1. Introduction
The Radical SAM enzymes –also referred to as the AdoMet radical enzymes – are a newly identified enzyme superfamily1 capable of catalyzing radical chemistry similar to, but more extensive than2, that performed by the AdoCbl-dependant enzymes2–7. The Radical SAM and AdoCbl-dependant enzymes have in common the 5'-dA• intermediate, a highly oxidizing and unstable radical intermediate that has never been directly observed, though its existence has been shown in both the Radical SAM and AdoCbl-dependant systems through the use of an allylic AdoMet analog8–11, and indirectly demonstrated in Radical SAM systems by incorporation of a radiolabel into the C5' position of the 5'-deoxyadenosine product12–14 and by its covalent addition to a substrate analog to form a C-adenosylated product15. AdoCbl enzymes produce 5'-dA• via homolytic cleavage of the Co-C5' bond of the corrin cofactor, whereas the Radical SAM enzymes generate the radical by reductive cleavage of a much simpler cofactor, AdoMet3,16. Comparison of the two enzyme families has led to the description of AdoMet by Baker and Frey as the “poor man’s adenosylcobalamin” on account of its relative simplicity and lower energetic cost of production3,17.
Radical SAM enzymes are interesting from an evolutionary perspective for this relative simplicity and other reasons. They require components that theoretically would have been available in the ancient world (reviewed in reference18), and although a primordial precursor of AdoCbl could also have existed19, it has been theorized that AdoMet preceded AdoCbl in the prebiotic world3. Radical SAM superfamily members are found spread across all of the kingdoms of life and catalyze a highly diverse set of reactions1,6,20, again implying an ancient origin with ample time over the course of evolution to diversify. Finally, members of this family are involved in fundamental biological processes that presumably evolved early on, such as metabolism, ribonucleotide reduction, and cofactor biosynthesis1,4,5.
As mentioned above, Radical SAM enzymes all catalyze radical chemistry and are united as a superfamily by their common mechanism of radical generation (reviewed in references1,4,5,7,21,22 (Figure 1a). The main hallmark of the superfamily, a conserved CX3CXϕC motif (ϕ = tyrosine, phenylalanine, histidine or tryptophan), coordinates an iron sulfur cluster that is instrumental in initiation of the radical reaction. Years of research have unambiguously shown that a [4Fe-4S]1+ cluster is the active iron-sulfur species in each of the Radical SAM systems23–25. The 4Fe-4S cluster, which is typically oxygen-sensitive, is ligated by the three cysteines of the conserved motif, leaving one iron unligated and therefore unique26. The second hallmark of the superfamily is a glycine-rich region involved in binding a molecule of AdoMet1,20. This bound AdoMet ligates the unique iron of the 4Fe-4S cluster26–30 and serves as the direct source of 5'-dA•26. Experiments have shown that the [4Fe-4S]1+ state of the cluster is the state competent for radical generation, likely via interaction with and reductive cleavage of AdoMet31,32. Though the detailed mechanism of this process is still unclear, it is thought to occur by electron transfer from an atom of the cluster to AdoMet through the AdoMet sulfur atom5,6,26–28,30,33–35. One important question surrounding this mechanism concerns which cluster atom, a sulfur or an iron, participates in this step, with density functional theory (DFT)36 studies recently focusing attention on iron as the likely mediator of electron transfer.
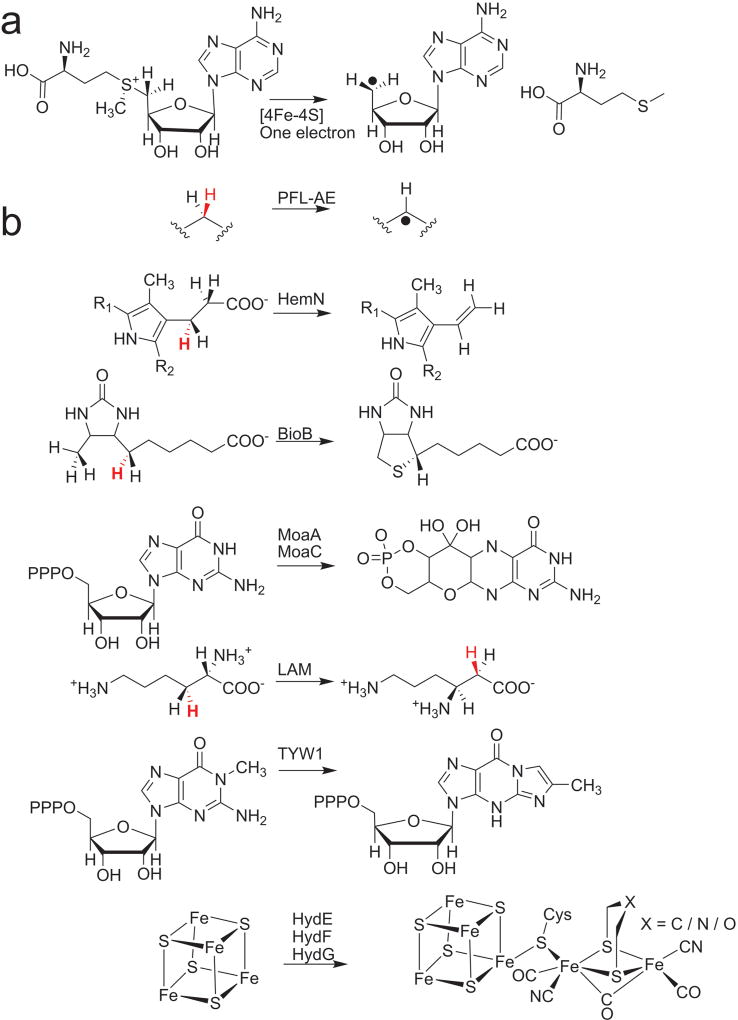
Figure 1. Radical SAM reactions.
(a) The general reaction each Radical SAM enzyme catalyzes to initiate radical chemistry. (b) Selected reactions catalyzed by the Radical SAM enzymes: PFL-AE, HemN (R1 and R2 in this reaction scheme correspond to the remainder of the coproporphyrinogen III tetrapyrrole macrocycle), BioB, MoaA/MoaC, LAM TYW1 and HydE/F/G. The hydrogen atom abstracted, if known, is shown in red type. Adapted from references5 and56.
Recently, the Radical SAM superfamily has been further expanded by the recent characterization of ThiC37–40. While studying thiamine pyrimidine biosynthesis, Downs et al found that a protein, ThiC, carries out Radical SAM chemistry but does not contain the conserved CX3CXϕC motif37,41. Although structures of the apo protein (with disordered cluster binding loop) are available39, comparison with other Radical SAM enzyme structures awaits crystallographic analysis of an iron-sulfur cluster bound form. Further, similar 4Fe-4S-dependent radicalization of SAM has been discovered in a new enzymatic fold with the characterization of Dph2, of the diphthamide biosynthetic pathway42–44. Although we will only discuss ThiC and Dph2 briefly in this review, these findings suggest that the predicted >2800 unique sequences6 already assigned to the CX3CXϕC-containing Radical SAM superfamily may need to make room for an unknown number of additional Radical SAM enzymes.
