Abstract
Mitochondrial NAD-dependent (IDH) and cytosolic NADP-dependent isocitrate dehydrogenases have been considered as candidates for the production of 2-oxoglutarate required by the glutamine synthetase/glutamate synthase cycle. The increase in IDH transcripts in leaf and root tissues, induced by nitrate or NH4+ resupply to short-term N-starved tobacco (Nicotiana tabacum) plants, suggested that this enzyme could play such a role. The leaf and root steady-state mRNA levels of citrate synthase, acotinase, IDH, and glutamine synthetase were found to respond similarly to nitrate, whereas those for cytosolic NADP-dependent isocitrate dehydrogenase and fumarase responded differently. This apparent coordination occurred only at the mRNA level, since activity and protein levels of certain corresponding enzymes were not altered. Roots and leaves were not affected to the same extent either by N starvation or nitrate addition, the roots showing smaller changes in N metabolite levels. After nitrate resupply, these organs showed different response kinetics with respect to mRNA and N metabolite levels, suggesting that under such conditions nitrate assimilation was preferentially carried out in the roots. The differential effects appeared to reflect the C/N status after N starvation, the response kinetics being associated with the nitrate assimilatory capacity of each organ, signaled either by nitrate status or by metabolite(s) associated with its metabolism.
Nitrate and NH4+ assimilation depends on C metabolism for energy, reducing power, and C skeletons. An important site for the production of critical organic acids, and perhaps ATP and reductant especially in nongreen tissues, is the mitochondria. In higher plants the major NH4+ assimilatory pathway is carried out in the plastids by the concerted action of GS and GOGAT. The GS/GOGAT cycle requires the input of an organic acid in the form of 2-OG to produce Glu. However, the presence of several different 2-OG-producing enzymes, as well as their isoenzymatic forms, means that this substrate is potentially synthesized in a number of subcellular compartments, including mitochondria, cytosol, and plastids. This has led to a poor understanding of how 2-OG is provided and how its provision is regulated for NH4+ assimilation.
In the literature it is often proposed that the 2-OG for the GS/GOGAT cycle is produced by either an isocitrate dehydrogenase (Gálvez and Gadal, 1995) or an Asp amino acid transferase (Lam et al., 1996). In the first case, there is a net synthesis of Glu, whereas in the second case, there is a production of Asp via an interconversion of C and amino compounds. Two isocitrate dehydrogenase enzymes can be distinguished that differ in their cofactor specificity: IDH (Lancien et al., 1998), a Krebs cycle enzyme that is restricted to the mitochondria, and ICDH. Several ICDH isoenzymes have been shown to be present in the plant, located in different subcellular compartments: the cytosol (Gálvez et al., 1996), chloroplasts (Gálvez et al., 1994), mitochondria (Gálvez et al., 1998), and peroxisomes (Yamazaki and Tolbert, 1970). Two hypotheses have been considered for the isocitrate dehydrogenase-dependent origin of 2-OG for N assimilation. Miflin and Lea (1980) suggested that this essential keto-acid is produced in the mitochondria by the IDH. However, according to Chen and Gadal (1990) citrate synthesized in the mitochondria is transferred to the cytosol where it is decarboxylated to 2-OG via the action of cytosolic ACO and ICDH (ICDH1). As yet, both hypotheses are poorly supported experimentally, but several observations have argued in favor of an ICDH1 origin. These include (a) a 2- to 3-fold higher export rate of citrate over 2-OG by isolated spinach mitochondria oxidizing malate (Hanning and Heldt, 1993), (b) an increase in ICDH1 transcript levels in NR mutants grown on high nitrate and the subsequent accumulation of 2-OG because of reduced nitrate assimilation (Scheible et al., 1997), and (c) an increase in ICDH1 transcript levels in detached potato leaves fed with nitrate (Fieuw et al., 1995). On the other hand, an attempt to show such a role for cytosolic ICDH using an antisense transgenic plant strategy has shown that a dramatic reduction of ICDH1 activity to less than 5% of the control plant level did not affect either plant growth and development or nitrogen metabolism under normal growth conditions (Kruse et al., 1998). To explain the lack of phenotype, these authors suggested that under such conditions either there was a contribution from other ICDH isoenzymatic activities or that the remaining ICDH1 activity was sufficient and that the rate of 2-OG synthesis was a nonlimiting step for N assimilation. Of course, it is possible that ICDH1 is not involved in the main pathway for 2-OG production for NH4+ assimilation.
The interaction between C and N metabolisms in higher plant cells is governed by many regulatory factors. This need to coordinate C and N metabolisms is reflected by the complex interplay between signals involving C metabolism, such as Suc and light, and those associated with N metabolism, such as nitrate (Crawford, 1995; Lam et al., 1996). It has become well established that nitrate (or N-containing compounds derived from nitrate metabolism) increases the expression of several genes encoding enzymes involved in N metabolism: NR (Pouteau et al., 1989), nitrate reductase, and (Rastogi et al., 1993), GS and GOGAT (Redinbaugh and Campbell, 1993). Recently, it has been shown that nitrate (Scheible et al., 1997) and/or sugars (Morcuende et al., 1998) also lead to a stimulation of organic acid biosynthesis.
The initial aim of this work was to investigate the role of an I(C)DH in 2-OG supply for amino acid synthesis during nitrate assimilation. The effect of N supplies (nitrate or ammonium) on I(C)DH transcript levels when supplied to short-term N-starved tobacco plants was carried out. It was found that only IDH was affected by both N sources in roots and leaves. The effect of nitrate resupply was chosen to see if the IDH changes were coordinated, at the transcript level, with other enzymes involved in N assimilation and 2-OG production. In this way it was found that nitrate supply led to a differential but coordinated response of several genes involved in N and C metabolisms in both roots and leaves. The analysis of metabolite levels was carried out to determine the signals involved in the observed simultaneous expression and to explain the observed differences between roots and leaves.
