Abstract.
Pyrethroid resistance has been detected in Triatoma infestans (Hemiptera: Reduviidae), which was atributed to target site insensitivity and increased oxidative metabolism of the insecticide by cytochrome P450s. Nicotinamide adenine dinucleotide phosphate (NADPH) cytochrome P450 reductase (CPR) plays an essential role in transferring electrons from NADPH to the P450-substrate complex. In this study, the full length CPR cDNA of T. infestans was isolated and gene expression was determined by quantitative polymerase chain reaction. The open reading frame is 2,046 bp long, encoding a protein of 682 amino acids. Amino acid sequence analysis indicates that the T. infestans CPR and the putative Rhodnius prolixus and Triatoma dimidiata CPRs present conserved ligand-binding domains. Congruent with a previous study of our laboratory, in which the expression of three cytochrome P450 genes (CYP4EM7, CYP3085B1, and CYP3092A6 genes) was induced by deltamethrin, the levels of T. infestans CPR mRNA were upregulated in the fat body of fifth instar nymphs after topical application of deltamethrin. Besides, as it was observed in the CYP4EM7 gene, it was detected overexpression of the CPR gene in the most resistant strain of T. infestans included in the study. These results suggest that CPR plays an essential role in P450-mediated resistance of T. infestans to insecticides.
INTRODUCTION
Chagas disease, also known as American trypanosomiasis, is caused by the parasite Trypanosoma cruzi, which is transmitted to humans by vectors of the subfamily Triatominae (Hemiptera: Reduviidae). The disease and its vectors are extensively distributed from southern United States of America to southern Argentina and Chile (latitude 42°N to latitude 46°S). Chagas disease is a serious public health problem in Latin America, where about eight million people are estimated to be infected with T. cruzi and more than 25 million people are at risk of contracting the infection.1,2
Triatoma infestans is the main vector of Chagas disease in the Southern Cone of Latin America between latitudes 10°S and 46°S, where it is primarily restricted to domestic and peridomestic environments. Pyrethroid insecticides have been the major means to control the vector populations. However, vector control has proven to be difficult because of the variability and extension of endemic areas, and the difficulties to implement sustained entomological vigilance to prevent the recovery of treated bug populations.3 In addition, during the past years, pyrethroid resistance in the vector insect has been reported as one of the main explanations of the unsatisfactory control observed.4 Analyses of genes involved in insecticide resistance in insect vectors of diseases as T. infestans are of considerable interest. Particularly, the importance of studying the expression patterns of genes involved in their resistance lies in its potential for exploitation in novel insect control strategies.
Results of a work focused on detecting the frequency of two knockdown resistance (Kdr) mutations in different T. infestans populations5 suggested that Kdr mutations and other insecticide resistance mechanism as enhanced metabolism and reduced penetrance would be involved in pyrethroid resistance in T. infestans.6,7 The single base-pair substitutions in the sodium channel gene can reduce the binding of pyrethroid insecticides in the target site of the voltage-gated sodium channel on the insect nervous system. However, in some insects as in the malaria vector Anopheles gambiae, pyrethroid resistance has been attributed to a combination of target site insensitivity and increased oxidative metabolism of the insecticide, catalyzed by cytochrome P450s.8
Cytochrome P450 monooxygenases (cytochrome P450s) are involved in the oxidative metabolism of various endogenous and exogenous substrates.9–11 They play a predominant role in the metabolism of insecticides, which often results in the development of insecticide resistance in insect populations.12 Increases of expression of cytochrome P450 genes at transcriptional level are often considered responsible for increasing the metabolism of insecticides and seems to be a common phenomenon in the evolution of resistance development in insects.13–17 In a previous work carried out in our laboratory, the cDNA sequences of three cytochrome P450 genes (CYP4EM7, CYP3085B1, and CYP3092A6) were identified in T. infestans. The mRNA levels of the cytochrome P450 genes identified were obtained from total RNA extracted from pools of fat body collected from individuals of different resistant and susceptible strains of T. infestans, and at different interval times after the topical application of the lethal doses 50% (LD50) of deltamethrin on the ventral abdomen of insects belonging to the different populations analyzed. The expression of the three cytochrome P450 genes isolated was induced by deltamethrin in the susceptible and resistant populations included in this study and overexpression of the CYP4EM7 gene was detected in the most resistant strain of T. infestans.18 The constitutively increased expression and induction of P450s are thought to be responsible for increased levels of detoxification of insecticides in insects and would be involved in the development of insecticide resistance.10,14,17,19–24 The results suggested that the CYP4EM7, CYP3085B1, and CYP3092A6 genes would be involved in the detoxification of deltamethrin and the evolution of resistance to this insecticide in T. infestans populations.
