Abstract.
Diagnostic tools for the detection of infection with Onchocerca volvulus are presently limited to microfilaria detection in skin biopsies and serological assessment using the Ov16 immunoglobulin G4 (IgG4) rapid test, both of which have limited sensitivity. We have investigated the diagnostic performance of a peptide enzyme-linked immunosorbent assay (ELISA) based on immunodominant linear epitopes previously discovered. Peptides that were used in these assays were designated O. volvulus motif peptides (OvMP): OvMP-1 (VSV-EPVTTQET-VSV), OvMP-2 (VSV-KDGEDK-VSV), OvMP-3 (VSV-QTSNLD-VSV), and the combination of the latter two, OvMP-23 (VSV-KDGEDK-VSV-QTSNLD-VSV). Sensitivity (O. volvulus infection), specificity (non-helminth infections), and cross-reactivity (helminth infections) were determined using several panels of clinical plasma isolates. OvMP-1 was found to be very sensitive (100%) and specific (98.7%), but showed substantial cross-reactivity with other helminths. Of the other peptides, OvMP-23 was the most promising peptide with a sensitivity of 92.7%, a specificity of 100%, and a cross-reactivity of 6%. It was also demonstrated that these peptides were immunoreactive to IgG but not IgG4, and there is no correlation with the Ov16 IgG4 status, making them promising candidates to complement this already available test. Combination of the Ov16 IgG4 rapid test and OvMP-23 peptide ELISA led to a sensitivity of 97.3% for the detection of O. volvulus infection, without compromising specificity and with minimal impact on cross-reactivity. The available results open the opportunity for a “clinical utility use case” discussion for improved O. volvulus epidemiological mapping.
BACKGROUND
One of the 20 communicable diseases listed on the World Health Organization (WHO) list of neglected tropical diseases is onchocerciasis (or river blindness), which is caused by infection with the filarial nematode Onchocerca volvulus.1–3 With at least 120 million people at risk to be infected, the majority of people suffering from this parasitic disease live in Africa.4,5 Recent updates on the Americas have reported that most of the last known areas of O. volvulus transmission have been freed of the parasite.6,7 However, challenges remain in the Amazonian focus straddling Venezuela and Brazil.8 In an effort to eliminate this disease, mass drug administration (MDA) programs with ivermectin have been ongoing for almost three decades.9,10 In 2016, the WHO released updated criteria for stopping MDA as a result of transmission interruption.11 The technical procedures and the corresponding cutoff values to confirm transmission interruption included the following: 1) screening pools of black flies by the polymerase chain reaction (PCR) for the DNA repeat sequence Ov150; minimal elimination value is < 1/2,000 Ov150-positive flies and 2) serological screening of school-aged children < 10 years of age for immunoglobulin G4 (IgG4) antibodies to the Ov16 antigen; elimination value is antibody prevalence < 0.1%. For the latter, a rapid-format test for the detection of IgG4 antibodies to the parasitic Ov16 antigen is available.12–17
We have previously performed a proteome-wide scanning of the O. volvulus proteome for the presence of linear epitopes.18 Of the list of 249 peptides that were found to contain immunodominant epitopes, more than half of them appeared to contain one of three epitope-forming motifs 1PxxTQE6, 1DGxDK5, and 1Qx(S/T)N(L/I)D6. In the study presented here, we have assessed the diagnostic performance of a peptide enzyme-linked immunosorbent assay (ELISA) using peptides containing these minimal epitopes and combinations thereof.
MATERIALS AND METHODS
Study samples.
All samples used in this study were de-identified before being provided and usage of these samples for research purposes was approved by an ethical committee or Institutional Review Board.
Plasma samples from O. volvulus–infected individuals were collected as part of a field study in Ghana. This study was undertaken in an onchocerciasis-endemic community located in Adansi South District along the Pra river basins in the Ashanti region of Ghana. Physical examinations were performed to identify those subjects having palpable nodules. Skin snips (biopsies) were then taken to determine the microfilarial (mf) load in the skin.19 Most subjects were participating in MDA programs. A total of 97 nodule-positive subjects who donated plasma samples were included.
In addition, a second set of 13 samples from O. volvulus–infected individuals, collected in Cameroon by Dr. Nutman, was obtained through the Filariasis Research Reagent Resource Center (FR3), Division of Microbiology and Infectious Diseases, National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH). Information on O. volvulus infection (number of microfilaria/mg skin and number of palpable nodules) was provided by FR3, along with demographic information (Table 1). All infected individuals had at least two palpable nodules and 25 mf/mg skin (microfilaridermia) as determined by skin snips. Sera were collected from clotted blood obtained by venipuncture.
Table 1.