MATERIALS AND METHODS
Plants and Growth Conditions
Tobacco (Nicotiana tabacum L. cv Xanthi) was grown aeroponically in a greenhouse under a 16-h day/8-h night photoperiod, with a supplementation by white fluorescent light to provide a PAR of 200 μmol photons m−2 s−1. The day/night temperature was 22°C/18°C. The plants were supplied every 3 d during 2 weeks with nutrient solution A containing the Murashige and Skoog macroelements, microelements, and Fe-EDTA (Murashige and Skoog, 1962). Then, the plants were treated with N-deficient solution B (solution A without mineral N source, K was supplied in the form of 10 mm KCl) for 4 d. Finally, 2 h after the beginning of the photoperiod, the plants were supplied with a solution consisting of solution B either with 10 mm potassium nitrate or 10 mm ammonium chloride added. Deveined leaves and roots were harvested at different times after the resupply of the N-containing solution. Plant tissues were fixed in liquid N2 and conserved at −80°C.
Isolation of RNA and Northern Analyses
Total RNA was extracted according to the method of Chirgwin et al. (1979) and Harpster et al. (1986). Northern analyses were carried out according to the standard procedures as described previously (Sambrook et al., 1989) using 20 μg of deveined leaf or root total RNA. DNA probes were generated, using the DNA fragments described in Table I, by random priming with the Nonaprimer kit (Pharmacia) and [32P]dCTP (Amersham). Specific 3′-noncoding region fragments were generated by PCR using Taq polymerase (Appligene, Illkirch, France) and the following oligonucleotide pairs: TGTCTGGGCAGACAAGAGG/TTGTAATTACGGACCTC and ATCCTGTAGCACAGAA/AGGATAGATACGCTA, for ICDH1 and mtICDH, respectively. Hybridization and wash conditions were as described previously (Gálvez et al., 1996) with final washes from 1× SSC and 0.2% SDS to 0.2× SSC and 0.2% SDS, depending on the origin of the probe.
Table I.
DNA probes used for northern analyses
| Name | Description | Source (Reference) |
|---|---|---|
| IDHa | 656-bp EcoRI-HindIII fragment | Tobacco (Lancien et al., 1998) |
| ICDH1 | 142-bp PCR fragment | Tobacco (Gálvez et al., 1996) |
| mtICDH | 186-bp PCR fragment | Tobacco (Gálvez et al., 1998) |
| CS | 400-bp EcoRI fragment | Tobacco (Landschütze et al., 1995) |
| ACO | 2,000-bp NotI-XhoI fragment | Tobacco (M. Surpili and B. Müller-Röber, unpublished data) |
| FUM | 18,000-bp BamHI fragment | Potato (Nast and Müller-Röber, 1996) |
| GS2 | 1,800-bp EcoRI fragment | Tobacco (Becker et al., 1992) |
| GS1 | 200-bp HindIII-EcoRI fragment | Tobacco (Dubois et al., 1996) |
| NR | 1,600-bp EcoRI fragment | Tobacco, nia2 (Calza et al., 1987) |
Enzyme Activities
Plant material was initially ground to a fine powder in liquid N2 using a mortar and pestle and then further ground in the appropriate extraction buffer with respect to the enzyme activity to be measured. After centrifugation at 20,000g, the supernatant gave the crude extract, which was used for subsequent enzyme activity measurements. ICDH activity was measured spectrophotometrically as the reduction of NADP at 340 nm, as described by Gálvez et al. (1996). Total GS was measured as the synthetase activity as described previously (O'Neal and Joy, 1973). GOGAT (Fd/NADH) activity was measured as described previously (Suzuki and Gadal, 1984) using Fd from spinach leaves or NADH. NR activity measurements were carried out in the presence of 5 mm EDTA according to the method of Ferrario-Méry et al. (1997).
Metabolite Measurements
Carbohydrates and organic acids were extracted with 1 m perchloric acid and amino acids were extracted with 3% sulfosalycilic acid, both from dry matter of roots and de-veined leaves. Suc, Fru, and Glc were measured in the soluble fraction using the Boerhinger Mannheim Suc/d-Glc/d-Fru test kit, following the manufacturer's instructions. The nitrate content was measured as described by Cataldo et al. (1975), and the ammonium content was measured as described by Rochat and Boutin (1989). Total amino acids were quantified by the method of Rosen (1957). Separation and analysis of amino acids were done by ion-exchange chromatography as in Rochat and Boutin (1989). Citrate and 2-OG were assayed as described previously (Bergmeyer, 1965).
SDS-PAGE and Western Analysis
Crude protein extracts (50 μg) were separated on 12% SDS-polyacrylamide gels according to the method of Laemmli (1970). For western analyses, protein transfer onto nitrocellulose membranes and immunodetection were performed as in Gálvez et al. (1996), using a 500-fold dilution of the 33% ammonium sulfate precipitate of the rabbit antiserum containing IDHa polyclonal antibodies (Lancien et al., 1998).