2. Unresolved questions in the Radical SAM enzyme field
Although we have learned a tremendous amount about Radical SAM enzymes in the last 10 years through the determination of the first few crystal structures and through elegant spectrostropic and biochemical studies, the majority of family members are still uncharacterized and several significant questions remain. Do members of the Radical SAM superfamily share a specific mechanism of control over the 5'-dA• intermediate, or does this vary along with the different reactions each individual enzyme catalyzes and substrates they bind? How similar will ThiC be to the other Radical SAM enzymes in terms of AdoMet binding and radical generation? In addition, the specific factors that govern AdoMet usage as cofactor or cosubstrate are still unclear, even with a range of Radical SAM structures in hand. Related to this, how one enzyme is capable of using two molecules of AdoMet during the course of one turnover is controversial. Finally, perhaps the most far-reaching unknown regarding the Radical SAM family concerns plasticity of the fold; for example, how is the same basic core fold modified to enable so many different types of chemistry, what are the detailed mechanisms of each enzyme after radical generation and how does this impact our understanding of the evolution of the superfamily? Many of these questions await better characterization of the individual family members; unfortunately, difficulties in reconstituting enzymatic activities of the majority of these enzymes in vitro is a fundamental problem plaguing the field that has yet to be fully resolved45–50.
Since the classification of these enzymes as a superfamily, researchers have elucidated key details of the radical generation processes, begun characterization of new Radical SAM enzymes, and published the first few crystal structures of superfamily members. This review will focus on the key aspects of the first series of Radical SAM structures in order to highlight the structural features of the superfamily and identify the main elements involved in substrate binding and catalysis.
3. Highlighted Radical SAM enzymes
The CX3CXϕC-containing Radical SAM structures now available represent a good cross section of the superfamily in terms of their diverse reactions and substrates47–49,51–57 (Figure 1). The methods used to solve each structure along with other relevant information are summarized in Table 1.
Table 1.
Radical SAM structures.
| PDB ID |
Resolution (Å) |
R / Rfree (%) | Space group |
Release date |
Source | Method | Ligands | #molec | |
|---|---|---|---|---|---|---|---|---|---|
| pept-PFL-AE | 3CB8 | 2.77 | 22.9 / 26.1 | P6122 | Oct-08 | E. coli | Fe MAD | 4Fe-4S, AdoMet, peptide substrate | 1 |
| PFL-AE | 3C8F | 2.25 | 22.4 / 29.8 | P3121 | Oct-08 | E. coli | Fe MAD | 4Fe-4S, Met | 1 |
| HemN | 1OLT | 2.07 | 15.5 / 18.7 | P63 | Dec-03 | E. coli | Fe MAD | 4Fe-4S, AdoMet | 1 |
| BioB | 1R30 | 3.4 | 25.6 / 30.0 | P3121 | Jan-04 | E. coli | Fe MAD | 4Fe-4S, AdoMet, DTB, 2Fe-2S | 2 |
| MoaA | 1TV7 | 2.8 | 18.9 / 21.1 | P212121 | Aug-04 | S. aureus | Fe MAD | 4Fe-4S | 2 |
| AdoMet-MoaA | 1TV8 | 2.2 | 21.4 / 24.1 | P212121 | Aug-04 | S. aureus | rigid body refinement | 4Fe-4S, AdoMet | 2 |
| 5'-GTP-MoaA | 2FB3 | 2.35 | 22.9 / 27.1 | P212121 | May-06 | S. aureus | rigid body refinement | 4Fe-4S,5'-dA, Met, 5'-GTP | 2 |
| R17/266/268AMoaA | 2FB2 | 2.25 | 19.6 / 23.7 | P212121 | May-06 | S. aureus | rigid body refinement | 4Fe-4S, AdoMet | 2 |
| LAM | 2A5H | 2.1 | 18.7 / 22.5 | C2 | Oct-05 | C. subterminale SB4 | Se MAD | 4Fe-4S, AdoMet, lysine, PLP | 4 |
| mjTYW1 | 2Z2U | 2.4 | 22.0 / 29.6 | C2 | Oct-07 | M. jannaschii | Se MAD | none | 1 |
| phTYW1 | 2YX0 | 2.21 | 19.4 / 24.2 | P212121 | Oct-07 | P. horikoshii | Se MAD | none | 1 |
| HydE | 3CIW | 1.35 | 14.3 / 17.7 | P212121 | Apr-08 | T. maritima | Fe SAD | 4Fe-4S, AdoHCys, | 1 |
| 2Fe2S-HydE | 3CIX | 1.70 | 14.7 / 18.3 | P212121 | Apr-08 | T. maritima | Fe SAD | 4Fe-4S, AdoHCys, thiocyanate, 2Fe-2S | 1 |
| AdoMet-HydE | 3IIZ | 1.62 | 14.0 / 18.4 | P212121 | Sep-09 | T. maritima | rigid body refinement | 4Fe-4S, AdoMet, 2Fe-2S | 1 |
| HydE with 5'-dA and Met | 3IIX | 1.25 | 14.0 / 16.6 | P212121 | Sep-09 | T. maritima | rigid body refinement | 4Fe-4S, 5'-dA, Met | 1 |
| ThiC | 3EPM | 2.79 | 18.5 / 24.4 | P21 | Oct-08 | C. crescentus | Se SAD | none | 2 |
| ThiC with substrate analog | 3EPN | 2.11 | 17.2 / 24.9 | P21 | Oct-08 | C. crescentus | rigid body refinement | imidazole ribonucleotide | 2 |
| ThiC with product | 3EPO | 2.10 | 15.6 / 22.7 | P21 | Oct-08 | C. crescentus | rigid body refinement | HMP-P | 2 |
| RimO | 2QGQ | 2.00 | 21.2 / 25.1 | P1 | Mar-10 | T. maritima | Se SAD | none | 8 |
3.1. Pyruvate formate-lyase activating enzyme (PFL-AE)
Along with lysine 2,3-aminomutase (LAM), PFL-AE is one of the most fully characterized Radical SAM enzyme. This enzyme is a member of the Radical SAM activase subfamily, all of which perform direct hydrogen atom abstraction from a target Glycyl Radical Enzyme, forming a catalytically essential glycyl radical (Figure 1b). PFL-AE itself forms a glycyl radical on G734 of E. coli pyruvate formate-lyase (PFL), activating it for homolytic cleavage of pyruvate to form acetyl-CoA and formate58–60. This reaction is very important under fermentative conditions where it serves as the organism’s source of acetyl-CoA61.
Recent structural characterization of PFL-AE has yielded two models of the 4Fe-4S bound form of the enzyme, in both the substrate-free and substrate-bound states55. The two structures were solved independently by iron-MAD techniques at 2.25 Å and 2.8 Å resolution, respectively (Table 1). The substrate-bound PFL-AE model includes the 4Fe-4S cluster, AdoMet and a seven-residue peptide substrate ordered in the active site. The particular peptide used in this study, RVSGYAV, corresponds to the seven residues of the PFL glycyl radical loop, and has been shown to be a decent substrate for PFL-AE (Km = 0.22 mM, Vmax = 11 nmol/min˙mg, compared to 1.4 µM and 54 nmol/min˙mg for PFL)14. The substrate-free model contains only the 4Fe-4S cluster and the methionyl moiety of a partially disordered AdoMet molecule. Any analysis of this model should take into account disorder in several of the loops near the active site of the substrate-free PFL-AE model, as well as the medium resolution of the substrate-bound form. The disorder observed in the substrate-free form is likely physiologically relevant, related to conformational flexibility of parts of the enzyme in the absence of substrate.
