Abstract
Background
Chrysanthemum is one kind of ornamental plant well-known and widely used in the world. However, its quality and production were severely affected by low temperature conditions in winter and early spring periods. Therefore, we used the RNA-Seq platform to perform a de novo transcriptome assembly to analyze chrysanthemum (Dendranthema grandiflorum) transcription response to low temperature.
Results
Using Illumina sequencing technology, a total of 86,444,237 high-quality clean reads and 93,837 unigenes were generated from four libraries: T01, controls; T02, 4 °C cold acclimation (CA) for 24 h; T03, − 4 °C freezing treatments for 4 h with prior CA; and T04, − 4 °C freezing treatments for 4 h without prior CA. In total, 7583 differentially expressed genes (DEGs) of 36,462 annotated unigenes were identified. We performed GO and KEGG pathway enrichment analyses, and excavated a group of important cold-responsive genes related to low temperature sensing and signal transduction, membrane lipid stability, reactive oxygen species (ROS) scavenging and osmoregulation. These genes encode many key proteins in plant biological processes, such as protein kinases, transcription factors, fatty acid desaturase, lipid-transfer proteins, antifreeze proteins, antioxidase and soluble sugars synthetases. We also verified expression levels of 10 DEGs using quantitative real-time polymerase chain reaction (qRT-PCR). In addition, we performed the determination of physiological indicators of chrysanthemum treated at low temperature, and the results were basically consistent with molecular sequencing results.
Conclusion
In summary, our study presents a genome-wide transcript profile of Dendranthema grandiflorum var. jinba and provides insights into the molecular mechanisms of D. grandiflorum in response to low temperature. These data contributes to our deeper relevant researches on cold tolerance and further exploring new candidate genes for chilling-tolerance and freezing-tolerance chrysanthemum molecular breeding.
Electronic supplementary material
The online version of this article (10.1186/s12864-018-4706-x) contains supplementary material, which is available to authorized users.
Keywords: Dendranthema grandiflorum, Low temperature stress, Cold acclimation, Transciptome sequence, Differentially expressed genes
Background
Low temperature is a common environmental stress factor, which seriously affects plant growth and becomes an important factors limiting plant distribution. Plants will make a series of physiological and biochemical changes as defense systems in response to chilling (0–15 °C) and freezing (< 0 °C) injury [1]. These cold resistance mechanisms involve biofilm system, osmotic regulators, protective enzyme systems, and fatty acids. For many temperate plant species, exposed to chilling temperature for a period of time could increase their tolerance to subsequent freezing conditions, and this process is called ‘cold acclimation (CA)’ [2]. Cold acclimation can regulate many cold-responsive genes differentially expressed, induce the establishment of cold resistance mechanism, and finally improve the adaptability of plants to low temperature [3].
Dendranthema grandiflorum var. jinba is one of the best-selling cut chrysanthemum varieties in China. In order to realize the annual production and supply of cut chrysanthemum, in most areas of China, factories need to use the facilities for heating in winter, which greatly increases the cost of production. Thus, understanding the mechanism of chilling and freezing stress responses and improving the cold tolerance of chrysanthemum by gene transfer are of great importance.
Chrysanthemum has large genomes and lacks genomic information on the basis of cold tolerance. With the development of next-generation sequencing (NGS) technology, large scale transcriptome data has become availavle in various species [4]. In recent years, Illumina RNA-Seq technology has been successfully applied to many plant species, such as Beta vulgaris [5], Spartina pectinata [6], Camellia sinensis [7], Lilium lancifolium [8], and Camellia japonica [9], for its high accuracy and sensitivity of gene discovery.
In our study, based on Illumina NGS technology, a fully characterized chrysanthemum transcriptome was represented. Physiological experiments and molecular sequencing analysis combined to explore the cold mechanism of chrysanthemum under chilling and freezing stresses. Moreover, we compare and analyze the physiological and molecular aspects of chrysanthemum with and without cold acclimation to explore the effect of acclimation on chrysanthemum. Lots of cold-induced genes were identified, and some of them were of great importance in cold-tolerance chrysanthemum molecular breeding.
Methods
Plant materials and low temperature treatments
Dendranthema grandiflorum var. jinba was used in this study. The buds raised from tissue-cultured seedlings were grown on MS medium (16 h photoperiod, 25 °C/22 °C day/night temperature) for twenty days. Then twenty-day old chrysanthemum seedlings were transferred to pots filled with a 1:1 mixture of peat and perlite, and acclimated for three days at normal condition. Then the four groups of seedlings were respectively subjected to following treatments: (T01) normal condition as control, (T02) 4 °C for 24 h, (T03) 4 °C for 24 h, followed by − 4 °C for 4 h, (T04) -4 °C for 4 h. Each treatment collected a mixture of three biologically replicated leaves, then samples rapidly frozen with liquid nitrogen and stored at − 80 °C. There were four samples in total used for RNA-Seq and differential expression analyses.
RNA preparation
Total RNA was extracted according to manufacturer’s instructions. 1% agarose gels was used for the monitoring of RNA degradation and contamination; the NanoPhotometer® spectrophotometer (IMPLEN, CA, USA) was used for check of RNA purity; Qubit® RNA Assay Kit was used for measurement of RNA concentration; and the RNA Nano 6000 Assay Kit of the Agilent Bioanalyzer 2100 system (Agilent Technologies, CA, USA) was used for assessing of RNA integrity.
Library preparation for transcriptome sequencing
A total amount of 3 μg RNA per sample was used as input material for the RNA sample preparations. Sequencing libraries were generated using NEBNext®Ultra™ RNA Library Prep Kit for Illumina® (NEB, USA) according to manufacturer’s recommendations. Briefly, mRNA was purified from total RNA by magnetic beads and cut randomly into short fragments by fragmentation buffer. First strand cDNA was synthesized by random hexamer primer and M-MuLV Reverse Transcriptase (RNase H-), using mRNA as a template. Second strand cDNA synthesis was subsequently performed using DNA Polymerase I and RNase H. The cDNA fragments of 150~ 200 bp in length were selected by AMPure XP beads, after end-repair and single nucleotide A (adenine) addition. Then, the cDNA library was obtained by PCR enrichment. At last, PCR products were purified and library quality was assessed on the Agilent Bioanalyzer 2100 system. The library preparations were sequenced on an Illumina Hiseq 4000 platform and paired-end reads were generated.
Transcriptome assembly and gene functional annotation
The clean data were obtained by removing reads containing adapter and low quality reads from raw data. Transcriptome assembly was performed using Trinity [10] based on clean data with high quality. The sequences of unigenes were compared with Nr (NCBI non-redundant), Pfam (Protein family), Swiss-Prot (a manually annotated and reviewed protein sequence database), GO (Gene Ontology), COG (Cluster of orthologous groups), KOG (euKaryotic Orthologous Groups) and eggNOG (evolutionary genealogy of genes: Non-supervised Orthologous Groups) databases by BLAST, and the KEGG (Kyoto Encyclopedia of Genes and Genomes) Orthology results were obtained by comparing with KEGG using KOBAS2.0 [11]. After predicting the amino acid sequence of unigenes, we use HMMER [12] to compare them with Pfam database to get the annotation information of unigenes.
Differential expression analysis
EdgeR program package was used for adjusting read counts of each sequenced library, and DEGseq (2010) R package was used for differential expression analysis of two samples. P value was adjusted using q value [13]. In this study, q value < 0.01 and |log2 (fold change)| > 1 were set as the threshold for significantly differential expression.
Validation of RNA-Seq data by qRT-PCR
Expression of Ten DEGs in chrysanthemum was analyzed by qRT-PCR with three replicates to verify the RNA-Seq data. The qRT-PCR was performed using the SsoFast EvaGreen supermix (Bio-Rad, Hercules, CA, USA) and Bio-Rad CFX96™ detection system. A final 10 μL qRT-PCR reaction mixture contained 5 uL SsoFast EvaGreen supermix, 1 uL diluted cDNA sample, and 150 nM primers. Relative expression levels were calculated by the 2−ΔΔCT method, and the Elongation Factor 1α (EF1α) gene (Forward primer: TTTTGGTATCTGGTCCTGGAG, Reverse primer: CCATTCAAGCGACAGACTCA) was used as a reference for quantitative expression analysis. The primers used in qRT-PCR are listed in Table 1.
Table 1.
Primers of qRT-PCR for validation of the selected DEGs
| Gene Name | Description | Primers |
|---|---|---|
| MEKK1 | Mitogen-activated protein kinase kinase kinase | F:CAATTCGCGGAACACCAATG |
| R:TTGAAACCGGGTCACTAACG | ||
| MYB-like | Myb-like DNA-binding domain | F:TGACCCTGATCCTGTGTTTG |
| R:GTCTACCTCTCCCAATTCATCTG | ||
| NAC90 | NAC domain-containing protein | F:TCCCACGACAAGAGAAACAAG |
| R:TGATCCCAATGGCTCGATTAC | ||
| FAD7 | omega-3 fatty acid desaturase | F:CTCCATTCCTTTCCACGGTAC |
| R:GTTATGGGTCCGCTTCAAATG | ||
| Hsp70 | Heat shock 70 kDa protein | F:CCAGCTCCACCTTGATACATC |
| R:GAAGATTGAGGATGCGATTGATG | ||
| LEA5 | Late embryogenesis abundant protein | F:AGAAAGTGTATCCGGAAGCG |
| R:CGTCAACTTGGCTTGTTTGG | ||
| LEA3 | Late embryogenesis abundant protein | F:GGGAAAGATAAGACAGGTGGG |
| R:GGAGTACCCATACCCAAAGC | ||
| P5CS | pyrroline-5-carboxylate synthetase | F:TTAGCAGGTCTTTGTGGGTG |
| R:GATGGGTGTTGAGAGGTAAAGG | ||
| ABF | ABA responsive element binding factor | F:CACCAAAGACTCCAATTCAACAG |
| R:CAGAGGGAAGAGGAAAGTGTTG | ||
| PP2C | Protein phosphatase 2C | F:TTGTCGCAGTTCTCATACTCC |
| R:TGATACGGCTTGTGGACTTG |
Determination of physiological indexes of chrysanthemum under low temperatures
Leaves of seedlings were used for determinations. Accumulation of H2O2 and O2− was detected by histochemical staining, referring to Wang et al. (2017) [14]. Relative electrolyte conductivity (EC) was also measured following Wang et al. (2017) [14]. Peroxidase (POD) and catalase (CAT) activities were respectively measured following Ranieri et al. (2000) [15] and Zhang et al. (2011) [16]. Proline and soluble sugar contents were determinated following Irigoyen et al. (1992) [17] and Wang et al. (2013) [18]. Each determinations included three biological and technical replicates.
Results
Transcriptome sequencing and assembly
To comprehensively investigate the transcriptome and gene expression profiles of D. grandiflorum under normal and low temperature stress, four cDNA sample from leaves of chrysanthemum seedlings were prepared and sequenced using Illumina HiSeq 4000 platform. An overview of the RNA-Seq reads derived from the four libraries was presented in Table 2. After the low-quality reads were removed, 25.69Gb clean reads were obtained with an average of 6.42Gb (21.5 million) reads for each sample, and the percentage of Q30 base in each sample was not less than 96.33% (Additional file 1: Figure S1).
