Abstract
Human vasorin (VASN) as a type I transmembrane protein, is a potential biomarker of hepatocellular carcinoma, which could expedite HepG2 cell proliferation and migration significantly in vitro. The ectodomain of VASN was proteolytically released to generate soluble VASN (sVASN), which was validated to be the active form. Among several monoclonal antibodies produced against sVASN, the clone V21 was found to bind with the recombinant human sVASN (rhsVASN) with the highest affinity and specificity, and also have inhibitory effects on proliferation and migration of HepG2 cells. Hence the phage-displayed peptide library was screened against the antibody V21. The positive phage clones were isolated and sequenced, and one unique consensus motifs was obtained. The result of sequence alignment showed that the conserved motif had similarity to VASN(Cys432-Cys441), embedded in the epidermal growth factor (EGF)-like domain. The synthetic mimotope peptide V21P1 and V21P2 were confirmed to bind with V21 and could compete with rhsVASN in ELISA assay. And they could also almost completely reverse the inhibitory effect of V21 on HepG2 migration and proliferation. Furthermore, the antibodies produced against V21P1 were able to bind not only with the peptide V21P1, but also with rhsVASN and the natural VASN from HepG2 cell. Our results showed that V21 seemed to be a functional antibody. The mimotopes toward V21 might mimic the functional domain of VASN, which would be helpful to exploit VASN functions and act as a candidate target for developing therapeutic antibodies against VASN.
Keywords: vasorin, hepatocellular carcinoma, inhibitory antibody, peptide library, mimotope
Introduction
Human vasorin (VASN), also known as Slit-like 2 protein (SLITL2), is a type I transmembrane glycoprotein, sharing similar structures with the proteins of SLIT family. VASN was initially identified as a secreted and cell-surface protein expressed in the cardiovascular system 1. In adult human tissues, VASN is expressed predominantly in aorta, moderately in kidney and placenta, and much less in other tissues, such as liver and brain. VASN has the capacity of binding with transforming growth factor β (TGFβ), and acts as a ligand trap of TGFβ. VASN modulates cellular responses to vascular lesion, including vessel repair and neointimal formation, by the mechanism of impeding TGFβ signaling.
In contrast to restricted expression in normal tissues, VASN was highly expressed in several types of tumor cells. VASN was detected in the indicated breast cancer cell lines 2 and seemed to be a novel breast cancer-related biomarker 3. Moreover, the presence of VASN in tumor samples from patients with triple-negative breast cancer seemed to be correlated with collective invasion and poor outcome 4. In human lung carcinoma cell line A549, overexpression of VASN interfered with TGFβ-mediated epithelial-to-mesenchymal transition (EMT), including the decrease in E-cadherin level and remodeling in actin cytoskeleton 2. The expression of VASN was also significantly increased in brain tumors, which protected the tumor cells from hypoxia- or TNF-α-induced apoptosis 5. We previously validated that VASN was also highly expressed in the cells and tissues of hepatocellular carcinoma (HCC), and at a high level in the serum samples of HCC patients. The cellular VASN seemed to be involved in proliferation, migration and apoptosis of HepG2 cells. When overexpressed in normal liver cell L02, VASN boosted cell proliferation and migration, and inhibited cell apoptosis 6. Meanwhile, exosomal VASN from HepG2 cells promoted migration of Human Umbilical Vein Endothelial Cells (HUVEC) 7. These reports suggest that VASN acts as a player in cancer pathogenesis, at least in part, through sequestering TGFβ. As is known, TGFβ has contextually pleiotropic activities, that is, it plays a role of a tumor suppressor during carcinoma progression, and on the contrary, it promotes the malignant progression and metastasis during cancer advance 8-10. Hence, the functions of VASN seemed to be sophisticated and remained to be well characterized.
VASN has two forms, the membrane-associated form and the soluble form 1. The ectodomain of VASN encompasses three known potential functional domains, ten tandem leucine-rich repeats (LRR), an epidermal growth factor (EGF)-like domain, and a fibronectin type III-like domain, which is proteolytically shedded by A Disintegrin and a Metalloprotease domain 17 (ADAM17) and secreted as soluble VASN (sVASN) 2. It is possible that besides TGFβ, sVASN might bind with multiple proteins, and so be endowed with corresponding versatile roles. Therefore, we hypothesize VASN is implicated in yet-to-be-identified signal pathways and exerts unknown TGFβ-independent roles.
The binding sites of protein-protein interaction are the molecular basis for protein functions, so profiling of these sites would facilitate the studies on relationship between structures and functions 11. Epitopes are valuable to unveil binding sites of protein interactions, and also to produce functional antibodies for probing or blocking the intact proteins 12. Diseases-related antigens and their epitopes played a primary role as biomarkers for immune diagnosis and therapy 13. Peptide libraries are molecular evolution in vitro, and a powerful tool for epitope mapping 14,15. Also, antibody-based strategies are useful in researches of protein functions and development of disease diagnosis and therapy 16,17. Screening peptide libraries against specific antibodies would throw a new light on researches of epitope profiles.
In this study, we try to address the dominant functional domains of VASN by biopanning of a random dodecapeptide library. The targets were two monoclonal antibodies, V20 and V21, with high affinity and specificity, and V21 antibody had inhibitory effects on VASN. After screening, two panels of mimetic peptides were obtained corresponding to the two antibodies, respectively. As expected, all of the peptide mimics had antigenity, and only the ones toward V21 might be functional.
Materials and methods
Cell culture
Human hepatocellular carcinoma cell line HepG2 (ATCC# HB-8065) was cultured in Dulbecco's Modified Eagle Medium (DMEM; Gibco, USA), supplemented with 10% fetal bovine serum (Hyclone, USA), 100 U/mL penicillin, and 100 μg/mL streptomycin. The cells were maintained in cell culture flasks (Costar, USA) in a humidified chamber with 5% CO2 at 37°C, and subcultured at a 1:3 ratio when the cells reached about 90% confluency.