Cytochrome P450s constitute a large superfamily of hem-containing monooxygenases. All P450 monooxygenation reactions occurring in the endoplasmic reticulum require electrons supplied by nicotinamide adenine dinucleotide phosphate (NADPH) cytochrome P450 reductase (CPR), a diflavin enzyme that contains flavin mononucleotide (FMN) and flavin adenine dinucleotide (FAD) cofactors that shuttle electrons from the reduced form of NADPH through a series of redox-coupled reactions to P450.25 In addition to cytochrome P450s, CPR also serves as the electron donor protein for several oxygenase enzymes found in the endoplasmic reticulum of most eukaryotic cells.26–29
Although multiple P450 genes have been found in the genomes of insects, typically only one CPR gene exists in each insect genome.30 Because the identification and analysis of P450 genes involved in insecticide resistance can be complex, it is important to analyze the CPR gene, which encodes for an enzyme essential for the activity of cytochrome P450s and is considered to be a vital part of P450-mediated insecticide resistance. The role of CPR in P450-mediated insecticide metabolism has been shown in some insects, but CPR has not been identified and characterized in T. infestans.31–33
In this study, we report the cDNA nucleotide sequence and deduced amino acid sequence for CPR in T. infestans, and we explore the expression pattern at the mRNA level of the CPR gene at different interval times after the application of deltamethrin. We also perform a comparative analysis of transcriptional expression of this gene in susceptible and resistant strains of T. infestans.
MATERIALS AND METHODS
Insects.
Four laboratory strains of T. infestans were analyzed. The insects from two of these strains were provided by the Centro de Referencia de Vectores of the Servicio Nacional de Chagas de Córdoba (Córdoba province, Argentina). One belonging to a colony susceptible to deltamethrin (Centro de Referencia de Vectores [CRV]-susceptible strain) that was originated in 2006 from insects collected in the locality of Chuña (Department of Ischilín, Córdoba province, Argentina) (30°28′S, 64°40′W) and other belonging to a second generation of a colony resistant to deltamethrin (CRV-resistant strain) that was originated from individuals collected in the locality of Mataral (Department of Santa Cruz, Bolivia) (18°06′S, 64°12′W). The insects from the other two strains were provided by the Centro de Investigaciones de Plagas e Insecticidas (Buenos Aires province, Argentina). They consisted of the first laboratory generation of a colony susceptible to deltamethrin (Centro de Investigaciones de Plagas e Insecticidas [CIP]-susceptible strain) that was originated from insects collected in the locality of Los Quirquinchos (25°07′S, 61°22′W) and of a colony resistant to deltamethrin (CIP-resistant strain) that was originated from specimens collected in the locality of La Esperanza (26°03′S, 60°27′W), both localities belonging to the Department of General Güemes (Chaco province, Argentina). The laboratory colonies were reared at 28°C ± 1°C at a relative humidity of 60–70% with a 6-hour light/18-hour dark cycle and fed once every 2 weeks on restrained chickens.
For the CPR cDNA sequence identification, fifth instar nymphs from the CRV-susceptible strain were fed after 7 days of moulting and their fat bodies were extracted 7 days later. For expression analysis of the CPR gene, fifth instar nymph fat bodies from the CRV-susceptible, CRV-resistant, CIP-susceptible, and CIP-resistant strains were extracted under the same conditions. Each sample was a pool of tissue from three specimens. Besides, fifth instar nymphs from the CRV-susceptible strain were fed after 7 days of moulting and 7 days later was performed a topical application of 1 μL of the LD50 of deltamethrin on the ventral abdomen of insects.18 Subsequently, pools of fat bodies from three fifth instar nymphs were collected at different time intervals after the application of insecticide. For control, topical applications with 1 μL of acetone were performed. In all cases, the topical application was carried out at the same time of day, and the tissue was dissected under aseptic conditions and stored in liquid nitrogen until used for RNA extraction.
Isolation of total RNA and cDNA synthesis.
Extraction of total RNA and cDNA synthesis were carried out as indicated in previous works.18,34–36 For CPR cDNA identification, total RNA was extracted from pools of fat bodies from five fifth instar nymphs and for quantitative polymerase chain reaction (qPCR), total RNA was isolated from pools of fat bodies from three fifth instar nymphs.
Amplification and sequencing of CPR gene.
Partial CPR cDNA sequence was amplified by the polymerase chain reaction (PCR), using unspecific primers (F3CPR and R3CPR) (Table 1) designed from the CPR gene coding region that was identified in the genome of Rhodnius prolixus (GenBank accession number ACPB03018078.1). The PCR was performed as described elsewhere.18,34–36 To obtain the complete open reading frame (ORF) of CPR, the rapid amplification of cDNA ends (RACE) method was performed by using the commercial kit ExactSTART Eukaryotic mRNA 5′- & 3′-RACE (Epicentre, Madison, WI).34 Specific primers for isolation of 3′ cDNA end and 5′ cDNA end are shown in Table 1. After the PCR products were electrophoresed, bands corresponding to the expected sizes were excised from the agarose gel and purified using QIAquick PCR Purification Kit (QIAGEN, Hilden, Germany). The PCR products were then cloned into the pCR4-TOPO TA cloning vector (Invitrogen, Carlsbad, CA) before being sequenced in an ABI 3130XL automated DNA sequencer (Applied Biosystems, Foster City, CA).
Table 1.