Table 2.
Overview of the sequencing and assembly
| Sample ID | Clean reads | Clean bases | Q30 | GC content | Mapped reads | Unique match |
|---|---|---|---|---|---|---|
| T01 | 22,802,541 | 6,774,398,078 | 96.33% | 43.62% | 73.40% | 51.43% |
| T02 | 21,839,313 | 6,501,753,240 | 96.52% | 42.93% | 73.77% | 49.89% |
| T03 | 20,478,749 | 6,077,198,988 | 96.78% | 42.90% | 73.10% | 47.49% |
| T04 | 21,323,634 | 6,339,923,198 | 96.68% | 42.88% | 71.71% | 49.06% |
With the Trinity program [10], a total of 199,754 transcripts were obtained from the clean reads with an N50 length of 1321 bp and a mean length of 878.8 bp. Among them, 93,837 unigenes were generated with an average length of 719.19 bp. The length distributions of transcripts and unigenes are given in Table 3. The clean data of each sample was compared with the transcript or unigene library. The proportion of mapped reads per library ranged from 71.71% to 73.77%, and unique mapped reads ranged from 47.49% to 51.43% (Table 2).
Table 3.
Length distribution of the transcripts and unigenes clustered from de novo assembly
| Length range | Transcript | Unigene |
|---|---|---|
| 200–300 | 40,988 (20.52%) | 30,475 |
| 300–500 | 44,661 (22.36%) | 23,753 |
| 500–1000 | 54,466 (27.27%) | 19,857 |
| 1000–2000 | 41,917 (20.98%) | 13,504 |
| 2000+ | 17,722 (8.87%) | 6248 |
| Total number | 199,754 | 93,837 |
| Total length | 175,543,436 | 67,486,480 |
| N50 length | 1321 | 1155 |
| Mean length | 878.8 | 719.19 |
Gene annotation and functional classification
To predict and analyze the function of the unigenes, we carried out functional annotation by using BLAST against multiple databases such as Nr, Swiss-Prot, GO, COG, KOG, KEGG, eggNOG, and Pfam. Of the 93,837 unigenes, 36,462 (38.86%) unigenes were successfully matched to homologous sequences in at least one of databases mentioned above. Among them, 10,226 (10.90%), 19,534 (20.82%), 12,584 (13.41%), 20,400 (21.74%), 24,137 (25.72%), 23,437 (24.98%), 33,201 (35.38%) and 35,370 (37.69%) unigenes were found in COG, GO, KEGG, KOG, Pfam, Swiss-Prot, eggNOG, and Nr databases, respectively. The Nr database produced the largest number of annotations. Compared with other species, chrysanthemum var. jinba showed the most matches to Vitis vinifera (3771), followed by Sesamum indicum (2623) and Coffea canephora (2230) (Fig. 1a).
Fig. 1.
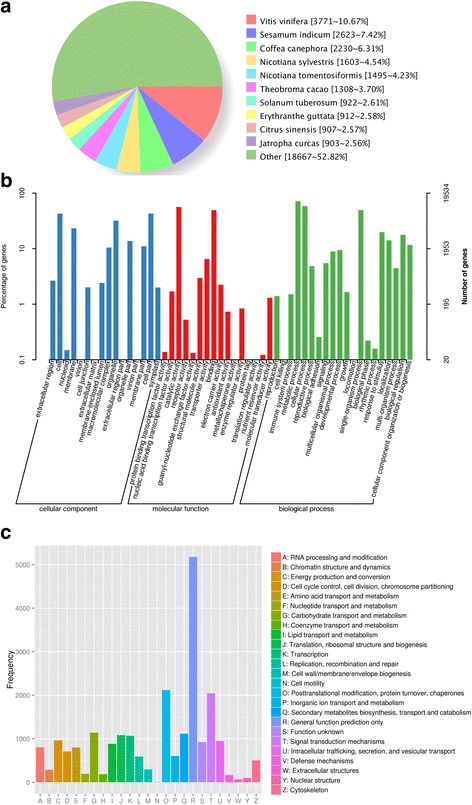
Functional annotation of assembled transcriptome. a Similarity of D. grandiflorum var. Jinba sequences with those of other species. b GO classification of the annotated unigenes. c KOG function classification of consensus sequence
GO assignments system was used to classify the possible functions of chrysanthemum genes. A total of 19,534 unigenes were successfully annotated and classified into 52 functional groups of three major GO categories (Cell Component (CC), Molecular Function (MF), and Biological Processes (BP)) (Fig. 1b). The top three GO terms for classified genes were ‘cell’ (8319), ‘cell part’(8319) and ‘organelle’ (6248) for cell component category; ‘catalytic activity’ (11,009), ‘binding’ (9622) and ‘transporter activity’ (1273) for molecular function; and ‘metabolic process’ (13,917), ‘cellular process’ (11,378) and ‘single-organism process’ (9623) for biological processes. In addition, genes involved in ‘membrane’, ‘response to stimulus’ and ‘biological regulation’ were also highly represent.
In total, 20,400 unigenes were grouped into 25 functional classifications based on KOG database (Fig. 1c). Among these classification, the largest group was ‘general function prediction only’ (5176), followed by ‘posttranslational modification, protein turnover, chaperones’ (2117), ‘signal transduction mechanisms’ (2046), ‘carbohydrate transport and metabolism’ (1140), ‘secondary metabolites biosynthesis, transport and catabolism’ (1115), ‘translation, ribosomal structure and biogenesis’ (1081), and ‘transcription’ (1067).
Analysis of potential differentially expressed genes (DEGs)
The clean reads from each sample were compared to the unigene library, and gene expression levels were estimated using RSEM (RNA-Seq by Expectation Maximization) according to the results of comparison. FPKM (fragments per kilobase of transcript per million fragments mapped) values were used to indicate the expression abundance of unigenes.
From three comparison, including CP1 (T01 vs T02), CP2 (T01 vs T03) and CP3 (T01 vs T04), a large number of DEGs were identified. The number of DEGs detected was as follows: CP1 3028 (1846 up- and 1182 down-regulated), CP2 3906 (2799 and 1107) and CP3 3245 (2270 and 975) (Fig. 2b). We found that more DEGs up regulated in treatment T03 which underwent CA than the treatment T04 without CA. Moreover, the venn diagram was used for analyses of unique and overlapping sets of DEGs in each treatment. As shown in Fig. 2a, there were 641 genes differentially expressed in all comparisons, indicating that these genes involved in the process of plants response to both chilling and freezing stresses, which were of great value for low temperature tolerance study. We also identified 1166, 1086 and 1324 DEGs expressed only in T02, T03 and T04, respectively. Thus it can be seen that there were 1166 genes only playing roles in the process of CA, not working in freezing period, and 1324 genes involved in response to rapid freezing. Of total 6557 DEGs, 4378 genes were successfully annotated with eight databases. The number of annotated DEGs in each treatment is shown in Table 4.
Fig. 2.
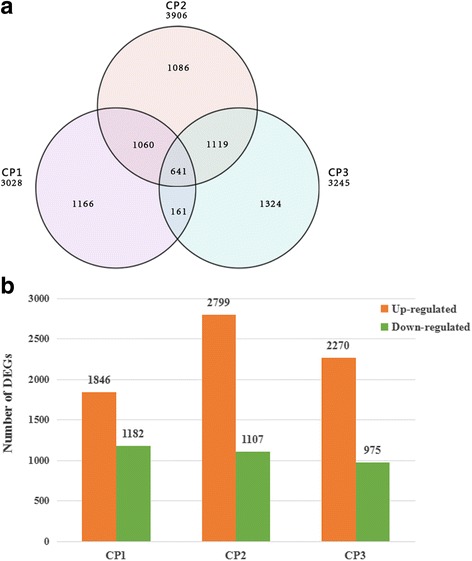
Venn diagram and histogram of DEGs during low temperature stresses. a Venn diagram showing DEGs expressed at each of the three low temperature treatments. b The numbers of DEGs identified in comparisons between pairs of libraries
Table 4.
DEGs annotation of D. grandiflorum var. Jinba
| CP1 | CP2 | CP3 | |
|---|---|---|---|
| Total annotated | 2280 | 2530 | 2166 |
| COG | 813 | 796 | 562 |
| GO | 1335 | 1390 | 1101 |
| KEGG | 827 | 832 | 596 |
| KOG | 1311 | 1332 | 990 |
| Pfam | 1825 | 1966 | 1627 |
| Swiss-Prot | 1654 | 1813 | 1534 |
| eggNOG | 2181 | 2400 | 2040 |
| Nr | 2257 | 2497 | 2140 |
KEGG pathway analysis for DEGs
KEGG annotation was used for DEGs from the above comparisons. In the CP1, 493 DEGs were assigned to the KEGG database involved 108 pathways; for CP2, 519 DEGs were assigned to 113 pathways; and for CP3, 386 DEGs assigned to 105 pathways. The most reliable top ten significantly enriched pathways of DEGs under low temperature treatments were represented as Table 5.
Table 5.