3.2. The oxygen-independent coproporphyrinogen III oxidase (HemN)
One of the two initial Radical SAM enzymes to be structurally characterized, HemN is one of the more recently biochemically characterized members of this superfamily. It catalyzes two oxidative decarboxylations of the propionate sidechains on rings A and B of coproporphyrinogen III to vinyl groups, yielding protoporphyrinogen IX, an important heme precursor (Figure 1)62. This reaction requires cleavage of two molecules of AdoMet46,63 and the action of an unidentified electron acceptor. Though HemN has been better characterized in recent years, full activity is still only obtained with the addition of cell free extract46,63,64. Presumably the addition of cell extract is necessary in order to provide the unknown electron acceptor, but the fact remains that the reaction requirements are currently incompletely defined, as is the case with several other Radical SAM systems4. However, this aspect of the HemN reaction – the enzyme’s inability to complete the reaction cycle in the absence of the electron acceptor – has made it amenable to spectroscopic characterization of radical reaction intermediates64.
The HemN structure was solved by iron-MAD techniques, refined to 2.07 Å resolution (Table 1), and contains the 4Fe-4S cluster, the cluster-bound AdoMet, and a poorly resolved additional molecule of AdoMet, termed SAM2, bound within the active site51. The possible role of SAM2, which could be physiologically relevant or an artifact of the crystallization conditions, is discussed in a later section. The actual substrate of this enzyme is not present in this model, and as mentioned above, the electron acceptor is also absent.
3.3. Biotin synthase (BioB)
Several Radical SAM enzymes catalyze sulfur insertion reactions, with BioB, LipA and MiaB serving as archetypes for this subfamily. BioB uses two molecules of AdoMet to insert a sulfur atom into positions C6 and C8 of dethiobiotin, forming the thiophane ring of biotin (Figure 1), while LipA catalyzes the sulfur insertion step that forms lipoic acid from octanoate, and MiaB thiomethylates isopentenyladenine, a step in the tRNA maturation pathway6. RimO is likely a new addition to this group, catalyzing the methylthiolation of a protein aspartate residue of the S12 protein57. Although the source of sulfur for biotin was a point of controversy for some time, there is now general agreement that a protein bound 2Fe-2S cluster is the source. This conclusion is based on 34S isotopic labeling studies65, the observed destruction of an Fe2S2 cluster that accompanies BioB turnover66–68, and a crystal structure of BioB that shows the 2Fe-2S cluster is close proximity to substrate dithiobiotin52. The requisite destruction of the 2Fe-2S cluster raised the issue of whether BioB is a suicide enzyme. In vivo studies suggest, however, that BioB does carry out multiple turnovers69, requiring that the cluster must be regenerated in vivo. In addition, recent in vitro studies have identified conditions under which BioB can undergo multiple turnovers, when Fe3+ and S2− are added to the reaction mix50. Components involved in reconstitution of this cluster in vivo have not yet been fully identified, though this is an area of ongoing research70.
The 3.4 Å resolution model of the BioB homodimer was solved by iron-MAD methods (Table 1). In the BioB structure, the 4Fe-4S cluster, AdoMet, the dethiobiotin substrate and a catalytically relevant 2Fe-2S cluster were observed bound to the active site. The resolution of the BioB structure is moderate, and as mentioned above, the components required for reconstitution of the 2Fe-2S cluster have not yet been fully identified70; however, this structure comprises all of the components required for catalysis and is therefore a complete model of the BioB pre-turnover Michaelis complex. Turnover is not observed only because the crystallization conditions lack the reductant necessary to reduce the [4Fe-4S] cluster to the 1+ oxidation state.
3.4. Molybdenum cofactor biosynthesis protein MoaA
One of the first steps of molybdenum cofactor biosynthesis is the rearrangement of guanosine-5'-triphosphate (5'-GTP) to form precursor Z (Figure 1b). Subsequent steps convert this intermediate to molybdopterin by forming the dithiolene group of molybdopterin that is responsible for coordination of molybdenum. Precursor Z differs from the intermediates of related pterin biosynthetic pathways in that C8 of the purine is retained and inserted between C2' and C3' of the ribose71,72. The formation of precursor Z is catalyzed by two enzymes, MoaA and MoaC, and though both structures have been solved, their specific roles during catalysis are presently unclear47,54,73. The identity of the actual substrate of each enzyme is also unknown, though 5'-GTP does bind to MoaA54. Finally, the question of whether MoaA and MoaC form a protein-protein complex is still unresolved.
Multiple structures of MoaA are available, each packing as a homodimer, with the essential 4Fe-4S cluster and an additional 4Fe-4S cluster at the C-terminus (Table 1). The MoaA structures solved include the “apo” (AdoMet-free) state, solved by iron-MAD, the AdoMet-bound state, solved by molecular replacement (MR) using the apo-MoaA structure as the search model, and the 5'-GTP-bound state with the AdoMet cleavage products in the AdoMet binding site, formed by soaking preformed AdoMet-bound crystals in a solution containing 5'-GTP and solved by MR using the MoaA-AdoMet complex structure47,54. A structure of the R17/266/268A triple mutant of MoaA has also been solved by MR with the AdoMet-bound MoaA model54. The second 4Fe-4S cluster seems to be involved in substrate binding, ligated at a unique iron by the 5'-GTP or, in the absence of 5'-GTP, dithiothreitol (DTT). ENDOR studies have confirmed an interaction between this second cluster and 5'-GTP via N1 of the purine ring74. Because the specific MoaA reaction has not been fully biochemically characterized, these structures may not represent a complete description of the components required for catalysis. MoaC may indeed form a complex with MoaA. Finally, because the structure of MoaA complexed with AdoMet and 5'-GTP was obtained by soaking and not cocrystallization, it could be argued that crystal contacts may have prevented a conformational change required for catalysis.
3.5. Lysine aminomutase (LAM)
Of all of the Radical SAM enzymes, LAM is currently the best characterized9,13,24,30,33,53,75–84. It catalyzes the migration of the amino group from C2 to C3 of lysine, using pyridoxal 5'-phosphate (PLP) to bind the substrate through the typical imine linkage (Figure 1). The reaction, essential components and mechanism of LAM have been very well characterized; therefore, many details of AdoMet cleavage by the Radical SAM enzymes have been provided by study of LAM84. For example, studies of LAM included demonstration of the allylic-5'-dA• intermediate using 3',4'-anhydro-AdoMet, a catalytically competent AdoMet analog9. At this point, LAM is the prototypical Radical SAM enzyme.
The structure of Clostridium subterminale 4Fe-4S-LAM in complex with AdoMet, substrate lysine and PLP was solved by iron-MAD techniques to 2.1 Å resolution53 (Table 1). The model is a homotetramer, with extensive dimerization and tetramerization interfaces. As was the case in the BioB crystallization conditions, turnover was prevented by omission of a reductant; therefore, this structure closely represents the Michaelis complex.