Sequences of all primers used in this study
| Name | Sequences | Function |
|---|---|---|
| Unspecific primers (for RT-PCR) | ||
| F3CPR | 5′-GACAATTGAAGGATCTAAAATGCG-3′ | Sense |
| R3CPR | 5′-CTTTCTGCATCCAAAATATAGG-3′ | Antisense |
| RACE primers (for 3′ cDNA isolation) | ||
| CPR3F | 5′-GTGTGGCTACATCATGGTTAGC-3′ | Sense |
| CPR3F1 | 5′-GACCTGGTACAGGTTTAGCTCC-3′ | Sense |
| PCR-Primer2 | 5′-TAGACTTAGAAATTAATACGACTCACTATAGGCGCGCCACCG-3′ | Antisense |
| RACE primers (for 5′ cDNA isolation) | ||
| PCR-Primer1 | 5′-TCATACACATACGATTTAGGTGACACTATAGAGCGGCCGCCTGCAGGAAA-3′ | Sense |
| CPR5R | 5′-TTAACTCCGAGTAGCGTGCC-3′ | Antisense |
| CPR5RN | 5′-CCGAGTAGCGTGCCAAACT-3′ | Antisense |
| Cloning | ||
| M13F | 5′-GTAAAACGACGGCCAG-3′ | Sense |
| M13R | 5′-CAGGAAACAGCTATGAC-3′ | Antisense |
| qPCR primers (for real time PCR) | ||
| CPRRTF | 5′-AACACAGATGAGGATTCGAGTAAAAA-3′ | Sense |
| CPRRTR | 5′-GTGTGCGTGGATTGGATGTTAT-3′ | Antisense |
| QβactinaF | 5′-CCCCTTTCAGTGAGGATCTTCA-3′ | Sense |
| QβactinaR | 5′-CGCCATCCTTCGATTGGA-3′ | Antisense |
PCR = polymerase chain reaction; qPCR = quantitative PCR; RACE = rapid amplification of cDNA end; RT-PCR = reverse transcription PCR.
cDNA sequence analysis.
The CPR cDNA sequence of T. infestans was compared with those of other insects deposited in GenBank using the “BLAST-N” tool available on the National Center for Biotechnology Information (NCBI) website (http://www.ncbi.nlm.nhi.gov) and the program Clustal W2 (http://www.ebi.ac.uk/Tools/clustalw2/index.html). The T. infestans amino acid CPR sequence was deduced from the corresponding cDNA using the translation tool from the ExPASy Proteomics website (http://www.expasy.org/tools/dna.html).
Phylogenetic relationship.
Phylogenetic analysis was conducted to investigate evolutionary relationships among the putative CPR protein identified in T. infestans and other selected sequences reported in the literature from Anopheles funestus (ABO77954.1), A. gambiae (AAO24765.1), Apis mellifera (XP_006569769.1, XP_016769012.1), Atta cephalotes (XP_012055902.1), Bactrocera dorsalis (NP_001306717.1), Bemisia tabaci (AGT15701.1), Bombus impatiens (XP_012246140.1, XP_012246141.1, XP_012246143.1), Bombus terrestris (XP_012173161.1), Cerapachys biroi (EZA61651.1), Cimex lectularius (NP_001303631.1), Culex quinquefasciatus (XP_001865801.1), Dendroctonus armandi (ALC78506.1), Dendroctonus ponderosae (AFI45002.1), Drosophila melanogaster (NP_477158.1, NP_723173.1, NP_001260128.1, NP_001260129.1), Drosophila mettleri (AAB48964.1), Fopius arisanus (XP_011306347.1, XP_011306348.1), Helicoverpa armigera (ADK25060.1), Laodelphax striatella (AHM93010.1), Monomorium pharaonis (XP_012541364.1), Musca domestica (NP_001273818.1), Nilaparvata lugens (AHB59865.1), Panonychus citri (AHZ12899.1), Papilio polytes (XP_013146229.1, XP_013146230.1), Papilio xuthus (XP_013182134.1), Pogonomyrmex barbatus (XP_011643152.1, XP_011643153.1), Spodoptera exigua (ADX95746.1), Spodoptera littoralis (AFP20584.1), Tenebrio molitor (AKZ17715.1), and Tribolium castaneum (XP_008197839.1). Multiple alignment of sequences was performed using the multiple alignment program Clustal W in MEGA version 6.06.37 Tree construction was performed using the maximum likelihood method, using MEGA version 6.06 software.37 The reliability of the trees was tested by the bootstrap procedure with 1,000 replications.
qPCR.
The transcript levels of the CPR gene identified in T. infestans were measured by using qPCR as described in previous works carried out in our laboratory.18,34–36 Specific primers (Table 1) and a Taqman probe were designed from the CPR cDNA sequence using Primer Express program (Applied Biosystems). The reaction conditions were of 15 minutes at 95°C, and 40 cycles of 15 seconds at 95°C and 60 seconds at 60°C. The relative copy number of CPR mRNA was calculated according to 2−ΔΔCT.38 The threshold cycle value difference ΔCT between CPR mRNA and β-actin mRNA of each reaction was used to normalize the level of total RNA.
Statistical analyses.
For the studies involving gene expression analysis by qPCR, two independent experiments were performed and data for each point were registered by triplicate to account for intra-experimental variation. Graphs and statistical tests were performed using GraphPad Prism version 5.00 for Windows (GraphPad Software, San Diego, CA). One way analysis of variance with Bonferroni posttest was used for comparisons. The results were presented as mean ± standard deviation and a P value < 0.05 was considered statistically significant.
RESULTS AND DISCUSSION
Identification and sequence analysis of CPR gene.