Demographic information of study populations used for determination of diagnostic performance
| Characteristic | Group | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Onchocerca volvulus–infected | Non-helminth–infected | Helminth-infected | |||||||||
| O. volvulus Cameroon | O. volvulus Ghana | HC southern Africa | HC Belgium | HIV | HCV | Dengue | Asthma | Wb | Bm | STH | |
| Origin | Cameroon | Ghana | Southern Africa | Belgium | USA | USA | Vietnam | USA | Sri Lanka (8) | Indonesia (Central Sulawesi) | Indonesia (Flores) |
| Tahiti (2) | |||||||||||
| Number of patients | 13 | 97 | 10 | 49 | 25 | 25 | 25 | 25 | 10 | 20 | 20 |
| Age, median (min–max) | 54 (20–72) | 47 (21–85) | 21 (17–47) | 40 (23–59) | n.a. | n.a. | 26 (4–67) | 46 (17–91) | 33 (13–48) | 19 (10–45) | 40 (25–75) |
| Gender, n (%) | |||||||||||
| Male | 6 (46) | 54 (56) | 7 (70) | 22 (45) | n.a. | n.a. | 10 (40) | 11 (44) | 6 (60) | 10 (50) | 2 (10) |
| Female | 7 (54) | 43 (44) | 3 (30) | 27 (55) | n.a. | n.a. | 15 (60) | 14 (56) | 3 (30) | 10 (50) | 18 (90) |
| Unknown | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (10) | 0 (0) | 0 (0) |
| Source* | FR3 | KCCR | TS | Janssen | MC | MC | DLS | BR | FR3 | UI | UI |
| Ov16 IgG4 positive, n | 12 | 66 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
Bm = Brugia malayi; HC = healthy control; HCV = hepatitis C virus; HIV = human immunodeficiency virus; IgG4 = immunoglobulin G4; n.a. = not available; STH = soil-transmitted helminths; Wb = Wuchereria bancrofti.
BR = Bioreclamation; DLS = Discovery Life Sciences; FR3 = Filariasis Research Reagent Resource Center; KCCR = Kumasi Center for Collaborative Research; MC = Mayo Clinic; TS = Tissue Solutions; UI = Universitas Indonesia.
For the non-helminth–infected control samples, demographic information is also provided in Table 1. A first healthy control sample set was composed of 10 serum samples from individuals from Southern Africa, collected in Food and Drug Administration-regulated donor centers in the USA, and was provided by Tissue Solutions Ltd. (Glasgow, Scotland). A second healthy control sample set was composed of 49 plasma samples from healthy individuals from Belgium.20–24 Furthermore, different sets of plasma samples were obtained from Discovery Life Sciences, Inc. (Los Osos, CA), Bioreclamation IVT (Baltimore, MD), and FR3 or Mayo Clinic (Jacksonville, FL).
For the cross-reactivity panels (non-Onchocerca helminth–infected individuals), a set of 10 samples from Wuchereria bancrofti–infected individuals, collected in Tahiti by Dr. Perolat or Sri Lanka (unknown collector), was obtained through the FR3, Division of Microbiology and Infectious Diseases, NIAID, NIH. Information on W. bancrofti infection (number of microfilaria/mL) was provided by FR3, along with demographic information (Table 1). Also, two sets of samples were collected in Indonesia. A first set was collected from individuals with Brugia malayi infection in central Sulawesi; a second set was collected in a region highly endemic for soil-transmitted helminths (STH) in Flores Island (Table 1).
Two additional groups were also included for which the helminth infection status is unknown. A first set of samples was collected from malaria-infected individuals from rural areas in Vietnam, which is endemic for STH and lymphatic filariasis, mainly B. malayi.25–29 Samples were provided by Discovery Life Sciences, Inc. A second set of samples was obtained from individuals in France (travelers and immigrants) who presented with symptoms of filariasis and who had a positive serology test against Acanthocheilonema viteae somatic antigens, which is used for screening of suspected clinical cases of human filarial infections (including W. bancrofti, B. malayi, Loa loa, O. volvulus, and Mansonella perstans). Samples were provided by Tissue Solutions Ltd. An overview of the patient demographics of both panels is provided in Table 2.
Table 2.
Demographic information of study populations with unknown helminth infection status
| Characteristic | Group | |
|---|---|---|
| Malaria | Filariasis | |
| Origin | Vietnam | France |
| No. of patients | 25 | 17 |
| Age, median (min–max) | 22 (18–40) | 44 (15–76) |
| Gender, n (%) | ||
| Male | 24 (96) | 10 (59) |
| Female | 1 (4) | 6 (35) |
| Unknown | 0 (0) | 1 (6) |
| Source* | DLS | TS |
| Ov16 IgG4 positive, n | 0 | 0 |
IgG4 = immunoglobulin G4.
DLS = Discovery Life Sciences; TS = Tissue Solutions.
Total IgG peptide ELISA.