Collection of cells, culture supernatants and exosomes
HepG2 cells were cultured in 25cm2 flasks. When the confluency reached 60-70%, the cells were washed by PBS twice and counted, and the medium was changed to fresh serum-free DMEM with or without TAPI-2 (Sigma) at the final concentration of 25 μmol/L 18. After 24h, the culture supernatants were collected and concentrated by ultrafilters (Millipore) with molecular weight cut-offs of 10,000. The exosomes were prepared from the concentrated supernatants using the ExoQuick-TCTM Exosome Precipitation Solution (System Biosciences, USA) as described by the manufacturer 7. Meanwhile, the cells were scraped and lysed by cold lysis buffer (Thermo Fisher) with protease inhibitor cocktail (Sigma).
Western Blot
The protein samples, such as the cell lysates, concentrated supernatants and exosomes, were subjected to 10% SDS-PAGE, then blotted onto polyvinylidene fluoride membrane (PVDF; Amersham Biosciences) using Mini Trans-Blot Modulate (Bio-Rad). After blocking for 1 h in 5% skim milk at room temperature, the membrane was probed with the primary antibodies, that is, anti-VASN antibody V20, anti-VASN (Cloud-Clone corporation), and serially diluted anti-V21P1 serum. The secondary antibody was horseradish peroxidase(HRP)-conjugated goat anti-mouse or anti-rabbit IgG. Finally, the blots were detected by the enhanced chemiluminescence (ECL) Western blot detection system (Pierce).
Sandwich ELISA
To quantify sVASN in the culture supernatants of HepG2, sandwich ELISA were performed using Human Vasorin ELISA Kit (Sino Biological Company), according to the manufacturer's instruction. In brief, 1.5×105 HepG2 cells were seeded on 6-well plates. After 24h culture in serum-free DMEM with or without 25 μmol/L TAPI-2, the supernatants were collected and centrifuged at 1000rpm to delete the cell debris. Sequentially, the supernatants were assayed and the concentration of sVASN was calculated according to the standard curve.
Random phage displayed peptide library and bacterial strains
Ph.D.-12TM Peptide Library Kit was purchased from New England Biolabs (Ipswich, MA, USA). The titer of the library is 1×1013 pfu/ml, and the complexity is on the order of 109. The host strain is E. coil ER2537.
Biopanning of binding peptides against anti-VASN antibodies
The dodecapeptide library was screened against the antibody V20 and V21, and three rounds of biopanning were carried out following the manufacturer's instruction. The stringency of selection was increased by lowering the target concentrations and raising the concentration of Tween-20 in the washing buffer stepwise. The enrichment was evaluated by output/input ratio 19 and P/N value 20. The P/N value was calculated by the numbers of phage colonies on the test well (P)/those on the control ones (N). The positive phage clones were screened by ELISA. The single-stranded DNA of phages was sequenced, the amino acid sequences were deduced, and sequence alignment was performed using DNAMAN and BLAST.
ELISA
To assess the binding of phage clones to the anti-VASN antibody V20 and V21, ELISA was carried out as described before 21,22. In brief, the ELISA strips (Costar) were coated with V20 or V21 at 0.5μg per well. The selected monoclonal phages (1×108 pfu) were added to each well in triplicate, and the plates were incubated at 37 °C for 1 h. After washed by TBS-0.05% Tween-20 for 5 times, the HRP-conjugated anti-M13 antibody (Amersham Biosciences) was added, and the plates were incubated at 37 °C for 1 h. The bound antibodies were detected using 3,3',5,5'-tetra-methyl-benzidine dihydrochloride (Sigma) as the substrate, and the color intensity was determined spectrophotometrically at 450 nm.
Competitive ELISA Assay
To assess specificity of phage clones binding with V20 and V21, competitive ELISA was carried out as described before 21,22. In brief, the ELISA strips (Costar) were coated with V20 or V21 at 0.2 μg per well. The selected monoclonal phages (1×108 pfu) were added to each well in triplicate with the recombinant human soluble VASN (rhsVASN; Novoprotein) at serial dilution, and the plates were incubated at 37 °C for 1 h. Then the binding phages were detected as mentioned afore. To analysis whether the peptide-BSA conjugates could interfere with the binding of VASN with V21, rhsVASN was coated at 0.5 μg per well, 80 ng/ml V21 was pre-incubated with the peptide-BSA proteins at various concentrations, and then added to the wells.
Transwell migration Assay
HepG2 motility were assayed using 12-well transwell plates (Corning) as described before 23. In brief, 1×105 cells were seeded on the upper chamber with a cell-permeable 8.0 μm membrane, and the lower chamber was filled with serum-free DMEM containing the antibodies with or without the peptide-BSA proteins. After 12h, cells on the upper surface of the membrane were removed using cotton swabs, and the cells that migrated to the bottom of the membrane were fixed with 4% paraformaldehyde in PBS and stained with 0.1% crystal violet solution. Cell micrographs were taken on bright field microscope equipped with a digital camera and the migratory cells were also counted.
Cell proliferation Assay
HepG2 were plated on 96-well plates at 3000 cells per well overnight. The medium was changed to fresh serum-free DMEM, and the mixtures of the antibodies and peptide-BSA proteins were added. After culture for 72h, CCK-8 assay was performed to detect cell proliferation.
Production of anti-mimic peptides sera
The mimic peptides were synthesized chemically and conjugated to Keyhole limpet hemocyanin (KLH). Female New Zealand White rabbits were first immunized by subcutaneously injecting them with 1 ml of the immunogen (0.25 mg of the peptide-KLH proteins in phosphate-buffered saline (PBS) mixed with complete Freund's adjuvant (Sigma)). Subsequent booster injections, i.e., 0.5 mg proteins in PBS emulsified in the rapid immune adjuvant (AbMax Biotechnology Co., Ltd), and were administered at 7-day intervals for 5 times.