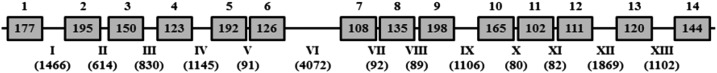
A fragment of 746 bp of CPR cDNA of T. infestans was amplified by reverse transcription PCR using unspecific primers designed from the CPR gene coding region that was identified in the genome of R. prolixus (GenBank accession number ACPB03018078.1). The procedure of rapid amplification of 3′ cDNA end (3′-RACE) allowed obtaining a segment of 452 bp, which overlapped with that fragment and contained the 3′ cDNA end of the gene. Subsequent rapid amplification of cDNA 5′ end (5′-RACE) resulted in a segment of 1,040 bp, which overlapped with the fragment of 746 bp and contained the 5′ cDNA end of the CPR gene. The comparative analysis of the cDNA fragments of T. infestans with cDNA sequences of the CPR gene of other insect species revealed that they were part of the CPR gene. A total of 2,049 bp of cDNA corresponding to CPR of T. infestans was sequenced, which comprises an ORF of 2,046 nucleotides encoding a protein of 682 amino acids and the stop codon thymine, adenine, and adenine (GenBank accession number KY350179). This cDNA allowed to identify in the genome of R. prolixus, in the reverse complementary chain between the nucleotide positions 12,036 and 26,719 of the Scaffold341 (GenBank accession number KQ034397.1), a total of 14,687 bp corresponding to the CPR gene that involve 14 exons encoding a protein of 682 amino acids, 13 introns, and the stop codon TAA (Figure 1). Moreover, in the present work, the deduced 682 amino acid sequence from the Transcriptome Shotgun Assembly of Triatoma dimidiata (GenBank accession number GECL01003769.1) identified as a “putative NADP-dependent flavoprotein reductase” (GenBank accession number JAP02355.1) was recognized as T. dimidiata CPR.
Figure 1.
Schematic representation of the CPR gene in Rhodnius prolixus. Exons are shown as shaded boxes and introns as horizontal lines. Exons and introns are labeled (5′ to 3′) with arabic and roman numerals, respectively. Their sizes in base pairs are indicated in the box (exons) and in parentheses (introns).
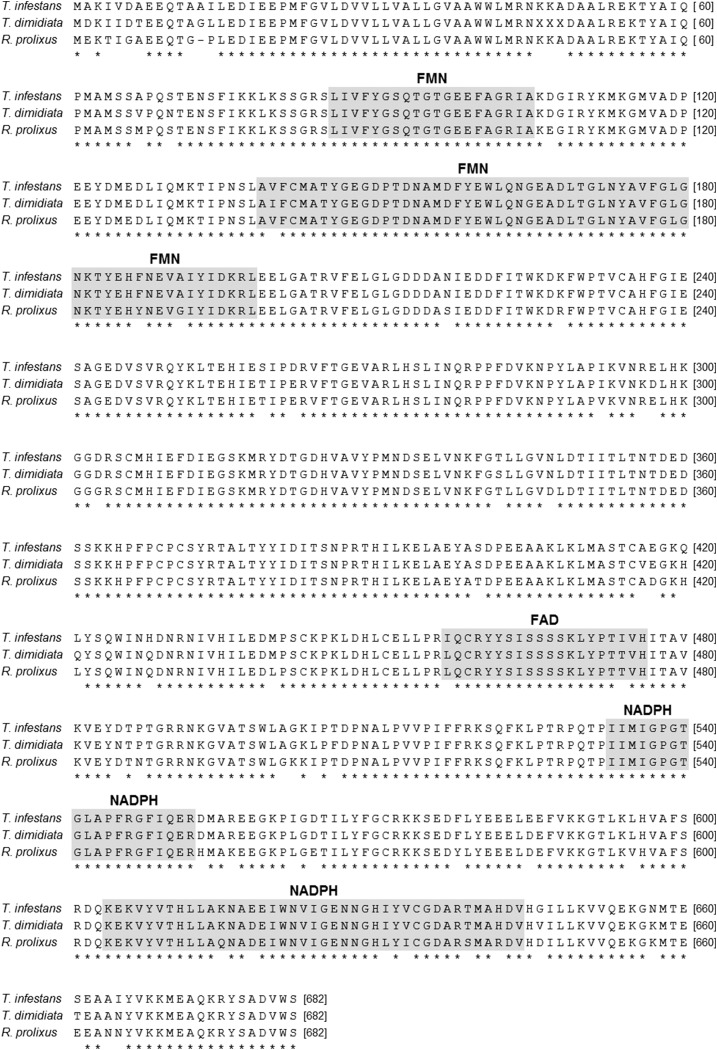
The alignment of the deduced amino acid sequences from CPR cDNA of the three Chagas disease vectors of the subfamily Triatominae (T. infestans, T. dimidiata, and R. prolixus) is shown in the Figure 2. All functional domains involved in the binding of cofactors FMN, FAD, and NADPH were identified in the predicted CPR protein primary structure.
Figure 2.
Alignment of amino acid sequences of cytochrome P450 reductases from Triatoma infestans, Triatoma dimidiata, and Rhodnius prolixus. Identical residues among the three sequences are marked by asterisks. The residues constituting the flavin mononucleotide (FMN), flavin adenine dinucleotide (FAD), and nicotinamide adenine dinucleotide phosphate (NADPH) binding sites are boxed.
The deduced amino acid sequence of CPR from T. infestans, T. dimidiata, and R. prolixus was compared with the corresponding sequences of other hemipteran CPRs. Sequence comparisons revealed that T. infestans CPR was most similar to CPR from T. dimidiata (95.31% identity), followed by R. prolixus (93.24%), C. lectularius (82.92%), L. striatella (77.12%), N. lugens (76.79%), and B. tabaci (74.44%).