Carboxy-terminally biotinylated synthetic peptides were synthesized by following standard procedures and purchased from PEPperPRINT GmbH (Heidelberg, Germany). For the determination of peptide-specific serum antibody levels, a peptide ELISA was developed and set up as follows. Streptavidin-Coated high capacity plates (Thermo Fisher Scientific, Breda, The Netherlands) were rinsed once with 200 μL phosphate buffered saline (PBS) + 0.05% Tween-20 (washing buffer). The plates were incubated with continuous shaking for 1 hour at room temperature with 100 μL of the selected biotinylated peptides, which were diluted at 1 μg/mL in PBS. In the “no peptide” control wells and positive control wells, PBS was added instead. The plates were rinsed three times with washing buffer. Then, the different wells were covered with 100 μL of human serum samples, diluted 200-fold in SuperBlock™ blocking buffer (Thermo Fisher Scientific). In “blank” control wells, SuperBlock blocking buffer was added instead, and in positive control wells, 6.25 ng/mL biotinylated Horse Radish Peroxidase in SuperBlock blocking buffer was added. The plate was incubated at room temperature for 1 hour. After incubation, a 5-fold rinsing cycle with washing buffer was performed. Then, the secondary antibody solution was added to each well. The solution contained an affinity-purified Donkey antihuman IgG (H+L) peroxidase conjugate (Jackson Immuno Research Europe Ltd., Newmarket, United Kingdom) diluted 1:10,000 in blocking solution. The reaction mixture was incubated at room temperature for 30 minutes. At the end of the incubation period, the plates were rinsed five times with washing buffer and treated with 100 μL 1-Step™ Ultra TMB-ELISA substrate solution (Thermo Fisher Scientific). After 10 minutes of incubation, the colorimetric reaction was stopped with 100 μL 1 N HCl. The plate was then read by using the SpectraMax Plus 384 microplate reader (Molecular Devices, Sunnyvale, CA) at a wavelength (λ) of 450 nm. For each sample, peptide-specific signals were corrected for the “no peptide” control signals to obtain the background-corrected optical density values. In cases where the peptide-specific signal was lower than the background, the result was adjusted to “0.”
IgG4 peptide ELISA.
Peptide ELISAs were performed as described earlier, except for the secondary antibody used, which was a mouse monoclonal HP6025 antihuman IgG4 (HRP) from Abcam (Cambridge, United Kingdom) and used after diluting 1:10,000 in blocking solution.
Onchocerciasis Ov16 IgG4 rapid test.
The presence of IgG4 antibodies against the O. volvulus antigen Ov16 was determined using the SD BIOLINE Onchocerciasis IgG4 test (Standard Diagnostics, Gyeonggi-do, Republic of Korea), according to the manufacturer’s instructions. Briefly, 10 μL of plasma was added to the round sample well on the lateral flow strip, immediately followed by the addition of four drops of assay diluent into the square assay diluent well. After 1 hour, the tests were scored. The tests were considered positive only when both the test and the control line were visible. Faint lines were also considered positive, as recommended by the manufacturer.
Onchocerciasis-selected peptides.
Based on the previous discovery of three immunodominant motifs and the epitope mapping results for these motifs, peptides were designed that contain the minimal epitope flanked both N- and C-terminally by a simple Val-Ser-Val linker (Table 3).18 For the nonessential amino acid residues in the epitope sequence, amino acid residues were chosen that showed optimal response in the epitope mapping experiments. Consequently, the epitope sequences used in the peptides were not present as such in any O. volvulus protein, but represented the optimized consensus epitope sequences of a set of cross-reacting epitopes. All peptides were designated O. volvulus motif peptides (for OvMP).
Table 3.
Peptides used in this study
| Peptide name | Sequence |
|---|---|
| OvMP-1 | VSV-EPVTTQET-VSV-Biotin |
| OvMP-2 | VSV-KDGEDK-VSV-Biotin |
| OvMP-3 | VSV-QTSNLD-VSV-Biotin |
| OvMP-23 | VSV-KDGEDK-VSV-QTSNLD-VSV-Biotin |
OvMP = Onchocerca volvulus motif peptides.
Statistical analysis.
For each peptide investigated, receiver operating characteristic (ROC) analysis was performed. Several sample sets from non-helminth–infected individuals were used as a control group (to determine specificity). These included healthy controls from both Belgium and South Africa, human immunodeficiency virus–infected or hepatitis C virus–infected individuals from the USA, dengue-infected individuals from Vietnam, and asthma patients from the USA. Two groups of confirmed (microfilaria positive, PCR positive, nodule positive, or Ov16 IgG4 positive) O. volvulus–infected individuals were used as the positive group (to determine sensitivity): one from Cameroon and one from Ghana. ROC analysis was performed using all non-helminth controls versus all O. volvulus positive samples and cutoffs were determined as the point with maximal Youden’s index ([Sensitivity + Specificity] − 1). Based on these cutoffs, sensitivity and specificity of each peptide ELISA were determined, as well as cross-reactivity with other helminths. This cross-reactivity was determined on sample sets from W. bancrofti-, B. malayi- and STH-infected individuals. All analyses were performed using GraphPad Prism 7 (GraphPad Software, Inc., La Jolla, CA). In the case of OvMP-23, the cutoff was defined as the point with maximal specificity, at the expense of a slight decrease of sensitivity.
For comparison of different groups, a two-tailed Mann–Whitney test was performed using GraphPad Prism 7. P value < 0.05 was considered significantly different.