3.6. Wye-base biosynthetic protein TYW1
A new putative Radical SAM enzyme, TYW1, was recently identified in a study aimed at isolating the enzymes involved in the biosynthesis of a specific class of modified tRNA base, the wye bases85,86. Wye bases have been found filling position 37, the position next to the 3'-position of the anticodon, in certian archaeal and eukaryotic tRNAs87,88. By stabilizing codon-anticodon interactions, these modified bases help to reinforce correct basepairing during translation89–91. An early step of the wye base biosynthetic pathway, formation of the tricyclic molecule 4-demethylwyosine from N-methylated guanosine base at position 37 of tRNAPhe (Figure 1b), is catalyzed by TYW186. In vivo down-regulation of the iron-sulfur cluster biosynthetic protein NFS1 in S. cerevisiae results in a reduction of wyobutosine synthesis, suggesting the requirement for an iron-sulfur cluster in catalysis86. Mutagenesis, crystallographic, and iron-sulfur cluster incorporation studies are consistent with assignment of TYW1 as a Radical SAM enzyme48,49,86; however, TYW1 activity has not yet been reconstituted in vitro and the source of the two carbons that are incorporated is unknown. Clearly, little is known about catalysis by this enzyme, and it may well require the involvement of additional proteins48,49.
Two groups have independently published structures of TYW1 from P. horokishii48 (phTYW1, 2.2 Å resolution) and M. jannaschii49 (mjTYW1, 2.4 Å resolution). Both structures were solved by Se-MAD techniques and neither contains the 4Fe-4S cluster, AdoMet, or substrate. TYW1 is a monomeric protein that, as demonstrated by cluster reconstitution experiments48,49, probably binds two FeS clusters; the 4Fe-4S cluster essential to 5'-dA• generation and a second cluster likely involved in substrate binding or activation. Low resolution anomalous difference maps collected at 1.54 Å indicate that TYW1 does bind two iron-sulfur clusters, although the data was apparently of too low quality to yield refined cluster-bound models. There is also disagreement as to whether the cluster at the second site is a 2Fe-2S or 4Fe-4S cluster48,49. Both structures have significant disorder, in particular near the active site, which may be attributed to the absence of cofactors, substrate, or unknown partner protein(s). In this review, we analyze the phTYW1 structure alone due to a lower amount of disorder than observed in the mjTYW1 structure. Both groups have modeled the tRNA substrate into the active site48,49, but no crystal structure is available of the complex.
3.7. [Fe-Fe] Hydrogenase Maturase Protein HydE
The Radical SAM enzyme HydE from Thermotoga maritima is one of three enzymes (HydE, HydF and HydG) recently identified as essential for maturation of the [FeFe] hydrogenase (HydA) from that organism92–94. Complex metallocluster bioassembly is a notoriously difficult field of study due to, among many things, instability of clusters during protein purification, the danger of incomplete cluster reconstitution, oxygen sensitivity and the existence of redundant assembly systems, which complicates identification of the specific components necessary95–97. Despite these difficulties, deconvolution of the specific functions of HydE, HydF and HydG has successfully begun. It is likely that HydE and HydG are responsible for formation of the binuclear iron subcluster of the HydA H cluster (Figure 1b)56,92,98,99, using HydF, a GTPase100, as a scaffold protein from which the subcluster can later be transferred to HydA101,102. Recent studies indicate that HydG catalyzes formation of the CO103 as well as the CN− ligands104. The involvement of Radical SAM proteins in the complicated hydrogenase maturation system highlights yet again the versatility of this ubiquitous superfamily.
Two very high-resolution structures of T. maritima HydE were solved independently by iron-SAD using anomalous signal from the enzyme’s reconsituted iron-sulfur clusters56. A structure of the 4Fe-4S form of the enzyme with bound S-adenosylhomocysteine (AdoHCys) was solved to 1.35 Ǻ resolution, while a structure of the enzyme with these ligands and an additional 2Fe-2S cluster and a bound thiocyanate molecule was refined to 1.7 Ǻ resolution56. Though the physiological relevance of this second cluster and the dithiocyanate is currently uncertain, it is this second structure that we discuss below. Additional structures were recently solved of the (a) AdoMet-bound and (b) the methionine- and 5'-dA bound forms of the enzyme, and used for QM/MM calculations to investigate the AdoMet cleavage mechanism in closer detail36. As noted above with respect to the HemN and MoaA structures, analysis of this structure must take into account the fact that the specific reaction catalyzed by HydE is unknown, as are the components necessary for its full activity.
4. Overall Fold
4.1. Radical SAM Core
Most Radical SAM enzymes share a common core fold responsible for radical generation to which additional protein structural elements can be added to tailor the enzyme to a particular substrate. As mentioned previously (here and elsewhere5,7,20,47,51–53,55,56,105,106), this core fold is comprised of six β/α motifs arranged in a manner that is reminiscent of a TIM barrel and is thought to be common to all of the superfamily members on the basis of structure-based sequence alignments20. It should be noted that despite the difference in cluster-binding cysteine motifs, ThiC also has a TIM barrel fold39, while Dph2 adopts a completely unrelated tertiary structure43. The TIM barrel fold is characterized by an eight-stranded, all-parallel β sheet made up of repeating β/α motifs, curved such that the eight strands form a barrel, surrounded on the outside by the eight helices. The Radical SAM core is similar to this fold, but contains only six of the eight β/α motifs, resulting in the formation of a “partial” or “3/4” TIM barrel (Figures 2 and 3). The helices of each β/α unit are located on one side of the sheet (the “outer” side of the barrel), while the active site is located on the opposite (or “inner”) side. The curvature of the core β sheet is typically less than that of a full barrel, except in the cases of BioB, HydE and ThiC, which do form a complete eight-stranded barrel. In the majority of the structures, the partial TIM barrel has less curvature (appearing “splayed”), reflecting that the orientation of the β/α motifs can vary slightly depending on the remaining protein fold. Protein elements outside of the core domain are involved in imparting specificity to the individual proteins, and provide surfaces for oligomerization as well.
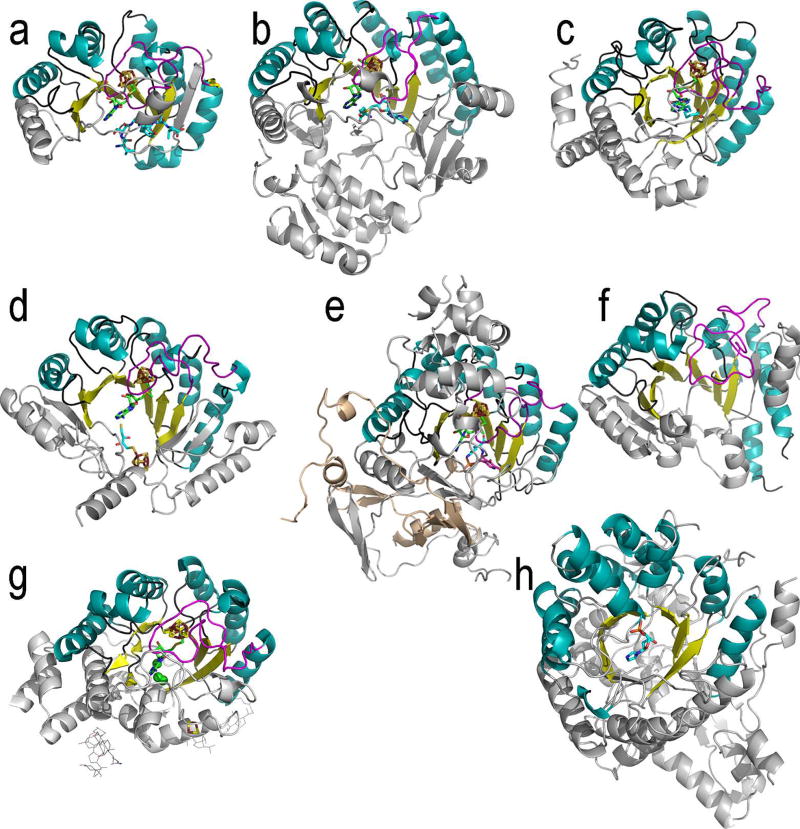
Figure 2. Ribbon diagrams of Radical SAM enzymes.