RESULTS
Effect of N Supply on Isocitrate Dehydrogenase Steady-State mRNA Levels
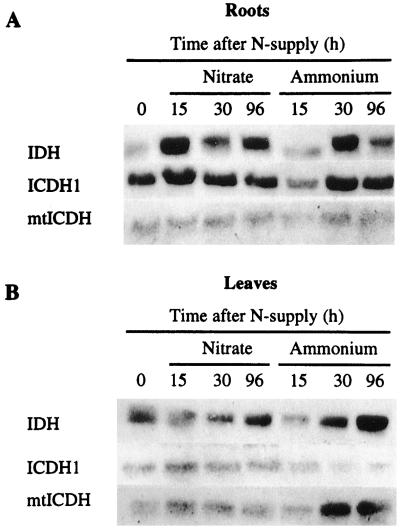
If a specific isocitrate dehydrogenase produces the 2-OG for NH4+ assimilation, it is possible that the addition of either nitrate or NH4+ to N-starved plants could affect the steady-state mRNA level of this enzyme. Therefore, mRNA levels of IDH and ICDH, taken at various times from roots and deveined leaves after the resupply of either 10 mm nitrate or 10 mm ammonium to aeroponically grown 4-d N-starved tobacco plants, were analyzed by northern blot. Figure 1 shows the effect of the above-mentioned treatments on the mRNA levels of IDH, ICDH1, and mtICDH. To distinguish the two different ICDH mRNAs, specific 3′-noncoding region probes were used as described in Methods. To investigate IDH expression a NtIDHa probe was used, which encodes the catalytic IDH subunit shown to be necessary for IDH activity (Lancien et al., 1998). In the experiment shown in Figure 1, nitrate supply led to a significant increase in IDHa mRNA steady-state levels only in the roots (similar changes have also been reported for IDHb and IDHc regulatory subunit mRNA; Lancien et al., 1998). This was seen in the roots and deveined leaves when NH4+ was given to the plants, although the response was slower in the roots when compared with nitrate addition. In contrast, nitrate did not affect ICDH1 and mtICDH mRNA levels (Fig. 1), whereas the addition of NH4+ gave rise to an increase in ICDH1 (only in the roots) and mtICDH (only in the leaves) transcript levels. Since only the IDH steady-state mRNA level was affected by the addition of the two different inorganic N supplies, IDH appeared to be a good candidate for the production of 2-OG with respect to N assimilation.
Figure 1.
Effect of nitrate and ammonium supply on isocitrate dehydrogenase transcripts in tobacco. Deveined leaves and roots were harvested after a 4-d N starvation (0 h) and after the addition of a N source (lanes 15, 30, and 96 h). mRNA was probed for IDH, ICDH1, and mtICDH. Twenty micrograms of total mRNA was loaded in each lane, which was checked by ethidium bromide staining.
If there is a major pathway leading to C skeleton production for N assimilation, the genes associated with this pathway could respond in a similar manner and be coordinated with N assimilation genes. Therefore, it was decided to investigate further the effect of nitrate resupply to the N-starved plants to see if similar changes to those observed for the IDH could be seen for enzymes associated with N assimilation and organic acid synthesis.
Differential but Coordinated Expression of Some Genes Encoding Enzymes Involved in C or N Metabolisms between Roots and Leaves during Nitrate Resupply
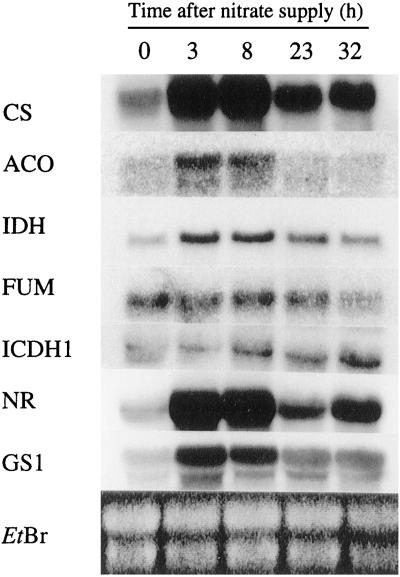
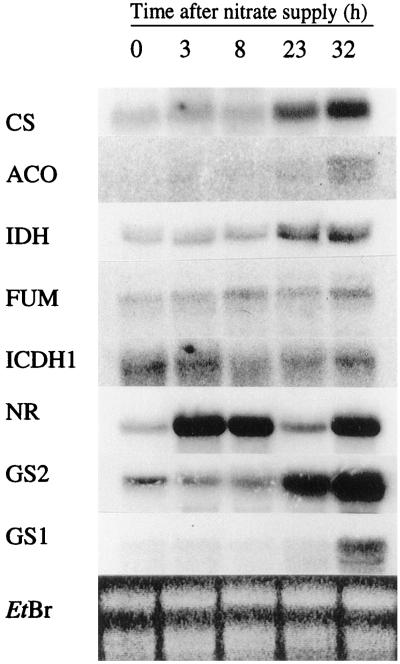
The effect of nitrate resupply to 4-d N-starved tobacco plants was investigated in roots (Fig. 2) and deveined leaves (Fig. 3) by northern analyses using DNA probes for N-assimilatory enzymes (NR and GS), certain Krebs cycle and putative organic acid pathway enzymes (IDH, CS, ACO, FUM, and ICDH1).
Figure 2.
Changes in steady-state transcript abundance in roots after nitrate resupply to 4-d N-starved tobacco plants. Northern analyses were performed at 0 h (after N starvation), and 3, 8, 23, and 32 h after nitrate resupply. An ethidium bromide (EtBr)-stained gel is presented to show loading in each lane. mRNAs were hybridized with probes for CS, ACO, IDH, FUM, ICDH1, NR, and GS1. Twenty micrograms of total mRNA was loaded in each lane.
Figure 3.
Changes in leaf steady-state transcript abundance in deveined leaves after nitrate resupply to 4-d-starved tobacco plants. Northern analyses were performed at 0 h (after N starvation), and 3, 8, 23, and 32 h after nitrate resupply. An ethidium bromide (EtBr)-stained gel is presented to show loading in each lane. mRNAs were hybridized with probes for CS, ACO, IDH, FUM, ICDH1, NR, and GS2 and GS1. Twenty micrograms of total mRNA was loaded in each lane.