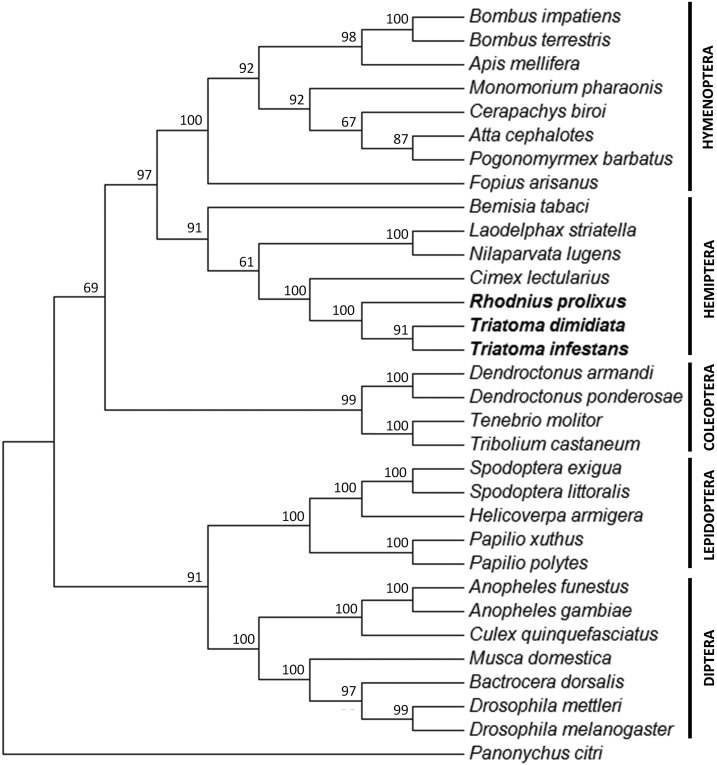
A phylogenetic analysis was performed based on the complete amino acid sequences of CPR of the hemipteran T. infestans, T. dimidiata, R. prolixus, C. lectularius, L. striatella, N. lugens, and B. tabaci, and of 24 representative species of the orders Diptera, Lepidoptera, Coleoptera, and Hymenoptera. As expected, insect CPRs from the same insect order were grouped together with significant bootstrap support (Figure 3). Within Hemiptera, T. infestans clustered with T. dimidiata with high bootstrap support (91%), as well as these two species with R. prolixus (100% bootstrap value). These three members of the subfamily Triatominae clustered with strong support with C. lectularius (100% bootstrap value), and N. lugens was also closely related to L. striatella (100% bootstrap value).
Figure 3.
Maximum likelihood phylogeny of Triatoma infestans, Triatoma dimidiata, and Rhodnius prolixus cytochrome P450 reductases (CPRs) deduced amino acid sequences (in bold), selected CPRs from other hemipteran species, and representative species of the orders Diptera, Lepidoptera, Coleoptera, and Hymenoptera. Bootstrap values next to nodes represent the percentage of 1,000 replicate trees that preserved the corresponding clade.
CPR gene expression.
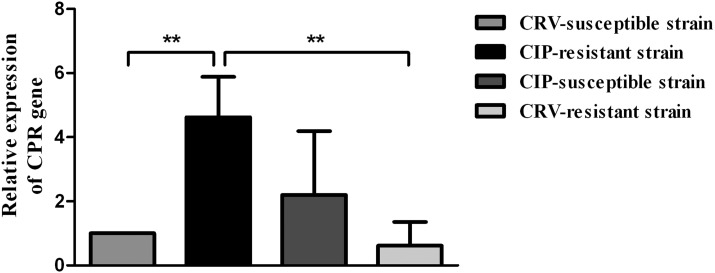
The expression at transcriptional level of the CPR gene was analyzed in two deltamethrin susceptible strains (CRV-susceptible and CIP-susceptible strains) and two deltamethrin-resistant strains (CRV-resistant and CIP-resistant strains). The CRV-resistant strain, which was originated from individuals collected in Mataral (Department of Santa Cruz) showed a lower resistance ratio (RR = 17.38) than the CIP-resistant strain (RR = 233.42) that was originated from specimens of La Esperanza (Department of General Güemes).39,40 Comparative analysis of transcriptional expression of the CPR gene of groups of fifth instar nymphs from the different strains considered for this study revealed that the mRNA levels of the CPR gene in individuals of the CIP-resistant strain were significantly higher than in insects of the CRV-susceptible and CRV-resistant strains (Figure 4). This result agrees with a recent finding in our laboratory of a high level of constitutive expression of a cytochrome P450 gene (CYP4EM7) in the CIP-resistant strain.18 However, the highly resistant individuals from the CIP-resistant strain did not show mRNA levels of the CPR gene significantly higher than in insects from the CIP-susceptible strain. The CIP-susceptible and CIP-resistant colonies were originated from specimens collected in two localities of the same geographical area, Los Quirquinchos and La Esperanza (Department of General Güemes), respectively. The constitutive expression of the CPR gene detected in the CIP-susceptible and CIP-resistant strains agrees with the high level of constitutive expression of the CYP4EM7 gene observed in both strains18 and would support the hypothesis stating that in this area could exist naturally tolerant populations to pyrethroids, explaining the presence of many localities with very low susceptibility.4
Figure 4.
