY, cat. no. ALX-380–014) and ABT-737 (Abbott Laboratories, Chicago, IL, cat. no. S1002). After 24 h the viability was analyzed using Cell Titer-Glo Luminescent Cell Viability Assay (Promega cat. no. 47571) and luminescence was analyzed with a Perkin Elmer Victor X 3 analyzer. The data shown represent at least 3 independent biological experiments. Data are expressed as the mean ± S.D. using the numbers of replicates reported in the figure legends. Statistical analysis was conducted using two-way ANOVA and unpaired Student's t test *p < 0.05; **p < 0.01; ***p < 0.001 with Prism (GraphPad) software. To analyze DNA fragmentation, 5 × 105 BJAB cells were treated overnight with ILZ-CD95L, lysed with Nicoletti buffer (0.1% sodium citrate, pH 7.4, 0.1% NP40, 9.65 mm NaCl, 200 μg/ml RNase A and 50 μg/ml propidium iodide). The sub-G1/0 nuclei were measured by flow cytometry (FACS Canto, Becton Dickinson) and analyzed with FlowJo software (https://www.flowjo.com/).
Western Blotting and Antibodies
For the stimulation kinetic, 5 × 105 BJAB control and PPM1G knockdown cells were treated with ILZ-CD95L for the indicated time and then lysed in RIPA buffer (150 mm NaCl, 20 mm Tris/HCl (pH 7.2), 0.05% SDS, 1.0% Triton X-100, 1% Deoxycholate and 5 mm EDTA) completed with Complete EDTA-free protease inhibitor mixture tablet and Phos-STOP phosphatase inhibitor mixture tablet (Roche Diagnostics). The lysates were cleared by centrifugation (14,800 RPM for 20 min at 4 °C).
For all Western blotting analyses, the lysates and bead bound proteins were treated with NUPAGE LDS sample buffer (Invitrogen cat. no. NP0008) under reducing conditions (50 mm TCEP, Thermo Fisher Scientific cat. no. 77720), separated by NuPAGE 4–12% Bis-Tris gradient gel (Invitrogen) using MOPS or MES buffer and blotted to nitrocellulose membranes (GE Healthcare Bio-Sciences). The membranes were blocked for 1 h at room temperature with PBST (PBS 0.02% TWEEN20) containing 5% blotting grade nonfat powdered milk.
The antibodies used for Western blotting were: anti-IKK alpha (Santa Cruz Biotechnology cat. no. sc-7606), anti-IKK beta (Santa Cruz Biotechnology cat. no. sc-8014), anti-IKK gamma (Santa Cruz Biotechnology cat. no. sc-8330), anti-CD95/FAS (Santa Cruz Biotechnology cat. no. sc-715), anti-Caspase 8 (clone C-15 (27)), anti-FADD (BD Biosciences cat. no. 610400) and anti-Caspase 3 (Cell Signaling Technology cat. no. 9665S), streptavidin HRP (GE Healthcare Life Sciences, Pittsburgh, Pennsylvania, cat. no. RPN1231VS), anti-β-tubulin (Sigma-Aldrich cat. no. T4026), anti-β-actin (Sigma-Aldrich cat. no. A5441), anti-IPO7C (Sigma-Aldrich cat. no. SAB4200152) and anti-IPO7N (Sigma-Aldrich cat. no. SAB4200153) antibodies. The anti-PPM1G antibody was a generous gift from Dr. Oliver Gruss (28). All primary antibodies were used 1:1000 diluted in PBS containing 3% BSA. Western blotting quantification was performed using ImageJ (https://imagej.nih.gov/ij/) and Image Lab 6.0 software (http://www.Bio-Rad.com/it-it/product/image-lab-software).
Experimental Design and Statistical Rationale
The experiments in this work were designed to answer two questions. First, we wanted to test whether the protein complex isolations performed with TIP or bTIP are more suited to identify PPIs when compared with single-step co-IP. To this end we chose the IKK-complex and the CD95/FAS-DISC, two multi-protein complexes for which the composition was largely evaluated before. Thus, we had a strong basis to determine the identification rates for specific binders. Single-step co-IP, TIP and/or bTIP experiments were performed using bait-specific and isotype control antibodies. Three biological replicates were performed for each experiment and the isolated proteins were analyzed after applying a two-hour LC gradient. MS-results were quantified, and potential specific binders were defined as proteins that showed a statistically significant (p value <0.01, student t test) enrichment (>4 fold enrichment) in the samples containing the specific bait antibodies. The second rationale was to test whether TIP can be used in discovery experiments using the CD95/FAS-DISC and the IKK-complex of stimulated and untreated primary human CD4+ T cells. In this case the proteins identified and quantified using specific and isotype-matched antibodies were compared with test the feasibility of TIP in a primary cell model: a single experiment was performed, and the results were compared with the results obtained with biological replicates in the cell line.
RESULTS
Principle of TIP
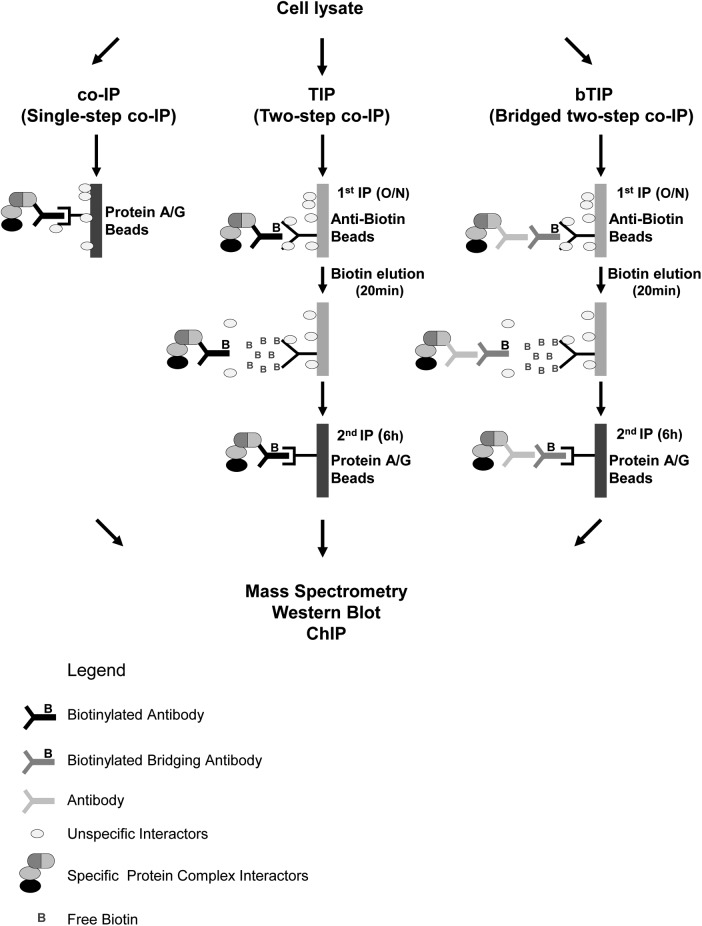
In contrast to conventional single-step co-IP (Fig. 1 left panel), TIP consists of two sequential coimmunoprecipitation steps targeting the endogenous protein(s) (Fig. 1, central panel). The first co-IP is performed with a biotinylated bait-specific antibody (biotin-ab), which is captured by anti-biotin beads. Alternatively, a biotinylated bridging ab can be combined with a native bait-specific ab (Fig. 1, right panel). Unlike avidin and streptavidin resins, the biotin-ab/anti-biotin-ab bond can be efficiently dissociated with free biotin, thereby releasing highly enriched complexes consisting of bait protein (bound to potential interaction partners) still attached to the biotinylated ab. To complete TIP, another affinity purification step is carried out with a standard protein A/G-resin that binds to the Fc-part of the abs (Fig. 1). TIP is applicable with several downstream applications, including the detection of PPIs by Western blot analysis or mass spectrometry and the detection of PNIs by qPCR.