As expected, nitrate given to the N-starved plants induced a rapid and transient increase of the NR steady-state mRNA level in both roots and leaves. These changes were considered as a reference for a nitrate-induced gene response. In the roots (Fig. 2), this response was also seen for GS1, encoding a cytosolic GS (Dubois et al., 1996), and the enzymes involved in the first decarboxylative step of the Krebs cycle: CS, ACO, and IDH. However, in the leaves (Fig. 3), the transcript levels of these enzymes, as well as GS2, appeared to be coordinated but showed slower response kinetics with respect to NR. This coordinated response was not observed for either the FUM or the ICDH1 (Figs. 2 and 3). These results show that there was a different response to nitrate supply between the roots and the leaves of N-starved tobacco plants affecting the steady-state mRNA levels of specific enzymes involved in N and C metabolisms. Although the responses were characterized by different kinetics between the two organs, there appeared to be a simultaneous response between the IDH and several other genes that could suggest a role for this enzyme in organic acid supply to N assimilation.
Therefore, it seemed that a regulatory control could occur to coordinate C skeleton partitioning for amino acid biosynthesis via the nitrate assimilatory pathway. To establish a possible correlation between gene expression and plant metabolic status of the different organs, an analysis of certain metabolite levels and enzymatic capacities was undertaken.
The Effect of N Starvation on Plant Metabolism
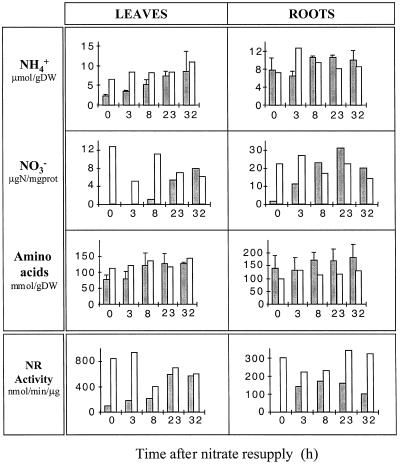
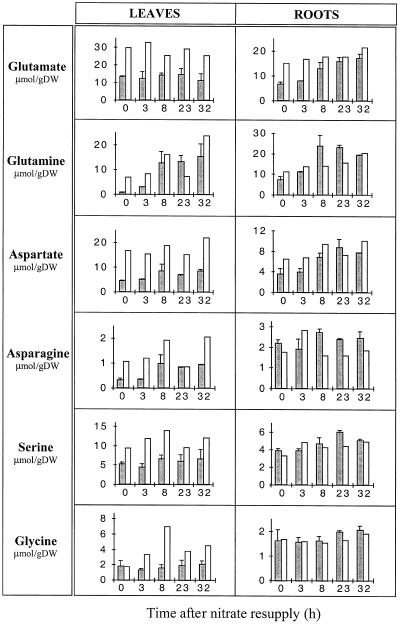
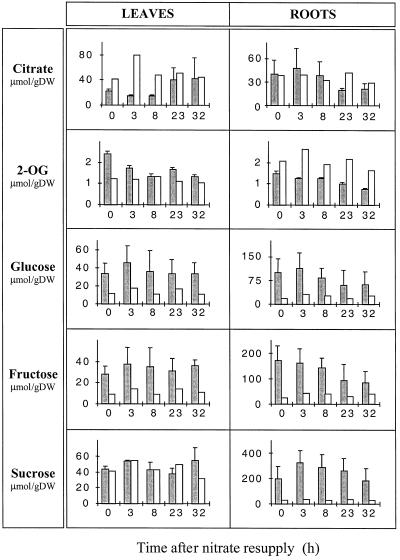
It is interesting to note that N starvation already affected the roots and the leaves to different extents with respect to N- and C-containing metabolites. As expected, the 4-d treatment caused a drastic decrease in stored endogenous nitrate in both leaves and roots (Fig. 4), leading to undetectable levels in the leaves, whereas a small amount remained in the roots. In general, leaves showed a larger decrease in the content of measured amino acids than roots. This was particularly pronounced for Gln, Asp, Asn, and Ser (compare leaves and roots in Fig. 5). On the other hand, the roots showed a greater increase in sugar levels, especially Suc and Fru (Fig. 6). N starvation also resulted in a differential response with respect to the 2-OG pool, since it increased in the leaves but decreased in the roots (Fig. 6).
Figure 4.
The effect of nitrate resupply on inorganic N (NH4+ and NO3−), amino acid content, and NR activity. Deveined leaves and roots were harvested from 4-d N-starved plants (0 h) and after nitrate resupply (3, 8, 23, 32 h), as shown by the shaded histograms. Control, nitrate-fed plants are shown by the nonshaded histograms. Maximal NR activities (nmol−1 h−1 μg−1 protein [prot]) were measured in the presence of EDTA. Values are the means of two independent experiments for nitrate supply experiments, whereas the controls are a single experimental series. Data from each individual experiment are the averages of material taken from four independent plants. DW, Dry weight.
Figure 5.
The effect of nitrate addition on the pool of major amino acids, Gln, Glu, Asp, Asn, Gly, and Ser in deveined leaves and roots. Results are shown relative to dry weight (DW). A calculation relative to total amino acid content did not change the observed amino acid variations. N-starved/nitrate-resupplied plants are shown by the shaded histograms; control nitrate-fed plants are depicted by the nonshaded histograms. Values are the means of two independent experiments for nitrate supply experiments, whereas the controls are single experimental series. Data from each individual experiment are the averages of material taken from four independent plants.
Figure 6.
The effect of nitrate addition on the pool of organic acids (citrate and 2-OG) and sugars (Glc, Fru, and Suc) in deveined leaves and roots. N-starved/nitrate-resupplied plants are shown by the shaded histograms; control nitrate-fed plants are depicted by the nonshaded histograms. Values are the means of two independent experiments for nitrate supply experiments, whereas the controls are single experiments. Data from each individual experiment are the averages of material taken from four independent plants. DW, Dry weight.