The most reliable top ten significantly enriched pathways of DEGs under low temperature treatments
| Pathway ID | Pathway | DEGs in pathway | All genes in pathway | P-value | |
|---|---|---|---|---|---|
| CP1 | ko03008 | Ribosome biogenesis in eukaryotes | 47 | 137 | 5.59E-13 |
| ko00196 | Photosynthesis - antenna proteins | 20 | 64 | 3.19E-09 | |
| ko00940 | Phenylpropanoid biosynthesis | 38 | 233 | 2.53E-07 | |
| ko00500 | Starch and sucrose metabolism | 42 | 301 | 4.29E-06 | |
| ko00052 | Galactose metabolism | 17 | 104 | 0.0005392 | |
| ko00400 | Phenylalanine, tyrosine and tryptophan biosynthesis | 13 | 78 | 0.0019789 | |
| ko00906 | Carotenoid biosynthesis | 8 | 38 | 0.0032878 | |
| ko00130 | Ubiquinone and other terpenoid-quinone biosynthesis | 10 | 66 | 0.0123906 | |
| ko00603 | Glycosphingolipid biosynthesis - globo series | 3 | 8 | 0.0132652 | |
| ko00360 | Phenylalanine metabolism | 17 | 143 | 0.0156871 | |
| CP2 | ko03008 | Ribosome biogenesis in eukaryotes | 31 | 137 | 4.55E-09 |
| ko00196 | Photosynthesis - antenna proteins | 20 | 64 | 7.76E-09 | |
| ko04626 | Plant-pathogen interaction | 45 | 272 | 5.72E-08 | |
| ko00940 | Phenylpropanoid biosynthesis | 29 | 233 | 0.0020767 | |
| ko00564 | Glycerophospholipid metabolism | 20 | 149 | 0.0042047 | |
| ko00360 | Phenylalanine metabolism | 19 | 143 | 0.0058143 | |
| ko00052 | Galactose metabolism | 15 | 104 | 0.0064176 | |
| ko00500 | Starch and sucrose metabolism | 33 | 301 | 0.0079662 | |
| ko04075 | Plant hormone signal transduction | 38 | 360 | 0.0084879 | |
| ko00740 | Riboflavin metabolism | 3 | 8 | 0.0152681 | |
| CP3 | ko00196 | Photosynthesis - antenna proteins | 28 | 64 | 0 |
| ko04626 | Plant-pathogen interaction | 33 | 272 | 5.61E-06 | |
| ko04075 | Plant hormone signal transduction | 39 | 360 | 1.24E-05 | |
| ko00860 | Porphyrin and chlorophyll metabolism | 14 | 77 | 4.17E-05 | |
| ko04712 | Circadian rhythm - plant | 11 | 55 | 0.0001138 | |
| ko00906 | Carotenoid biosynthesis | 8 | 38 | 0.0006843 | |
| ko00195 | Photosynthesis | 15 | 114 | 0.0009170 | |
| ko00740 | Riboflavin metabolism | 3 | 8 | 0.0067278 | |
| ko00591 | Linoleic acid metabolism | 6 | 36 | 0.0106150 | |
| ko00564 | Glycerophospholipid metabolism | 14 | 149 | 0.0256548 |
Integrate these three comparisons, the major pathways involved in the cold response mechanism were ‘Starch and sucrose metabolism’, ‘Galactose metabolism’, ‘Glycerophospholipid metabolism’, ‘Biosynthesis of unsaturated fatty acids’, ‘Plant hormone signal transduction’ and ‘Plant-pathogen interaction’ (Additional file 1: Figure S2). As shown in Table 6, for first four biological metabolism and synthesis pathways, the number of DEGs in CP1 was more than CP2 or CP3; but for last two signal transduction related pathways, DEGs number of CP1 was less than CP2 or CP3.
Table 6.
DEG statistics in five major pathways
| #Pathways | CP1 | CP2 | CP3 |
|---|---|---|---|
| Starch and sucrose metabolism | 42 | 33 | 22 |
| Galactose metabolism | 17 | 15 | 6 |
| Glycerophospholipid metabolism | 14 | 20 | 14 |
| Biosynthesis of unsaturated fatty acids | 7 | 9 | 1 |
| Plant hormone signal transduction | 29 | 38 | 39 |
| Plant-pathogen interaction | 19 | 45 | 33 |
Protein kinase (PK) and transcription factors (TFs) responding to low temperature
A total of 96, 131, and 129 DEGs encoding protein kinases belonging to 23 major families were found in CP1, CP2 and CP3 treatments, respectively. Among them, 51 (14 up- and 37 down-regulated), 71 (50 and 21) and 80 (71 and 9) receptor-like kinases (RLKs) were identified. And there were 4 (2 up- and 2 down-regulated), 8 (7 and 1) and 10 (10 and 0) genes of CP1, CP2 and CP3 involved in the mitogen-activated protein kinase (MAPK) cascade. We found 3, 12 and 9 DEGs of calcium-dependent protein kinase (CDPK) family and 7, 5 and 4 DEGs of CBL-interacting protein kinase (CIPK) family were all up regulated in CP1, CP2 and CP3. Moreover, we identified 13, 12 and 11 protein phosphatase (PP) differentially expressed in three treatments. The details of these PK and PP genes are shown in Additional file 2: Table S1. These results indicated that protein phosphorylation may be induced by low temperature in chrysanthemum. In addition, it was interesting that there were more up-regulated PK in CP2 and CP3 than CP1 (Fig. 3a).
Fig. 3.
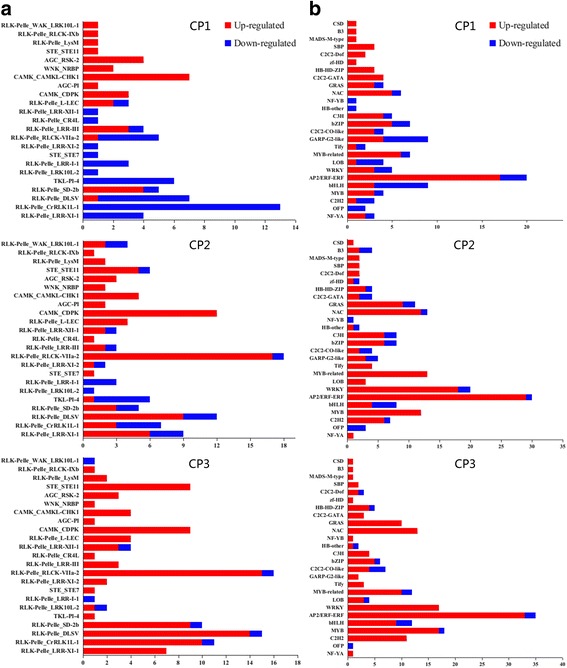
Differentially expressed protein kinases a and transcription factors b responsive to low temperature. Within each bar, number of up- and down-regulated genes is shown in red and blue, respectively
In this study, totally 123 (84 up- and 39 down-regulated), 190 (157 and 33) and 193 (172 and 21) transcription factor (TF) genes were identified in CP1, CP2 and CP3. Obviously, the number of up- regulated expressed transcription TFs in CP2 and CP3 was greater than that of CP1 (Fig. 3b). A large number of TF families were involved in low temperature response. Generally, the major TF families were AP2/ERF, WRKY, bHLH, MYB, MYB-related, and NAC. In treatment CP1, the AP2/ERF family was the largest group (18%), followed by the bHLH family (8%) and the GARP-G2-like family (6%); for CP2, the top three family were AP2/ERF (17%), WRKY (12%) and MYB (7%); and for CP3, the top three were AP2/ERF (20%), WRKY (10%), MYB (10%) (Additional file 1: Figure S3). Of AP2/ERF genes, 17, 29 and 33 were up-regulated in CP1, CP2 and CP3, respectively. And the members of WRKY and MYB families were more active in freezing condition rather than in chilling condition.
Gene related to the membrane system and osmoregulation
The increase of membrane lipids unsaturation level can increase the fluidity of membrane, accordingly improving the cold resistance of plants [19]. In our study, we identified 12 lipid-transfer proteins (LTPs) and 7 fatty acid desaturase (FADs) genes. Among them, 5 LTPs and 3 FADs were up-regulated in CP1, 7 LTPs and 5 FADs up-regulalted in CP2, and only 4 LTPs and 1 FADs were up-regulated in CP3. The details of genes related to the biofilm system are shown in Table 7.
Table 7.
Gene related to the biofilm system in response to low temperature
| Gene ID | CP1 log2FC | CP1 regulated | CP2 log2FC | CP2 regulated | CP3 log2FC | CP3 regulated | Gene description | |
|---|---|---|---|---|---|---|---|---|
| LTPs | c98772.graph_c0 | 2.06 | up | 2.26 | up | 0.38 | normal | Non-specific lipid transfer protein |
| c94507.graph_c0 | 0.43 | normal | −0.12 | normal | 1.13 | up | Non-specific lipid transfer protein | |
| c90293.graph_c0 | 4.24 | up | 4.30 | up | 1.47 | normal | Non-specific lipid-transfer protein-like protein | |
| c86130.graph_c0 | 4.39 | up | 4.66 | up | 1.53 | normal | Lipid transfer-like protein | |
| c96305.graph_c0 | 1.50 | normal | 2.41 | up | 1.28 | normal | Non-specific lipid-transfer protein-like protein | |
| c93927.graph_c1 | 3.17 | normal | 3.48 | up | – | – | Probable non-specific lipid-transfer protein-like protein | |
| c111292.graph_c1 | 3.44 | up | 3.97 | up | 1.36 | up | Non-specific lipid-transfer protein-like protein | |
| c95472.graph_c0 | 0.93 | normal | 1.38 | normal | 2.16 | up | Non-specific lipid-transfer protein | |
| c45235.graph_c0 | 3.55 | up | 2.77 | normal | 3.83 | up | Putative lipid-transfer protein | |
| c91182.graph_c0 | 0.78 | normal | 0.86 | normal | −1.50 | down | Lipid transfer protein EARLI 1 | |
| c92863.graph_c0 | 2.98 | normal | 3.81 | up | – | – | Non-specific lipid-transfer protein-like protein | |
| c95124.graph_c0 | −0.95 | normal | −1.41 | normal | −3.08 | down | Lipid transfer protein EARLI 1 | |
| FADs | c103089.graph_c1 | 3.38 | up | 2.95 | up | −0.09 | normal | Omega-3 fatty acid desaturase |
| c98666.graph_c0 | 3.35 | up | 2.63 | up | 0.11 | normal | Omega-3 fatty acid desaturase | |
| c86611.graph_c0 | 1.84 | up | 1.59 | normal | 0.21 | normal | Omega-3 fatty acid desaturase | |
| c87665.graph_c0 | 1.21 | normal | 2.50 | up | 0.56 | normal | Omega-6 fatty acid desaturase | |
| c87665.graph_c1 | 1.02 | normal | 2.44 | up | 0.88 | normal | Omega-6 fatty acid desaturase | |
| c86525.graph_c0 | 0.64 | normal | 2.02 | up | −0.58 | normal | Omega-6 fatty acid desaturase | |
| c106808.graph_c0 | −0.07 | normal | 0.82 | normal | 1.26 | up | Omega-6 fatty acid desaturase | |
| PLDs | c105153.graph_c0 | 0.03 | normal | 0.95 | normal | 1.10 | up | Phospholipase D alpha 1 |
| c112648.graph_c0 | 3.42 | up | 3.67 | up | 1.43 | up | Phospholipase D alpha 1 | |
| c115302.graph_c0 | 3.50 | up | 4.51 | up | 0.49 | normal | Phospholipase D p1 | |
| c98821.graph_c0 | 2.89 | up | 3.39 | up | 0.97 | normal | Phospholipase D alpha 1 | |
| c114556.graph_c0 | −2.12 | down | −3.02 | down | −0.19 | normal | phospholipase D Z-like | |
| c113287.graph_c0 | −0.20 | normal | 2.22 | up | 0.71 | normal | Phospholipase D beta 1 | |
| SPLA2 | c116204.graph_c1 | −0.83 | normal | 1.78 | normal | 1.59 | up | Triacylglycerol lipase SDP1 |
| c96544.graph_c0 | 1.75 | up | 0.35 | normal | 2.09 | up | Phospholipase A2-alpha | |
| c105515.graph_c0 | 2.17 | up | 1.65 | normal | 0.94 | normal | Patatin-like phospholipase | |
| POD | c96977.graph_c0 | – | – | 3.80 | up | 2.84 | up | Peroxidase N1 |
| c103682.graph_c0 | 4.29 | up | 3.85 | up | 4.27 | up | Peroxidase 17 | |
| c106645.graph_c0 | 3.36 | up | 3.94 | up | 1.70 | normal | Cationic peroxidase 1 | |
| c113544.graph_c0 | −2.75 | down | −0.11 | normal | −0.22 | normal | Peroxidase N1 | |
| c100631.graph_c0 | – | – | 3.97 | up | 4.86 | up | Lignin-forming anionic peroxidase | |
| c114296.graph_c0 | −0.51 | normal | −1.66 | normal | −2.82 | down | Peroxidase 64 | |
| c90653.graph_c0 | −3.06 | down | −0.21 | normal | 0.24 | normal | Peroxidase (Precursor) | |
| c98404.graph_c0 | 0.17 | normal | 0.26 | normal | −2.07 | down | Lignin-forming anionic peroxidase (Precursor) | |
| c99288.graph_c0 | −2.62 | down | −2.34 | down | −0.69 | normal | Peroxidase 12 |
According to the pathway enrichment analysis, ‘starch and sucrose metabolism’ (ko00500), ‘galactose metabolism’ (ko00052), ‘arginine and proline metabolism’ (ko00330) involved plenty of genes that regulating and relieving the osmotic stress caused by cold-evoked dehydration. In the treatment CP1, 7 starch-degrading genes, 4 cellulose-degrading genes, 3 sucrose-phosphate synthase (SPS) genes, 2 trehalose phosphate synthase (TPS) genes, 3 trehalose phosphatase (TPP) genes, 3 galactinol synthase genes, 4 raffinose synthase gene and 1 pyrroline-5-carboxylate synthetase (P5CS) gene were up-regulated; an equivalent order for CP2 were 7, 3, 3, 2, 3, 3, 4 and 1; for CP3 were 4, 1, 0, 0, 2, 1, 3 and 0. Evidently, under freezing conditions, activities of starch and cellulose degradation and above soluble sugars syntheses were decreased in chrysanthemum without cold acclimation (T04). The details of genes related to osmotic regulation system are shown in Table 8.
