Abstract.
Sporadic cases of Tricostrongylosis are reported in humans. Diagnosis of enteric Trichostrongylus relies primarily on coproscopic analysis but morphological identification is difficult because of similarity among nematode species. The method is time consuming and requires some expertise. To overcome these limitations, we developed a molecular approach by real-time polymerase chain reaction (PCR) to provide a rapid, specific, and sensitive tool to detect Trichostrongylus spp. in human feces. We designed primers and probe specific for Trichostrongylus rDNA region 5.8S and internal transcribed spacer 2. Three Italian family clusters were analyzed and DNA sequencing was performed to confirm real-time PCR results comparing with known GenBank sequence data. Sequence analysis showed ≥ 99% identity to Trichostrongylus colubriformis and Trichostrongylus axei. This study provides a molecular methodology suitable for fast and specific detection of Trichostrongylus in fecal specimens and to distinguish the zoonotic species.
Trichostrongylus spp. are worldwide helminth parasites primarily of herbivores. Human trichostrongylosis cases have been sporadically reported from many countries.1 In Europe, the infection is known to be present mostly in goats and sheep while accidental infections are reported in humans, generally in temperate and rural areas.2–5 Indeed, human infections seem to coincide with close contact with farm animals through ingestion of food or water contaminated with feces of the host.6 Infected people can develop gastrointestinal symptoms of different intensities, although most subjects are asymptomatic and present eosinophilia as the only finding. Recently, we published data on Trichostrongylus infection in four family clusters,7 the second case of the two reports that has been described in Italy overall.8 Diagnosis of trichostrongylosis was performed as routinely by finding the eggs in fecal samples or by agar plate culture (APC). However, this methodology has limitations as the egg morphology of Trichostrongylus and hookworm species are relatively similar, making it difficult to differentiate them. In addition, the culturing process is very laborious and time consuming. Most investigations have showed molecular methods as valuable tools to detect and differentiate Trichostongylus spp. Some polymerase chain reaction (PCR)-based and nested-PCR were developed for epidemiologic studies, using as amplification target the ribosomal DNA internal transcribed spacer 1 and/or 2 region (ITS1-ITS2).1,5,9 To our knowledge, there is only one study reporting the real-time PCR technique to identify and quantify Trichostrongylus colubriformis larvae (L3)/worms in sheep farm.10 For the intrinsic features of real-time PCR, such as higher specificity, sensitivity, speed and lower contamination risk than conventional PCR,11–16 we decided to assess a specific real-time PCR to detect Trichostrongylus spp. DNA in human fecal samples. We conducted a preliminary study analyzing data obtained from samples of Italian family clusters previously described in our center.7 We analyzed the rDNA region 5.8S and ITS2 as established genetic markers for parasite of interest. The specificity of results was confirmed by comparing DNA sequences obtained using the specific real-time PCR to known sequence data (Genbank) for Trichostrongylus spp.
Fecal samples were collected in accordance with the tenets of the Declaration of Helsinki and with the informed consent obtained from three Italian family clusters of Trichostrongylus infection diagnosed at the Center for Tropical Diseases in Negrar (Verona, Italy) between 2014 and 2016 as previously described.7 Fecal samples were also collected from farm goats (N = 4) in two out of three family clusters, and they were used to set up the real-time PCR. A total of 11 human fecal specimens and four goats feces were collected according to the routine procedure of our laboratory. In detail, 200 mg of feces were stored at −20°C overnight in a solution of phosphate buffered saline 1X with 2% of polyvinylpolypyrrolidone (Sigma-Aldrich, Milan, Italy). In each sample, Phocine Herpes Virus type-1 (PhHV-1, kindly provided by Dr. Pas S., ErasmusMC, Department of Virology, Rotterdam, The Netherlands) was added within the S.T.A.R. buffer (Roche Diagnostic, Monza, Italy), serving as an internal control for the isolation and amplification steps. Before the start of the DNA extraction, all the samples were thawed and then boiled for 10 minutes at 100°C. The DNA was extracted using MagnaPureLC.2 instrument (Roche Diagnostic), following