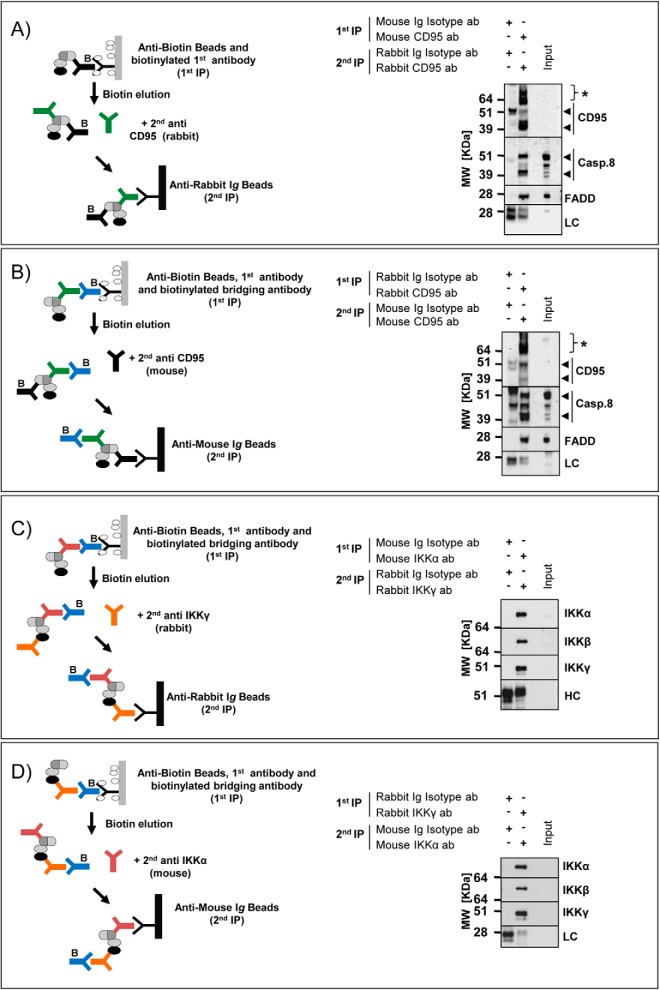
Fig. 1.
Schematic Representation of TIP and bTIP. Overview of the TIP and bTIP procedures: the cell lysate can be prepared from unmodified cells. It is incubated with a biotinylated antibody (ab) recognizing the bait protein (TIP) or with a primary ab followed by a biotinylated bridging secondary ab (bTIP); the first affinity purification is performed with anti-biotin agarose. The complexes are released by addition of free biotin, recaptured with protein A/G Sepharose and eluted for downstream applications (O/n = over night).
TIP is Highly Selective and Facilitates the Identification of Complex-Associated Proteins Using Monoclonal and Polyclonal Antibodies
To optimize and benchmark the TIP protocol, we used the well-known CD95/FAS “death inducing signaling complex” (DISC). First, we biotinylated the anti-CD95/FAS ab, and to rule out a reduced activity because of this modification, we compared its affinity with the nonbiotinylated ab by flow cytometry. Because the surface binding of both was similar, we concluded that the biotinylation had no impact on antibody function (supplemental Fig. S1A). We chose buffer conditions recently published (29) and tested the optimal precipitation timing for the first and second coimmunoprecipitation step. The association of known DISC-components such as CD95/FAS, caspase-8 (Casp.8) and FADD (18) was maximal when performing the anti-biotin IP overnight (14 h) (supplemental Fig. S1B). This is also the time frame frequently published for this complex (29, 30). Under these conditions, the anti-biotin resin precipitated the complex at levels comparable to protein A/G beads (supplemental Fig. S1C). The silver gel analysis suggested that substituting the affinity resin did not result in decreased protein background (supplemental Fig. S1D). Importantly, the anti-biotin resin enabled an almost quantitative elution of the complexes already 5 min after the addition of free biotin (supplemental Fig. S1E). We also determined the optimal time frame for the second coimmunoprecipitation step and found that caspase-8 (Casp.8) and FADD were efficiently re-precipitated after six hours (supplemental Fig. S1F). Therefore the final TIP protocol for this complex consists of a first purification overnight and a second precipitation for six hours. To ensure a quantitative release of the complexes two applications of the eluent for 10 min were applied before the second precipitation step. We used these conditions also for the isolation of the IKK-complex and obtained comparable results (supplemental Fig. S1G–S1I).
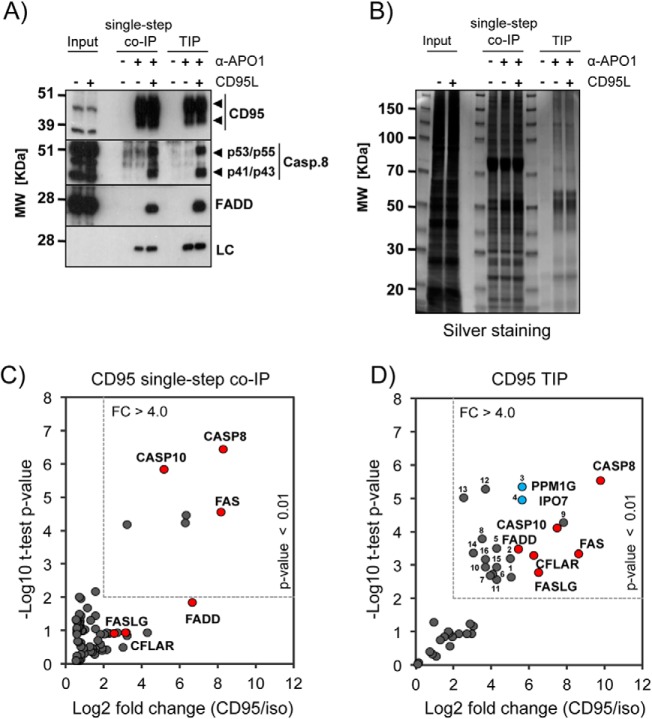
Next, we performed TIP and single-step co-IP head-to head for the CD95/FAS-DISC using the optimized conditions described above. Both methods resulted in equal levels of specifically bound proteins such as CD95/FAS, caspase-8 (Casp.8) and FADD, as shown by Western blot analysis (Fig. 2A). However, the protein pattern obtained by silver staining of a 1D SDS-PAGE suggested that the sample complexity was significantly reduced in the TIP samples (Fig. 2B). We next tested whether the marked increase in sample purity also influenced the identification and quantification of known DISC components by MS. The statistical analysis of the label free quantification (LFQ)-values indicated that the strong increase in specificity achieved by TIP supported the identification of specific complex binders (supplemental Table S1). We set the significance threshold for potential complex binders to a p value of <0.01 and an enrichment-factor of >4-fold in the anti-CD95/FAS containing samples when compared with the isotype control. Using these conservative settings, the single-step co-IP resulted in the significant enrichment of six proteins. Three of these (CASP8, CASP10, and FAS) were already described to be CD95/FAS-DISC components. When analyzing the data applying a less stringent threshold (p value of < 0.05), FADD was also identified as CD95/FAS binder whereas CFLAR and FASLG showed no significant enrichment (Fig. 2C, supplemental Table S2A). In contrast, the statistical analyses of the TIP samples revealed an enrichment of all six known DISC core components (FAS, CASP8, CASP10, FADD, CFLAR, and FASLG) (18) in the anti-CD95/FAS samples with high confidence (Fig. 2D, supplemental Table S2B). Interestingly, we also obtained significant enrichment for 16 additional proteins (supplemental Table S2B and S2C for detailed description of the identified proteins), including PPM1G and IPO7 that we later confirmed to interact specifically with CD95/FAS (Fig. 2D, blue dots; see below for validation).
Fig. 2.