Table 8.
Gene related to osmotic regulation system in response to low temperature
| Gene ID | CP1 log2FC | CP1 regulated | CP2 log2FC | CP2 regulated | CP3 log2FC | CP3 regulated | Gene description | |
|---|---|---|---|---|---|---|---|---|
| Starch-degrading genes | c106047.graph_c0 | 1.82 | normal | 2.10 | up | 0.57 | normal | Beta-amylase |
| c111660.graph_c0 | 4.57 | up | 4.51 | up | 4.83 | up | Beta-amylase | |
| c98302.graph_c0 | 3.25 | up | 3.61 | up | 1.87 | up | Beta-amylase | |
| c111420.graph_c0 | 2.83 | up | 3.22 | up | 1.81 | up | Beta-amylase | |
| c111420.graph_c1 | 2.83 | up | 3.22 | up | 1.58 | up | Beta-amylase | |
| c102389.graph_c0 | 2.43 | up | 1.99 | up | 0.97 | normal | Probable alpha-amylase | |
| c116071.graph_c0 | 2.01 | up | 1.94 | normal | −0.90 | normal | Alpha-amylase | |
| c116145.graph_c0 | 1.93 | up | 2.06 | up | 0.02 | normal | Alpha-amylase | |
| Cellulose-degrading genes | c111098.graph_c0 | 2.59 | up | 2.32 | up | 1.37 | normal | Beta glucosidase 41 isoform 3 |
| c91317.graph_c0 | 2.74 | up | 2.02 | normal | 0.98 | normal | Raucaffricine-O-beta-D- glucosidase | |
| c108565.graph_c1 | 1.88 | up | 1.43 | normal | 0.26 | normal | lysosomal beta glucosidase-like | |
| c96136.graph_c0 | 2.96 | up | 2.90 | up | 0.33 | normal | Beta-glucosidase 18 | |
| c106631.graph_c0 | −2.07 | down | −1.52 | normal | 0.23 | normal | Strictosidine-O-beta-D- glucosidase | |
| c104722.graph_c1 | −2.20 | down | −2.06 | normal | −0.81 | normal | Probable beta-D-xylosidase 5 | |
| c93973.graph_c0 | −3.35 | down | −1.34 | normal | −0.06 | normal | lysosomal beta glucosidase-like isoform X1 | |
| c98430.graph_c0 | −1.82 | down | −1.98 | normal | −0.44 | normal | Beta-glucosidase 18 | |
| c98430.graph_c1 | −2.07 | down | −2.41 | down | −0.82 | normal | beta-glucosidase 18 isoform X1 | |
| c104480.graph_c0 | 0.12 | normal | 2.23 | up | 0.56 | normal | Probable beta-D-xylosidase 5 | |
| c112478.graph_c0 | 1.49 | normal | 1.12 | normal | 1.12 | up | Beta-glucosidase 11 | |
| c114139.graph_c0 | −1.23 | normal | −1.96 | normal | −1.23 | down | Beta-glucosidase 18 | |
| SPS | c114821.graph_c1 | 3.09 | up | 3.44 | up | 0.34 | normal | sucrose phosphate synthase |
| c114715.graph_c0 | 2.56 | up | 2.48 | up | −0.88 | normal | sucrose phosphate synthase | |
| c114821.graph_c0 | 3.67 | up | 4.00 | up | 0.92 | normal | sucrose phosphate synthase | |
| SS | c103323.graph_c0 | −1.84 | normal | −1.53 | normal | 1.11 | up | Sucrose synthase 3 |
| TPS | c102854.graph_c0 | −4.99 | down | −2.01 | normal | −1.08 | normal | Probable alpha,alpha-trehalose- phosphate synthase |
| c104136.graph_c0 | −4.61 | down | −1.25 | normal | −0.91 | normal | Probable alpha,alpha-trehalose- phosphate synthase |
|
| c115624.graph_c0 | −2.17 | down | −0.23 | normal | −0.63 | normal | trehalose-6-phosphate synthase | |
| c92114.graph_c0 | 2.38 | normal | 3.25 | up | – | – | trehalose-7-phosphate synthase | |
| c112854.graph_c0 | −3.03 | down | −1.39 | normal | −1.34 | down | Probable alpha,alpha-trehalose- phosphate synthase |
|
| c113270.graph_c0 | −2.41 | down | 1.08 | normal | 0.44 | normal | Probable alpha,alpha-trehalose- phosphate synthase |
|
| c93906.graph_c0 | −4.25 | down | −2.27 | normal | −0.82 | normal | Probable alpha,alpha-trehalose- phosphate synthase |
|
| c99330.graph_c0 | 3.86 | up | 3.89 | up | – | – | Alpha,alpha-trehalose- phosphate synthase |
|
| c107046.graph_c1 | −1.62 | normal | −2.72 | down | −0.88 | normal | trehalose 6-phosphate synthase | |
| c108202.graph_c0 | −3.46 | down | 0.56 | normal | −0.17 | normal | Probable alpha,alpha-trehalose- phosphate synthase |
|
| c94647.graph_c0 | 2.41 | up | 1.49 | normal | – | – | Alpha,alpha-trehalose- phosphate synthase |
|
| c86304.graph_c0 | −3.60 | down | −1.52 | normal | −0.57 | normal | Probable alpha,alpha-trehalose- phosphate synthase |
|
| TPP | c110653.graph_c0 | 3.26 | up | 5.87 | up | 5.00 | up | trehalose-phosphate phosphatase |
| c116316.graph_c0 | 1.99 | up | 2.15 | up | 1.00 | normal | trehalose-phosphate phosphatase | |
| c104330.graph_c0 | 3.07 | up | 3.06 | up | 2.73 | up | trehalose-phosphate phosphatase | |
| Raffinose synthase genes | c80434.graph_c0 | 2.58 | up | 3.34 | up | 1.41 | normal | Galactinol synthase |
| c98797.graph_c0 | 4.00 | up | 5.46 | up | 1.90 | up | Galactinol synthase | |
| c81587.graph_c0 | 2.44 | up | 2.55 | up | 0.97 | normal | Galactinol synthase | |
| c115834.graph_c0 | 4.05 | up | 7.49 | up | 4.99 | up | Probable galactinol--sucrose galactosyltransferase | |
| c115556.graph_c0 | −4.49 | down | 1.01 | normal | 1.97 | up | Probable galactinol--sucrose galactosyltransferase | |
| galM | c98143.graph_c0 | −1.78 | down | −1.26 | normal | −0.44 | normal | Aldose 1-epimerase |
| galA | c110075.graph_c0 | −2.00 | down | −2.54 | down | −0.43 | normal | Alpha-galactosidase |
| P5CS | c110924.graph_c0 | 2.76 | up | 2.95 | up | 0.74 | normal | pyrroline-5-carboxylate synthetase |
Moreover, we found a group of cold-related proteins, including cold-regulated (COR), cold shock and heat shock proteins (HSPs), which were invovled in the low temperature stress response process. Among them, HSPs accounted for a large proportion. And we also identified some anti-freezing proteins (AFPs) [7], such as beta-1,3-glucanase proteins (GLPs), thaumatin-like proteins (TLPs), polygalacturonase-inhibiting proteins (PGIPs) and late-embryogenesis-abundant proteins (LEAs). In total, there were 14, 17 and 13 AFPs-related genes up-regulated in CP1, CP2 and CP3, respectively. These genes are shown as Table 9.
Table 9.