the protocol DNA_I_Blood_Cells_High performance_II, using the DNA isolation kit I (Roche Diagnostic) with a final elution volume of 100 µL. In the first step, we aligned all known sequences available in Genbank of Trichostrongylus spp. rDNA region 5.8S and ITS2 using the multiple sequence alignment program of ClustalW (http://www.genome.jp/tools/clustalw/) (Supplemental Figure 1). Then, PrimerExpress®Softwarev3.0.1 (Thermo Fisher Scientific, Milan, Italy) was used to design primers and probe. We chose the primers/probe combination resulting in a Trichostrongylus spp. amplicon size of 76 bp (Supplemental Table 1) and having the forward primer overlapping the NC1 region5 as further guarantee of conserved region for Trichostrongylus spp. (Figure 1). To perform TaqMan real-time PCR, we evaluated the optimal amounts of primers/probe by preparing a dilution series to determine the minimum concentrations giving the maximum ∆Rn (normalized reporter). For both primers and probe, 300 nM yielded the highest endpoint fluorescence and the lowest threshold cycle (Ct). Amplification was carried out with the cycling protocol: 95°C for 3 minutes, and 40 cycles each of 95°C for 15 seconds, 60°C for 30 seconds, and 72°C for 30 seconds. For all experiments, reactions were performed in triplicate on CFX96 thermalcycler systems (Bio-Rad Laboratories, Milan, Italy). The PCR set up was performed using goats DNA; a negative control containing no template DNA was also verified and Ct value ≥ 40 was defined as a negative result. As control for PCR inhibitors and amplification quality, the PhHV-1 control DNA was amplified with the appropriate primers/probe mix (Supplemental Table 1)17 in the same reaction as Trichostrongylus spp. in multiplex PCR. In brief, amplification reactions for all the real-time PCRs were performed in 25 µL volumes containing PCR buffer (SsoAdvanced universal probes supermix, Bio-Rad Laboratories), 2.5 µg of Bovine Serum Albumin (BSA) (Sigma-Aldrich), 80 nM of each PhHV-1 specific primers, 200 nM of PhHV-1 CY5-BHQ2 labeled probe, 5 µL of DNA sample, and 300 nM of Trichostrongylus spp. primers/probe set. The amplification of individual samples was considered to be hampered by inhibitory factors if the expected Ct value of 32 in the PhHV was increased by more than three cycles. The PhHV analysis showed no significant reduction in Ct values as a result of the newly procedure. For Trichostrongylus specific target, DNA loads were arbitrally categorized into the following intensity groups: low (38 ≥ Ct < 40), moderate, and high (Ct ≤ 37.9). The developed methodology was applied to all the 11 extracted DNA and the results were compared with the coproparasitological and cultures data available from three family clusters (Table 1). In cluster 1 (N = 2) (corresponding to cluster 3 in our previous study7), real-time PCR analysis was positive (median Ct 35.97) for patient 1 and so were APC and coproparasitological exam. Patient 2 resulted positive to microscopic fecal examination and negative to real-time PCR, possibly due to different dates of specimen collection for analysis. Microscopy and real-time PCR analyses were negative 15 days after albendazole (400 mg twice a day for 10 days) treatment of both patients. Fecal examinations, culture, and real-time PCR resulted negative at diagnosis for all members of cluster 2 (N = 2) (corresponding to cluster 2 in our previous study7), who recovered after presumptive treatment of Trichostrongylus infection. Regarding cluster 3 (corresponding to cluster 4 in our previous study7), specimens were isolated from all patients presumed to be affected in outbreak belong to Trichostrongylus and treated for that. Only patient 1 resulted positive exclusively to microscopy. The most likely explanation for the lack of amplification of Trichostrongylus DNA could be related to the scanty number of eggs (only one egg observed after an extensive microscopic examination of entire specimens) and the possible absence of eggs in 200 mg samples processed specifically for real-time PCR analysis. Indeed, the probability to harvest the parasite is highest in larger amount of sample material. To confirm Trichostrongylus spp. identity, Sanger sequencing analysis was performed on all DNA samples (human and goat). Conventional PCR was conducted using primers forward JhTsp and reverse Tricho-R5 resulting in a 228 bp fragment size encompassing the 76 bp fragment (Figure 1). The reaction was carried out with AmpliTaqGold (Thermo Fisher Scientific) in a 25 µL volume containing 2.5 µg of BSA (Sigma-Aldrich) and 5 µL of DNA sample and