Comparison of Single-Step co-IP and TIP. A, Representative Western blotting for the CD95/FAS-complex isolations: 1 × 107 BJAB cells were used to perform single-step co-IP and TIP from untreated (−) and CD95L stimulated (+) cells (LC: biotinylated mouse IgG light chain; arrowheads: specific signals). B, Silver staining of a representative SDS-PAGE for the CD95/FAS isolations described above. C, Scatter plot showing the enrichment analysis for CD95/FAS single-step co-IP versus the isotype control samples (three replicates each). The dotted square indicates the cutoff used to define significant enrichment that was set to a p value <0.01 (student t test) and a fold change (FC) >4-fold. Known interacting proteins are colored in red and labeled with their gene name. For the full analysis see supplemental Tables S1 and S2A. D, Scatter plot showing the enrichment analysis for CD95/FAS-TIP versus the isotype control samples (three replicates each). The cutoff for significant enrichment was defined as in (C). For the detailed analysis see supplemental Tables S1 and S2B. Known interacting proteins are colored in red and labeled with their gene name; the blue dots represent PPM1G and IPO7; the identified proteins are numbered and described in supplemental Table S2C.
To test whether TIP was applicable using other antibodies and to validate its broad applicability, we also isolated IKKα- and caspase-8 containing protein complexes. We used Western blotting analyses to investigate the recovery of known complex components compared with the single-step co-IP. We found that TIP performed with a monoclonal anti-IKKα ab resulted in only a minor decrease of the signals for specific IKK-complex binding proteins (IKKα and IKKγ) (16, 31) (supplemental Fig. S2A). Also here, 1D SDS-PAGE analysis suggested a considerable decrease of total protein content in the TIP samples (supplemental Fig. S2B). Similar results were obtained by isolating caspase-8 associated proteins using a biotinylated polyclonal goat ab (supplemental Fig. S2C and S2D).
In summary, the high specificity of TIP facilitates the identification of specific interactors by MS analysis. Our results further indicate that TIP can be performed with mono- and polyclonal antibodies. Thus, TIP represents a powerful method for the biochemical isolation and characterization of stable protein complexes from native cells.
TIP with Bridging Antibodies
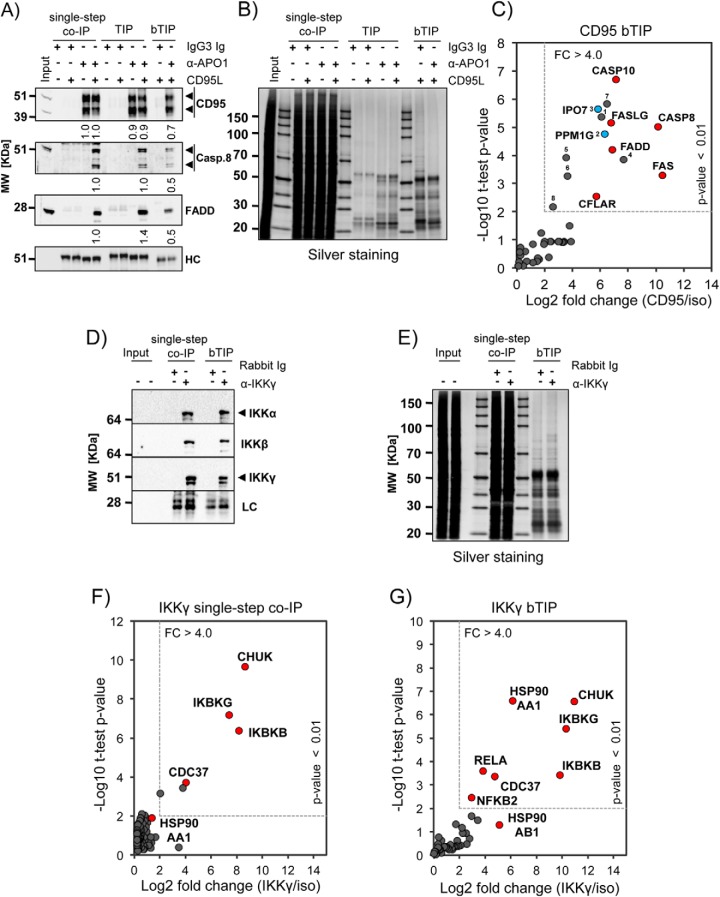
Although we demonstrated above the usefulness of TIP for decreasing the background contaminants and improving the detection of specific interactors, biotinylated abs are unfortunately not commonly available and biotinylation may reduce their functionality in some cases. Therefore, we tested, whether it was possible to utilize biotinylated (secondary) bridging abs to perform the TIP protocol. We used recombinant oligoclonal abs to standardize the experimental conditions and to avoid batch-to-batch differences of polyclonal secondary abs. Compared with the single-step co-IP and TIP, bridged TIP (bTIP) resulted in about 50% decrease of signal intensity for specifically CD95/FAS-associated proteins such as caspase-8 (Casp.8) and FADD as shown by quantitative Western blot analysis (Fig. 3A). However, as already observed for TIP, bTIP led to a remarkable reduction of sample complexity when compared with the single-step co-IP as shown by 1D SDS-PAGE (Fig. 3B) and MS1 base peak chromatograms (supplemental Fig. S3A–S3C). This improved the identification of CD95/FAS-associated proteins by MS (Fig. 3C, supplemental Table S1 and supplemental Table S3A). Comparable with TIP, MS analyses of the anti-CD95/FAS bTIP samples identified and quantified all known major DISC components (FAS, CASP8, CASP10, FADD, CFLAR, and FASLG). In addition we found significant enrichment of eight additional proteins, including PPM1G and IPO7 (p value >0.01; Fig. 3C, blue dots and supplemental Table S3B for detailed description of the proteins identified by TIP).
Fig. 3.
Comparison of Single-Step co-IP, TIP and bTIP. A, CD95/FAS-DISC isolations from 1 × 107 BJAB cells was performed with anti-CD95/FAS ab (α-APO1) using single-step co-IP, TIP and bTIP. Shown is the Western blot analysis for the indicated proteins (HC: mouse IgG heavy chain). The densitometric analyses of the Western blotting signal intensities were normalized to the single-step co-IP (co-IP) samples. (Means and standard deviation (n = 3): CD95(-): co-IP 1.0 ± 0; TIP 0.9 ± 0.2; CD95(+): co-IP 1.0 ± 0; TIP 0.9 ± 0.1; bTIP: 0.7 ± 0.1 | Casp.8: co-IP 1.0 ± 0; TIP 1.0 ± 0.1; bTIP; 0.5 ± 0.1 | FADD: co-IP 1.0 ± 0; TIP 1.4 ± 0.1; bTIP: 0.5 ± 0; arrowheads: specific signals). B, Silver staining of a representative SDS-PAGE for the CD95/FAS-complex isolations described in (A). C, Scatter plot showing the enrichment analysis for CD95/FAS-bTIP versus the isotype control samples. The dotted square indicates the cutoff used to define significant enrichment that was set to a p value <0.01 (student t test) and a fold change (FC) >4-fold (see supplemental Tables S1 and S3A). The known interacting proteins are colored in red and labeled with their gene name. The blue dots represent PPM1G and IPO7; the identified proteins are numbered and described in supplemental Table S3B. D, Shown is a representative Western blotting of IKK-complex isolations from 1 × 107 BJAB cells. These were performed with an anti-IKKγ ab either by single-step co-IP or by bTIP (LC: rabbit Ig light chain; arrowheads: specific signals). E, Silver staining of a representative SDS-PAGE for the IKK-complex isolations described in (D). F, Scatter plot showing the enrichment analysis for IKKγ single-step co-IP versus the isotype control samples. The dotted square indicates the cutoff used to define significant enrichment that was set to a p value <0.01 (student t test) and a fold change (FC) >4-fold. The known interacting proteins are colored in red and labeled with their gene name. For the detailed analysis see supplemental Tables S4 and S5A. G, Scatter plot showing the enrichment analysis for IKKγ-bTIP versus the isotype control samples. The analysis was performed as described in (F). For the detailed MS results and statistical analyses see supplemental Tables S4 and S5B.