Genes encoding cold-related proteins and anti-freezing proteins
| Gene ID | CP1 log2FC | CP1 regulated | CP2 log2FC | CP2 regulated | CP3 log2FC | CP3 regulated | Gene description | |
|---|---|---|---|---|---|---|---|---|
| COR | c98333.graph_c1 | – | – | 4.07 | up | 4.47 | up | putative cold-inducible protein |
| c97739.graph_c0 | 3.41 | up | 4.12 | up | 1.01 | normal | cold-responsive protein | |
| c99607.graph_c0 | 3.15 | up | 1.50 | normal | 3.31 | up | Cold regulated gene 27 | |
| Cold shock proteins | c113528.graph_c0 | 2.83 | up | 2.73 | up | 0.01 | normal | Cold shock protein 1 |
| c94983.graph_c0 | 2.47 | up | 2.52 | up | – | – | Cold shock protein 1 | |
| c99900.graph_c0 | 1.78 | up | 1.19 | normal | 0.37 | normal | Cold shock domain-containing protein 3 | |
| AFPs | c99499.graph_c0 | −1.26 | normal | −2.87 | down | −0.40 | normal | beta-1,3-glucanase |
| c106137.graph_c0 | 0.85 | normal | 2.24 | up | 1.83 | up | beta-1,3-glucanase | |
| c101035.graph_c0 | −2.55 | down | −3.21 | down | 0.55 | normal | chitinase 2-like | |
| c82667.graph_c0 | 4.46 | up | 8.71 | up | 5.59 | up | Thaumatin-like protein | |
| c82207.graph_c0 | −3.00 | down | −1.66 | normal | − 1.40 | normal | Thaumatin-like protein | |
| c107884.graph_c0 | 3.81 | up | 4.24 | up | 2.17 | up | Thaumatin-like protein 1 | |
| c100473.graph_c1 | −2.06 | down | −1.79 | normal | −0.31 | normal | Thaumatin-like protein 1a | |
| c97008.graph_c0 | −3.44 | down | −2.88 | normal | −1.77 | normal | Thaumatin-like protein 1 | |
| c96509.graph_c0 | 5.01 | up | 4.94 | up | 0.41 | normal | polygalacturonase-inhibiting protein 4 | |
| c92070.graph_c0 | 7.56 | up | 8.54 | up | – | – | Late embryogenesis abundant protein | |
| c70366.graph_c0 | 5.37 | up | 5.74 | up | 0.05 | normal | Late embryogenesis abundant protein | |
| c58403.graph_c0 | 4.52 | up | 5.52 | up | 0.24 | normal | Late embryogenesis abundant protein | |
| c105364.graph_c0 | 1.15 | normal | 7.32 | up | 6.28 | up | Late embryogenesis abundant protein | |
| c114597.graph_c0 | 0.27 | normal | 1.97 | up | 2.85 | up | Late embryogenesis abundant protein | |
| c105750.graph_c0 | 2.76 | up | 2.12 | normal | – | – | Late embryogenesis abundant protein | |
| c100124.graph_c0 | 2.77 | up | 2.27 | up | −0.52 | normal | Late embryogenesis abundant protein | |
| c110317.graph_c0 | 1.76 | up | 3.51 | up | 2.82 | up | Late embryogenesis abundant protein | |
| c94818.graph_c0 | – | – | 10.01 | up | 9.79 | up | Late embryogenesis abundant protein | |
| c78821.graph_c0 | 3.23 | up | 7.04 | up | 6.42 | up | Late embryogenesis abundant protein | |
| c102635.graph_c0 | 2.79 | up | 1.32 | normal | −0.25 | normal | Late embryogenesis abundant protein | |
| c108163.graph_c0 | 3.73 | up | 4.59 | up | 2.68 | up | Late embryogenesis abundant protein | |
| c112429.graph_c0 | 1.99 | up | 3.51 | up | 2.45 | up | Late embryogenesis abundant protein | |
| c96108.graph_c0 | 1.09 | normal | 6.29 | up | 5.22 | up | Late embryogenesis abundant protein | |
| c114782.graph_c0 | 0.56 | normal | 1.61 | normal | 2.22 | up | Late embryogenesis abundant protein | |
| c108048.graph_c2 | 6.00 | up | 6.95 | up | 4.39 | up | Late embryogenesis abundant protein | |
| HSPs | c108834.graph_c0 | −0.67 | normal | 4.29 | up | 3.33 | up | Heat shock protein |
| c100907.graph_c0 | 0.01 | normal | 2.85 | up | 2.48 | up | Heat shock protein | |
| c120647.graph_c0 | – | – | – | – | 3.32 | up | Heat shock protein 70 | |
| c104201.graph_c0 | 1.36 | normal | 2.04 | up | −0.07 | normal | Heat shock 70 kDa protein | |
| c113668.graph_c0 | −1.70 | normal | −2.63 | down | −0.89 | normal | Heat shock 70 kDa protein | |
| c97547.graph_c1 | 4.43 | up | 5.32 | up | 1.26 | up | Heat shock 70 kDa protein | |
| c86961.graph_c0 | −0.15 | normal | 2.23 | up | 1.37 | up | Heat shock cognate 70 kDa protein | |
| c82373.graph_c0 | – | – | 3.05 | up | – | – | Heat shock cognate 70 kDa protein | |
| c108184.graph_c0 | 0.93 | normal | 2.56 | up | 1.01 | normal | Heat shock cognate 70 kDa protein | |
| c107360.graph_c0 | 0.74 | normal | 2.23 | up | 1.51 | up | Heat shock factor protein HSF24 | |
| c98633.graph_c0 | 0.03 | normal | 1.28 | normal | 1.10 | up | Heat shock cognate 70 kDa protein | |
| c88835.graph_c0 | −1.57 | normal | 2.03 | up | 1.90 | up | Heat shock cognate 70 kDa protein | |
| c114383.graph_c0 | 0.88 | normal | 3.24 | up | 2.47 | up | Heat shock cognate 70 kDa protein | |
| c114642.graph_c0 | 1.03 | normal | 2.34 | up | −0.50 | normal | Heat shock 70 kDa protein | |
| c106065.graph_c0 | 2.03 | up | 2.13 | normal | 0.51 | normal | Heat shock cognate protein | |
| c112665.graph_c0 | 0.39 | normal | 3.17 | up | 1.91 | up | Heat shock cognate 70 kDa protein | |
| c95742.graph_c0 | 3.89 | up | 3.90 | up | −0.30 | normal | Heat shock 70 kDa protein | |
| c111753.graph_c0 | −0.02 | normal | 2.82 | up | 1.95 | up | Heat shock cognate 70 kDa protein | |
| c81956.graph_c0 | −1.60 | normal | −2.41 | down | −1.84 | down | Stromal 70 kDa heat shock-related protein | |
| c97818.graph_c0 | −2.43 | down | −2.06 | down | −0.93 | normal | 15.4 kDa class V heat shock protein | |
| c91820.graph_c0 | −1.35 | normal | −2.66 | down | −1.93 | down | Stromal 70 kDa heat shock-related protein | |
| c96991.graph_c0 | −2.12 | down | −2.42 | down | −2.23 | down | Stromal 70 kDa heat shock-related protein | |
| c113713.graph_c1 | −0.33 | normal | −1.35 | normal | 1.65 | up | 22.7 kDa class IV heat shock protein | |
| c102754.graph_c0 | 3.00 | up | 3.17 | up | 0.01 | normal | Heat shock cognate protein 80 | |
| c112282.graph_c0 | 4.18 | up | 4.97 | up | 1.27 | up | Heat shock cognate 70 kDa protein | |
| c115107.graph_c2 | 3.11 | up | 3.26 | up | 0.16 | normal | Heat shock protein 90–2 | |
| c102959.graph_c0 | 3.21 | up | 3.31 | up | −0.06 | normal | Heat shock 70 kDa protein | |
| c112881.graph_c0 | 2.46 | up | 2.55 | up | −0.20 | normal | Stromal 70 kDa heat shock-related protein | |
| c58792.graph_c0 | – | – | 4.51 | up | 3.93 | up | Small heat shock protein | |
| c115107.graph_c0 | 1.87 | up | 1.89 | normal | 0.12 | normal | Heat shock protein 90–2 | |
| c109478.graph_c0 | 0.06 | normal | 2.11 | up | 1.02 | normal | Heat shock cognate 70 kDa protein | |
| c99010.graph_c0 | −1.34 | normal | −2.22 | down | −0.64 | normal | 15.7 kDa heat shock protein |
Plant hormone signal transduction pathway
A total of 68 DEGs were invovled in several plant hormone signal transduction pathways, such as auxin, cytokinin (CK), gibberellin (GA), abscisic acid (ABA), ethylelne (ET), brassinosteroid (BR), jasmonic acid (JA) and salicylic acid (SA) in our study. Among them, auxin, ABA, ET and JA pathways were predominatly induced. In the auxin signaling pathway, there were 9, 10 and 9 genes up-regulated in T02, T03 and T04; in the CK pathway, the number of up-regulated genes in three treatments was 4, 3 and 2; in the ET pathway, the number was 2, 2 and 7; and in the JA pathway, the number was 1, 5 and 7. In ABA signaling pathway, totally 4 ABA receptor genes, 1 ABA-responsive element binding factor (ABF) gene, 4 serine/threonine-protein kinase SRK2 (SnRK2) genes, and 6 protein phosphatase 2C (PP2C) genes were differentially expressed during low temperature conditions. Unexpectedly, under the chilling condition, there was no gene differentially expressed in the SA signaling pathway, while one gene up-regulated under freezing conditions. The above results suggested that plant hormone played important roles in response to low temperature stress in chrysanthemum.
Verification of DEGs using qRT-PCR
To further verify the expression levels of genes obtained from Illumina RNA-Seq platform, we have selected ten genes induced by low temperature to perform qRT-PCR, including genes encoding protein kinases, transcription factors, fatty acid desaturase, heat shock protein, late-embryogenesis-abundant protein, pyrroline-5-carboxylate synthetase, ABA-responsive element binding factor and polygalacturonase-inhibiting protein. The results of experiment showed a strong correlation with the RNA-Seq data (Fig. 4).
Fig. 4.
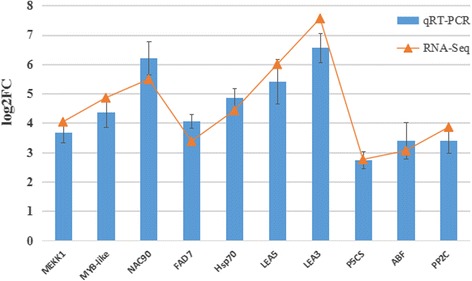
The relative expression levels of ten DEGs identified in the comparison CP1 between RNA-Seq and qRT-PCR. The genes relative expression levels were determined by 2−ΔΔCT as expressed, and were normalized to the expression level of EF1α
Analyses of phenotypic and physiological changes of chrysanthemum at different low temperatures.
As shown in Fig. 5a, the phenotypes of chrysanthemum seedlings under chilling stress (4 °C) was not significantly different from those under normal conditions, but only the youngest leaves of individual seedlings appeared dying. And under freezing stress (− 4 °C), it was clear that the seedlings without prior CA (T04) were more wilted than those with CA (T03). Under low temperature, the membrane permeability of chrysanthemum leaves increased and electrolyte leakage occurred. The degree of electrolyte extravasation was higher in chrysanthemum under freezing conditions than that under the chilling condition (Fig. 5c). And the electrolyte conductivity of T03 was lower than T04, suggesting that CA could increase the tolerance of chrysanthemum to freezing stress.
Fig. 5.
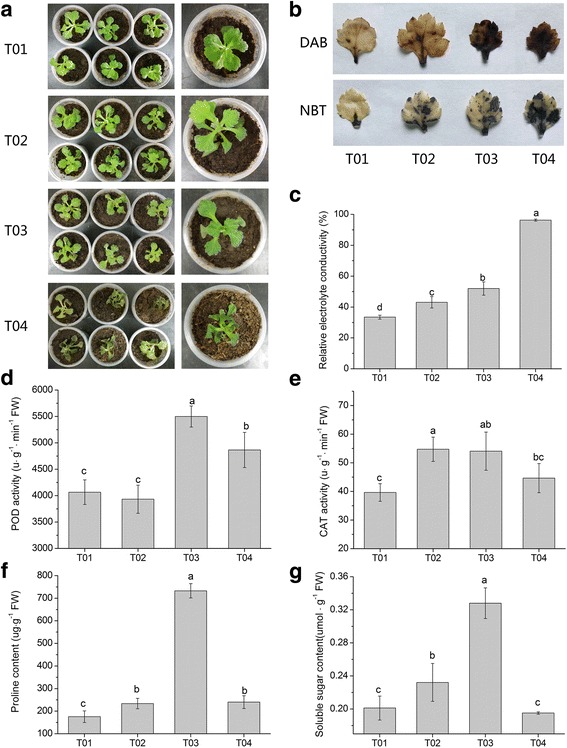
Phenotypic and physiological changes of chrysanthemum at different low temperatures. a Phenotypic comparison of chrysanthemum in four different treatments. b Histochemical staining with DAB and NBT for assessing the accumulation of H2O2 and O2−, respectively, under low temperatures. c-g Analysis of relative electrolyte conductivity c, POD activity d, CAT activity e, proline content f, and soluble sugar g under low temperatures. Data represent means and standard errors of three replicates. The different letters above the columns indicate significant (P < 0.05) differences according to Duncan’s multiple range test
In order to further explore the oxidative damage of chrysantmeum seedlings, we carried out histochemical staining experiments on the leaves. As a whole, chrysanthemum leaves were subjected to more severe oxidative stress under the conditions of freezing than chilling; and under freezing conditions, seedling with prior CA (T03) accumulated less H2O2 and O2− than seedlings without CA (T04) (Fig. 5b). Furthermore, we determined activities of antioxidant enzymes (POD and CAT) in chrysanthemum. Under chilling stress, POD activity of seedlings was not significantly different than normal condition, but CAT activity increased obviously. Under freezing conditions, both POD and CAT activities significantly increased, and activities of T03 were remarkably higher than T04 (Fig. 5d, e).