RESULTS AND DISCUSSION
IgG antibodies against peptides with immunodominant motifs.
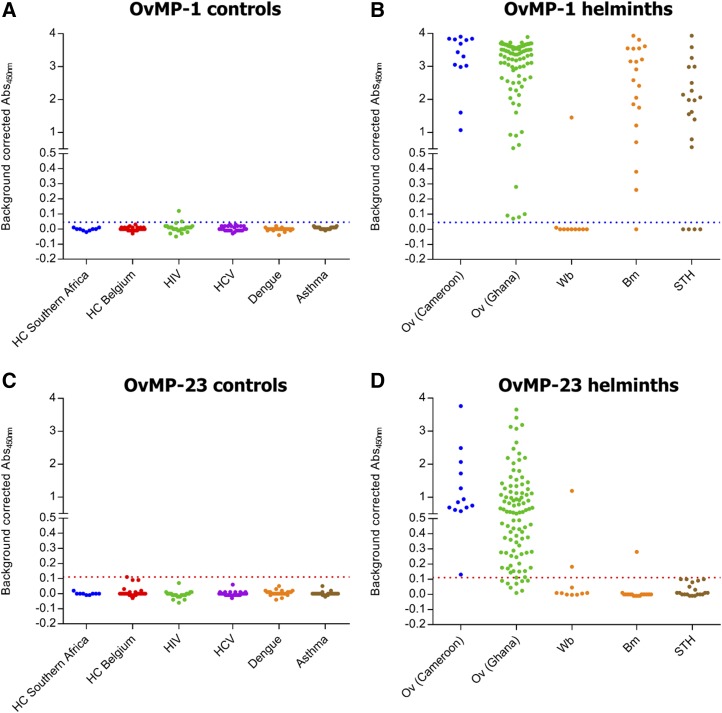
The peptides containing consensus immunodominant linear epitope motifs from O. volvulus were used to set up peptide ELISA and the diagnostic performance of these peptides was assessed. A total of 110 samples from O. volvulus–infected individuals were used to determine sensitivity, whereas 159 samples from non-helminth–infected individuals were used for the determination of specificity. To assess cross-reactivity with other helminths, 50 samples from W. bancrofti, B. malayi, or STH-infected individuals were used. Table 4 summarizes the performance characteristics of the three peptides that were investigated in this study. As an example, individual data of all OvMP-1 data are presented in Figure 1A and B.
Table 4.
Performance characteristics of Onchocerca volvulus minimal epitope containing peptides
| OvMP-1 | OvMP-2 | OvMP-3 | OvMP-23 | |
|---|---|---|---|---|
| Cut-off* | 0.045 | 0.025 | 0.015 | 0.11 |
| Sensitivity (%) | 100.0 | 99.1 | 89.1 | 92.7 |
| Specificity (%) | 98.7 | 95.6 | 88.6 | 100.0 |
| Cross-reactivity (%) | 72.0 | 20.0 | 10.0 | 6.0 |
OvMP = O. volvulus motif peptides.
Background-corrected absorbance.
Figure 1.
Assessment of immune response against OvMP-1 and OvMP-23. Immunoreactivity against OvMP-1 (A, B) and OvMP-23 (C, D) was determined in control groups (A, C) and in helminth-infected individuals (B, D). Dotted lines indicate the cutoff determined by receiver operating characteristic analysis (blue for OvMP-1, red for OvMP-23). All samples used in this graph are described in Table 1. OvMP = Onchocerca volvulus motif peptides. This figure appears in color at www.ajtmh.org.
The peptide OvMP-1 appears to be very specific (98.7%) and to give a positive response in all (100%) O. volvulus–infected individuals. However, also in most other helminth-infected individuals, a positive response is observed (72%). This high cross-reactivity is mostly apparent for B. malayi and STH, while only a few samples from W. bancrofti–infected individuals appear to react with this peptide.
The peptides OvMP-2 and OvMP-3 showed sensitivities of 99.1% and 89.1%, respectively. The specificity of the assays was 95.6% and 88.6% for OvMP-2 and OvMP-3, respectively. A substantial cross-reactivity with other helminths of 20% was observed for OvMP-2, whereas for peptide OvMP-3 this was 10%.
Combination of epitopes in a single peptide results in improved diagnostic characteristics.
To improve further on sensitivity and specificity, a combined epitope peptide was constructed where the motif sequences from OvMP-2 and OvMP-3 were linked to each other, separated by a VSV linker. This peptide (called OvMP-23) was also used in a peptide ELISA and its performance characteristics were determined, similarly as described earlier (Table 4, Figure 1C and D). As specificity and minimal cross-reactivity are of high importance, a cutoff has been defined for this peptide that corresponds to 100% specificity. This combined epitope peptide appears to have superior sensitivity and specificity characteristics compared with the individual motifs (92.7% and 100%, respectively) and only 6% of the 50 non-Onchocerca–, but helminth-infected individuals were seropositive for this peptide. The data presented here clearly demonstrate that combining two linear epitopes is a successful approach to optimize the sensitivity and specificity of the test.