To test whether bTIP could also be applied with polyclonal antibodies (pAbs), we isolated the IKK-complex described above by using a rabbit pAb recognizing IKKγ/NEMO (IKBKG). Western blots analysis indicated only a slight decrease of signal intensity for the specifically associated proteins IKKα (CHUK) and IKKβ (IKBKB) in the bTIP samples when compared with the single-step co-IP (Fig. 3d). However, presumably coprecipitated contaminant proteins were strongly reduced in the bTIP samples (Fig. 3e). We compared bTIP with single-step co-IP by MS. Known complex binders such as CHUK, IKBKB, IKBKG, and CDC37 (32) were significantly enriched in both anti-IKKγ isolations. Moreover, bTIP resulted in the identification of three additional proteins, namely HSP90AA, RELA, and NFKB2 (Fig. 3F and 3G, supplemental Tables S4 and S5). These proteins were already found to interact with the IKK-complex in a comprehensive TAP-approach (32). These results indicated that the identification of specific binders by MS was more efficient in the bTIP preparations when compared with the samples isolated by single-step co-IP. We can conclude that TIP can also be performed with biotinylated bridging antibodies.
Application of TIP to ChIP
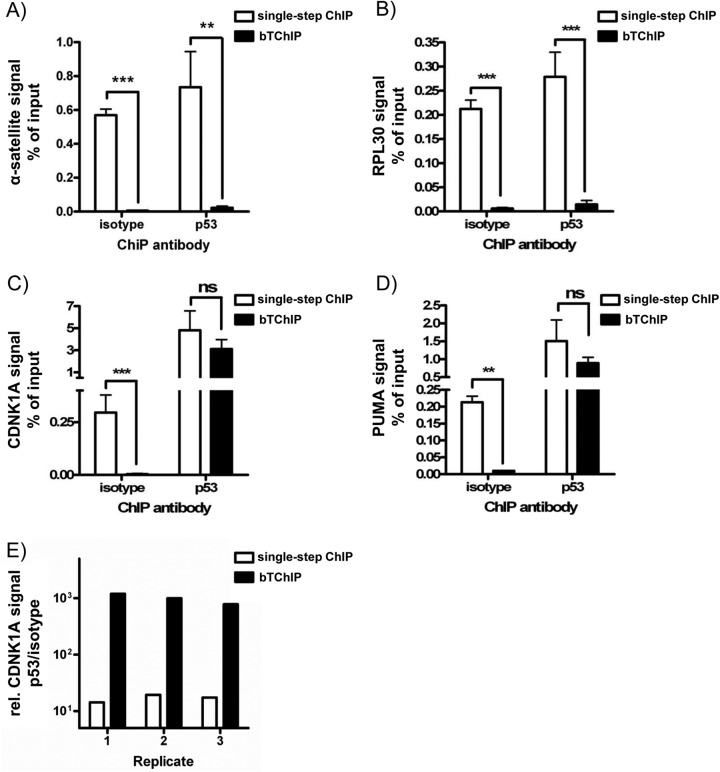
To investigate if the TIP approach was applicable for other antibody-based assays and to get a quantitative estimate of the increased specificity, TIP was adapted to ChIP. To induce p53 activation, the cells were treated with the topoisomerase-I inhibitor SN38 and chromatin was coimmunoprecipitated with anti-p53 (DO-I) or isotype control ab. A bridged two-step chromatin immunoprecipitation (bTChIP) was performed as described above and compared with conventional single-step ChIP. The isolated DNAs were quantified by qPCRs for promoter regions of p53 target genes (CDNK1A and PUMA) or unrelated chromatin areas (RPL30 and α-satellite). In these experiments, bTChIP with both abs (anti-p53 and isotype control) resulted in more than 94% signal reduction for nonspecific α-satellite repeats and RPL30 (Fig. 4A and 4B). The slight decrease of signals for promoter regions of known p53 target genes such as CDKN1A and PUMA was vastly over-compensated by 95% loss of nonspecific signal with the control ab (Fig. 4C and 4D). This led to an impressive increase of the p53/isotype signal ratios for CDKN1A and PUMA promoter regions from 17 ± 2 to 991 ± 121 and from 7 ± 3 to 100 ± 20 (average±S.E.), respectively (Fig. 4E). In summary, we can conclude that bTChIP represents an improved alternative to conventional ChIP protocols.
Fig. 4.
TIP Can be Adapted to Chromatin Immunoprecipitation. 5 × 106 HCT116 cells were treated with SN38 for 4 h. Then chromatin was prepared and precipitated using an isotype control ab (MOPC21) or an anti-p53 ab (DO-I) by either standard single-step ChIP or bTChIP. The bead-associated chromatin was determined by qPCR for: A, α-satellite sequences, B, RPL30 promoter, C, CDNK1A promoter, and D, PUMA promoter. Average and S.E. of 3 independent experiments are shown (**p < 0.01; ***p < 0.001; student t test; ns: not significant). E, Comparison of signal-to noise ratio (anti-p53 to the isotype control) upon single-step ChIP and bTChIP. Shown are 3 biological replicates using CDNK1A primers (logarithmic scale).
Sequential Protein Complex Isolation with Two Different Antibodies
We further tested whether a modified TIP protocol would enable the purification of protein complexes using two abs with different specificities either against the same bait or against two different proteins of the complex. This would be useful to distinguish protein complexes that share a common member or if antibody cross-reactivity is an important issue. In a first application we tested whether the TIP protocol could be performed with two abs recognizing different epitopes of CD95/FAS. To avoid artifacts, we validated that the resins used for the second precipitations were not binding the abs used for the initial isolation step (supplemental Fig. S4A and S4B). Then we purified proteins associated with ligand-stimulated CD95/FAS with a mouse ab followed by a re-precipitation of the complex with a CD95/FAS-specific rabbit ab conjugated to an anti-rabbit resin (Fig. 5A left panel). Next, we reversed the sequence and used first a CD95/FAS-specific rabbit ab and biotinylated bridging abs followed by the re-precipitation with a mouse ab and anti-mouse resin (Fig. 5B left panel). The Western blotting analyses indicated that it was possible to isolate interacting proteins such as caspase-8 (Casp.8) and FADD by performing the sequential co-IPs in both ways (Fig. 5A and 5B right panels). Next, we purified the IKK-complex, and instead of utilizing two abs recognizing one bait protein, we sequentially precipitated two different proteins of the complex. We first isolated the IKK-complex with a mouse ab recognizing IKKα using biotinylated bridging abs and then re-precipitated with a IKKγ-specific rabbit ab coupled to anti-rabbit beads (Fig. 5C left panel) and vice versa (Fig. 5D left panel). Also, in these experiments we were able to isolate the core components of the IKK-complex and got similar results by changing the order of the bait proteins (Fig. 5C and 5D, right panels). Thus, we provide proof of concept that this variation of TIP could be adopted to purify protein complexes with two abs of different specificities.
Fig. 5.
TIP for a sequential protein complex purification with two different antibodies. A, CD95/FAS-complex was isolated from 5 × 107 CD95L stimulated (+) BJAB cells using a mouse biotinylated anti-CD95/FAS (α-APO1) ab and coimmunoprecipitated with anti-biotin beads. After the elution, the supernatant was incubated with a rabbit anti-CD95/FAS ab (sc-715) and anti-rabbit beads. Western blot analysis for the indicated proteins was performed (LC: rabbit Ig light chain; arrowheads: specific signals; asterisks: nonspecific signals). B, CD95/FAS-complex was isolated with the same preparation of the cell lysates as above using a rabbit anti-CD95/FAS ab (sc-715) in combination with a goat biotinylated anti-rabbit secondary ab and precipitated with anti-biotin beads. After the elution, the supernatant was incubated with a mouse anti-CD95/FAS ab (α-APO1) and the complex was purified with anti-mouse IgG beads. Western blot analysis for the indicated proteins was performed (LC: mouse IgG light chain; arrowheads: specific signals; asterisks: nonspecific signals). C, IKK-complex was isolated from 5 × 107 BJAB cells using a mouse anti-IKKα and a biotinylated goat anti-mouse secondary ab and precipitated with anti-biotin beads. After the elution, the supernatant was incubated with a rabbit anti-IKKγ ab and the complex was isolated with anti-rabbit beads. Western blot analysis for the indicated proteins was performed (HC: rabbit Ig heavy chain). D, The IKK-complex was isolated with the same preparation of the cell lysates as above using a rabbit anti-IKKγ ab and a biotinylated goat anti-rabbit secondary ab. After the elution, the IKK-complex was re-isolated with a anti-IKKα ab and anti-mouse beads. Western blot analysis for the indicated proteins was performed (LC: mouse IgG light chain).