Statistical analysis
Prism 6 (GraphPad Software) was used for statistical analysis. Data were tested for significance using unpaired Student t-test for two sets of data, and one-way ANOVA or two-way ANOVA for multiple comparison. Results with P value <0.05 were considered statistically significant. Data are presented as means ± standard deviation.
Results
Soluble VASN present in the conditioned medium of HepG2 cells
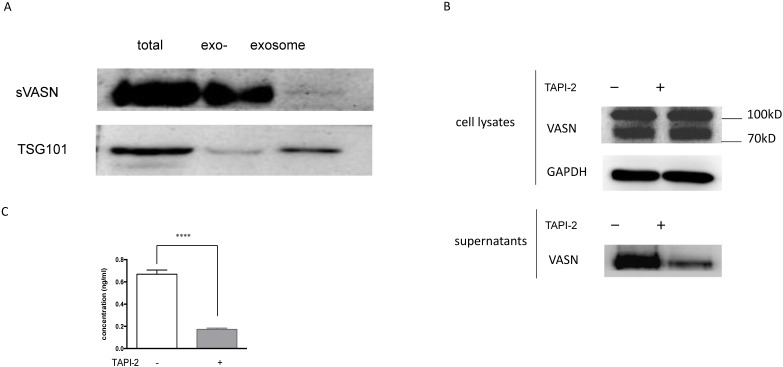
As we reported before, VASN was expressed highly in the hepatocellular carcinoma HepG2 cells and tissues, and might be a potential biomarker 6. Moreover, VASN was found to exist in HepG2-derived exosomes 7. To detect whether sVASN was released from HepG2 cells into the culture medium, Western blot was performed. After cultured in the serum-free medium with or without ADAM17 inhibitor TAPI-2 for 24h, the supernatant of HepG2 cells was collected and concentrated by ultrafiltration. Then exosomes were precipitated and purified, and meanwhile, the exosome-depleted supernatants were obtained. The result of Western blot showed that HepG2 cells could secrete VASN protein into the culture supernatant, corresponding to high expression in the cells (Fig. 1A). However, Western blot assay of exosomes showed that the exosomal VASN from the same volume supernatant was too little to be detected, in which TSG101 was taken as the gel loading control of exosomes 24. To further validate whether the soluble VASN was cleaved by ADAM17, TAPI-2 was added to the culture medium to inhibit ADAM17 activity. The result of Western blot showed that TAPI-2 had little effects on VASN expression in the cells, while the soluble VASN was much less in the supernatant of TAPI-2-treated HepG2 than that of the untreated ones (Fig. 1B). It meant that TAPI-2 impeded release of VASN ectodomain quite significantly, and so ADAM17 was vital for cleavage. Soluble VASN in the culture supernatants was also assayed by sandwich ELISA. The result showed that sVASN in HepG2 supernatant decreased more than threefold after TAPI-2 addition (Fig. 1C). These results suggested that VASN existed mainly in the soluble form in the supernatant of HepG2 cells.
Figure 1.
Soluble VASN existed mainly in soluble form in the culture supernatant of HepG2 cells. (A) Western blot assay of VASN in the culture supernatant. The supernatants were collected and concentrated, exosomes from the same volume of the supernatant were purified, and the same volume of the exosome-depleted supernatant were collected by the same way. TSG101 was taken as the gel loading control of exosomes. exo-: exosome-depleted supernatant. (B) Western blot of VASN in the cell lysates and culture supernatants with or without addition of TAPI-2. (C) Sandwich ELISA assay. Soluble VASN in 100 μl of the culture supernatants with or without addition of TAPI-2. **** P value < 0.0001 compared to the supernatant untreated by TAPI-2 by an unpaired Student t-test.
VASN Antibody V21 attenuating proliferation and migration of HepG2 cells
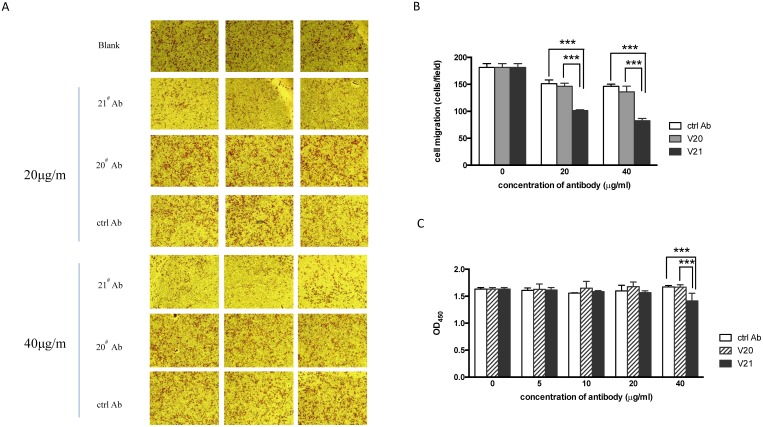
We previously validated that cellular VASN was involved in proliferation and migration of HepG2 6. Here, we tried to explore whether sVASN in the supernatant had the similar role. We prepared several clones of monoclonal antibodies against rhsVASN. Among them, the clone V20 and V21 showed the highest affinity with rhsVASN (data not shown). Both V20 and V21 were also able to bind with native VASN protein of HepG2 cells in Western blot assay (Figs. 1A, B). Sequentially, VASN antibodies were added to the medium of HepG2 at the indicated concentrations, and Transwell and CCK-8 assays were carried out. The results showed that migration of HepG2 was inhibited notably by V21 antibody in a dose-dependent manner, but not by V20 antibody or the control antibody (Figs. 2A, B). Meantime, V21 antibody also hindered HepG2 proliferation (Fig. 2C). It was suggested V21 was an inhibitory antibody, and sVASN had effects on proliferation and migration of HepG2. The antibody V20 might exert little functions as indicated in the above-mentioned assays (Fig. 2).
Figure 2.