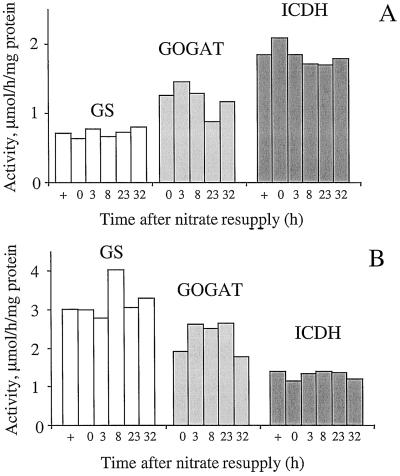
In both roots and leaves, N starvation led to a decrease in NR activity (Fig. 4) but no significant changes were seen in either the in vitro total GS and ICDH activities (Fig. 7, compare + and 0).
Figure 7.
The effect of nitrate addition on GS, Fd-dependent GOGAT (GOGAT), and ICDH activities in root (A) and deveined leaf (B) extracts. Values are the average of two independent experiments. + corresponds to activities from control plants grown on nitrate and harvested at 0 h. Data from each individual experiment are the average of material taken from four independent plants.
A Differential Effect of Nitrate Supply on N Metabolism between Leaves and Roots
Nitrate resupply after N starvation led to a rapid accumulation of nitrate in the roots, which was mirrored by an increase in NR activity (Fig. 4). The induced changes allowed the roots to attain the control plant levels during the first 8 h after nitrate supply. These changes were accompanied by similar rapid increases in measured amino acid levels, including Glu, Gln, and Asp (Fig. 5). Gln reached and exceeded the control level 8 h after resupply. No significant changes were observed in GS and Fd-GOGAT activities (Fig. 7A); however, there was an increase in NADH-GOGAT activity from 10 to 390 nmol NADH oxidized h−1 mg−1 protein, reaching 30% of the total GOGAT capacity 32 h after nitrate addition.
In the leaves the differential kinetic response seen at the mRNA level with respect to the roots was also observed at the N metabolism level. Nitrate supply led to a much slower accumulation of nitrate, which like the NR activity, increased to reach the control plant level only 23 h after resupply (Fig. 4). Similar kinetics were also observed for the NH4+ content (Fig. 4) and to a lesser extent for the Gln content, both of which returned to the control plant levels (Fig. 5). However, the other measured amino acids did not increase significantly, staying close to the low N-starved levels (Fig. 5). As in the roots, the in vitro GS/GOGAT cycle capacity was not modified (Fig. 7B) and an NADH-GOGAT activity was induced to give an activity of 164 nmol NADPH oxidized h−1 mg−1 protein 32 h after the nitrate addition (10% of the total leaf GOGAT activity).
A Differential Effect of Nitrate Supply on C Metabolism between Leaves and Roots
The resupply of N-starved plants with nitrate also led to a differential response between the roots and the leaves with respect to organic acid levels and to a lesser extent with respect to sugar content (Fig. 6). After nitrate supply, the citrate content slowly increased, whereas 2-OG levels decreased in the leaf tissues, both reaching the control plant levels. However, in the roots citrate and 2-OG contents decreased to levels lower than the control plants. Nitrate addition also led to a decrease in Glc and Fru levels in both roots and leaves; in all cases the level remained higher than in the control plants. A differential effect between the roots and leaves was observed for the Suc content that stayed at the nonstarved level in the leaves but remained higher than the control level in the roots. IDH protein content (Fig. 8) and total ICDH activity (Fig. 7) were unchanged after nitrate resupply, staying at the control plant level in both leaves and roots.
Figure 8.
The effect of nitrate resupply on IDH protein content in roots (A) and leaves (B). Antibodies raised against recombinant IDHa protein were used to detect IDH in 50 μg of crude protein extracts. Control, Nitrate-grown plant extracts. Resupply corresponds to 4-d N-starved plants re-fed with 10 mm nitrate.
DISCUSSION
2-OG Production for N Assimilation
The hypothesis that cytosolic ICDH plays an important role in the production of C skeletons for NH4+ assimilation (Chen and Gadal, 1990) has recently been supported indirectly by several observations. These include an increase in ICDH1 transcript levels and 2-OG levels by the addition of nitrate to NR tobacco mutants (Scheible et al., 1997) and increased ICDH1 transcript levels in detached potato leaves fed with nitrate (Fieuw et al., 1995). However, in the first case (Scheible et al., 1997), the ICDH activity was not measured, and in the second example, the changes in transcript levels could not be correlated with the measured ICDH activity (Fieuw et al., 1995). In this work the addition of 10 mm nitrate to 4-d N-starved tobacco plants did not give rise to significant increases in leaf ICDH1 transcript levels (Figs. 1 and 2), although root ICDH1 mRNA occasionally showed a slow increase (Fig. 2). However, in both organs total ICDH activity did not change after nitrate resupply (Fig. 7). The apparent contradiction with the above-mentioned works could be explained by the different experimental models/conditions used.
The comparison of results obtained using slow-growing NR mutants containing high nitrate and low Gln levels (Scheibel et al., 1997), nutrient-fed detached leaves (Fieuw et al., 1995), and N-starved plants containing low nitrate and low Gln levels is probably imprudent. However, in our case it is not possible to say that ICDH1 does not play a role in N metabolism, because it was also seen that our N starvation and N resupply conditions did not alter the in vitro GS and GOGAT enzymatic capacities (Fig. 7). An absence of any modification in the GS/GOGAT cycle capacity could be explained by the fact that nitrate assimilation only produces a small fraction of the total NH4+ assimilated by a leaf (Keys et al., 1978). Although it appeared that photorespiration was reduced by N starvation (as judged by the changes in Gly, Ser, and NH4+ levels after the treatment), it was possible that sufficient activity remained to justify the stability of leaf GS/GOGAT capacity. The absence of an increase in ICDH1 activity could also reflect the fact that 2-OG production is a nonlimiting step for the GS/GOGAT cycle, as suggested by the absence of a phenotype in antisense potato (Kruse et al., 1998) and tobacco (S. Gálvez and M. Hodges, unpublished observations) severely inhibited in ICDH1 activity.