IgG4 response against OvMP-1 and OvMP-23.
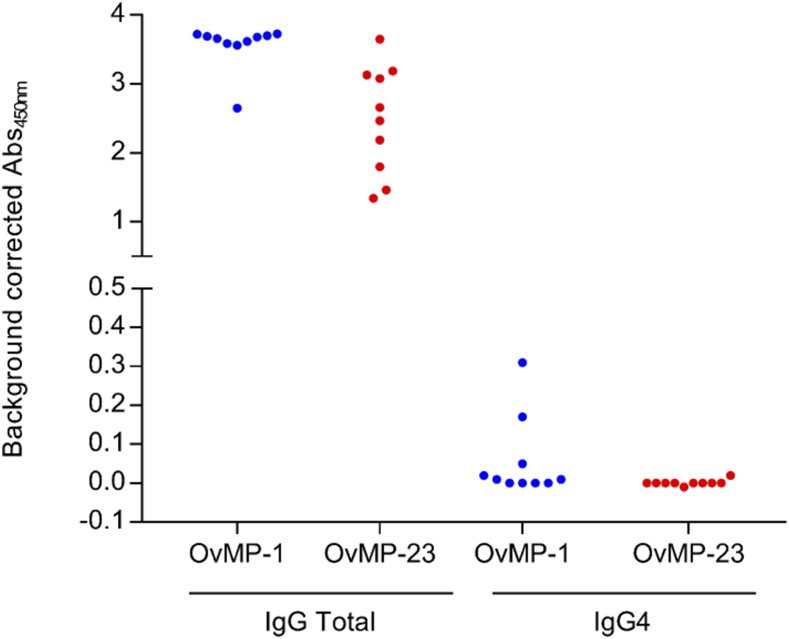
As it is well-known that the immune response against several parasitic antigens is dominated by an IgG4 response, we have measured IgG4 antibodies against OvMP-1 and OvMP-23 in a selection of samples that have a very strong total IgG response against these peptides (Figure 2). Of the 10 samples evaluated, only two appeared to have detectable IgG4 levels (i.e., background-corrected absorbance > 0.1) that recognize OvMP-1, whereas for OvMP-23 no positive signals were detected. This result indicates that the response against these peptides is dominated by non-IgG4 responses, as was already observed before.18 Here again, IgG1 and IgG3 are considered to be the main isotypes.
Figure 2.
Total IgG and IgG4 isotype response against OvMP-1 and OvMP-23. Immunoreactivity against OvMP-1 (blue) and OvMP-23 (red) was determined in 10 Onchocerca volvulus–infected individuals using either total IgG detection antibody or IgG4-specific detection antibody. IgG = immunoglobulin G; OvMP = O. volvulus motif peptides. This figure appears in color at www.ajtmh.org.
Evaluation of the peptides in sample sets from individuals with unknown helminth infection status.
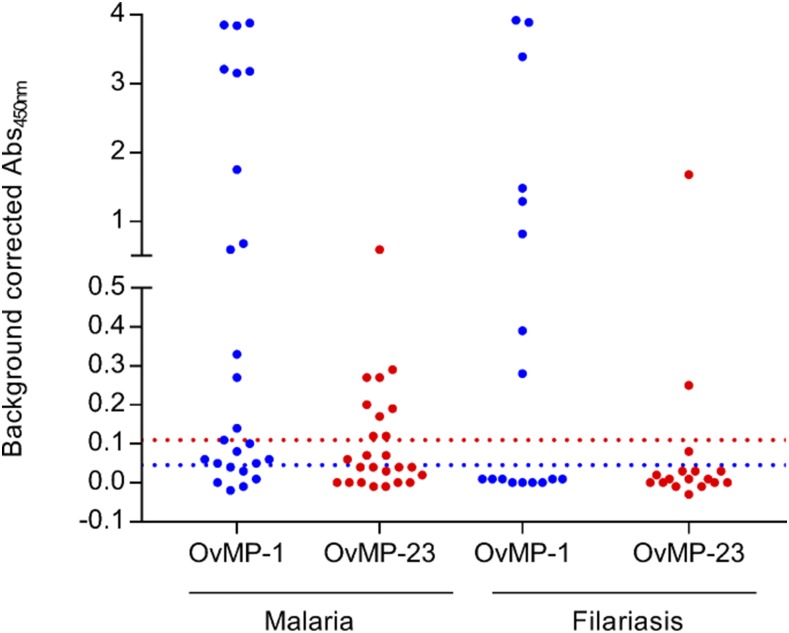
In addition to the groups described earlier that had a well-documented helminth infection or exposure status, we also assessed the response against OvMP-1 and OvMP-23 in sample sets with limited or no helminth-related information (Figure 3). In the first set of 25 malaria-infected samples from Vietnam, 19 samples were positive for OvMP-1. As O. volvulus is not endemic in Vietnam, exposure to or infection with another infectious agent must be responsible for this immune response. Given the high prevalence of B. malayi and STH in this country and the fact that we showed earlier that B. malayi and/or STH clearly cross-react with OvMP-1, it is reasonable to assume that infection with or exposure to B. malayi or STH is responsible for the observed reactivity. However, as Plasmodium falciparum, the causative agent of malaria, is known to express a protein called the interspersed repeat antigen that contains highly immunogenic tandem repeat sequences that perfectly match with the motif in OvMP-1, it cannot be excluded that the reactivity toward OvMP-1 is caused by the malaria infection.30–32 Also, nine of these samples tested positive for OvMP-23, which might potentially be caused by cross-reactivity with B. malayi or exposure to other filaria species. At this point, it is not clear what is causing the reactivity of these samples to this peptide.