The antibody V21 could inhibit migration and proliferation of HepG2 cells. The representative photographs of cell migration in transwell assay were taken (A), and the migratory cells were analyzed quantitively (B). ctrl Ab: the isotype control antibody. (C) The cell proliferation was detected by CCK-8 assay. Data are means ± SD. *** P value < 0.001 compared to the V21 treated groups by two-way ANOVA.
Screening mimotopes of VASN against the antibodies by peptide library
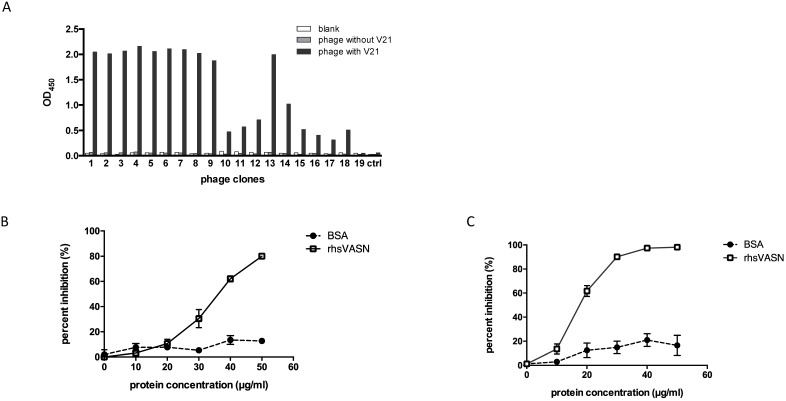
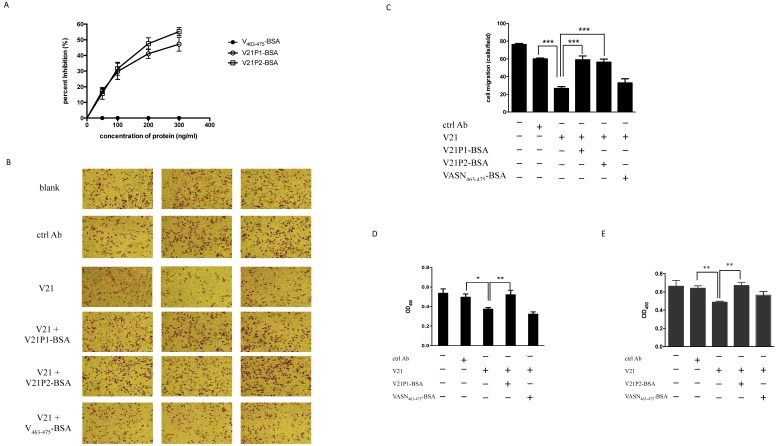
As is known, the extracellular region of VASN comprises three putative functional domains, which implies potential functions 1. However, up to now, little is known about the dominant motifs of VASN and their functions. For epitope profiles of proteins were always the interaction sites of protein-proteins, we tried to map epitopes of VASN. Phage-displayed random dodecapeptide library was screened against the antibody V21. The enrichment extent was monitored by output/input ratio and P/N value. After 3 rounds of biopanning, the specific phage clones were enriched (Table 1). The binding of isolated phage monoclones with the antibodies was assayed by ELISA. The results showed that 18 of 19 clones were able to bind specifically with V21 antibody (Fig. 3A). Then the insert peptides of positive phages were sequenced. The sequencing result showed that 5 sequences toward V21 were obtained, and the result of sequence alignment by DNAMAN and BLAST indicated that these sequences had one unique conserved motif, which had moderate similarity with the amino acid residues 432-441 of VASN (Tables 2). VASN(C432-C441) located in the epidermal growth factor (EGF)-like domain, and Cys432 and Cys441 formed a disulfide bond. Interestingly, the peptide SLTLPPFTGLAG, named V21P1, also has some similarity with SLTLGIEPVSPTS, the residues 463-475 of VASN, which was identified as a B-cell epitope by mass spectrometry combined with a multiengine search approach 25. Furthermore, competitive ELISA was carried out to assess the specificity of binding between the positive phage clones and the antibodies. V21 was coated, and two selected positive phages (clone 6 and 1, named as V21P1 and V21P2) were simultaneously added with serially diluted rhsVASN, taking BSA as the negative control (Figs. 3B and 3C). The rhsVASN could attenuate the binding of phages with antibodies dose-dependently, and the inhibition percentages were more than 90% at the final concentration of 50 μg/ml. The results suggested that the phage clones V21P1 and V21P2 bound V21 with high affinity and specificity, and the insert peptides of phages might be mimotopes of VASN.
Table 1.
The enrichment procedure of peptide library screening against V21.
| Round | coated-Ab (μg/well) | input (pfu) |
output of sample well (pfu) | output of control well (pfu) | P/N value |
|---|---|---|---|---|---|
| 1 | 1.5 | 4.7×1010 | 4.36×105 | 4.6×104 | 9 |
| 2 | 1.0 | 1.67×1010 | 1.1×108 | 2.5×105 | 440 |
| 3 | 0.5 | 1.16×1010 | 1.81×109 | 1.19×106 | 1521 |
Figure 3.
Affinity and specificity of isolated phage clones against the VASN antibodies. ELISA assays of isolated phage clones binding with V21 (A). ctrl: peptide library, blank: without phages. Competitive ELISA assays of isolated phage clones V21P1 (B) and V21P2 (C) binding with V21, competed by rhsVASN.
Table 2.
Sequence alignment of peptide inserts binding with V21
| phage clone No. | sequencea | frequency |
|---|---|---|
| 1 | TQFSITPPFTGS | 2 |
| 3 | SHWFTGSDAGP | 1 |
| 4 | EYYGFTGLHSPH | 1 |
| 5 | HPFPPSFTGKAS | 1 |
| 6 | SLTLPPFTGLAG | 9 |
| consensus motif | PPFTGL(S) | |
| VASN(432-441) | CPEGFTGLYC |
a The consensus motifs were shown in italic letters.