TIP Enables Discovery Proteomics in Primary Human CD4+ T Cells
Currently most protein interaction studies are performed in expandable and genetically accessible (tumor-) cells that allow to perform multiple biological replicates and ectopic protein expression. Unfortunately, these properties are not shared by most of the primary human cells.
As a proof of principle, we used TIP to isolate the IKK- and CD95/FAS-complexes from naïve and activated human CD4+ T cells, respectively. We obtained human CD4+ T cells from whole blood and performed TIP with anti-IKKα 1 day after the isolation (supplemental Fig. S5A). MS analyses were performed with 3-hour LC gradients, and we considered only proteins identified with two or more unique peptides. Also, in the primary cell system we validated known complex proteins such as IKKα and IKKγ by Western blot analysis (supplemental Fig. S5B). By MS analysis and MaxQuant assisted data processing the known core components of the IKK-complex were specifically enriched in the anti-IKKα samples (supplemental Table S6). The orthogonal comparison with the isotype control samples resulted in 14 and 18 proteins that were only present in the IKKα-TIP of resting and PMA/ionomycin stimulated cells, respectively (supplemental Fig. S5C). Except for HSP90AA1, all known IKK-complex core components (IKBKB, IKBKG, CHUK, HSP90AB1 and CDC37) were only found in the stimulated or nonstimulated IKKα-TIP samples, confirming the high specificity and sensitivity of TIP. Of note, although it was not passing our stringent criteria excluding all proteins quantified in the control, the LFQ value of HSP90AA1 was more than 16 fold higher in the IKKα-TIP compared with the isotype control (supplemental Table S6).
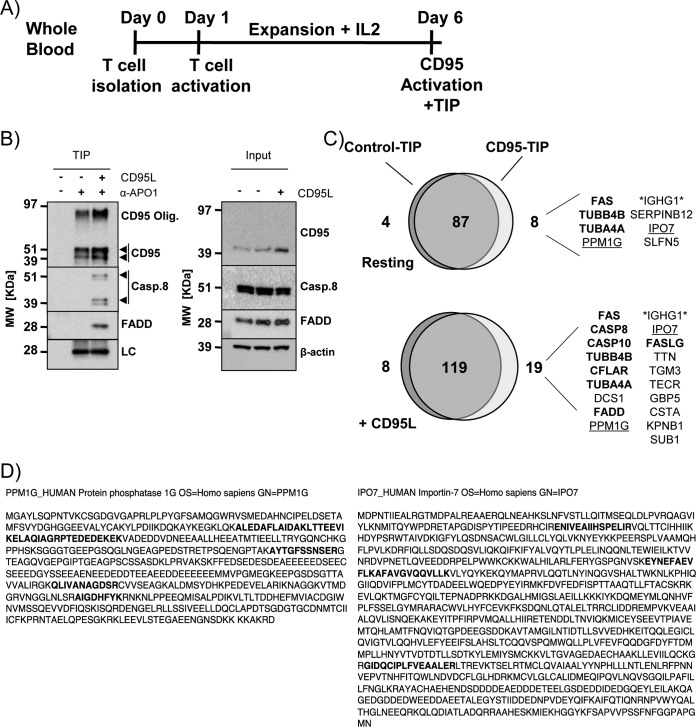
We next performed TIP for CD95/FAS-associated proteins in primary CD4+ T cells from healthy human donors activated and expanded for 6 days in vitro (Fig. 6A). We identified the bait protein (CD95/FAS) and all previously described DISC core components such as CASP8, CASP10, FASLG, CFLAR, and FADD in the CD95/FAS-TIP sample obtained from CD95L stimulated cells (Fig. 6B and 6C). Supporting the published constitutive interaction of CD95/FAS and the tubulin cytoskeleton (33), alpha and beta tubulin isoforms were identified in a stimulation independent manner (Fig. 6C, supplemental Table S7). Interestingly, importin 7 (IPO7) and protein phosphatase 1G (PPM1G) were also identified to be associated with CD95/FAS in primary CD4+ T cells (Fig. 6C and 6D). In summary these data indicate that TIP enabled the identification of PPIs in primary human CD4+ T cells.
Fig. 6.
TIP for CD95/FAS-complex purification from primary human CD4+ T cells. A, Scheme describing the treatment of primary human CD4+ T cells before CD95/FAS-TIP isolation using 3 × 107 untreated (-) or CD95L stimulated (+) cells. B, Western blot analysis for the indicated proteins isolated from nonstimulated (-) and CD95L stimulated (+) primary human CD4+ T cells by CD95/FAS TIP (LC: biotinylated mouse IgG light chain; arrowheads: specific signals). C, Venn diagram of the proteins identified and quantified by MaxQuant analysis comparing the isotype control-TIP and anti-CD95/FAS-TIP in resting and CD95L-treated samples. The list shows the proteins selectively enriched in the anti-CD95/FAS samples (ordered by LFQ values). The proteins in bold represent published complex components (for the MS results see supplemental Table S7). D, Sequence coverage of the peptides identified for PPM1G (left) and IPO7 (right). Bold letters indicate the peptides identified by LC-MS/MS.
PPM1G Knockdown Sensitizes Cells to CD95-Induced Apoptosis
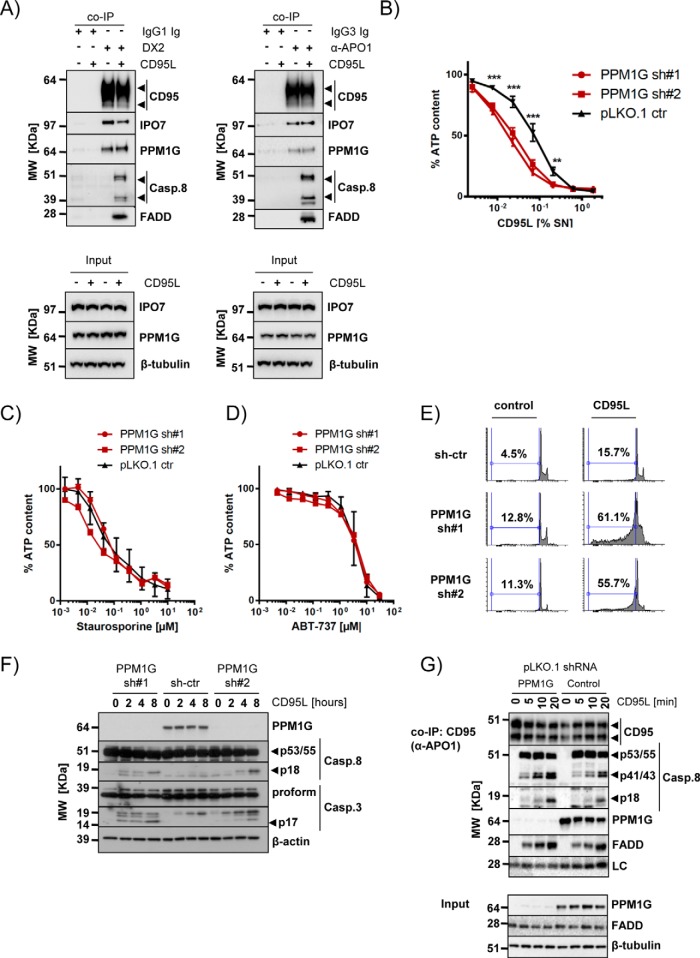
Besides the well-known interaction partners (CASP8, CASP10, FASLG, CFLAR, and FADD), IPO7 and PPM1G were the only proteins found to be enriched in the anti-CD95/FAS TIPs performed with the tumor cell line that were also identified in primary CD4+ T cells. We verified the interaction of IPO7 and PPM1G with CD95/FAS under native conditions using two different antibodies (Fig. 7A). This ruled out an unspecific cross-reactivity of the anti-APO1 ab used in the preparative approach. Because PPM1G showed higher sequence coverage in the MS analyses of the samples obtained from primary CD4+ T cells (Fig. 6D), we decided to investigate its function in the CD95/FAS signal transduction. Upon binding of CD95L/FASL to its cognate receptor, the DISC is formed. The DISC is an activation platform for the so-called initiator caspases (caspase-8 and -10) that then cleave and activate effector caspases (e.g. caspase-3 and -7). This caspase cascade ultimately leads to the induction of apoptotic cell death. The binding of PPM1G to CD95/FAS was detected in various cellular systems (supplemental Fig. S6A–S6C). Down-regulation of PPM1G resulted in a 3–10 fold increase of CD95L sensitivity in several cell lines (Fig. 7B and supplemental Fig. S6D). Differently, we have not observed any sensitization to other apoptosis inducers such as staurosporine or the Bcl2/XL inhibitor ABT-737 (Fig. 7C–7D). The higher sensitivity for CD95L was because of an increase of apoptotic cell death as demonstrated by flow cytometry (Fig. 7E) and Western blot analysis for the activation of caspases triggered by CD95/FAS-stimulation. In cells knocked down for PPM1G expression, DNA fragmentation was also detected in absence of CD95L, indicating a mild increase of basal apoptosis in these cells that was not evident by Western blot analysis (Fig. 7F and supplemental Fig. S6E). However, the active p18 subunit of caspase-8 (Casp.8) was observed two to four hours after CD95L treatment in cells lacking PPM1G, whereas it accumulated significantly less in control cells. The same applied for the fully processed active p17 subunit of caspase-3 (Casp.3) (Fig. 7F and supplemental Fig. S6E). Furthermore, we noted a faster accumulation of the cleaved form of both caspase-8 isoforms (p43/41) in the CD95/FAS-DISC formed by PPM1G silenced cells. This was paralleled by an increase of the fully activated caspase-8 (p18 subunit) in the DISC of cells lacking PPM1G (Fig. 7G, supplemental Fig. S6F and S6G) and interestingly, in H9 cells we have also detected an increased amounts of noncleaved caspase-8 (supplemental Fig. S6F and S6G). In conclusion these results suggest a higher activity of the CD95/FAS-DISC in absence of PPM1G indicating its inhibitory role in CD95/FAS-induced apoptosis.