Figure 3.
Assessment of immune response against OvMP-1 and OvMP-23 in individuals with unknown helminth infection status. Immunoreactivity against OvMP-1 (blue) and OvMP-23 (red) was determined in malaria-infected individuals from rural Vietnam and in suspected filariasis patients (travelers and immigrants) in France. The blue dotted line indicates the cutoff determined by ROC analysis for OvMP-1, and the red dotted line indicates the cutoff determined by ROC analysis for OvMP-23. All samples used in this graph are described in Table 2. OvMP = Onchocerca volvulus motif peptides; ROC = receiver operating characteristic. This figure appears in color at www.ajtmh.org.
In the second set of 17 filariasis patients, eight were positive for OvMP-1, whereas only two tested positive for OvMP-23. These data suggest that OvMP-1 identifies a proportion of helminth (filariasis)-infected individuals. As discussed earlier, OvMP-1 shows some reactivity to W. bancrofti infection, which might be reflected in the data of these filariasis samples. OvMP-23, on the other hand, only reacts in a limited number of filariasis patients. Based on the data obtained in the well-characterized sample sets described earlier, we can now state that the nine samples that do not respond to OvMP-1 are most likely from W. bancrofti–infected individuals, whereas the six samples that only respond to OvMP-1 are from B. malayi–infected individuals and the samples that respond to both peptides are from O. volvulus–infected individuals. However, as no specific information is available for these individuals, we are unable to confirm these conclusions.
IgG antibodies against peptides are not related to Ov16 IgG4 status.
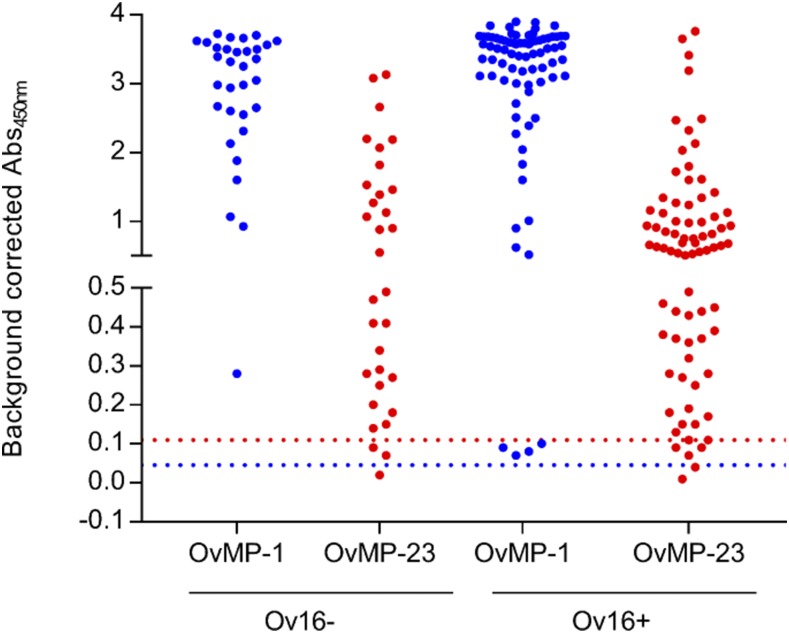
All samples that were used for the assessment of the diagnostic performance of the different peptides were also tested with the Ov16 IgG4 rapid test. All non-Onchocerca–infected samples were Ov16 negative, except for one sample from an asthma patient (from the USA). In the Onchocerca-infected individuals, 78 of a total of 110 samples were positive for Ov16 IgG4, corresponding to a sensitivity of 70.9%, which is comparable to previous reports.12–15,33 We have grouped samples based on their Ov16 IgG4 status and investigated whether there was a correlation between Ov16 IgG4 and peptide serology (Figure 4). The results clearly demonstrate that peptide serology and Ov16 IgG4 serology can be used as complementary markers for Onchocerca infection. In the sample set investigated in this study, 29 of 32 Ov16 negative samples had antibodies against OvMP-23, resulting in a total sensitivity of 97.3% for the combined Ov16 and peptide serology test. This is also reflected in the fact that for both OvMP-1 and OvMP-23, there is no statistical difference between the Ov16-positive and Ov16-negative groups (P value > 0.05).