Shown are the top view of each monomer with the 4Fe-4S cluster, AdoMet and substrate, if present, in stick representation: (a) PFL-AE, (b) HemN, (c) BioB, (d) MoaA, (e) LAM, (f) phTYW1 (g) HydE and (h) ThiC. The Radical SAM core domain is colored as follows: helices, teal; strands, yellow; loops, dark grey; cluster-binding loop harboring the CX3CXϕC motif, magenta. The AdoMet, 4Fe-4S cluster and substrate atoms are colored as follows: iron, ruby; sulfur, gold; AdoMet carbons, green; substrate carbons, teal; oxygen, red, nitrogen, blue. Protein elements outside the core are colored light grey.
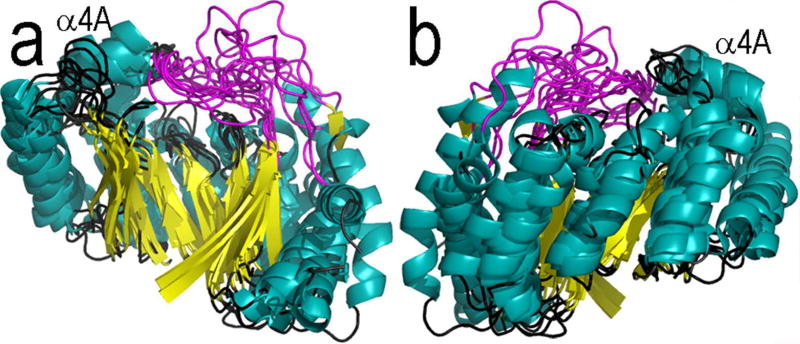
Figure 3. Superposition of seven Radical SAM core domains, colored as in Figure 1.
Here are shown two side views of the Radical SAM cores of PFL-AE, HemN, BioB, MoaA, LAM, TYW1, and HydE, from (a) the front and (b) the back.
The lack of closure of the partial barrel results in exposure of one face of the β sheet, which forms what is here referred to as the barrel’s lateral opening. This opening can be “covered” or “plugged” by protein elements outside of the Radical SAM core, typically from the C-terminal region with some minor contributions from the N-terminus or another molecule (Figure 2). It is within this lateral opening that the active site resides. The essential 4Fe-4S cluster and AdoMet bind via loops emanating from core β strands. Except in the case of ThiC, the cluster binding loop follows β1 and harbors the canonical CX3CXϕC motif (Figures 2 – 4). Binding of the 4Fe-4S cluster and AdoMet is discussed in more detail in the following sections.
Figure 4. Structure based sequence alignment of the Radical SAM enzymes.
The seven sequences, including both structurally characterized TYW1 sequences, are aligned with the main secondary structural elements labeled above the alignment. The Radical SAM core is highlighted in yellow (strands) and teal (helices). Residues of interest are colored as follows: the CX3CXϕC motif cysteines are in red; residues that contact AdoMet (or the TYW1 residues that are expected to contact AdoMet based on an analysis of the structures) in green; residues that contact the substrate, blue; residues that contact a cofactor, pink; and residues that contact both AdoMet and the substrate, orange. For clarity and with the exception of PFL-AE, each sequence was truncated as follows: HemN (33 – 331), BioB (23 – 313), MoaA (1 – 285), LAM (96 – 381), phTYW1 (30 – 342), mjTYW1 (30 – 311) and HydE (35 – 335). Residues of the four motifs (see text) are boxed in red and identified as follows: red stars correspond to the cysteines of the cluster-binding loop; red circles, the GGE motif; red triangles, the GxIxGxxE motif; and red squares, the conserved structural motif. A previously published alignment20 was used as a starting point for this alignment. It was then adjusted manually to reflect the exact structural elements of each enzyme and, in the case of the GxIxGxxE motif, to align the residues involved in conserved hydrogen bonding networks.
Superimposition of the Radical SAM core from multiple enzymes shows that the structure of this domain is very highly conserved (Figure 3). The core domain is defined here as beginning at the N-terminus of the strand that leads into the cluster-binding loop (PFL-AE R21, HemN L53, BioB Q41, MoaA D15, LAM R116, phTYW1 C71 and E58 of HydE) and ending at the C-terminus of the sixth strand (PFL-AE P200, HemN N241, BioB M223, MoaA E195, LAM Q291, phTYW1 A273 and HydE T85), at which point the structures begin to diverge. The first section of the core, the first strand and the cluster-binding loop, differ the most (Figure 3). The remaining protein fold shows very little variation, with strands β2 – β6 and the helices that follow them almost identically positioned in each structure. The helices of the core show more variation than the strands, and one helix of note, helix α4A (Figure 3), is missing in the LAM structure (Figure 4). Helix α4A, which is located on the loop connecting β4 and α4, is positioned along the top of the partial barrel, with the helix C-terminus pointing towards the cluster. Finally, the structures begin to diverge at strand β6 and the loop following it in order to appropriately accommodate substrate.
4.2. Protein elements outside of the Radical SAM core
The Radical SAM core is typically located at or near the N-terminus of the molecule, and the length and sequence of the C-terminal region is highly divergent between the different Radical SAM subfamilies. While the C-terminal regions contribute to substrate specificity, explaining some of the observed differences between subfamilies, a significant portion of the substrate binding site is provided by the core itself. Although it is simpler to think of these enzymes as containing separate domains, with the substrate binding site at the interface of those domains, examination of the Radical SAM structures (Figure 2) shows that this is not necessarily true. HemN does appear to have a separate C-terminal domain (Figure 2b), but PFL-AE, BioB, MoaA, LAM and HydE each contain additional C-terminal (and N-terminal) protein elements that simply extend or complete the core domain. For example, in BioB and HydE, two β strands are added to complete a (β/α)8 barrel. These additional non-core regions can complete the binding site of substrate or an additional cofactor (such as the extra FeS clusters of BioB, MoaA and HydE) and/or form oligomerization surfaces as in the HemN, BioB, MoaA and LAM structures. See the Supporting Information and Figures S1–S7 for a detailed discussion of the overall fold of each enzyme.
5. The FeS cluster
5.1. Location of the 4Fe-4S cluster binding site
The most recognized Radical SAM enzyme feature is the highly conserved CX3CXϕC motif, which binds an anaerobically stable, site-differentiated 4Fe-4S cluster1. The unique iron of the cluster is coordinated by the methionyl moiety of a bound molecule of AdoMet26–28.