Relative expression (mRNA) of the CPR gene in the fat body of Triatoma infestans nymphs from the CRV-susceptible, CRV-resistant, CIP-susceptible, and CIP-resistant strains. The error bars represent the standard deviation of the mean. Two asterisks indicate significant difference between the mean of insects from the CIP-resistant strain and the mean of the CRV-susceptible and CRV-resistant strains at P < 0.01.
Although substantially less than in the CIP-resistant strain, resistance to deltamethrin was detected in the CRV-resistant strain originated from specimens collected in the locality of Mataral (Bolivia). However, the transcript level of the CPR gene detected in individuals of the CRV-resistant strain was not significantly higher than in individuals of the CRV-susceptible strain (Figure 4). Therefore, as it was also observed for the CYP4EM7 gene, the constitutive expression of the CPR gene in the CRV-resistant strain was not significant.18 This may arise in response to different insecticides applied in the places of origin of each population and/or genetic variations between Argentinian and Bolivian populations of T. infestans. Vector control of Chagas disease in Argentina is mainly based on the application of deltamethrin, whereas in Bolivia other pyrethroids are more frequently used. Differences in treatments with insecticides could have selected different mechanisms that confer resistance in the insect vector.4
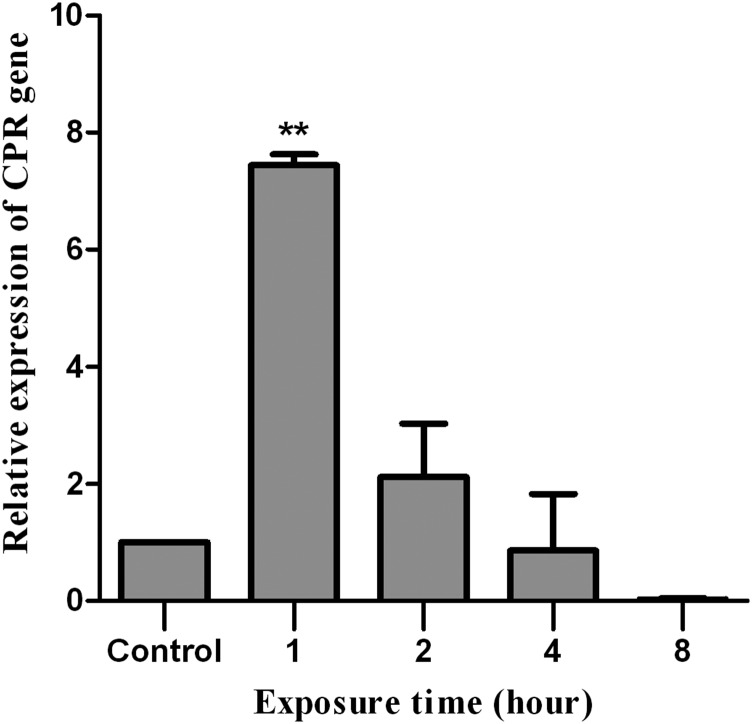
The mRNA levels of the CPR gene were also determined at different interval times after the application of deltamethrin in fifth instar nymphs from the CRV-susceptible strain (Figure 5). The results show that the levels of CPR mRNAs were increased significantly (7-fold) 1 hour after insecticide application in relation to those detected in individuals not exposed to deltamethrin (control). Coincidently, the expression at transcriptional level of the three cytochrome P450 genes previously isolated (CYP4EM7, CYP3085B1, and CYP3092A6) was induced by deltamethrin in the CRV-susceptible, CRV-resistant, CIP-susceptible, and CIP-resistant strains.18 Such an induction could lead to an elevated tolerance to these insecticides, which consequently contributes to a difficulty in controlling T. infestans populations in the field. Because the CPR gene encodes for an enzyme essential for the activity of cytochrome P450s and considered to be a vital part of P450-mediated insecticide resistance, the overexpression of the CPR gene detected in the CIP-resistant strain and the deltamethrin-inducibility of that gene in the CRV-susceptible strain is consistent with the hypothesis that postulates the involvement of P450 genes in the development of resistance to pyrethroid insecticide in T. infestans.
Figure 5.
Relative expression (mRNA) of the CPR gene in the fat body of Triatoma infestans nymphs of the CRV-susceptible strain at different times after treatment with deltamethrin at LD50 concentration. The error bars represent the standard deviation of the mean. Two asterisks on the standard error bar indicate significant difference between the mean of the treatment with the insecticide and the mean of the control at P < 0.01.
CONCLUSIONS
This study is the first report of the CPR gene in triatomines. Consistent with the overexpression of the CYP4EM7 gene in the CIP-resistant strain of T. infestans and the deltamethrin-inducibility of the CYP4EM7, CYP3085B1, and CYP3092A6 genes in all the strains analyzed in a previous work,18 it was observed overexpression of the CPR gene in the CIP-resistant strain and induction of that gene in the CRV-susceptible strain. These results suggest that the P450-mediated metabolism detoxification of xenobiotics might be an important mechanism for deltamethrin resistance in T. infestans.
Acknowledgments:
We thank the Centro de Referencia de Vectores, Servicio Nacional de Chagas de Córdoba (Córdoba, Argentina) and the Centro de Investigaciones de Plagas e Insecticidas (Buenos Aires, Argentina) for providing insects used in our studies.