Fig. 7.
Validation of IPO7 and PPM1G interaction with CD95/FAS and PPM1G function. A, Copurification of PPM1G and IPO7 with CD95/FAS using two different anti-CD95/FAS antibodies (DX2 and α-APO1) under native expression conditions (arrowheads: specific signals). B, PPM1G knockdown sensitizes cells to CD95/FAS-induced cell death. BJAB cells were knocked down with two independent PPM1G shRNAs and treated for 24 h with a CD95L as indicated. Viability was determined by cell titer glo assay. Shown are average and S.D. of eight independent experiments (**p < 0.01; ***p < 0.001; ANOVA). C–D, PPM1G knockdown does not sensitize BJAB cells for Staurosporine (C) or ABT-737 (D). BJAB cells were knocked down as in (B) and treated for 24 h with the indicated drugs at different dilutions. Viability was determined by cell titer glo assay. Shown are average and S.D. of four independent biological experiments. E, DNA fragmentation of BJAB cells silenced for PPM1G expression was analyzed by flow cytometry 16 h after CD95L addition. F, The processing of caspases upon CD95L stimulation of the cells described in (E) was investigated. Shown is a representative Western blot analysis for the indicated proteins (arrowheads: specific signals). G, Analysis of the CD95/FAS-DISC formed by 2 × 107 BJAB cells transduced with control or PPM1G shRNAs. The cells were stimulated with CD95L for the indicated timeframes and CD95/FAS was precipitated using α-APO1 ab and shown is a representative Western blot analysis for the indicated proteins. The total lysate is shown in the lower part (LC: mouse IgG light chain; arrowheads: specific signals).
DISCUSSION
TIP combines the advantages of conventional single-step co-IP and TAP (34) for the purification of stable protein complexes and overcomes several limitations associated with each of these methods. In contrast to TAP (35), the TIP and bTIP approaches do not require the transgenic expression of the bait protein and thus can be performed in virtually all cell systems, including primary human cells. The opportunity to work under native expression conditions also reduces the risk for nonspecific interactions resulting from alterations in protein homeostasis (36) or unintended activation of pathways such as apoptosis induction triggered by forced expression of CD95/FAS or caspase-8 (37–39).
TIP and bTIP largely reduced nonspecific binders using abs covering different monoclonal mouse IgG isotypes as well as commercially available pAbs. Interestingly, in TIP applications rabbit pAbs were as effective and specific as mAbs.
When compared with the single-step co-IP, TIP and bTIP resulted in the identification of more known complex components in the CD95/FAS-DISC and the IKK-complex. Indeed, TIP and bTIP resulted in the significant enrichment of all so far known and validated core components of the CD95/FAS-DISC formed by this cell type (18). The same was obtained for the purification of the IKK-complex with bTIP. In a comprehensive TAP-approach published before (32), the TAP-tagged IKKγ/NEMO only identified the core components of the complex (CHUK, IKBKB, IKBKG, CDC37, HSP90AA1 and HSP90AB1). In our bTIP experiments we also identified these proteins as IKKγ/NEMO interactors without the need of bait protein overexpression. With a p value of 0.051, HSP90AB1 was the only known interactor not unambiguously identified by bTIP. Even more, we identified with RELA and NFKB2 two other known proteins of the IKK-complex that were not found before when IKKγ/NEMO was used as bait-protein for TAP (32). We want to emphasize that the quality of the abs used for the experiments is of highest importance to avoid inefficient precipitation or abundant nonspecific interactions that are because of poor affinities or cross-reactivity of many commercially available abs (40, 41). Fortunately, this problem is known and currently there is a strong tendency of companies and researchers to shift toward high quality reagents such as recombinant or rabbit monoclonal abs. To circumvent problems with cross-reactivity, we demonstrated that TIP could also be used to precipitate protein complexes with two different abs. This facilitates a stringent purification of a complex even with two less specific abs and opens the opportunity to use two abs recognizing two independent proteins in the same complex.
We also demonstrated that TIP is not only suited to investigate PPIs but can also be adapted to chromatin immunoprecipitation. Combined with ORGANIC (42) it may be used to perform high quality sequential purifications of chromatin bound transcription factors under native conditions, which was until now not compatible with the re-ChIP protocol (43). We also expect that the increased specificity obtained by bTChIP will facilitate ChIP sequencing approaches.
TIP resulted in the identification of IPO7 and PPM1G as CD95/FAS-binding proteins in primary human CD4+ T cells. PPM1G was initially described to bind and dephosphorylate metabotrobic glutamate receptor 3 (44), regulate Cajal body localization (28), histone phosphorylation (45), p21 stability (46), alternative splicing (47), p53 signaling (48), transcription elongation (49), and protein translation (50, 51). Here we found this protein as constitutively interacting with the death receptor CD95/FAS. However, the exact molecular mechanism of how PPM1G depletion leads to the specific sensitization for CD95L remains to be investigated.
In conclusion we showed that TIP and bTIP could be powerful tools to elucidate the composition of stable protein complexes even in primary cells. Therefore, we envision a broad spectrum of applications for TIP and especially bTIP in affinity based discovery proteomics.
Data Availability
The data obtained are accessible at the EMBL-EBI, URL: https://www.ebi.ac.uk/pride/archive/projects/PXD006700.
Supplementary Material
Acknowledgments
We thank Dr. Oliver Gruss for kindly providing the PPM1G antiserum and Dr. Martin Leverkus and Dr. Valeria Coppola for helpful discussions during the preparation of the manuscript. Many thanks to Ramona Mayer for sample preparation for MS analyses.