consisted of an initial heating at 95°C for 10 minutes, followed by 40 amplification cycles of 95° for 15 seconds, 58°C for 40 seconds, and 72°C for 1 minutes, with a final elongation at 72°C for 10 minutes. PCR products were resolved on a 1.5% agarose gel stained with runSafe (Eppendorf, Milan, Italy) and analyzed on runVIEW system (Cleaver Scientific, Rugby, United Kingdom) (data not shown). Then, the PCR products after purification by ExoSap-IT (Thermo Fisher Scientific) were sequenced bidirectionally for more accuracy using BigDye terminator sequencing_3.1 kit (Thermo Fisher Scientific) on an ABI Prism 3500 sequencer (Thermo Fisher Scientific), following the manufacturer’s instructions. The sequences obtained were aligned and compared with known sequence data for Trichostrongylus spp. in the GenBank database using Sequencing Analysisv6 Software (Thermo Fisher Scientific), BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE_TYPE=BlastSearch), and ClustalW programs. Aligning was done in two steps: first, an automatic aligning phase and second, a manual adjustment of multiple alignments. Query cover and identity score percentage in 5.8S and ITS2 region of Italian nematodes were from 99% to 100% for T. colubriformis and Trichostrongylus axei from GenBank (KR011272.1; KR011271.1; KR002111.1; KR002110.1; KT215388.1; AB908960.1; AB908959.1; AB908958.1; JQ889794.1; JF680985.1; HQ844229.1; HQ389232.1; AY439026.1; KT215384.1) (Figure 2). The sequence results confirmed that each amplicon was derived from the appropriate rDNA region. In addition, the sequence analysis confirmed that the human infection for cluster 1 and 2 was due to the ingestion of vegetables fertilized with goat manure. Interestingly, DNA sequences from patient 1 (Samples 1, 2) and Goat2 (both coming from same cluster 1) presented the same variation C > T (KR011272.1 c.383C > T).
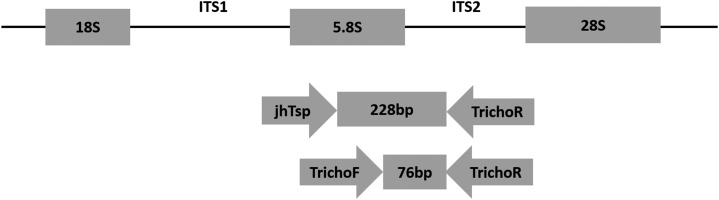
Figure 1.
Schematic representation of part of the rDNA transcriptional unit and relative locations of primers (Tricho-F, Tricho-R and JhTsp) used in this study. The region harboring 5.8S and internal transcribed spacer 2 region (ITS2) rDNA was amplified with Tricho-F and Tricho-R by real-time polymerase chain reaction and with JhTsp and Tricho-R for DNA sequencing.
Table 1.
Laboratory analyses results for Trichostrongylus spp. in fecal samples of three family clusters
| Cluster | Patient | APC | Coproparassitological exam | Real-time PCR |
|---|---|---|---|---|
| 1 | Patient 1 | Positive | Positive* | Positive |
| Patient 1 (treated) | NA | Negative | Negative | |
| Patient 2 | NA | NA | Negative | |
| Patient 2 | NA | Positive* | NA | |
| Patient 2 (treated) | NA | Negative | Negative | |
| 2 | Patient 1 | Negative | Negative | Negative |
| Patient 2 | Negative | Negative | Negative | |
| 3 | Patient 1 | Negative | Positive† | Negative |
| Patient 2 | Negative | Negative | Negative | |
| Patient 3 | Negative | Negative | Negative | |
| Patient 4 | Negative | Negative | Negative |
APC = agar plate culture; NA = not available; PCR = polymerase chain reaction.
1–2 eggs detected per slide.
Only one egg detected.
Figure 2.
Nucleotide sequences alignment of 5.8S-ITS2 region using ClustalW. The locations of forward primer JhTsp and reverse primer Tricho-R are underlined in each line. Asterisks indicate identical nucleotides. ITS2 = internal transcribed spacer 2 region.
In the present study, a specific real-time PCR method for detection of Trichostrogylus spp. in human fecal samples was developed. The primer/probe set for Trichostrongylus was designed to amplify an internal region harboring the 5.8S and ITS2, as a genetic marker for the detection and differentiation of Trichostrongylus.1,5,9 We evaluated the specificity of our molecular approach by direct sequencing of all amplicons from human and goat samples. We can conclude that for the diagnosis of this parasite infection, microscopic direct examination continued to be mandatory. Therefore, the developed molecular procedure combined with microscopy would allow a more accurate screening in humans and animals for this disease. Further studies to determine the limit of detection including larger cohort of samples are required to improve this method. Taken together, our results show that the present real-time PCR could be a suitable, rapid approach for the diagnosis of tricostrongylosis.
Supplementary Material
Supplemental Table and Figure.
Acknowledgments:
We would like to thank Monica Degani and Stefano Tais for their contributions to this study.
Note: Supplemental table and figure appear at www.ajtmh.org.