Figure 4.
Immune response against OvMP-1 and OvMP-23 according to Ov16 IgG4 status. Immunoreactivity against OvMP-1 (blue) and OvMP-23 (red) was determined in Onchocerca volvulus–infected individuals who were grouped according to their Ov16 IgG4 status. The blue dotted line indicates the cutoff determined by ROC analysis for OvMP-1, and the red dotted line indicates the cutoff determined by ROC analysis for OvMP-23. IgG4 = immunoglobulin G4; OvMP = O. volvulus motif peptides; ROC = receiver operating characteristic. This figure appears in color at www.ajtmh.org.
CONCLUSION
We have developed peptide ELISAs based on three consensus immunodominant linear epitope motifs from O. volvulus. The assay based on OvMP-1 not only has excellent sensitivity for O. volvulus, but also cross-reacts substantially with other helminths. Further investigation using clinical samples from well-characterized helminth-infected and/or exposed individuals will be needed to have a clear picture on which helminths cross-react in this assay. The assay based on OvMP-23 has been shown to have a specificity of 100% with a sensitivity of 92% and minimal cross-reactivity with other helminths. When used in conjunction with the Ov16 IgG4 test, a total sensitivity of 97.3% was obtained. The available results open the opportunity for a “clinical utility use case” discussion for improved O. volvulus epidemiological mapping.
Acknowledgments:
We thank the Filariasis Research Reagent Resource Center (FR3) at the Smith College, Northampton, USA, for providing serum samples from Onchocerca volvulus–infected individuals under a contract from the National Institute of Allergy and Infectious Diseases, USA. We also acknowledge Prof. Maria Yazdanbakhsh for providing plasma samples of Brugia malayi– and STH-infected individuals from Indonesia. We thank Janssen Biobank for logistic support, Liesbeth Van Wesenbeeck and Hanne Meeuws for scientific discussions, Will Colon for critically reading the manuscript, and Benny Baeten and Marc Engelen from Janssen Global Public Health for programmatic support.
REFERENCES
- 1.Holmes P; WHO Strategic and Advisory Group on Neglected Tropical Diseases , 2014. Neglected tropical diseases in the post-2015 health agenda. Lancet 383: 1803. [DOI] [PubMed] [Google Scholar]
- 2.Hotez PJ, Brindley PJ, Bethony JM, King CH, Pearce EJ, Jacobson J, 2008. Helminth infections: the great neglected tropical diseases. J Clin Invest 118: 1311–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.WHO , 2017. Report of the Tenth Meeting of the WHO Strategic and Technical Advisory Group for Neglected Tropical Diseases. Geneva, Switzerland: WHO Press. [Google Scholar]
- 4.Enk CD, 2006. Onchocerciasis—river blindness. Clin Dermatol 24: 176–180. [DOI] [PubMed] [Google Scholar]
- 5.Borup LH, Peters JS, Sartori CR, 2003. Onchocerciasis (river blindness). Cutis 72: 297–302. [PubMed] [Google Scholar]
- 6.Rodriguez-Perez MA, et al. 2015. Elimination of onchocerciasis from Mexico. PLoS Negl Trop Dis 9: e0003922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Richards F, Jr, et al. 2015. One hundred years after its discovery in Guatemala by Rodolfo Robles, Onchocerca volvulus transmission has been eliminated from the central endemic zone. Am J Trop Med Hyg 93: 1295–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Botto C, et al. 2016. Evidence of suppression of onchocerciasis transmission in the Venezuelan Amazonian focus. Parasit Vectors 9: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dull HB, Meredith SE, 1998. The Mectizan Donation Programme—a 10-year report. Ann Trop Med Parasitol 92 (Suppl 1): S69–S71. [PubMed] [Google Scholar]
- 10.Evans DS, Unnasch TR, Richards FO, 2015. Onchocerciasis and lymphatic filariasis elimination in Africa: it’s about time. Lancet 385: 2151–2152. [DOI] [PubMed] [Google Scholar]
- 11.WHO , 2016. Guidelines for Stopping Mass Drug Administration and Verifying Elimination of Human onchocerciasis: Criteria and Procedures. Geneva, Switzerland: World Health Organization. [PubMed] [Google Scholar]
- 12.Lipner EM, Dembele N, Souleymane S, Alley WS, Prevots DR, Toe L, Boatin B, Weil GJ, Nutman TB, 2006. Field applicability of a rapid-format anti-Ov-16 antibody test for the assessment of onchocerciasis control measures in regions of endemicity. J Infect Dis 194: 216–221. [DOI] [PubMed] [Google Scholar]
- 13.Weil GJ, Steel C, Liftis F, Li BW, Mearns G, Lobos E, Nutman TB, 2000. A rapid-format antibody card test for diagnosis of onchocerciasis. J Infect Dis 182: 1796–1799. [DOI] [PubMed] [Google Scholar]