Moreover, we measured the changes in the osmoregulation substances of chrysanthemum at low temperatures. Under chilling stress, both proline and soluble sugar contents of chrysanthemum were increased. And under freezing condition, proline and soluble sugar contents of T03 were substantially increased, whereas T04 was low in proline and soluble sugar contents (Fig. 5f). The result indicated that CA can induce chrysanthemum osmotic regulation systems to work better under freezing conditions.
Discussion
Low temperature is an important stress factor for plants. It can not only limit the widespread distribution of plants, but also seriously affect the growth and development of them, and cause a series of adverse metabolic reactions [9]. However, ‘jinba’ exactly is a kind of cutting-chrysanthemum which is difficult to mass-produce in winter with normal form. And now, the data available on the molecular basis of chrysanthemum response to cold stress is not limited any longer with the development of high-throughput sequencing.
Based on the quantitative statistics of DEGs in each comparison, we found a lot of genes induced by low temperature and the majority of them involved in up-regulation rather than down-regulation (Fig. 2b). From Fig. 2a, we found that some genes may only be induced by freezing and function on protecting chrysanthemum from freezing damage. As expected, more genes were up-regulated in T03 than T04 (Fig. 2b), supporting the statement that CA process may contribute to plants obtaining increased tolerance to freezing conditions [20].
KEGG analysis
As shown in Table 6, when exposed to chilling conditions, DEGs of chrysanthemum associated with biosynthesis and metabolism pathways were enriched more than freezing conditions, suggesting that chilling stress could induce more biosynthesis and metabolism genes than freezing. However, the pathways of ‘Plant-pathogen interaction’ and ‘Plant hormone signal transduction’ were greater enriched in freezing condition than chilling. Plant-pathogen interaction pathway involves lots of sensing and signaling genes related to Ca2+ signal transducion pathway and MAPK cascade, it demonstrated that more genes participated in the signal sensing and transduction pathways were induced by freezing than chilling stress.
In addition, under freezing stress, whether biosynthesis and metabolism related pathways or signal related pathways in CP2 were greater enriched than CP3. It indicated that CA can relieve the inhibition of biosynthesis and metabolism by freezing injury, and strengthen the response of signal transduction pathway to freezing injury.
Low temperature signal transduction
For surviving in low temperature environment, plants developed a series of complex signaling pathways, and the activities of which are modulated by adding or removing phosphate groups [21]. Lots of protein kinases and protein phosphatases participate in the processes of phosphorylation and de-phosphorylation, such as RLKs, MAPKs, CDPKs, CIPKs, protein phosphatase P (PPP), protein phosphatase M (PPM), and protein tyrosine phosphatase (PTP) [22]. Larges of previous experiments implicated that RLKs work on hormone signaling events in response to environmental stimuli [23–26]. In our experiment, lots of RLKs were found both in all treatments, and up-regulated RLKs in freezing stress were more than in chilling stress (Additional file 2: Table S1). The MAPK cascade activated by reactive oxygen species mediates cold stress signaling in Arabidopsis [27]. Plants over-expressing MKK2 increased freezing tolerance, whereas mkk2 null mutant plants were hypersensitive to cold stress [27]. Overexpression of OsMAPK5 conferred cold tolerance in rice seedlings [28]. We respectively identified 2, 8, 10 MAPKs genes up-regulated in CP1, CP2 and CP3, suggesting that MAPK cascade played a significant role in the low temperature stress, especially in freezing stress in chrysanthemum. In addition, protein phosphatase has been shown to be crucial components of MAPKs cascade [21]. Relevantly, several PPP, PPM and PTP differentially expressed in each treatment were identified. CDPKs and CIPKs are two key kinases of the Ca2+-signaling pathway, which is known to be involved in responses to cold stimuli [29, 30]. And it has been reported that OsCIPK03, OsCIPK12, OsCDPK7 and OsCDPK13 played important roles in conferring cold tolerance in rice [31–33]. In this study, 3, 12, 9 CDPK and 7, 5, 4 CIPK genes were differentially expressed in three treatments, respectively, and all of them were up-regulated. These genes may provide excellent genetic resources for breeding engineering of low-temperature tolerance chrysanthemum.
The different expression of genes involving in the signal cascade mechanism can affect the expression of genes participating in the formation of plant hormones such as ABA, ET, SA and JA. And then, theses hormones may amplify the cascades or initiate some new signaling pathways [34]. In this experiment, ABA, ET and JA are top three responsive hormone signal transduction pathways in chrysanthemum, and there were numbers of DEGs involved in them. ABA is a plant hormone extensively involved in environmental stresses. ABA-responsive element (ABRE) and dehydration-responsive element (DRE) were two cis-acting elements involved in ABA-mediated gene expression [21]. Besides DEGs encoding three key proteins (ABA receptor, PP2C and SnRK2) of ABA signaling pathway were identified, another one ABF and 11 DRE-binding factors (DREB) were obtained in our study (Additional file 3: Table S2). Both ABF and DREB can activate the down-stream genes expression in response to cold stress [35]. This indicated that low temperature-induced expression of chrysanthemum appeared to be ABA dependent.
Lipid molecules are also involved in signal transduction during low temperature stress. Phosphatidic acid (one kind of membranous secondary messenger molecule) can be produced by phospholipase D (PLD) [36]. Li et al. have shown that the cold-induced freezing tolerance of Atpldd T-DNA knock-out mutant Arabidopsis was impaired, whereas that of AtPLDd-overexpression plants was enhanced [37]. In our study, we totally found 6 PLD DEGs, 3, 4 and 2 of which represented up-regulation in CP1, CP2 and CP3, respectively. The up-regulation amplitude of PLD genes in CP2 was higher than CP3. This indicated that PLD was closely related to low-temperature tolerance of chrysanthemum, and CA might contribute to improve freezing tolerance of chrysanthemum.
TFs responding to low temperature
When subjected to low temperature stress, TFs could be activated through a series of signal transduction pathways. Then activated TFs bind specifically to the corresponding cis-acting element to activate the expressions of various downstream resistance-related genes, thereby enhancing the cold tolerance of plants [38]. AP2/ERF, WRKY, MYB-related, MYB, NAC were five major TF classes identified as DEGs in response to low temperature stress. And some members of above TF families have been identified as regulators involved in low temperature stress responses [39–41]. Among various reported TFs, DREBs/CBFs played pivotal roles in improving low temperature tolerance of plants [34]. Overexpression of Arabidopsis DREBs enhanced chilling or freezing tolerance in many plant species, and overexpression DREB1 of several plant species in transgenic Arabidopsis also conferred tolerance to freezing stress [34]. In our study, a total of 10 DREBs were up-regulated under chilling or freezing stress. Interestingly, there were only 3 WRKY and 3 MYB TFs up-regulated in the chilling treatment, whereas respectively about 17 were up-regulated in freezing conditions. It can be inferred that WRKY and MYB TFs may play vital roles in response to the freezing stress. Moreover, some other TF families were also found to be differentially expressed in our study, such as bHLH, bZIP, C2H2, GRAS and C3H. These results suggested that a large number of TFs were involved in the regulation mechanism of chrysanthemum in response to low temperature stress through different pathways.
Cold resistant genes associated with the biofilm system
When the temperature drops, phase changes firstly occurs in the biofilm membrane lipid, which leads to the increase of membrane permeability and the leakage of electrolyte. Physiologically, Fig. 5c showed the chrysanthemum biofilm system suffered more severe persecution under freezing conditions, especially T04 chrysanthemum. Therefore, we explored the molecular mechanisms of biological membranes. Increasing the proportion of unsaturated fatty acids in membrane lipids is propitious to membrane fluidity, thus decreasing the phase changes temperature of the membrane lipids [42]. Therefore, the content of membrane fatty acid and the fluidity of membrane are closely related to the cold resistance of plants. In molecular terms, we found a total of 12 genes encoding LTPs and 7 genes encoding FADs were differentially expressed from three libraries (CP1, CP2, and CP3). Among them, 7 FADs genes included omega-6 and omega-3 fatty acid desaturases genes, which are mainly involved in processes of unsaturated fatty acids (linoleic acid and α-linolenic acid) biosynthesis. And previous reports showed that increasing omega-3 desaturase genes expression could enhanced resistance of different plants to cold stress [43, 44]. Meanwhile, PLD and secretory phospholipase A2 (SPLA2) were also two favorable factors of membrane fluidity [45, 46]. Relevantly, we found numbers of DEGs encoding PLD and SPLA2 in three libraries. These results indicated that chrysanthemum may be able to tolerant low temperatures by enhancing the stabilization of cell membrane. In general, genes encoding LTPs, FADs, PLDs and SPLA2 were obviously much more active in chrysanthemum with CA (T03) than in chrysanthemum without CA (T04) (Table 7), suggesting that chrysanthemum acquired freezing tolerance from CA process.
The damage of cell membrane system under low temperature stress may be related to membrane lipid peroxidation induced by free radical and ROS. Membrane lipid peroxidation product malondialdehyde (MDA) conbined with protein can cause membrane protein denaturation, so membrane lipid peroxidation is also an important reason leading to the decrease of membrane lipid fluidity [47]. Fig. 5b showed that chrysanthemum was subjected to more severe reactive oxygen injury under freezing stress than chilling stress, and T04 was more serious than T03. The increase of contents or activities of protective enzymes is beneficial to maintain the balance between ROS production and removal. Under the same freezing condition, activities of POD and CAT enzymes of T03 was higher than that of T04, and more genes encoding peroxidase represented up-regulated in T03, suggesting that CA process induced chrysanthemum to establish a better protective enzyme system to improve their ability to resist frost damage.