Footnotes
* This work was supported by the FIRB RBAP10KJC5_004 grant from the Ministry of Education, University and Research and by RF-2011-02349985 grant from the Ministry of Health.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- Co-IP
- coimmunoprecipitation
- ChIP
- chromatin immunoprecipitation
- TAP
- tandem affinity purification
- PPI
- protein-protein interaction
- PNI
- protein-nucleic acid interaction
- TIP
- two-step coimmunoprecipitation
- bTIP
- bridged two-step coimmunoprecipitation
- bTChIP
- bridged two-step chromatin immunoprecipitation
- LC/MS
- liquid chromatography and mass spectrometry
- LFQ
- label free quantification
- FC
- fold change
- Ig(s)
- immunoglobulin(-s)
- ab(s)
- antibody(-ies)
- mAb(s)
- monoclonal antibody(-ies)
- pAb(s)
- polyclonal antibody(-ies)
- qPCR
- quantitative polymerase chain reaction
- PMA
- phorbol 12-myristate 13-acetate
- PAGE
- polyacrylamide gel electrophoresis
- CD95(L)
- cluster of differentiation 95 (ligand)
- IKK
- Inhibitor of kappa B Kinase
- DISC
- death inducing signaling complex
- APO-1
- apoptosis antigen 1
- FADD
- FAS-associated protein with death domain.
REFERENCES
- 1. Nilsson T., Mann M., Aebersold R., Yates J. R. 3rd, Bairoch A., and Bergeron J. J. (2010) Mass spectrometry in high-throughput proteomics: ready for the big time. Nat. Methods 7, 681–685 [DOI] [PubMed] [Google Scholar]
- 2. Gingras A. C., Gstaiger M., Raught B., and Aebersold R. (2007) Analysis of protein complexes using mass spectrometry. Nat. Rev. Molecular Cell Biology 8, 645–654 [DOI] [PubMed] [Google Scholar]
- 3. Warnken U., Schleich K., Schnolzer M., and Lavrik I. (2013) Quantification of high-molecular weight protein platforms by AQUA mass spectrometry as exemplified for the CD95 death-inducing signaling complex (DISC). Cells 2, 476–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Meyer K., and Selbach M. (2015) Quantitative affinity purification mass spectrometry: a versatile technology to study protein-protein interactions. Front. Gen. 6, 237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Boldt K., Gloeckner C. J., Texier Y., von Zweydorf F., and Ueffing M. (2014) Applying SILAC for the differential analysis of protein complexes. Methods Mol. Biol. 1188, 177–190 [DOI] [PubMed] [Google Scholar]
- 6. Selbach M., and Mann M. (2006) Protein interaction screening by quantitative immunoprecipitation combined with knockdown (QUICK). Nat. Methods 3, 981–983 [DOI] [PubMed] [Google Scholar]
- 7. Rigaut G., Shevchenko A., Rutz B., Wilm M., Mann M., and Seraphin B. (1999) A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17, 1030–1032 [DOI] [PubMed] [Google Scholar]
- 8. Bigenzahn J. W., Fauster A., Rebsamen M., Kandasamy R. K., Scorzoni S., Vladimer G. I., Muller A. C., Gstaiger M., Zuber J., Bennett K. L., and Superti-Furga G. (2016) An Inducible Retroviral Expression System for Tandem Affinity Purification Mass-Spectrometry-Based Proteomics Identifies Mixed Lineage Kinase Domain-like Protein (MLKL) as an Heat Shock Protein 90 (HSP90) Client. Mol. Cell. Proteomics 15, 1139–1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Krogan N. J., Cagney G., Yu H., Zhong G., Guo X., Ignatchenko A., Li J., Pu S., Datta N., Tikuisis A. P., Punna T., Peregrin-Alvarez J. M., Shales M., Zhang X., Davey M., Robinson M. D., Paccanaro A., Bray J. E., Sheung A., Beattie B., Richards D. P., Canadien V., Lalev A., Mena F., Wong P., Starostine A., Canete M. M., Vlasblom J., Wu S., Orsi C., Collins S. R., Chandran S., Haw R., Rilstone J. J., Gandi K., Thompson N. J., Musso G., St Onge P., Ghanny S., Lam M. H., Butland G., Altaf-Ul A. M., Kanaya S., Shilatifard A., O'Shea E., Weissman J. S., Ingles C. J., Hughes T. R., Parkinson J., Gerstein M., Wodak S. J., Emili A., and Greenblatt J. F. (2006) Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 440, 637–643 [DOI] [PubMed] [Google Scholar]
- 10. Wang J., Rao S., Chu J., Shen X., Levasseur D. N., Theunissen T. W., and Orkin S. H. (2006) A protein interaction network for pluripotency of embryonic stem cells. Nature 444, 364–368 [DOI] [PubMed] [Google Scholar]
- 11. Burckstummer T., Bennett K. L., Preradovic A., Schutze G., Hantschel O., Superti-Furga G., and Bauch A. (2006) An efficient tandem affinity purification procedure for interaction proteomics in mammalian cells. Nat. Methods 3, 1013–1019 [DOI] [PubMed] [Google Scholar]
- 12. Dalvai M., Loehr J., Jacquet K., Huard C. C., Roques C., Herst P., Cote J., and Doyon Y. (2015) A Scalable Genome-Editing-Based Approach for Mapping Multiprotein Complexes in Human Cells. Cell Reports 13, 621–633 [DOI] [PubMed] [Google Scholar]
- 13. Forler D., Kocher T., Rode M., Gentzel M., Izaurralde E., and Wilm M. (2003) An efficient protein complex purification method for functional proteomics in higher eukaryotes. Nat. Biotechnol. 21, 89–92 [DOI] [PubMed] [Google Scholar]
- 14. Li Y. (2010) Commonly used tag combinations for tandem affinity purification. Biotechnol. Appl. Biochem. 55, 73–83 [DOI] [PubMed] [Google Scholar]
- 15. Zhang X., Guo C., Chen Y., Shulha H. P., Schnetz M. P., LaFramboise T., Bartels C. F., Markowitz S., Weng Z., Scacheri P. C., and Wang Z. (2008) Epitope tagging of endogenous proteins for genome-wide ChIP-chip studies. Nat. Methods 5, 163–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hinz M., and Scheidereit C. (2014) The IkappaB kinase complex in NF-kappaB regulation and beyond. EMBO Reports 15, 46–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Strasser A., Jost P. J., and Nagata S. (2009) The many roles of FAS receptor signaling in the immune system. Immunity 30, 180–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lavrik I. N., and Krammer P. H. (2012) Regulation of CD95/Fas signaling at the DISC. Cell Death Differentiation 19, 36–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Baldwin A. S. (2012) Regulation of cell death and autophagy by IKK and NF-kappaB: critical mechanisms in immune function and cancer. Immunol. Rev. 246, 327–345 [DOI] [PubMed] [Google Scholar]
- 20. Trauth B. C., Klas C., Peters A. M., Matzku S., Moller P., Falk W., Debatin K. M., and Krammer P. H. (1989) Monoclonal antibody-mediated tumor regression by induction of apoptosis. Science 245, 301–305 [DOI] [PubMed] [Google Scholar]
- 21. Preobrazhensky S. N., and Bahler D. W. (2008) Optimization of flow cytometric measurement of ZAP-70 in chronic lymphocytic leukemia. Cytometry 74, 118–127 [DOI] [PubMed] [Google Scholar]
- 22. Cox J., and Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 23. Cox J., Neuhauser N., Michalski A., Scheltema R. A., Olsen J. V., and Mann M. (2011) Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805 [DOI] [PubMed] [Google Scholar]
- 24. Tyanova S., Temu T., and Sinitcyn P. (2016) The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Methods 13, 731–740 [DOI] [PubMed] [Google Scholar]