- 14.Steel C, Golden A, Stevens E, Yokobe L, Domingo GJ, de Los Santos T, Nutman TB, 2015. Rapid point-of-contact tool for mapping and integrated surveillance of Wuchereria bancrofti and Onchocerca volvulus infection. Clin Vaccine Immunol 22: 896–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Golden A, et al. 2013. Extended result reading window in lateral flow tests detecting exposure to Onchocerca volvulus: a new technology to improve epidemiological surveillance tools. PLoS One 8: e69231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lavebratt C, Dalhammar G, Adamafio NA, Nykanen-Dejerud U, Mingarini K, Ingemarsson K, Opoku N, Akuffo HO, 1994. A simple dot blot assay adaptable for field use in the diagnosis of onchocerciasis: preparation of an adult worm antigen fraction which enhances sensitivity and specificity. Trans R Soc Trop Med Hyg 88: 303–306. [DOI] [PubMed] [Google Scholar]
- 17.Chandrashekar R, Ogunrinade AF, Weil GJ, 1996. Use of recombinant Onchocerca volvulus antigens for diagnosis and surveillance of human onchocerciasis. Trop Med Int Health 1: 575–580. [DOI] [PubMed] [Google Scholar]
- 18.Lagatie O, Van Dorst B, Stuyver LJ, 2017. Identification of three immunodominant motifs with atypical isotype profile scattered over the Onchocerca volvulus proteome. PLoS Negl Trop Dis 11: e0005330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Debrah AY, et al. 2015. Doxycycline leads to sterility and enhanced killing of female Onchocerca volvulus worms in an area with persistent microfilaridermia after repeated ivermectin treatment: a randomized, placebo-controlled, double-blind trial. Clin Infect Dis 61: 517–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stuyver LJ, Verbeke T, Van Loy T, Van Gulck E, Tritsmans L, 2013. An antibody response to human polyomavirus 15-mer peptides is highly abundant in healthy human subjects. Virol J 10: 192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van Loy T, Thys K, Tritsmans L, Stuyver LJ, 2013. Quasispecies analysis of JC virus DNA present in urine of healthy subjects. PLoS One 8: e70950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lagatie O, Van Loy T, Tritsmans L, Stuyver LJ, 2014. Circulating human microRNAs are not linked to JC polyomavirus serology or urinary viral load in healthy subjects. Virol J 11: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lagatie O, Van Loy T, Tritsmans L, Stuyver LJ, 2014. Antibodies reacting with JCPyV_VP2 _167-15mer as a novel serological marker for JC polyomavirus infection. Virol J 11: 174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lagatie O, Van Loy T, Tritsmans L, Stuyver LJ, 2014. Viral miRNAs in plasma and urine divulge JC polyomavirus infection. Virol J 11: 158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van der Hoek W, De NV, Konradsen F, Cam PD, Hoa NT, Toan ND, Cong le D, 2003. Current status of soil-transmitted helminths in Vietnam. Southeast Asian J Trop Med Public Health 34 (Suppl 1): 1–11. [PubMed] [Google Scholar]
- 26.Hung BK, De NV, Duyet le V, Chai JY, 2016. Prevalence of soil-transmitted helminths and molecular clarification of hookworm species in ethnic Ede primary schoolchildren in Dak Lak Province, southern Vietnam. Korean J Parasitol 54: 471–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meyrowitsch DW, Nguyen DT, Hoang TH, Nguyen TD, Michael E, 1998. A review of the present status of lymphatic filariasis in Vietnam. Acta Trop 70: 335–347. [DOI] [PubMed] [Google Scholar]
- 28.Noordin R, Shenoy RK, Lim BH, Ramachandran CP, 2013. Filarial worms in southeast Asia. Lim YAL, Vythilingam I, eds. Parasites and Their Vectors: A Special Focus on Southeast Asia Vienna, Austria: Springer-Verlag Wien, 33–56. [Google Scholar]
- 29.Rebollo MP, et al. 2015. Shrinking the lymphatic filariasis map of Ethiopia: reassessing the population at risk through nationwide mapping. PLoS Negl Trop Dis 9: e0004172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stahl HD, Crewther PE, Anders RF, Kemp DJ, 1987. Structure of the FIRA gene of Plasmodium falciparum. Mol Biol Med 4: 199–211. [PubMed] [Google Scholar]
- 31.Stahl HD, Crewther PE, Anders RF, Brown GV, Coppel RL, Bianco AE, Mitchell GF, Kemp DJ, 1985. Interspersed blocks of repetitive and charged amino acids in a dominant immunogen of Plasmodium falciparum. Proc Natl Acad Sci USA 82: 543–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaur P, Sharma P, Kumar A, Chauhan VS, 1990. Synthetic, immunological and structural studies on repeat unit peptides of Plasmodium falciparum antigens. Int J Pept Protein Res 36: 515–521. [DOI] [PubMed] [Google Scholar]
- 33.Winthrop KL, et al. 2006. The reliability of anterior segment lesions as indicators of onchocercal eye disease in Guatemala. Am J Trop Med Hyg 75: 1058–1062. [PubMed] [Google Scholar]