At the same time, we also biopanned the peptide library against V20 antibody. After 3 rounds of biopanning, the specific phage clones were enriched (Supplementary Table S1). All of 10 isolated clones were able to bind specifically with V20 in ELISA (Supplementary Fig. S1), and 7 sequences was obtained (Supplementary Table S2). The results of sequence alignment showed that the sequences toward V20 could be grouped into three clusters, which were completely different from those of V21. The result of competitive ELISA showed that binding of the clone 4 and 9 (named as V20P1 and V20P2) with V20 could be inhibited by rhsVASN in a dose-dependent manner, and the inhibition percentages were more than 90% (Supplementary Fig. S2). The results suggested that the insert peptides of phages might mimic some epitope of VASN, other than that against V21.
Affinity and specificity of mimotope peptides
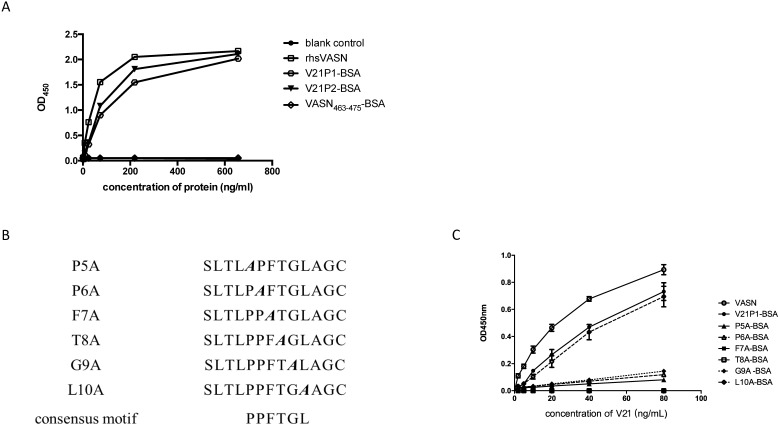
To assess whether they were binding sites for V21, the mimotope peptides were synthesized and conjugated to BSA, named as V21P1-BSA (SLTLPPFTGLAG) and V21P2-BSA (TQFSITPPFTGS). The residues VASN(Ser463-Ser475), was prepared as VASN463-475-BSA and taken as the control. ELISA were performed to detect the affinity and specificity of the peptide-BSA proteins binding to V21. The result of ELISA showed that the serial-diluted V21P1-BSA and V21P2-BSA were able to bind with V21, similarly to rhsVASN (Fig. 4A). In contrast, VASN463-475-BSA couldn't bind with V21. It suggested that the mimotope peptides might have high affinity with the antibodies, mimicking the antigenicity of sVASN protein. Meanwhile, we also assayed binding of the mimic peptides towards V20. The proteins, named as V20P1-BSA (SFYIEAQGDRLH) and V20P2-BSA (LPGYYSEAGNWR), could bind with V20 with high affinity (Supplementary Fig. S3).
Figure 4.
The synthetic mimotope peptides could interfere with the inhibitory effects of V21 on HepG2 cells. (A) Affinity of synthetic mimotope peptides binding to V21.rhsVASN was taken as the positive control, VASN463-475-BSA as the negative control, and no-protein as the blank control. (B) The sequences of mutant peptides. The alanine mutant sites were shown in bold and italic. (C) Affinity of synthetic peptide mutants binding to V21. rhsVASN and V21P1 were taken as the positive control.
Moreover, we tried to identify the key amino acids of the mimic peptides by the means like alanine scanning 22. Six mutant peptides, with single alanine mutation of the consensus motif in V21P1, were synthesized (Fig. 4B). ELISA were carried out to assay the interaction between the mutants and V21. The result showed that the mutant L10A almost completely retained the binding ability to V21, while F7A and T8A couldn't bind with V21, and P5A, P6A and G9A kept little binding ability (Fig. 4C). It suggested that the consensus residues were critical amino acids in the mimotope, except Leu10.
Mimotope peptides attenuating inhibitory activity of V21 antibody
We had validated that the antibody V21 could inhibit proliferation and, migration of HepG2 cells (Fig. 2), and V21P1 and V21P2 seemed to be the peptide mimics of V21 binding site. Hence, we hypothesized the two peptides might affect the effects of V21. At first, competitive ELISA was carried out to assay whether the two peptides could interfere with binding between rhsVASN and V21. The result showed that V21P1-BSA and V21P2-BSA could inhibit rhsVASN binding to V21 to a certain extent, and the percentage inhibition was 47.2% and 55.3%, respectively, while the control VASN463-475-BSA had no inhibition effects (Fig. 5A). It suggested that the peptide V21P1 and V21P2 might mimic the binding site of VASN toward V21.
Figure 5.
The mimic peptides were able to attenuate inhibitory activity of V21 antibody. (A) Competitive ELISA. The rhsVASN was coated, and V21 was pre-incubated with the peptide-BSA conjugates at various concentrations, and then added to the wells. (B, C) Transwell assay of cell migration. 20 μg/ml V21 was added with or without 10 μg/ml peptide-BSA proteins. The representative photographs of cell migration were taken (B), and the migratory cells were quantitatively analyzed (C). The cell proliferation assay was performed to analyze the effects of V21P1-BSA (D) and V21P2-BSA (E) mixed with V21 on HepG2 proliferation. Data are means ± SD. * P value < 0.05, ** P value < 0.01 and *** P value <0.001 compared to the V21 treated groups by one-way ANOVA.
Furthermore, the peptide-BSA proteins and V21 were pre-mixed and co-incubated, then added into the medium of HepG2 cells for further culture for the indicated time. Transwell assay and CCK8 assay were preform to analyze cell migration and proliferation. The result of transwell assay showed that both V21P1-BSA and V21P2-BSA could almost completely reverse the inhibitory effects of V21 on HepG2 migration, while VASN463-475-BSA couldn't (Figs. 5B, C). Meanwhile, they could also rescue proliferation of HepG2, antagonizing the inhibition of V21, as suspected (Figs. 5D, E). It seemed that V21P1 and V21P2 could mimic the cognate binding sites in the inhibitory antibody, which might represent the dominant functional domain of sVASN.