REFERENCES
- 1.Rassi A, Rassi A, Jr, Marin-Neto JA, 2010. Chagas disease. Lancet 375: 1388–1402. [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization , 2014. Chagas Disease (American trypanosomiasis) World Health Organ Fact Sheet 340 Available at: http://who.int/mediacentre/factsheets/fs340/en/.
- 3.Tarleton R, Gürtler RE, Urbina JA, Ramsey J, Viotti R, 2014. Chagas disease and the London declaration on neglected tropical diseases. PLoS Negl Trop Dis 8: e3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mougabure-Cueto G, Picollo MI, 2015. Insecticide resistance in vector Chagas disease: evolution, mechanisms and management. Acta Trop 149: 70–85. [DOI] [PubMed] [Google Scholar]
- 5.Sierra I, Capriotti N, Fronza G, Mougabure-Cueto G, Ons S, 2016. Kdr mutations in Triatoma infestans from the Gran Chaco are distributed in two differentiated foci: implications for pyrethroid resistance management. Acta Trop 158: 208–213. [DOI] [PubMed] [Google Scholar]
- 6.Santo Orihuela PL, Vassena CV, Zerba EN, Picollo MI, 2008. Relative contribution of monooxygenase and esterase to pyrethroid resistance in Triatoma infestans (Hemiptera: Reduviidae) from Argentina and Bolivia. J Med Entomol 45: 298–306. [DOI] [PubMed] [Google Scholar]
- 7.Pedrini N, Mijailovsky SJ, Girotti JR, Stariolo R, Cardozo RM, Gentile A, Juárez MP, 2009. Control of pyrethroid-resistant Chagas disease vectors with entomopathogenic fungi. PLoS Negl Trop Dis 3: e434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nikou D, Ranson H, Hemingway J, 2003. An adult-specific CYP6 P450 gene is overexpressed in a pyrethroid-resistant strain of the malaria vector, Anopheles gambiae. Gene 318: 91–102. [DOI] [PubMed] [Google Scholar]
- 9.Gilbert LI, 2004. Halloween genes encode P450 enzymes that mediate steroid hormone biosynthesis in Drosophila melanogaster. Mol Cell Endocrinol 215: 1–10. [DOI] [PubMed] [Google Scholar]
- 10.Feyereisen R, 2005. Insect cytochrome P450. Gilbert LI, Latrou K, Gill SS, eds. Comprehensive Molecular Insect Science, Vol. 4. Oxford, United Kingdom: Elsevier, 1–77. [Google Scholar]
- 11.Rewitz KF, O’Connor MB, Gilbert LI, 2007. Molecular evolution of the insect Halloween family of cytochrome P450s: phylogeny, gene organization and functional conservation. Insect Biochem Mol Biol 37: 741–753. [DOI] [PubMed] [Google Scholar]
- 12.Zhou XJ, Ma CX, Li M, Sheng CF, Liu HX, Qiu XH, 2010. CYP9A12 and CYP9A17 in the cotton bollworm Helicoverpa armigera: sequence similarity, expression profile and xenobiotic response. Pest Manag Sci 66: 65–73. [DOI] [PubMed] [Google Scholar]
- 13.Carino FA, Koener JP, Plapp FW, Jr, Feyereisen R, 1992. Expression of the cytochrome P450 gene CYP6A1 in the housefly, Musca domestica Mullin CA, Scott JG, eds. Molecular Mechanisms of Insecticide Resistance: Diversity among Insects, Vol. 5. ACS Symposium Series. Washington, DC: ACS, 31–40. [Google Scholar]
- 14.Carino FA, Koener JF, Plapp FW, Jr, Feyereisen R, 1994. Constitutive over expression of the cytochrome P450 gene CYP6A1 in a house fly strain with metabolic resistance to insecticides. Insect Biochem Mol Biol 24: 411–418. [DOI] [PubMed] [Google Scholar]
- 15.Liu N, Scott JG, 1997. Phenobarbital induction of CYP6D1 is due to a trans acting factor on autosome 2 in house flies, Musca domestica. Insect Mol Biol 6: 77–81. [DOI] [PubMed] [Google Scholar]
- 16.Liu N, Scott JG, 1998. Increased transcription of CYP6D1 causes cytochrome P450-mediated insecticide resistance in house fly. Insect Mol Biol 28: 531–535. [DOI] [PubMed] [Google Scholar]
- 17.Li X, Schuler MA, Berenbaum MR, 2007. Molecular mechanisms of metabolic resistance to synthetic and natural xenobiotics. Annu Rev Entomol 52: 231–253. [DOI] [PubMed] [Google Scholar]
- 18.Grosso CG, Blariza MJ, Mougabure-Cueto G, Picollo MI, García BA, 2016. Identification of three cytochrome P450 genes in the Chagas’ disease vector Triatoma infestans: expression analysis in deltamethrin susceptible and resistant populations. Infect Genet Evol 44: 459–470. [DOI] [PubMed] [Google Scholar]
- 19.Terriere LC, 1983. Enzyme induction, gene amplification, and insect resistance to insecticides. Georghiou GP, Satio T, eds. Pest Resistance to Pesticides New York, NY: Plenum Press, 265–297. [Google Scholar]
- 20.Terriere LC, 1984. Induction of detoxification enzymes in insects. Annu Rev Entomol 29: 71–88. [DOI] [PubMed] [Google Scholar]