It has already been well documented that nitrate addition can induce an increase in NR, nitrite reductase, GS, and Fd-GOGAT transcript levels (see the introduction). On the other hand, NH4+ has been shown to decrease NR levels (Hoff et al., 1994) and increase only the transcript levels of enzymes involved in its assimilation (e.g. GS; Hirel et al., 1987). Therefore, it is interesting that the addition of either nitrate or NH4+ to N-starved tobacco plants led to an increase in IDH transcript levels, both in roots and leaves. Although, NH4+ did produce an increase in mtICDH and ICDH1 mRNA levels in the leaves and roots, respectively. These observations concerning IDH could reflect a role in 2-OG production for N assimilation. However, as with total GS activity, the increase in IDH mRNA levels was not mirrored by an increase in IDH protein levels, as judged by western analyses (Fig. 8). This shows that the nitrate-induced effect, presumably at the transcriptional level, does not necessarily lead to a similar response at the protein/activity level, perhaps reflecting a control at the translational level.
Further indications for a possible role of IDH in nitrate assimilation are seen by the coordinated changes in steady-state transcript levels between IDH, NR and GS1 (in the roots), and GS2 (in the leaves). This could be because of the important role of Krebs cycle activity in general cell functioning, in which case it might be predicted that all Krebs cycle genes would be modified. However, this was not observed because only CS, mitochondrial ACO, and IDH mRNA levels were equally affected, whereas FUM transcripts were unaffected by the addition of nitrate, this occurring in both the roots and the leaves. From these results, it is clear that nitrate addition exerts a control on certain Krebs cycle enzyme genes in a manner that suggests the induction of an organic acid production pathway for N assimilation. Recently, using NR mutants, PEP carboxylase, and CS mRNA levels and organic acid content were shown to increase when nitrate accumulates (Scheible et al., 1997), and it was proposed that nitrate induced organic acid production for nitrate assimilation via the ICDH1 pathway. However, in this work IDH was not investigated.
Differential Response between the Roots and Leaves
The experimental conditions used in this work provoked a differential response between the roots and the leaves both during the N-starvation period and after the addition of nitrate. First, the withdrawal of N supply led to less-pronounced changes in N-containing metabolite levels in the roots than in the leaves; this was seen for nitrate, NH4+, and amino acid contents (Figs. 4 and 5). It seemed that leaf N metabolism was down-regulated and export of N-containing compounds to the roots was up-regulated by the N stress. As a consequence, such changes would favor root growth under “N-limiting” conditions. It has already been shown that a moderate N deficiency stimulates root growth (Agren and Ingestad, 1987). It has been suggested that under such conditions root growth would be selectively stimulated by a regulation of shoot/root allocation and that roots would recruit more amino acids, which have been shown to cycle between the leaves and the roots (Cooper and Clarkson, 1989). Under our experimental conditions the root contained higher sugar levels both after and during the N-starvation period. Such changes could reflect the requirement of an increased Krebs cycle activity to sustain cell metabolic functions, including organic acid synthesis for N-metabolism. The decrease in the root 2-OG level could be explained by an increased demand for C skeletons for N assimilation. On the other hand, the increase in leaf 2-OG levels after N starvation might reflect the observed decrease in nitrate assimilatory capacity. Therefore, the level of 2-OG seems to reflect the changes in N assimilatory capacity and C/N status in the different organs. Perhaps, the 2-OG content, alone or in association with a N compound like the Gln, allows the cell to sense the C/N status and signal the need to regulate/coordinate C and N metabolic pathways. Such a signaling pathway is well documented in bacteria in which Gln and 2-OG levels regulate the so-called two-component system (Jiang et al., 1997).
The addition of nitrate to the N-starved plants also produced a differential effect on the roots and leaves. This could be explained by a preferential assimilation of the newly supplied nitrate in the roots. Under normal growth conditions, nitrate is mainly exported to the leaves to be assimilated but it appears that under our conditions the transport of nitrate from the roots could be either inhibited or drastically reduced. The proposed stimulation of root nitrate assimilation is suggested by the observed rapid increase in NR activity and amino acid levels (especially Gln) and the decrease in sugar and 2-OG levels. It is possible that during this time the root exports Gln to the leaves, thus explaining why the leaf Gln content increased, whereas there was no significant change in nitrate accumulation, NR activity, and the content of the other measured amino acids. The changes in leaf metabolite levels after nitrate addition suggest a much slower up-regulation in leaf N metabolism with respect to that measured in the root.
On the whole, the kinetics of mRNA accumulation and the up-regulation of nitrate assimilation capacity seem to be similar. Therefore, in both organs the effect on the observed transcript levels (except for leaf NR) could be explained by a similar signaling pathway involving either nitrate directly or a nitrate assimilation derived metabolite. In the roots nitrate accumulated and appeared to be metabolized more rapidly than in the leaves. However, since IDH transcript levels were found also to respond to NH4+ supply in both roots and leaves, it could be that a downstream signal linked to nitrate assimilation might be involved to control the observed coordination.
In the leaves only the NR was rapidly affected by the addition of nitrate (mRNA and enzymatic capacity). This could be because of a specific signaling pathway between the roots and the leaves. NR is known to be responsive to cytokinins (Vincentz et al., 1993), and recent advances have identified a related action of nitrate and cytokinins in the metabolic cross-talk between roots and leaves. A response regulator homologous to the bacterial two-component system has been isolated in maize leaves and has been assumed to be involved in the early response and signal transduction pathway involved in the inorganic N supply transduction, mediated by cytokinins (Sakakibara et al., 1998). Perhaps, such a signaling pathway could have triggered the rapid leaf NR response.
In conclusion, it is suggested that IDH could play an important role in the production of 2-OG for N assimilation. This is based on the following observations: (a) IDH mRNA levels were increased by the addition of either nitrate or NH4+ to N-starved tobacco plants; (b) there was a nitrate-associated coordinated induction in the steady-state mRNA levels of IDH with mitochondrial CS and ACO, as well as with NR (in the roots) and GS; and (c) the mRNA induction kinetics were closely related to the accumulation of nitrate and the metabolic changes associated with N assimilation in each organ examined.