In addition, under low temperature stress, the genes encoding antifreeze proteins and molecular chaperones altered to provide protection to plants. LEAs are known for its aggressive response to dehydration; however, its function in freezing tolerance has been confirmed [48]. A total of 16 LEA protein genes among DEGs were identified in our study. Some cold-responsive genes encoded molecular chaperones like HSP70s and HSP90s. A Brassica napus gene hsp90 contributed to freezing tolerance by stabilizing proteins against freeze-induced denaturation [49]. There were 9, 21 and 14 genes encoding HSPs identified in DEGs of CP1, CP2 and CP3, speculating that HSPs chaperones played important roles in resistance to low temperature stresses in chrysanthemum.
Cold resistant genes associated with osmotic regulation system
Osmotic adjustment is a regulation of plants when dehydration stress evocked by low temperature occurs. As shown in Fig. 5g, low temperature induced the accumulation of soluble sugar in chrysanthemum. Soluble sugar, as an osmotic regulator and an anti dehydrating agent, can reduce cell water potential and enhance water holding capacity [50]. Sugar can provide carbon source and substrate to induce other related cold resistance and physiological and biochemical process, contributing to the enhancement of cold resistance; it also can avoid the protein coagulating caused by the low temperature, to further improve the cold resistance of plant [51, 52]. Significant changes in expressions of the key enzyme genes involved in starch and cellulose degrading, soluble sugars synthesis, and galactose degradation. α−/β-amylase and β-glucosidase were respectively involved in the degradation of starch and cellulose, and the genes encoding these enzymes showed different degrees of rise. Meanwhile, the genes involved in the processes of sucrose, trehalose and raffinose synthesis were also up regulated, which encoding sucrose-phosphate synthase, sucrose synthase, trehalose phosphate synthase, trehalose phosphatase, galactinol synthase, and raffinose synthase. The previous study reported that overexpression of sucrose-phosphate synthase promoted sucrose levels and increased the degree of cold tolerance in transgenic Arabidopsis [53]. What’s more, we found aldose 1-epimerase (galM), which acts on the degradation of galactose, and alpha-galactosidase (galA), which hydrolyze substances containing galactoside bonds, were down regulated in T02 or T03. It’s beneficial to maintain the stability of soluble sugar content.
Low temperature also induced the accumulation of proline in chrysanthemum (Fig. 5f). When plants are subjected to cold stress, proline can not only reduce cell water potential, but also promote protein hydration, thus playing a certain role in cells protection [54]. P5CS, which participates in the synthesis of proline, was only found to be up-regulated in T02 and T03. PRODH, which participates in the degradation of poline, was only represented up-regulated in T04. It suggests that T03 has a better capacity of maintaining high levels of proline than T04, which is one of reasons why T03 is more resistant to freezing than T04.
Conclusion
Our study presents a genome-wide transcript profile of Dendranthema grandiflorum var. jinba, which supported by physiological tests, and provides insights into the molecular mechanisms of D. grandiflorum in response to low temperature. Analyses of physiological and molecular data showed that CA could lead to the establishment of cold resistance mechanism of chrysanthemum, and finally improve the adaptability of chrysanthemum to freezing. And we explored a series of potential chilling-resistant and antifreeze genes associated with low temperature signal transduction, biofilm system and osmotic regulation system, which can serve as candidate genes for cold-tolerance molecular breeding project of chrysanthemum.
Additional files
Figure S1. Composition of raw reads in the four RNA libraries. Figure S2. Functional classification and pathway assignment of DEGs by KEGG. The y-axis indicates the name of the KEGG pathways. The x-axis indicates the percentage of the number of annotated DEGs under the pathway in total number of DEGs in all pathways. Figure S3. Transcription factor families occupied proportion in Dendranthema grandiflorum DEGs. (DOC 1620 kb)
Table S1. Differentially expressed protein kinases and protein phosphatases genes involved in low temperature responses. (XLS 74 kb)
Table S2. Genes involved in ABA signaling pathway and DREBs in response to low temperature. (XLS 27 kb)
Acknowledgments
Funding
The RNAseq analysis was supported by National Natural Science Foundation of China (31770742). The funding body had no role in the design of the study, the interpretation of data and in writing the manuscript.
Availability of data and materials
The raw sequencing data have been submitted to the NCBI Sequence Read Archive database with accession number GSE102413.
Abbreviations
- ABA
Abscisic acid
- ABF
ABA-responsive element binding factor
- ABRE
ABA-responsive element
- AFPs
Anti-freezing proteins
- BP
Biological Processes
- BR
Brassinosteroid
- CA
Cold acclimation
- CAT
Catalase
- CC
Cell Component
- CDPK
Calcium-dependent protein kinase
- CIPK
CBL-interacting protein kinase
- CK
Cytokinin
- COG
Cluster of orthologous groups
- COR
Cold-regulated gene
- DEGs
Differentially expressed genes
- DRE
Dehydration-responsive element
- DREB
DRE-binding factors
- EC
Electrolyte conductivity
- eggNOG
Evolutionary genealogy of genes: Non-supervised Orthologous Groups
- ET
Ethylelne
- FADs
Fatty acid desaturase
- FPKM
Fragments per kilobase of transcript per million fragments mapped
- GA
Gibberellin
- galA
Alpha-galactosidase
- galM
Aldose 1-epimerase
- GLPs
Beta-1,3-glucanase proteins
- GO
Gene Ontology
- HSPs
Heat shock proteins
- JA
Jasmonic acid
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- KOG
euKaryotic Orthologous Groups
- LEAs
Late-embryogenesis-abundant proteins
- LTPs
Lipid-transfer proteins
- MAPK
Mitogen-activated protein kinase
- MDA
Malondialdehyde
- MF
Molecular Function
- NGS
Next-generation sequencing
- Nr
NCBI non-redundant
- P5CS
Pyrroline-5-carboxylate synthetase
- Pfam
Protein family
- PGIPs
Polygalacturonase-inhibiting proteins
- PK
Protein kinase
- PLD
Phospholipase D
- POD
Peroxidase
- PP
Protein phosphatase
- PP2C
Protein phosphatase 2C
- PPM
Protein phosphatase M
- PPP
Protein phosphatase P
- PTP
Protein tyrosine phosphatase
- qRT-PCR
Quantitative real-time polymerase chain reaction
- RLKs
Receptor-like kinases
- ROS
Reactive oxygen species
- RSEM
RNA-Seq by Expectation Maximization
- SA
Salicylic acid
- SnRK2
Serine/threonine-protein kinase
- SPLA2
Phospholipase A2
- SPS
Sucrose-phosphate synthase
- TFs
Transcription factors
- TLPs
Thaumatin-like proteins
- TPP
Trehalose phosphatase
- TPS
Trehalose phosphate synthase
Authors’ contributions
KW and QLL conceived and designed the experiments. KW, ZB, BJ, FZ and QYL performed the experiments. KW, QLL, YP, LZ, YJ and GL analyzed and interpreted the sequence data. KW wrote the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Chrysanthemum materials of this study were provided by plant tissue culture room of Sichuan Agricultural Uniersity. No field permission was necessary to collect the plant samples in this study. This experimental research on chrysanthemum complied with institutional, national, or international guidelines.
Competing interests
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s12864-018-4706-x) contains supplementary material, which is available to authorized users.
Contributor Information
Ke Wang, Email: wkarol@sina.com.
Zhen-yu Bai, Email: nickcutter78@gmail.com.
Qian-yu Liang, Email: 415893783@qq.com.
Qing-lin Liu, Phone: +86-28-86290881, Email: qinglinliu@126.com.
Lei Zhang, Email: Leizhang1985@126.com.
Yuan-zhi Pan, Email: yangliu126126@126.com.
Guang-li Liu, Email: Guangliliu12@126.com.
Bei-bei Jiang, Email: bbj1231@126.com.
Fan Zhang, Email: wuyinhuan@outlook.com.
Yin Jia, Email: jinyin83@126.com.
References
- 1.Han J, Thamilarasan SK, Natarajan S, Park JI, Chung MY, Nou IS. De novo assembly and transcriptome analysis of bulb onion (Allium cepa L.) during cold acclimation using contrasting genotypes. PLoS One. 2016;11:e0161987. doi: 10.1371/journal.pone.0161987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu J, Dong CH, Zhu JK. Interplay between cold-responsive gene regulation, metabolism and RNA processing during plant cold acclimation. Curr Opin Plant Biol. 2007;10:290–295. doi: 10.1016/j.pbi.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 3.Chinnusamy V, Zhu J, Zhu JK. Cold stress regulation of gene expression in plants. Trends Plant Sci. 2007;12:444–451. doi: 10.1016/j.tplants.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 4.Qu Y, Zhou A, Zhang X, Tang H, Liang M, Han H, et al. De novo transcriptome sequencing of low temperature-treated phlox subulata and analysis of the genes involved in cold stress. Int J Mol Sci. 2015;16:9732–9748. doi: 10.3390/ijms16059732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moliterni VMC, Paris R, Onofri C, Orrù L, Cattivelli L, Pacifico D, et al. Early transcriptional changes in Beta vulgaris in response to low temperature. Planta. 2015;242:187–201. doi: 10.1007/s00425-015-2299-z. [DOI] [PubMed] [Google Scholar]
- 6.Nah G, Lee M, Kim DS, Rayburn AL, Voigt T, Lee DK. Transcriptome analysis of spartina pectinata in response to freezing stress. PLoS One. 2016;11:e0152294. doi: 10.1371/journal.pone.0152294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang XC, Zhao QY, Ma CL, Zhang ZH, Cao HL, Kong YM, et al. Global transcripome profiles of Camellia sinensis during cold acclimation. BMC Genomics. 2013;14:415. doi: 10.1186/1471-2164-14-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang J, Yang Y, Liu X, Huang J, Wang Q. Gu J, et al. transcriptome profiling of the cold response and signaling pathways in Lilium lancifolium. BMC Genomics. 2014;15:203. doi: 10.1186/1471-2164-15-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Q, Lei S, Du K, Li L, Pang X, Wang Z, et al. RNA-seq based transcriptomic analysis uncovers α-linolenic acid and jasmonic acid biosynthesis pathways respond to cold acclimation in Camellia japonica. Sci Rep. 2016;6:36463. doi: 10.1038/srep36463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xie C, Mao X, Huang J, Ding Y, Wu J, Dong S, et al. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011;39:W316–W322. doi: 10.1093/nar/gkr483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eddy SR. Profile hidden Markov models. Bioinformatics Italic. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 14.Wang K, Zhong M, Wu YH, Bai ZY, Liang QY, Liu QL, et al. Overexpression of a chrysanthemum transcription factor gene DgNAC1 improves the salinity tolerance in chrysanthemum. Plant Cell Rep. 2017;36:571–581. doi: 10.1007/s00299-017-2103-6. [DOI] [PubMed] [Google Scholar]