Immunogenicity of the mimotope peptides
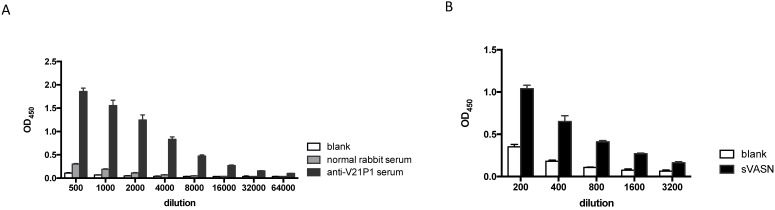
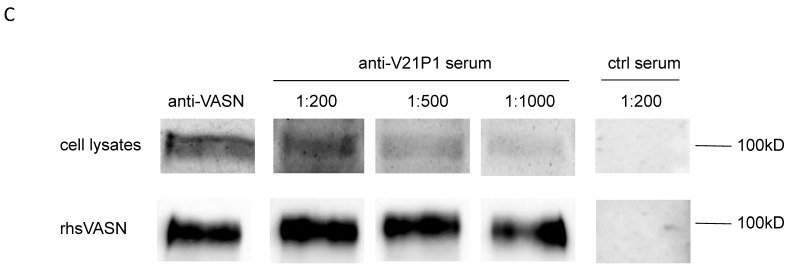
The mimic peptide V21P1 was synthesized and conjugated to KLH, and polyclonal antibodies against the peptides were prepared by immunizing rabbits. The titer of anti-sera was detected by ELISA. The result showed that the titer of the anti-serum against V21P1 was 1:64,000 (Fig. 6A). Moreover, to detect whether the anti-sera had cross-activity with rhsVASN, ELISA and Western Blot assay were performed. The result of ELISA showed that titer of the anti-serum against rhsVASN was 1:3,200 (Fig. 6B). Meanwhile, it was also showed that the anti-sera were able to bind with VASN from HepG2 cell lysates and rhsVASN specifically (Fig. 6C) and the titer was 1:1,000. It seemed the mimotope peptides could act as an immunogen to induce not only immune response against itself, but also cross-reaction against VASN.
Figure 6.
Immunogenicity of the mimotope peptide V21P1. (A) ELISA assay of anti-sera binding with V21P1. The peptide V21P1 were coated at 5μg per well. The antiserum was obtained from the immunized rabbits and serially two-fold diluted. Results are expressed as OD at 450 nm. Normal rabbit serum was taken as the negative control. blank: no serum. (B) ELISA assay of the anti-serum cross-reactivity with rhsVASN protein. The protein rhsVASN were coated at 0.5 μg per well. blank: non-coated. (C) Western Blot assay of the anti-serum cross-reactivity with natural VASN in HepG2 cell lysates or rhsVASN protein, taking anti-VASN antibody (Cloud-Clone corporation) as the positive control. ctrl serum: normal rabbit serum.
Discussion
Up to now, it was showed that human VASN protein might play a role in cancer pathogenesis, such as tumor growth, survival, metastasis and angiogenesis 2,4,6,7. In our previous study, it was revealed that VASN was correlated with HCC progression, as well as a candidate biomarker 6. As is known, VASN has two forms, and the soluble form might be biologically active 2. We found that the level of soluble VASN was much more highly in HCC patient sera than that in the healthy control, except that VASN was expressed more highly on HCC tissue slides than hepatitis liver tissue slides. Furthermore, the secretome compartments of some cancer cells consisted of VASN, which was sensitive to RPI-1 and dasatinib treatments, and could be a biomarker for measuring the effectiveness of cancer therapies 26. These results suggested that soluble VASN might be a potential target for cancer therapy. As VASN was found expressed highly in some solid tumors, it could be targeted, at least, as a topical therapy. However, the precise functions of sVASN and the underlined molecular mechanisms remained to be addressed. Hence, we tried to profile the functional motifs of sVASN, and to explored whether they were involved in HCC pathogenesis.
In our previous work, VASN ectodomain was expressed in E. coli and purified, and then several monoclonal antibodies against rhsVASN were generated (data not shown). Among them, V20 and V21 had relatively high affinity and specificity, and could bind with native VASN protein. In the present study, for the first time, we found V21 had inhibitory capacity on proliferation and migration of HepG2, by attenuating functions of VASN (Fig. 2). A panel of peptides toward V21 were identified by peptide library screening and share a consensus motif, sharing 4 amino- acid residues in common with VASN(Cys432-Cys441) (Table 2). We hypothesized that VASN(Cys432-Cys441) might consist of one protein interface hot spot of sVASN, and the 4 residues seemed to be the primary ones. We try to explore the key residues of the mimotope in depth. By the means of alanine mutation, we found that five out of six consensus residues were almost crucial to the affinity of the mimotope to V21, which seemed that the mimotope was exactly the corresponding epitope against V21. A mimotope is defined as a molecular, always a peptide, which mimics the structure of a natural epitope, which might cause an antibody response similar to the one elicited by the epitope 27. Hence, the anti-serum was prepared against V21P1, and proved to be able to bind with rhsVASN and the natural protein in HepG2. It was validated that V21P1 was the mimotope of VASN not only with antigenicity but also with immunogenicity. Moreover, for mimotopes bear the critical cognate binding sites of intact proteins, they are useful for mapping the functional motifs 28,29. As the results showed here, the mimotope peptides could act as a candidate target for generation of VASN antagonists, such as blocking antibodies and small molecule inhibitors.