- 21.Kasai S, Weerashinghe IS, Shono T, Yamakawa M, 2000. Molecular cloning, nucleotide sequence, and gene expression of a cytochrome P450 (CYP6F1) from the pyrethroid-resistant mosquito, Culex quinquefasciatus say. Insect Biochem Mol Biol 30: 163–171. [DOI] [PubMed] [Google Scholar]
- 22.Daborn PJ, et al. 2002. A single P450 allele associated with insecticide resistance in Drosophila. Science 297: 2253–2256. [DOI] [PubMed] [Google Scholar]
- 23.Daborn PJ, Lumb C, Boey A, Wong W, Ffrench-Constant RH, Batterham P, 2007. Evaluating the insecticide resistance potential of eight Drosophila melanogaster cytlchrome P450 genes by transgenic over-expression. Insect Biochem Mol Biol 37: 512–519. [DOI] [PubMed] [Google Scholar]
- 24.Zhu F, Li T, Zhang L, Liu N, 2008. Co-up-regulation of three P450 genes in response to permethrin exposure in permethrin resistant house flies, Musca domestica. BMC Physiol 8: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paine MJI, Scrutton NS, Munro AW, Roberts GCK, Wolf CR, 2004. Electron transfer partners of cytochrome P450. Ortiz de Montellano PR, ed. Cytochromes P450: Stucture, Mechanism and Biochemistry New York, NY: Kluwer Academic, 115–148. [Google Scholar]
- 26.Ono T, Ozasa S, Hasegawa F, Imai Y, 1977. Involvement of NADPH-cytochrome c reductase in the rat liver squalene epoxidase system. Biochim Biophys Acta 486: 401–407. [PubMed] [Google Scholar]
- 27.Schenkman JB, Jansson I, 1999. Interactions between cytochrome P450 monooxygenases and cytochrome b5. Drug Metab Rev 31: 351–364. [DOI] [PubMed] [Google Scholar]
- 28.Nishino H, Ishibashi T, 2000. Evidence for requirement of NADPH-cytochrome P450 oxidoreductase in the microsomal NADPH-sterol delta7-reductase system. Arch Biochem Biophys 374: 293–298. [DOI] [PubMed] [Google Scholar]
- 29.Wang J, Ortiz de Montellano PR, 2003. The binding sites on human heme oxygenase-1 for cytochrome P450 reductase and biliverdin reductase. J Biol Chem 278: 20069–20076. [DOI] [PubMed] [Google Scholar]
- 30.Nelson D, 2009. The cytochrome P450 homepage. Hum Genomics 4: 59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lycett GJ, McLaughlin LA, Ranson H, Hemingway J, Kafatos FC, Loukeris TG, Paine MJ, 2006. Anopheles gambiae P450 reductase is highly expressed in oenocytes and in vivo knockdown increases permethrin susceptibility. Insect Mol Biol 15: 321–327. [DOI] [PubMed] [Google Scholar]
- 32.Sarapusit S, Pethuan S, Rongnoparut P, 2010. Mosquito NADPH-cytochrome P450 oxidoreductase: kinetics and role of phenylalanine amino acid substitutions at Leu86 and Leu219 in CYP6AA3-mediated deltamethrin metabolism. Arch Insect Biochem Physiol 73: 232–244. [DOI] [PubMed] [Google Scholar]
- 33.Zhu F, Sams S, Moural T, Haynes KF, Potter MF, Palli SR, 2012. RNA interference of NADPH-cytochrome P450 reductase results in reduced insecticide resistance in the bed bug, Cimex lectularius. PLoS One 7: e31037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blariza MJ, Soria NW, Torres AG, Grosso CG, García BA, 2014. cDNA isolation and characterization of two vitellogenin genes in the Chagas’ disease vector Triatoma infestans (Hemiptera, Reduviidae). Gene 543: 118–124. [DOI] [PubMed] [Google Scholar]
- 35.Blariza MJ, Leyria J, Canavoso LE, Soria NW, García BA, 2016. Dynamics of expression of two vitellogenin genes in the Chagas’ disease vector Triatoma infestans: analysis throughout pre-vitellogenesis and vitellogenesis. Acta Trop 156: 100–107. [DOI] [PubMed] [Google Scholar]
- 36.Blariza MJ, Grosso CG, García BA, 2017. Silencing of two vitellogenin genes inhibits oviposition in the Chagas’ disease vector Triatoma infestans (Hemiptera: Reduviidae). Am J Trop Med Hyg 97: 477–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S, 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Livak KJ, Schmittgen TD, 2001. Análisis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 39.Toloza A, Germano M, Mougabure Cueto G, Vassena C, Zerba E, Picollo MI, 2008. Differential patterns of insecticide resistance in eggs and first instars of Triatoma infestans (Hemiptera: Reduviidae) from Argentina and Bolivia. J Med Entomol 45: 421–426. [DOI] [PubMed] [Google Scholar]
- 40.Germano M, Picollo MI, Spillmann C, Mougabure Cueto G, 2014. Fenitrothion: an alternative insecticide for the control of deltamethrin-resistant populations of Triatoma infestans in northern Argentina. Med Vet Entomol 28: 21–25. [DOI] [PubMed] [Google Scholar]