The 4Fe-4S cluster is bound at the top (C-terminal end) of the Radical SAM partial β-barrel core domain, located above the lateral opening discussed above (Figures 2 and 3). The CX3CXϕC motif resides on the loop following the first β strand of the Radical SAM core (termed the cluster-binding loop). The length of the cluster-binding loop varies from enzyme to enzyme, depending on the spacing of the β/α motifs surrounding it. PFL-AE and LAM have cluster-binding loops of similar length and orientation, but those of the other enzymes differ widely (Figure 3). The loop winds around the cluster from “behind” to the front of the cluster, towards the lateral opening of the Radical SAM core. Residues following the CX3CXϕC motif are therefore located near the substrate. This loop may also provide part of the interaction surface for binding of the physiological reductant (see below).
5.2. The environment surrounding the cluster
The nature of the chemistry catalyzed by the Radical SAM enzymes suggests that the 4Fe-4S cluster should be fully protected from solvent, and this seclusion is for the most part observed in all of the structures (with the exception of MoaA, which may undergo further conformational changes in the presence of additional reaction components). The cluster is typically located approximately 7–10 Å from the nearest protein surface, which is formed opposite the lateral opening by the cluster-binding loop. Because the 4Fe-4S cluster binds at the C-termini of the core β strands but inside the ring of helices, it is buried by the loops at the top of the partial barrel. Conserved protein elements involved in burial of the cluster include helix α4A and the aromatic residue of the CX3CXϕC motif. This aromatic residue is observed in all Radical SAM enzymes except the aRNR activase. Helix α4A is structurally conserved in all of the Radical SAM enzymes with an interesting variation in LAM, where this helix is not part of the Radical SAM core; rather, it is replaced by a helix contributed by the N-terminal helical domain of this enzyme. Finally, several of the enzymes, PFL-AE and HemN in particular, have additional long loops immediately following the Radical SAM core that fold over and presumably help bury the cluster.
5.3. Interactions between the 4Fe-4S cluster and AdoMet
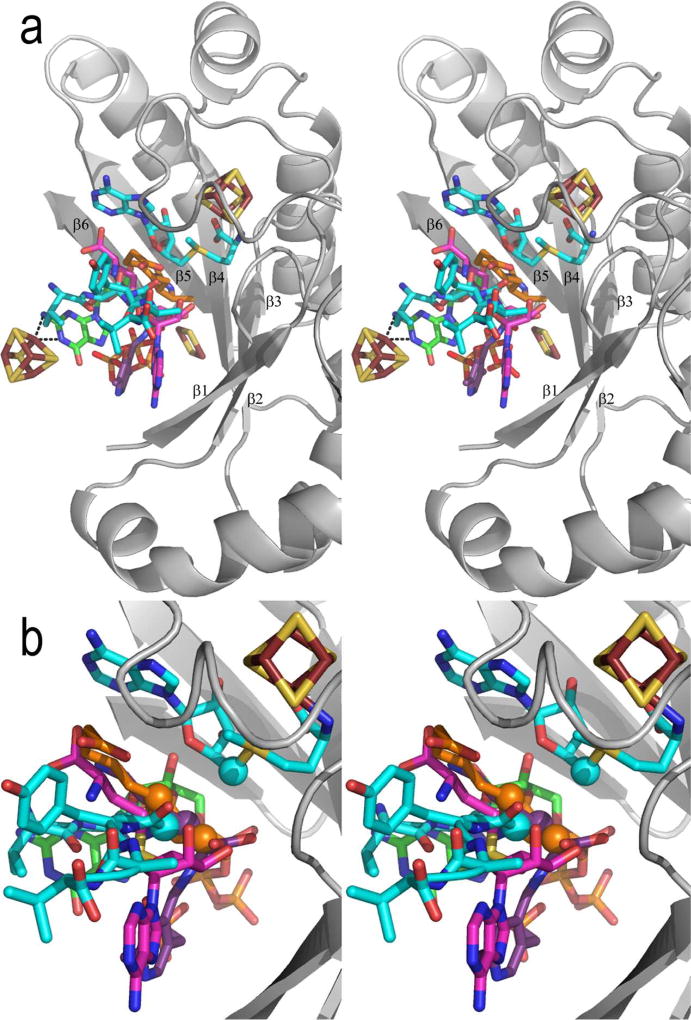
These Radical SAM enzyme crystal structures confirmed the results of several spectroscopic studies that showed that AdoMet binds directly to the 4Fe-4S cluster26–30,34 through its amino nitrogen and carboxylate group. Ligation to the cluster presumably helps to secure AdoMet in the binding site and properly position it for radical generation. Superimposition of six AdoMet-bound 4Fe-4S clusters reveals similar positioning with some variation (Figure 5); the biggest differences being in the position of the methyl group of AdoMet (Table 2). Some of this variation may be artifactual, due to differences in the resolution of the structures (1.62 to 3.4 Å) or discrepancies in refinement procedures. Other variation may reflect the manner in which protein side chains in each enzyme contact the AdoMet. Overall, the postion of the sulfur of AdoMet fairly constant, consistent with the theory that electron transfer occurs between an atom of the cluster and the AdoMet sulfur5,27,28,30,34,36. Interestingly, the average distance between the AdoMet sulfur and the closest Fe of the cluster is shorter (3.4 Å) than the average distance between the AdoMet sulfur and the closest sulfur (3.8 Å), in agreement with recent DFT calculations that point to Fe as the likely mediator of electron transfer36. In the following section, the AdoMet binding site is examined in more detail to understand more fully how the enzymes ligate AdoMet in order to produce 5'dA•.
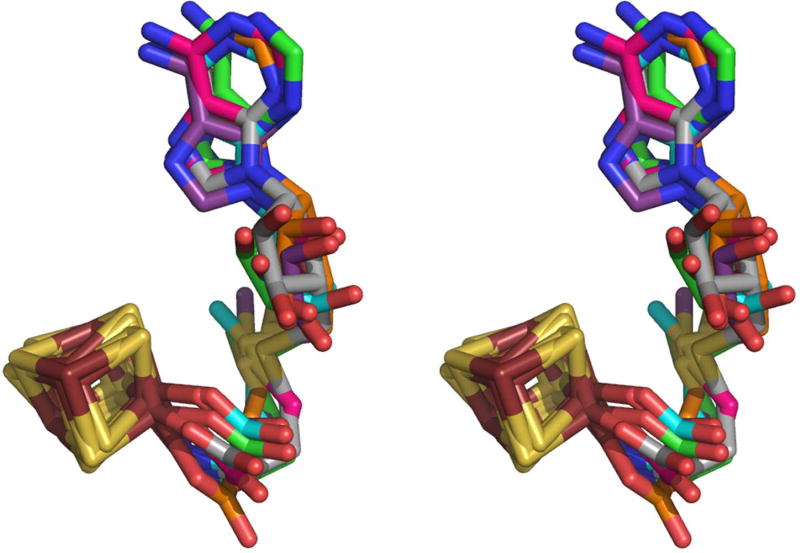
Figure 5. Overlay of 4Fe-4S clusters and bound AdoMet from six Radical SAM enzymes, shown in stereoview.
Colors are as follows: iron, ruby; sulfur, gold; oxygen, red; nitrogen, blue. In order to distinguish the six enzymes, the AdoMet carbons of each enzyme are colored as follows: teal, PFL-AE; magenta, HemN; orange, BioB; green, MoaA; purple, LAM; grey, HydE.