In addition, we found that the mimotope V21P1 had the same 4 amino-acid residues at the N-terminus as those of VASN(Ser463-Ser475), the reported motif 25. Nevertheless, the 4 amino-acid residues were not the conserved ones embedded in V21P1, which were always thought as the key residues of the epitope. The synthetic peptide VASN(Ser463-Ser475) couldn't bind with V21 in ELISA assay or hinder binding of rhsVASN to V21 in competitive ELISA assay (Figs. 4A,5A). Moreover, VASN(Ser463-Ser475) was not able to interfere with V21 inhibitory effects on HepG2 (Fig. 5B-5E). In view of the results, we speculated that V21P1 and VASN(Ser463-Ser475) were two different B-cell epitopes of VASN, and possibly adjacent in three-dimensional conformation. V21P1 might mimic one conformational epitope, while VASN(Ser463-Ser475) was a linear one. It seemed that the two epitopes were involved in different functions.
Otherwise, we also obtained the mimotopes toward V20, which had more than one conserved motif and differed from that toward V21. And V20 was proved to have no obvious effects on proliferation and migration of HepG2 (Fig. 2). These results suggested that V20 and V21 recognized distinct epitopes. As is known, proteins always don't exist as a straight chain, but fold into the tertiary structure. Therefore, the natural epitopes are always discontinuous in the primary structure, that is, they are conformational epitopes 30. It meant that the identified peptides toward V20 were mimotopes of one conformational epitopes of VASN, which could mimic the structure of the natural epitope. Maybe the motifs toward V20 were correlated with other unknown functions of VASN. Further investigations needed to be done.
In summary, the present work mapped the epitope profiles of human VASN protein for the first time. The mimotopes might be representatives of VASN functional motifs, which would benefit the detailed research of VASN functions and relationship between structure and functions. The corresponding motif in VASN, as well as the mimotopes, might act as candidate targets of VASN antagonists for developing novel anti-HCC therapies. In addition, the antibody V21 was able to be one lead molecule of VASN inhibitors.
Supplementary Material
Supplementary figures and tables.
Acknowledgments
We thank the staff of Institute of Military Cognitive and Brain Sciences, Academy of Military Medical Sciences, for their assistance. This study was funded by the National Natural Science Foundation of China funded by the Ministry of Science and Technology of China (Grant number 81572846 to S.L., and 31570817 to J.L.).
Abbreviations
- VASN
vasorin
- sVASN
soluble VASN
- SLITL2
Slit-like 2 protein
- TGFβ
transforming growth factor β
- EMT
epithelial-to-mesenchymal transition
- HCC
hepatocellular carcinoma
- HUVEC
Human Umbilical Vein Endothelial Cells
- LRR
leucine-rich repeats (LRR)
- EGF
epidermal growth factor
- ADAM17
A Disintegrin and a Metalloprotease domain 17
- PVDF
polyvinylidene fluoride membrane
- HRP
horseradish peroxidase
- ECL
enhanced chemiluminescence.
References
- 1.Ikeda Y, Imai Y, Kumagai H. et al. Vasorin, a transforming growth factor beta-binding protein expressed in vascular smooth muscle cells, modulates the arterial response to injury in vivo. Proc Natl Acad Sci USA. 2004;101(29):10732–7. doi: 10.1073/pnas.0404117101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Malapeira J, Esselens C, Bech-Serra JJ. et al. ADAM17 (TACE) regulates TGFβ signaling through the cleavage of vasorin. Oncogene. 2011;30(16):1912–22. doi: 10.1038/onc.2010.565. doi: 10.1038/onc.2010.565. [DOI] [PubMed] [Google Scholar]
- 3.Zhang Y, Zhang J, Liu Z. et al. A network-based approach to identify disease-associated gene modules through integrating DNA methylation and gene expression. Biochem Biophys Res Commun. 2015;465(3):437–42. doi: 10.1016/j.bbrc.2015.08.033. doi: 10.1016/ j.bbrc.2015.08.033. [DOI] [PubMed] [Google Scholar]
- 4.Westcott JM, Prechtl AM, Maine EA. et al. An epigenetically distinct breast cancer cell subpopulation promotes collective invasion. J Clin Invest. 2015;125(5):1927–43. doi: 10.1172/JCI77767. doi:10.1172/ JCI77767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choksi S, Lin Y, Pobezinskaya Y. et al. A HIF-1 target, ATIA, protects cells from apoptosis by modulating the mitochondrial thioredoxin, TRX2. Mol Cell. 2011;42(5):597–609. doi: 10.1016/j.molcel.2011.03.030. doi: 10.1016/j.molcel. 2011. 03. 030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li S, Li H, Yang X. et al. Vasorin is a potential serum biomarker and drug target of hepatocarcinoma screened by subtractive-EMSA-SELEX to clinic patient serum. Oncotarget. 2015;6(12):10045–59. doi: 10.18632/oncotarget.3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang A, Dong J, Li S. et al. Exosomal transfer of vasorin expressed in hepatocellular carcinoma cells promotes migration of human umbilical vein endothelial cells. Int J Biol Sci. 2015;11(8):961–9. doi: 10.7150/ijbs.11943. doi: 10.7150/ ijbs.11943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muraoka-Cook RS, Dumont N, Arteaga CL. Dual role of transforming growth factor beta in mammary tumorigenesis and metastatic progression. Clin Cancer Res. 2005;11(2 Pt 2):937s–43s. [PubMed] [Google Scholar]