- 25. Vizcaino J. A., Csordas A., Del-Toro N., Dianes J. A., Griss J., Lavidas I., Mayer G., Perez-Riverol Y., Reisinger F., Ternent T., Xu Q. W., Wang R., and Hermjakob H. (2016) 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 44, 11033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kaeser M. D., and Iggo R. D. (2002) Chromatin immunoprecipitation analysis fails to support the latency model for regulation of p53 DNA binding activity in vivo. Proc. Natl. Acad. Sci. U.S.A. 99, 95–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Scaffidi C., Medema J. P., Krammer P. H., and Peter M. E. (1997) FLICE is predominantly expressed as two functionally active isoforms, caspase-8/a and caspase-8/b. J. Biol. Chem. 272, 26953–26958 [DOI] [PubMed] [Google Scholar]
- 28. Petri S., Grimmler M., Over S., Fischer U., and Gruss O. J. (2007) Dephosphorylation of survival motor neurons (SMN) by PPM1G/PP2Cgamma governs Cajal body localization and stability of the SMN complex. J. Cell Biol. 179, 451–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pearson J. S., Giogha C., Ong S. Y., Kennedy C. L., Kelly M., Robinson K. S., Lung T. W., Mansell A., Riedmaier P., Oates C. V., Zaid A., Muhlen S., Crepin V. F., Marches O., Ang C. S., Williamson N. A., O'Reilly L. A., Bankovacki A., Nachbur U., Infusini G., Webb A. I., Silke J., Strasser A., Frankel G., and Hartland E. L. (2013) A type III effector antagonizes death receptor signalling during bacterial gut infection. Nature 501, 247–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cullen S. P., Henry C. M., Kearney C. J., Logue S. E., Feoktistova M., Tynan G. A., Lavelle E. C., Leverkus M., and Martin S. J. (2013) Fas/CD95-induced chemokines can serve as “find-me” signals for apoptotic cells. Mol. Cell 49, 1034–1048 [DOI] [PubMed] [Google Scholar]
- 31. Kovalenko A., and Wallach D. (2006) If the prophet does not come to the mountain: dynamics of signaling complexes in NF-kappaB activation. Mol. Cell 22, 433–436 [DOI] [PubMed] [Google Scholar]
- 32. Bouwmeester T., Bauch A., Ruffner H., Angrand P. O., Bergamini G., Croughton K., Cruciat C., Eberhard D., Gagneur J., Ghidelli S., Hopf C., Huhse B., Mangano R., Michon A. M., Schirle M., Schlegl J., Schwab M., Stein M. A., Bauer A., Casari G., Drewes G., Gavin A. C., Jackson D. B., Joberty G., Neubauer G., Rick J., Kuster B., and Superti-Furga G. (2004) A physical and functional map of the human TNF-alpha/NF-kappa B signal transduction pathway. Nat. Cell Biol. 6, 97–105 [DOI] [PubMed] [Google Scholar]
- 33. Doma E., Chakrabandhu K., and Hueber A. O. (2010) A novel role of microtubular cytoskeleton in the dynamics of caspase-dependent Fas/CD95 death receptor complexes during apoptosis. FEBS Lett. 584, 1033–1040 [DOI] [PubMed] [Google Scholar]
- 34. Carneiro D. G., Clarke T., Davies C. C., and Bailey D. (2016) Identifying novel protein interactions: Proteomic methods, optimisation approaches and data analysis pipelines. Methods 95, 46–54 [DOI] [PubMed] [Google Scholar]
- 35. Puig O., Caspary F., Rigaut G., Rutz B., Bouveret E., Bragado-Nilsson E., Wilm M., and Seraphin B. (2001) The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods 24, 218–229 [DOI] [PubMed] [Google Scholar]
- 36. Gibson T. J., Seiler M., and Veitia R. A. (2013) The transience of transient overexpression. Nat. Methods 10, 715–721 [DOI] [PubMed] [Google Scholar]
- 37. Boldin M. P., Mett I. L., Varfolomeev E. E., Chumakov I., Shemer-Avni Y., Camonis J. H., and Wallach D. (1995) Self-association of the “death domains” of the p55 tumor necrosis factor (TNF) receptor and Fas/APO1 prompts signaling for TNF and Fas/APO1 effects. J. Biol. Chem. 270, 387–391 [DOI] [PubMed] [Google Scholar]
- 38. Chinnaiyan A. M., O'Rourke K., Tewari M., and Dixit V. M. (1995) FADD, a novel death domain-containing protein, interacts with the death domain of Fas and initiates apoptosis. Cell 81, 505–512 [DOI] [PubMed] [Google Scholar]
- 39. Muzio M., Chinnaiyan A. M., Kischkel F. C., O'Rourke K., Shevchenko A., Ni J., Scaffidi C., Bretz J. D., Zhang M., Gentz R., Mann M., Krammer P. H., Peter M. E., and Dixit V. M. (1996) FLICE, a novel FADD-homologous ICE/CED-3-like protease, is recruited to the CD95 (Fas/APO-1) death–inducing signaling complex. Cell 85, 817–827 [DOI] [PubMed] [Google Scholar]
- 40. Bradbury A., and Pluckthun A. (2015) Reproducibility: Standardize antibodies used in research. Nature 518, 27–29 [DOI] [PubMed] [Google Scholar]
- 41. Baker M. (2015) Reproducibility crisis: Blame it on the antibodies. Nature 521, 274–276 [DOI] [PubMed] [Google Scholar]
- 42. Kasinathan S., Orsi G. A., Zentner G. E., Ahmad K., and Henikoff S. (2014) High-resolution mapping of transcription factor binding sites on native chromatin. Nat. Methods 11, 203–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Furlan-Magaril M., Rincon-Arano H., and Recillas-Targa F. (2009) Sequential chromatin immunoprecipitation protocol: ChIP-reChIP. Methods Mol. Biol. 543, 253–266 [DOI] [PubMed] [Google Scholar]
- 44. Flajolet M., Rakhilin S., Wang H., Starkova N., Nuangchamnong N., Nairn A. C., and Greengard P. (2003) Protein phosphatase 2C binds selectively to and dephosphorylates metabotropic glutamate receptor 3. Proc. Natl. Acad. Sci. U.S.A. 100, 16006–16011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kimura H., Takizawa N., Allemand E., Hori T., Iborra F. J., Nozaki N., Muraki M., Hagiwara M., Krainer A. R., Fukagawa T., and Okawa K. (2006) A novel histone exchange factor, protein phosphatase 2Cgamma, mediates the exchange and dephosphorylation of H2A-H2B. J. Cell Biol. 175, 389–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Suh E. J., Kim Y. J., and Kim S. H. (2009) Protein phosphatase 2Cgamma regulates the level of p21Cip1/WAF1 by Akt signaling. Biochem. Biophys. Res. Commun. 386, 467–470 [DOI] [PubMed] [Google Scholar]
- 47. Allemand E., Hastings M. L., Murray M. V., Myers M. P., and Krainer A. R. (2007) Alternative splicing regulation by interaction of phosphatase PP2Cgamma with nucleic acid-binding protein YB-1. Nat. Structural Mol. Biol. 14, 630–638 [DOI] [PubMed] [Google Scholar]
- 48. Khoronenkova S. V., Dianova I. I., Ternette N., Kessler B. M., Parsons J. L., and Dianov G. L. (2012) ATM-dependent downregulation of USP7/HAUSP by PPM1G activates p53 response to DNA damage. Mol. Cell 45, 801–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. McNamara R. P., McCann J. L., Gudipaty S. A., and D'Orso I. (2013) Transcription factors mediate the enzymatic disassembly of promoter-bound 7SK snRNP to locally recruit P-TEFb for transcription elongation. Cell Reports 5, 1256–1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Liu J., Stevens P. D., Eshleman N. E., and Gao T. (2013) Protein phosphatase PPM1G regulates protein translation and cell growth by dephosphorylating 4E binding protein 1 (4E-BP1). J. Biol. Chem. 288, 23225–23233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Xu K., Wang L., Feng W., Feng Y., and Shu H. K. (2016) Phosphatidylinositol-3 kinase-dependent translational regulation of Id1 involves the PPM1G phosphatase. Oncogene 35, 5807–5816 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data obtained are accessible at the EMBL-EBI, URL: https://www.ebi.ac.uk/pride/archive/projects/PXD006700.