ACKNOWLEDGMENTS
The authors would like to thank Dr. Bernd Müller-Röber (Max-Planck-Institut für Molekulare Pflanzenphysiologie, Golm, Germany) and Dr. Christian Meyer (Institut National de la Recherche Agronomique [INRA], Versailles, France) for making available certain cDNA probes, Dr Akira Suzuki (INRA) for his help in measuring GOGAT activities, and R. Boyer (Institut de Biotechnologie des Plantes, Université Paris-Sud, Orsay, France) for his excellent photographic skills.
Abbreviations:
- 2-OG
2-oxoglutarate
- ACO
aconitase
- CS
citrate synthase
- FUM
fumarase
- GS(1/2)
Gln synthetase (cytosolic/chloroplastic)
- ICDH(1)
NADP-dependent isocitrate dehydrogenase (cytosolic)
- IDH
NAD-dependent isocitrate dehydrogenase
- mtICDH
mitochondrial ICDH
- NR
nitrate reductase
LITERATURE CITED
- Agren GI, Ingestad T. Root:shoot ratio is a balance between nitrogen productivity and photosynthesis. Plant Cell Environ. 1987;10:579–586. [Google Scholar]
- Becker TW, Caboche M, Carrayol E, Hirel B. Nucleotide sequence of the tobacco cDNA encoding plastidic glutamine synthetase and light inducibility, organ specificity and diurnal rythmicity in the expression of the corresponding gene of tobacco and tomato. Physiol Plant. 1992;99:241–248. doi: 10.1007/BF00023384. [DOI] [PubMed] [Google Scholar]
- Bergmeyer HU (1965) Citrate, Malate, α ketoglutarate. In HU Bergmeyer, ed, Methods of Enzymatic Analysis. Academic Press, New York, pp 318–334
- Calza R, Huttner E, Vincentz M, Rouze P, Galangau F, Vaucheret H, Cherel I, Meyer C, Kronenberger J, Caboche M. Cloning of DNA fragments complementary to tobacco nitrate reductase mRNA and encoding epitopes common to the nitrate reductase from higher plants. Mol Gen Genet. 1987;209:552–562. doi: 10.1007/BF00331162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cataldo DSA, Haroon M, Schrader LE, Youngs VL. Rapid colorimetric determination of nitrate in plant tissue by nitration of salicylic acid. Commun Soil Sci Plant Anal. 1975;72:248–253. [Google Scholar]
- Chen R-D, Gadal P. Do the mitochondria provide 2-oxoglutarate needed for glutamate synthesis in higher plant chloroplasts? Plant Physiol Biochem. 1990;28:141–145. [Google Scholar]
- Chirgwin JM, Przybyla AE, MacDonald RJ, Rutter WJ. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979;18:5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Cooper HD, Clarkson DT. Cycling of amino-nitrogen and other nutrients between shoots and roots in cereals: a possible mechanism integrating shoot and root in the regulation of nutrient uptake. J Exp Bot. 1989;40:753–762. [Google Scholar]
- Crawford NM. Nitrate: nutrient and signal for plant growth. Plant Cell. 1995;7:859–868. doi: 10.1105/tpc.7.7.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois F, Brugière N, Sangwan RS, Hirel B. Localization of tobacco cytosolic glutamine synthetase enzymes and the corresponding transcripts show organ- and cell-specific patterns of protein synthesis and gene expression. Plant Mol Biol. 1996;31:803–817. doi: 10.1007/BF00019468. [DOI] [PubMed] [Google Scholar]
- Ferrario-Méry S, Thibaud M-C, Betsche T, Valadier M-H, Foyer CH. Modulation of carbon and nitrogen metabolism, and of nitrate reductase, in untransformed and transformed Nicotiana plumbaginifolia during CO2 enrichment of plants grown in pots and in hydroponic cultures. Planta. 1997;202:510–520. [Google Scholar]
- Fieuw S, Müller-Röber B, Galvez S, Willmitzer L. Cloning and expression analysis of the cytosolic NADP-dependent isocitrate dehydrogenase from potato. Plant Physiol. 1995;107:905–913. doi: 10.1104/pp.107.3.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gálvez S, Bismuth E, Sarda C, Gadal P. Purification and characterization of chloroplastic NADP-isocitrate dehydrogenase from mixotrophic tobacco cells. Plant Physiol. 1994;105:593–600. doi: 10.1104/pp.105.2.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gálvez S, Gadal P. On the function of the NADP-dependent isocitrate dehydrogenase isoenzymes in living organisms. Plant Sci. 1995;105:1–14. [Google Scholar]
- Gálvez S, Hodges M, Decottignies P, Bismuth E, Lancien M, Sangwan RS, Dubois F, LeMaréchal P, Crétin C, Gadal P. Identification of a tobacco cDNA encoding a cytosolic NADP-isocitrate dehydrogenase. Plant Mol Biol. 1996;30:307–320. doi: 10.1007/BF00020116. [DOI] [PubMed] [Google Scholar]
- Gálvez S, Roche O, Bismuth E, Brown S, Gadal P, Hodges M. Mitochondrial localisation of a NADP-dependent isocitrate dehydrogenase isoenzyme using the green fluorescent protein as a marker. Proc Natl Acad Sci USA. 1998;95:7813–7818. doi: 10.1073/pnas.95.13.7813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanning I, Heldt HW. On the function of mitochondrial metabolism during photosynthesis in spinach (Spinacia oleracea) leaves. Partitioning between respiration and export of redox equivalents and precursors for nitrate assimilation products. Plant Physiol. 1993;103:1147–1154. doi: 10.1104/pp.103.4.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harpster MH, Taylor WC. Maize phosphoenolpyruvate carboxylate: cloning and characterization of mRNAs encoding isozymic forms. J Biol Chem. 1986;261:6132–6136. [PubMed] [Google Scholar]