- 15.Ranieri A, Petacco F, Castagna A, Soldatini GF. Redox state and peroxidase system in sunflower plants exposed to ozone. Plant Sci. 2000;159:159–167. doi: 10.1016/S0168-9452(00)00352-6. [DOI] [PubMed] [Google Scholar]
- 16.Zhang L, Xi D, Luo L, Meng F, Li Y, Wu CA, et al. Cotton GhMPK2 is involved in multiple signaling pathways and mediates defense responses to pathogen infection and oxidative stress. FEBS J. 2011;278:1367–1378. doi: 10.1111/j.1742-4658.2011.08056.x. [DOI] [PubMed] [Google Scholar]
- 17.Irigoyen JJ, Emerich DW, Sanchez-Diaz M. Water stress induced changes in concentrations of proline and total soluble sugars in nodulated alfalfa (Medicago sativa) plants. Physiol Plant. 1992;84:55–60. doi: 10.1111/j.1399-3054.1992.tb08764.x. [DOI] [Google Scholar]
- 18.Wang C, Deng P, Chen L, Wang X, Ma H, Hu W. A wheat WRKY transcription factor TaWRKY10 confers tolerance to multiple abiotic stresses in transgenic tobacco. PLoS One. 2013;8:e65120. doi: 10.1371/journal.pone.0065120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li B, Takahashi D, Kawamura Y, Uemura M. Comparison of plasma membrane proteomic changes of Arabidopsis suspension-cultured cells (T87 line) after cold and ABA treatment in association with freezing tolerance development. Plant Cell Physiol. 2012;53:543–554. doi: 10.1093/pcp/pcs010. [DOI] [PubMed] [Google Scholar]
- 20.Chen L, Fan J, Hu L, Hu Z, Xie Y, Zhang Y, et al. A transcriptomic analysis of bermudagrass (Cynodon dactylon) provides novel insights into the basis of low temperature tolerance. BMC Plant Biol. 2015;15:216. doi: 10.1186/s12870-015-0598-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pareek A, Sopory SK, Bohnert HJ. Govindjee. Abiotic stress adaptation in plants: physiological, molecular and genomic foundation. Berlin: Springer; 2010. [Google Scholar]
- 22.Denu JM, Stuckey JA, Saper MA, Dixon JE. Form and function in protein dephosphorylation. Cell. 1996;87:361–364. doi: 10.1016/S0092-8674(00)81356-2. [DOI] [PubMed] [Google Scholar]
- 23.Johnson KL, Ingram GC. Sending the right signals: regulating receptor kinase activity. Curr Opin Plant Biol. 2005;8:648–656. doi: 10.1016/j.pbi.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 24.Morris ER, Walker JC. Receptor-like protein kinase: the keys to response. Curr Opin Plant Biol. 2003;6:339–342. doi: 10.1016/S1369-5266(03)00055-4. [DOI] [PubMed] [Google Scholar]
- 25.Tichtinsky G, Vanoosthuyse V, Cock JM, Gaude T. Making inroads into plant receptor kinase signaling pathways. Trends Plant Sci. 2003;8:231–237. doi: 10.1016/S1360-1385(03)00062-1. [DOI] [PubMed] [Google Scholar]
- 26.Haubrick LL, Assmann SM. Brassinosteroids and plant function: some clues, more puzzles. Plant Cell Environ. 2006;29:446–457. doi: 10.1111/j.1365-3040.2005.01481.x. [DOI] [PubMed] [Google Scholar]
- 27.Teige M, Scheikl E, Eulgem T, Doczi R, Ichimura K, Shinozaki K, et al. The MKK2 pathway mediates cold and salt stress signaling in Arabidopsis. Mol Cell. 2004;15:141–152. doi: 10.1016/j.molcel.2004.06.023. [DOI] [PubMed] [Google Scholar]
- 28.Xiong L, Yang Y. Disease resistance and abiotic stress tolerance in rice are inversely modulated by an abscisic acid-inducible mitogen-activated protein kinase. Plant Cell. 2003;15:745–759. doi: 10.1105/tpc.008714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheong YH, Kim KN, Pandey GK, Gupta R, Grant JJ, Luan S. CBL1, a calcium sensor that differentially regulates salt, drought and cold responses in Arabidopsis. Plant Cell. 2003;15:1833–1845. doi: 10.1105/tpc.012393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim KN, Cheong YH, Grant JJ, Pandey GK, Luan S. CIPK3, a calcium sensor-associated protein kinase that regulates abscisic acid and cold signal transduction in Arabidopsis. Plant Cell. 2003;15:411–423. doi: 10.1105/tpc.006858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saijo Y, Hata S, Kyozuka J, Shimamoto K, Izui K. Overexpression of a single Ca2+-dependent protein kinase confers both cold and salt/drought tolerance on rice plants. Plant J. 2000;23:319–327. doi: 10.1046/j.1365-313x.2000.00787.x. [DOI] [PubMed] [Google Scholar]
- 32.Saijo Y, Kinoshita N, Ishiyama K, Hata S, Kyozuka J, Hayakawa T, et al. A Ca2+-dependent protein kinase that endows rice plants with cold- and salt-stress tolerance functions in vascular bundles. Plant Cell Physiol. 2001;42:1228–1233. doi: 10.1093/pcp/pce158. [DOI] [PubMed] [Google Scholar]
- 33.Xiang Y, Huang Y, Xiong L. Characterization of stressresponsive CIPK genes in rice for stress tolerance improvement. Plant Physiol. 2007;144:1416–1428. doi: 10.1104/pp.107.101295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yadav SK. Cold stress tolerance mechanisms in plants. A review. Agron Sustain Dev. 2010;30:515–527. doi: 10.1051/agro/2009050. [DOI] [Google Scholar]
- 35.Thomashow MF. Arabidopsis thaliana as a model for studying mechanisms of plant cold tolerance. Cold Spring Harbor Monograph Archive. 1994;27:807–834. [Google Scholar]
- 36.Meijer HJ, Munnik T. Phospholipid-based signaling in plants. Annu Rev Plant Biol. 2003;54:265–306. doi: 10.1146/annurev.arplant.54.031902.134748. [DOI] [PubMed] [Google Scholar]
- 37.Li W, Li M, Zhang W, Welti R, Wang X. The plasma membrane-bound phospholipase Ddelta enhances freezing tolerance in Arabidopsis thaliana. Nat Biotechnol. 2004;22:427–433. doi: 10.1038/nbt949. [DOI] [PubMed] [Google Scholar]
- 38.Kasuga M, Liu Q, Miura S, Yamaguchishinozaki K, Shinozaki K. Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Novartis Found Symp. 1999;17:287. doi: 10.1038/7036. [DOI] [PubMed] [Google Scholar]
- 39.Hu H, You J, Fang Y, Zhu X, Qi Z, Xiong L. Characterization of transcription factor gene SNAC2 conferring cold and salt tolerance in rice. Plant Mol Biol. 2008;67:169–181. doi: 10.1007/s11103-008-9309-5. [DOI] [PubMed] [Google Scholar]
- 40.Ito Y. Functional analysis of rice DREB1/CBF-type transcription factors involved in cold-responsive gene expression in transgenic rice. Plant Cell Physiol. 2006;47:141–153. doi: 10.1093/pcp/pci230. [DOI] [PubMed] [Google Scholar]
- 41.Vannini C, Locatelli F, Bracale M, Magnani E, Marsoni M, Osnato M, et al. Overexpression of the rice Osmyb4 gene increases chilling and freezing tolerance of Arabidopsis thaliana plants. Plant J. 2004;37:115–127. doi: 10.1046/j.1365-313X.2003.01938.x. [DOI] [PubMed] [Google Scholar]
- 42.Zhou Z, Wang MJ, Zhao ST, Hu JJ, Lu MZ. Changes in freezing tolerance in hybrid poplar caused by up- and down-regulation of PtFAD2 gene expression. Transgenic Res. 2010;19:647–654. doi: 10.1007/s11248-009-9349-x. [DOI] [PubMed] [Google Scholar]
- 43.Domínguez T, Hernández ML, Pennycooke JC, Jiménez P, Martínez-Rivas JM, Sanz C, et al. Increasing ω-3 desaturase expression in tomato results in altered aroma profile and enhanced resistance to cold stress. Plant Physiol. 2010;153:655–665. doi: 10.1104/pp.110.154815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kodama H, Hamada T, Horiguchi G, Nishimura M, Iba K. Genetic enhancement of cold tolerance by expression of a gene for chloroplast [omega]-3 fatty acid desaturase in transgenic tobacco. Plant Physiol. 1994;105:601–605. doi: 10.1104/pp.105.2.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gardiner J, Marc J. Phospholipases may play multiple roles in anisotropic plant cell growth. Protoplasma. 2013;250:391–395. doi: 10.1007/s00709-012-0377-7. [DOI] [PubMed] [Google Scholar]
- 46.Yang X, Sheng W, He Y, Cui J, Haidekker MA, Sun GY, et al. Secretory phospholipase A2 type III enhances alpha-secretase-dependent amyloid precursor protein processing through alterations in membrane fluidity. J Lipid Res. 2010;51:957–966. doi: 10.1194/jlr.M002287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peng XN, Yi ZL, Jiang JX. Progress in the study of cold resistance in plant. Biotechnol Bull. 2007;4:15–18. [Google Scholar]
- 48.Sasaki K, Christov NK, Tsuda S, Imai R. Identification of a novel LEA protein involved in freezing tolerance in wheat. Plant Cell Physiol. 2014;55:136–147. doi: 10.1093/pcp/pct164. [DOI] [PubMed] [Google Scholar]
- 49.Krishna P, Sacco M, Cherutti JF, Hill S. Cold-induced accumulation of hsp 90 transcripts in Brasscia napus. Plant Physiol. 1995;107:915–923. doi: 10.1104/pp.107.3.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ma YY, Xiao X, Zhang WN. Research progress of plants under cold stress. J. Anhui. Agric Sci. 2012;40:7007–7008. [Google Scholar]
- 51.Liu WY, Liu JX, Han B, Zhang R. Determination of reduced sugar content in potatoes. Agriculture & Technology. 2013;1(26):33. [Google Scholar]
- 52.Xu CX. Research progress on the mechanism of improving plant cold hardiness. Acta Ecol Sin. 2012;32:7966–7980. doi: 10.5846/stxb201106260945. [DOI] [Google Scholar]
- 53.Liu WY. Plant stress and genes. Bei Jing: Beijing Institute of Technology Press; 2015. [Google Scholar]
- 54.Liu HM, Wen YH, Wang SK, You Y. A review of plant cold resistance. Northern Horticulture. 2003;6:14–15. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Composition of raw reads in the four RNA libraries. Figure S2. Functional classification and pathway assignment of DEGs by KEGG. The y-axis indicates the name of the KEGG pathways. The x-axis indicates the percentage of the number of annotated DEGs under the pathway in total number of DEGs in all pathways. Figure S3. Transcription factor families occupied proportion in Dendranthema grandiflorum DEGs. (DOC 1620 kb)
Table S1. Differentially expressed protein kinases and protein phosphatases genes involved in low temperature responses. (XLS 74 kb)
Table S2. Genes involved in ABA signaling pathway and DREBs in response to low temperature. (XLS 27 kb)
Data Availability Statement
The raw sequencing data have been submitted to the NCBI Sequence Read Archive database with accession number GSE102413.