Table 2.
4Fe-4S cluster to AdoMet distances in Å.
| Fe – AdoMet N | Fe – AdoMet O | Fe – AdoMet C | Fe – AdoMet S | Fe – AdoMet CH3 |
S – AdoMet S | |
|---|---|---|---|---|---|---|
| pept-PFL-AE | 2.2 | 2.2 | 3.0 | 3.2 | 3.5 | 3.9 |
| HemNa | 2.6 | 2.2 | 3.1 | 3.5 | 4.4 | 3.6 |
| BioB | 2.4 | 2.5 | 3.0 | 4.0 | 5.4 | 4.2 |
| AdoMet-MoaA | 2.3 | 2.0 | 2.9 | 3.2 | 4.4 | 3.4 |
| LAM | 2.0 | 2.0 | 3.0 | 3.1 | 4.5 | 3.9 |
| HydE | 2.3 | 2.3 | 3.1 | 3.4 | N/A | 3.8 / 4.1 |
| Spectroscopic studies of PFL-AE | --------- | --------- | 3.3 | --------- | 4–5 | --------- |
The structure HemN contains a mixture of AdoMet enantiomers. The physiologically relevant enantiomer was used to obtain the values in this table and to prepare all figures.
6. AdoMet binding
6.1. AdoMet conformation
The conformation of AdoMet observed bound to the Radical SAM enzymes is similar in each structure (Figure 5). The molecule always adopts an anti conformation at the glycosidic bond in these enzymes, but varies in terms of the puckering of ribose (3'-endo in the majority of the enzymes, with PFL-AE and MoaA as the exceptions). In terms of the position of the AdoMet methionine with respect to the 5'-dA moiety, AdoMet can assume a variety of conformations, either “folded” or “extended” based on the torsional angle at atoms O4'-C4'-C5'-SB (referred to as the ψ torsional angle)107. The lowest energy extended conformation of AdoMet has a ψ torsional angle of ~180°, compared to <90° for folded conformations. In contrast to the majority of protein structures with bound AdoMet, all of the structures of Radical SAM enzymes bind AdoMet in the folded conformation, with the sulfonium located close to O4' of the ribose ring. HemN is the outlier of the group on this point, binding AdoMet with a ψ torsional angle of −112.80°, compared to −64.29° (PFL-AE), −48.75° (BioB), −73.96° (MoaA), −60.42° (LAM), and −80.78° (HydE). The implications of the conformation of AdoMet bound by an enzyme are unclear, though it has been observed that when AdoMet is bound with the purpose of serving a reactive role (transfer of one of the groups bound to the sulfonium), it adopts an extended conformation, and when bound as an activator or substrate, as in MetJ and AdoMet decarboxylase, it binds without a preference for the extended ψ torsional angles107. The overall conformation AdoMet adopts when bound to a Radical SAM enzyme is likely important to allow correct positioning of the AdoMet sulfur atom and enable electron transfer from the cluster to that sulfur atom (Figure 5, discussed above).
The anchoring of AdoMet by an iron-sulfur cluster via the methionyl amino and carboxyl atoms had, of course, never been observed in a protein before structural characterization of a Radical SAM enzyme, all of which employ a unique structural motif for binding AdoMet.
6.2. General properties of the AdoMet binding site
In all of the Radical SAM enzyme structures discussed here47,51–56, AdoMet is observed bound across the top of the partial or full TIM barrel in a binding site made up of residues from each of the core β strands (Figures 4 and 6). The binding site is hydrophilic and provides residues for several specific interactions with AdoMet (see below). Although little conservation is observed across the superfamily, many of the residues involved in binding AdoMet and/or substrate are highly to fully conserved within individual Radical SAM subfamilies, such as the BioB protein subfamily.
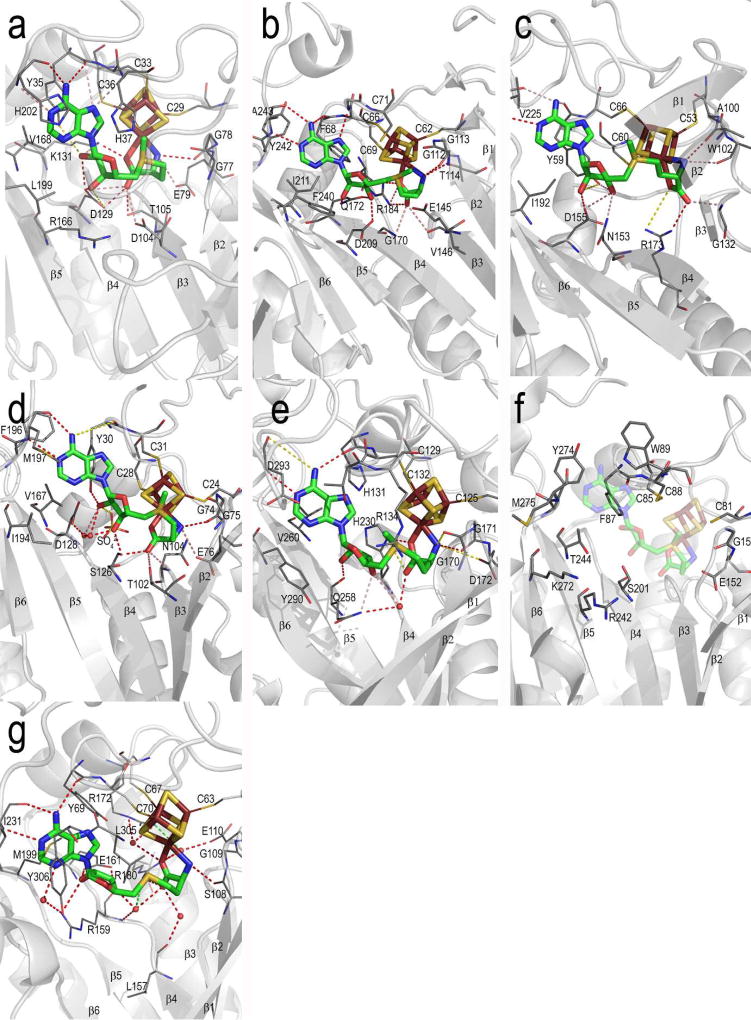
Figure 6. Details of Radical SAM enzyme AdoMet binding site.
The protein backbone is shown as grey cartoons, with AdoMet and the 4Fe-4S cluster shown in sticks (AdoMet, green carbons; iron, ruby; sulfur, gold) and core β strands labeled 1 – 6. Protein sidechains that interact with AdoMet are shown as lines with carbons colored dark grey. Hydrogen bonding contacts are shown as red (within 3.2 Å distance), green (3.2 – 3.7 Å) or yellow (more than 3.7 Å) dashed lines. Shown in this figure are the AdoMet binding sites of (a) PFL-AE (b) HemN (c) BioB (d) MoaA (e) LAM (f) the putative AdoMet binding site of phTYW1, shown with the superimposed 4Fe-4S cluster and AdoMet of the MoaA structure and (g) HydE. See Figure S8 for these images in stereoview.