REFERENCES
- 1.Phosuk I, Intapan PM, Sanpool O, Janwan P, Thanchomnang T, Sawanyawisuth K, Morakote N, Maleewong W, 2013. Molecular evidence of Trichostrongylus colubriformis and Trichostrongylus axei infections in humans from Thailand and Lao PDR. Am J Trop Med Hyg 89: 376–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lattes S, Ferte H, Delaunay P, Depaquit J, Vassallo M, Vittier M, Kokcha S, Coulibaly E, Marty P, 2011. Trichostrongylus colubriformis nematode infections in humans, France. Emerg Infect Dis 17: 1301–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thibert JB, Guiguen C, Gangneux JP, 2006. Human trichostrongyloidosis: case report and microscopic difficulties to identify ankylostomidae eggs [in French]. Ann Biol Clin (Paris) 64: 281–285. [PubMed] [Google Scholar]
- 4.Wall EC, Bhatnagar N, Watson J, Doherty T, 2011. An unusual case of hypereosinophilia and abdominal pain: an outbreak of Trichostrongylus imported from New Zealand. J Travel Med 18: 59–60. [DOI] [PubMed] [Google Scholar]
- 5.Yong TS, Lee JH, Sim S, Lee J, Min DY, Chai JY, Eom KS, Sohn WM, Lee SH, Rim HJ, 2007. Differential diagnosis of Trichostrongylus and hookworm eggs via PCR using ITS-1 sequence. Korean J Parasitol 45: 69–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.The Centers for Disease Control and Prevention , 2016. Trichostrongylosis. Available at: https://www.cdc.gov/dpdx/trichostrongylosis/index.html. Accessed July 20, 2017.
- 7.Buonfrate D, Angheben A, Gobbi F, Mistretta M, Degani M, Bisoffi Z, 2017. Four clusters of Trichostrongylus infection diagnosed in a single center, in Italy. Infection 45: 233–236. [DOI] [PubMed] [Google Scholar]
- 8.Cancrini G, Boemi G, Iori A, Corselli A, 1982. Human infestations by Trichostrongylus axei, T. capricola and T. vitrinus: 1st report in Italy [in Italian]. Parassitologia 24: 145–149. [PubMed] [Google Scholar]
- 9.Sato M, Yoonuan T, Sanguankiat S, Nuamtanong S, Pongvongsa T, Phimmayoi I, Phanhanan V, Boupha B, Moji K, Waikagul J, 2011. Short report: human Trichostrongylus colubriformis infection in a rural village in Laos. Am J Trop Med Hyg 84: 52–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Milhes M, et al. , 2017. A real-time PCR approach to identify anthelmintic-resistant nematodes in sheep farms. Parasitol Res 116: 909–920. [DOI] [PubMed] [Google Scholar]
- 11.Amar CF, East CL, Gray J, Iturriza-Gomara M, Maclure EA, McLauchlin J, 2007. Detection by PCR of eight groups of enteric pathogens in 4,627 faecal samples: re-examination of the English case-control Infectious Intestinal Disease Study (1993–1996). Eur J Clin Microbiol Infect Dis 26: 311–323. [DOI] [PubMed] [Google Scholar]
- 12.Bruijnesteijn van Coppenraet LE, Wallinga JA, Ruijs GJ, Bruins MJ, Verweij JJ, 2009. Parasitological diagnosis combining an internally controlled real-time PCR assay for the detection of four protozoa in stool samples with a testing algorithm for microscopy. Clin Microbiol Infect 15: 869–874. [DOI] [PubMed] [Google Scholar]
- 13.Gunson RN, Bennett S, Maclean A, Carman WF, 2008. Using multiplex real time PCR in order to streamline a routine diagnostic service. J Clin Virol 43: 372–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu YT, 2008. A technological update of molecular diagnostics for infectious diseases. Infect Disord Drug Targets 8: 183–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muldrew KL, 2009. Molecular diagnostics of infectious diseases. Curr Opin Pediatr 21: 102–111. [DOI] [PubMed] [Google Scholar]
- 16.Verweij JJ, Blange RA, Templeton K, Schinkel J, Brienen EA, van Rooyen MA, van Lieshout L, Polderman AM, 2004. Simultaneous detection of Entamoeba histolytica, Giardia lamblia, and Cryptosporidium parvum in fecal samples by using multiplex real-time PCR. J Clin Microbiol 42: 1220–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Niesters HG, 2002. Clinical virology in real time. J Clin Virol 25 (Suppl 3): S3–S12. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table and Figure.