- 9.Massagué J. TGFbeta in Cancer. Cell. 2008;134(2):215–30. doi: 10.1016/j.cell.2008.07.001. doi:10.1016/j.cell. 2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Massagué J. TGFβ signalling in context. Nat Rev Mol Cell Biol. 2012;13(10):616–30. doi: 10.1038/nrm3434. doi: 10.1038/nrm3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bienstock RJ. Computational drug design targeting protein-protein interactions. Curr Pharm Des. 2012;18(9):1240–54. doi: 10.2174/138161212799436449. [DOI] [PubMed] [Google Scholar]
- 12.González-Ruiz D, Gohlke H. Targeting protein-protein interactions with small molecules: challenges and perspectives for computational binding epitope detection and ligand finding. Curr Med Chem. 2006;13(22):2607–25. doi: 10.2174/092986706778201530. [DOI] [PubMed] [Google Scholar]
- 13.Meeusen EN, Lim E, Mathivanan S. Secreted tumor antigens -immune biomarkers for diagnosis and therapy. Proteomics; 2017. doi: 10.1002/pmic. 201600442. [DOI] [PubMed] [Google Scholar]
- 14.Yaffe MB, Smerdon SJ. The use of in vitro peptide-library screens in the analysis of phosphoserine/threonine-binding domain structure and function. Annu Rev Biophys Biomol Struct. 2004;33:225–44. doi: 10.1146/annurev.biophys.33.110502.133346. [DOI] [PubMed] [Google Scholar]
- 15.Kokoszka ME, Kay BK. Mapping protein-protein interactions with phage-displayed combinatorial peptide libraries and alanine scanning. Methods Mol Biol. 2015;1248:173–88. doi: 10.1007/978-1-4939-2020-4_12. doi: 10.1007/978-1-4939-2020-4_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu Y, Ling S, Hegde AM. et al. Using reverse-phase protein arrays as pharmacodynamic assays for functional proteomics, biomarker discovery, and drug development in cancer. Semin Oncol. 2016;43(4):476–83. doi: 10.1053/j.seminoncol.2016.06.005. doi: 10.1053/ j. seminoncol.2016.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beghein E, Gettemans J. Nanobody Technology: A Versatile Toolkit for Microscopic Imaging, Protein-Protein Interaction Analysis, and Protein Function Exploration. Front Immunol. 2017;8:771.. doi: 10.3389/fimmu.2017.00771. doi: 10.3389/fimmu.2017.00771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fréour T, Jarry A, Bach-Ngohou K. et al. TACE inhibition amplifies TNF-alpha-mediated colonic epithelial barrier disruption. Int J Mol Med. 2009;23(1):41–8. [PubMed] [Google Scholar]
- 19.Mao J, Shi X, Wu YB, Identification of Specific Hydroxyapatite Binding Heptapeptide by Phage Display and Its Nucleation Effect. Materials (Basel); 2016. p. 9. (8). pii: E700. doi: 10.3390/ma9080700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dong J, Liu C, Zhang J. et al. Selection of novel nickel-binding peptides from flagella displayed secondary peptide library. Chem Biol Drug Des. 2006;68(2):107–12. doi: 10.1111/j.1747-0285.2006.00421.x. [DOI] [PubMed] [Google Scholar]
- 21.Gao YP, Dong J, Zhang X. et al. A novel peptide can mimic extracellular fibrinogen-binding protein to block the activation of complement system. Cell Biochem Biophys. 2013;66(3):753–7. doi: 10.1007/s12013-013-9520-0. doi:10.1007/ s12013-013-9520-0. [DOI] [PubMed] [Google Scholar]
- 22.Dong J, Gao Y, Liu Y. et al. The protective antibodies induced by a novel epitope of human TNF-alpha could suppress the development of collagen-induced arthritis. PLoS One. 2010;5(1):e8920.. doi: 10.1371/journal.pone.0008920. doi: 10.1371/journal. pone.0008920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guerra AD, Rose WE, Hematti P. et al. Minocycline modulates NFκB phosphorylation and enhances antimicrobial activity against Staphylococcus aureus in mesenchymal stromal/stem cells. Stem Cell Res Ther. 2017;8(1):171.. doi: 10.1186/s13287-017-0623-1. doi:10.1186/s13287-017-0623-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greening DW, Xu R, Ji H, Tauro BJ, Simpson RJ. A protocol for exosome isolation and characterization: evaluation of ultracentrifugation, density-gradient separation, and immunoaffinity capture methods. Methods Mol Biol. 2015;1295:179–209. doi: 10.1007/978-1-4939-2550-6_15. doi: 10.1007/978-1-4939-2550-6_15. [DOI] [PubMed] [Google Scholar]
- 25.Marcilla M, Alvarez I, Ramos-Fernández A. et al. Comparative Analysis of the Endogenous Peptidomes Displayed by HLA-B*27 and Mamu-B*08: Two MHC Class I Alleles Associated with Elite Control of HIV/SIV Infection. J Proteome Res. 2016;15(3):1059–69. doi: 10.1021/acs.jproteome.5b01146. doi:10.1021/acs. jproteome.5b01146. [DOI] [PubMed] [Google Scholar]
- 26.Caccia D, Zanetti DL, Miccichè F. et al. Secretome compartment is a valuable source of biomarkers for cancer-relevant pathways. J Proteome Res. 2011;10(9):4196–207. doi: 10.1021/pr200344n. [DOI] [PubMed] [Google Scholar]
- 27.Huang J, He B, Zhou P. Mimotope-based prediction of B-cell epitopes. Methods Mol Biol. 2014;1184:237–43. doi: 10.1007/978-1-4939-1115-8_13. doi: 10.1007/978-1-4939-1115-8_13. [DOI] [PubMed] [Google Scholar]
- 28.Toyoda T, Masunaga K, Ohtsu Y. et al. Antibody-scanning and epitope-tagging methods; molecular mapping of proteins using antibodies. Curr Protein Pept Sci. 2000;1(3):303–8. doi: 10.2174/1389203003381360. [DOI] [PubMed] [Google Scholar]
- 29.Fuchs S, Kasher R, Balass M. et al. The binding site of acetylcholine receptor: from synthetic peptides to solution and crystal structure. Ann N Y Acad Sci. 2003;998:93–100. doi: 10.1196/annals.1254.011. [DOI] [PubMed] [Google Scholar]
- 30.Goldsby R, Kindt TJ, Osborne BA, et al. Antigens (Chapter 3) In: Immunology fifth., editor. New York: W. H. Freeman and Company; 2003. pp. 57–75. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures and tables.