- Hirel B, Bouet C, King B, Layzell D, Jacobs F, Verma DP. Glutamine synthetase genes are regulated by ammonia provided externally or by symbiotic nitrogen fixation. EMBO J. 1987;6:1167–1171. doi: 10.1002/j.1460-2075.1987.tb02350.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoff T, Truong H-N, Caboche M. The use of mutants and transgenic plants to study nitrate assimilation. Plant Cell Environ. 1994;17:489–506. [Google Scholar]
- Jiang P, Zucker P, Atkinson MR, Kamberov ES, Tirasophon W, Chandran P, Schefke BR, Ninfa AJ. Structure/function analysis of the PII signal transduction protein of Escherichia coli: genetic separation of interactions with protein receptors. J Bacteriol. 1997;179:4342–4353. doi: 10.1128/jb.179.13.4342-4353.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keys AJ, Bird IF, Cornelius MJ, Lea PJ, Wallsgrove RM, Miflin BJ. Photorespiratory nitrogen cycle. Nature. 1978;275:741–743. [Google Scholar]
- Kruse A, Fieuw S, Heineke D, Müller-Röber B. Antisense inhibition of cytosolic NADP-dependent isocitrate dehydrogenase in transgenic potato plants. Planta. 1998;205:82–91. [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of the bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lam HM, Coshigano K, Oliveira I, Melo-Oliveira R, Coruzzi G. The molecular genetics of nitrogen assimilation into amino acids in higher plants. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:569–593. doi: 10.1146/annurev.arplant.47.1.569. [DOI] [PubMed] [Google Scholar]
- Lancien M, Gadal P, Hodges M. Molecular characterization of higher plant NAD-dependent isocitrate dehydrogenase: evidence for a heteromeric structure by the complementation of yeast mutants. Plant J. 1998;16:325–333. doi: 10.1046/j.1365-313x.1998.00305.x. [DOI] [PubMed] [Google Scholar]
- Landschütze V, Müller-Röber B, Willmitzer L. Mitochondrial citrate synthase from potato: predominant expression in mature leaves and young flower buds. Planta. 1995;196:756–764. [PubMed] [Google Scholar]
- Miflin BJ, Lea PJ (1980) Ammonia assimilation. In BJ Miflin, ed, The Biochemistry of Plants, Vol 5. Academic Press, New York, pp 169–202
- Morcuende R, Krapp A, Hurry V, Stitt M. Sucrose feeding leads to increased rates of nitrate assimilation, increased rates of 2-oxoglutarte synthesis, and increased synthesis of a wide spectrum of amino acids in tobacco leaves. Planta. 1998;206:394–409. [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco cultures. Physiol Plant. 1962;15:473–479. [Google Scholar]
- Nast G, Müller-Röber B. Molecular characterization of potato fumurate hydratase and functional expression in Escherichia coli. Plant Physiol. 1996;112:1219–1227. doi: 10.1104/pp.112.3.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neal D, Joy KW. Localization of glutamate synthetase in chloroplasts. Nat New Biol. 1973;246:61–62. doi: 10.1038/newbio246061a0. [DOI] [PubMed] [Google Scholar]
- Pouteau S, Chérel I, Vaucheret H, Caboche M. Nitrate reductase mRNA regulation in Nicotiana plumbaginifolia nitrase reductase-deficient mutants. Plant Cell. 1989;1:1111–1120. doi: 10.1105/tpc.1.11.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastogi R, Back E, Schneiderbauer A, Bowsher CG, Moffatt B, Rothstein SJ. A 330-bp region in the spinach nitrite reductase gene promoter directs nitrate-inducible tissue-specific expression in transgenic tobacco. Plant J. 1993;4:317–326. [Google Scholar]
- Redinbaugh MG, Campbell WH. Glutamine synthetase and ferredoxin-dependent glutamate synthase expression in the maize (Zea mays) root primary response to nitrate. Plant Physiol. 1993;101:1249–1255. doi: 10.1104/pp.101.4.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochat C, Boutin J-P. Carbohydrates and nitrogenous compounds change in the hull and in the seed during the pod development of pea. Plant Physiol Biochem. 1989;202:510–521. [Google Scholar]
- Rosen H. A modified ninhidrin colorimetric analysis for amino acids. Arch Biochem Biophys. 1957;67:10–15. doi: 10.1016/0003-9861(57)90241-2. [DOI] [PubMed] [Google Scholar]
- Sakakibara H, Suzuki M, Takei K, Deji A, Taniguchi M, Sugiyama T. A response-regulator homologue possibly involved in nitrogen signal transduction mediated by cytokinin in maize. Plant J. 1998;14:337–344. doi: 10.1046/j.1365-313x.1998.00134.x. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning. A Laboratory Manual. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Scheible WR, Gonzalez-Fontes A, Lauerer M, Müller-Röber B, Caboche M, Stitt M. Accumulation of nitrate acts as a signal to induce organic acid metabolism and repress starch metabolism in tobacco. Plant Cell. 1997;9:783–798. doi: 10.1105/tpc.9.5.783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A, Gadal P. Glutamate synthase: physicochemical and functional properties of different forms in higher plants and in other organisms. Physiol Veg. 1984;22:471–486. [Google Scholar]
- Vincentz M, Moureaux T, Leydecker M-T, Vaucheret H, Caboche M. Regulation of nitrate and nitrite reductase expression in Nicotiana plumbaginifolia leaves by nitrogen and carbon metabolites. Plant J. 1993;3:315–324. doi: 10.1111/j.1365-313x.1993.tb00183.x. [DOI] [PubMed] [Google Scholar]
- Yamazaki RK, Tolbert NE. Enzymic characterization of leaf peroxisomes. J Biol Chem. 1970;245:5137–5144. [PubMed] [Google Scholar]