As discussed above, the chemistry performed by the Radical SAM enzymes suggests that the active site would require burial within a protected protein environment during catalysis to prevent quenching of the radical. This sequestration of AdoMet from solvent is indeed observed in the BioB, LAM, HydE and substrate-bound PFL-AE structures52,53,55,56. The HemN and MoaA structures do not provide a complete view of the substrate-bound forms of those enzymes (Table 1 and above), and as one would expect, their active sites are solvent exposed47,51,54. Further investigation of these two enzymes, and structural characterization of more of the Radical SAM superfamily members, will clarify this aspect of catalysis.
6.3. Overall description of the AdoMet binding site
Low sequence conservation within the Radical SAM superfamily may arise in part from the location of the AdoMet binding site in these proteins. Because AdoMet binds across the top of the β barrel-like sheet, the residues of the binding site originate from loops following each β strand, and are therefore dispersed throughout the primary sequence (Figure 4). This dispersion hinders identification of a specific AdoMet binding motif beyond the previously identified glycine-rich region1, or GGE motif20. Low sequence similarities notwithstanding, comparison of available CX3CXϕC-containing Radical SAM structures shows that they employ remarkably similar modes of interaction with AdoMet (Figure 6, see Figure S8 for stereoviews of the AdoMet binding site). Examination of the dendrogram visualization of the Radical SAM core domains constructed by Sofia et al.1 demonstrates that the Radical SAM structures we now have are distantly related within the superfamily and represent a good cross-section of the sequence space covered by the individual members. Therefore, the structural comparison conducted here allows us to provide a general description of AdoMet binding in this superfamily that is likely applicable to all members that are not in the ThiC-like subclass (Figure 4). TYW1 is excluded from the following discussion as AdoMet is not observed bound in the active site.
6.3.1. The AdoMet methionyl moiety
The methionyl carboxylate and amino groups of AdoMet ligate the unique iron of the Radical SAM 4Fe-4S cluster26–28 in order to help anchor and orient the molecule for electron transfer, and possibly to alter the redox potential of the 4Fe-4S cluster and/or AdoMet82,84. In each structure, the amino group is observed binding close to the C-terminus of strand β2, making hydrogen bonds with residues of the “GGE motif” – also termed the “glycine rich region” – located on that strand (Figures 4, 6, and Figure S8; PFL-AE G77, G78, E79; HemN G112, G113, T114; BioB A100, A101, W102; MoaA G74, G75, E76; LAM G170, G171, D172; HydE S108, G109, E110). The hydrogen bonds provided by this motif are the only interactions observed to the amino group, and ensure the correct orientation of this part of AdoMet in the active site, as well as proper coordination of the unique iron by the methionyl moiety. In contrast, much more variation is observed with respect to contacts with the AdoMet carboxylate by the Radical SAM enzymes. In fact, it is this particular interaction with AdoMet that varies the most between the individual enzymes in terms of location within the primary sequence of the residues involved, with residues originating from β3, the loop following β3, β4 and α4 (Figures 4, 6, and S8). The majority of the enzymes use a conserved arginine, lysine or histidine (PFL-AE K131; HemN R184; BioB R173; LAM H230; HydE R180) to bind and orient the carboxylate, while the MoaA binding site provides threonine and serine hydroxyls for hydrogen bonding (MoaA T102, S126). Positioning of this portion of AdoMet is further fine tuned by interactions with backbone atoms (PFL-AE T105; HemN sV146 and G170; BioB G132; MoaA N104), additional sidechain atoms (PFL-AE D104 and D129) or, via protein-bound water molecules (Q258 of LAM; HydE E110, L157, R159, and R172). These variations in binding likely yield the somewhat different conformations of the AdoMet methionine in the structures discussed here (Figure 6 and S8).
6.3.2. The AdoMet ribose
Some variation in the Radical SAM enzymes is also evident in the details of their interactions with the AdoMet ribose. Residues responsible for interacting with the ribose originate mainly from strands β4 and β5 (Figures 4, 6, and S8). Though in some of the structures the hydroxyl of the CX3CXϕC motif tyrosine (PFL-AE Y35; BioB Y59; MoaA Y30) is within 4 Å of a ribose hydroxyl, it is unlikely that this residue plays a major role in ribose binding. This residue is not fully conserved (though it is always aromatic) and appears to be most important in terms of adenine binding (see below) and protection of the cluster from solvent. The ribose hydroxyls can interact with charged or polar protein sidechains directly (PFL-AE D129; HemN Q172, D209; BioB N153, D155; MoaA S126; LAM Q258; HydE R159, E161) or via protein-bound water molecules (MoaA D128; HydE R159). Finally, with the exception of both BioB and HydE, these Radical SAM enzymes have a highly conserved charged or polar residue within strand β5 that interacts with or is located very near the AdoMet ribose and/or substrate (PFL-AE R166; HemN D209; MoaA N165; LAM Q258). Within each enzyme subfamily, that residue is positioned close to the ribose hydroxyls and C5' of AdoMet, as well as to the substrate. Moreover, many Radical SAM enzyme subfamily members, not just those with known structure, appear to have a similarly conserved residue at the same place in β5 (data not shown). The high conservation and position of that residue in the structures suggests an important role during catalysis such as in the orientation of 5'dA• and/or substrate for proper hydrogen atom abstraction. Recent mutagenesis studies on BioB are consistent with this idea108.
6.3.3. The AdoMet adenine moiety
When bound to a Radical SAM enzyme, the adenine moiety rests against 3–5 hydrophobic residue sidechains and is specifically recognized by 3–4 hydrogen bonds made to protein backbone atoms. This portion of the binding site is made up mainly by residues from β strands 5 and 6, with contributions from the CX3CXϕC motif of the cluster-binding loop (Figures 4, 6, and S8). The interactions with adenine are maintained in the superfamily and are more easily predicted than other interactions, though enzyme evolution has resulted in a variety of compensatory mutations, particularly at those residues involved in protein backbone to AdoMet hydrogen bonds. Hydrophobic interactions are invariably provided by three specific regions of the primary structure (Figures 4, 6, and S8): (1) the conserved aromatic residue of the CX3CXϕC motif (Y35 in PFL-AE; F68 in HemN; Y59 in BioB; Y30 of MoaA; H131 of LAM; Y69 of HydE), (2) a residue from β5, located at position 2 of a conserved motif utilized in part for adenine binding, termed the GxIxGxxE motif (V168 of PFL-AE; I211 of HemN; I192 of BioB; V167 of MoaA; V260 of LAM; M199 of HydE) and (3) one or two residues located at the end of β6 that form a conserved structural motif (L199, H202 of PFL-AE; F240, A243 of HemN; N222,V225 of BioB; I194, M197 of MoaA; Y290, D293 of LAM; I231 of HydE). Hydrogen bonds are made to atoms N1 and N6 of AdoMet by backbone atoms. These backbone hydrogen bonds are made by the aromatic residue of the CX3CXϕC motif and by a residue from the adenine-binding structural motif of β6 (H202 of PFL-AE; A243 of HemN; V225 of BioB; M197 of MoaA; D293 of LAM; I231 of HydE). Lastly, N7 of AdoMet is in some cases hydrogen bonded by a backbone atom from the residue in the CX3CXϕC motif (PFL-AE Y35, H37; HemN G70).
6.4. AdoMet binding motifs in the Radical SAM superfamily
The solution of a Radical SAM structure was crucial for identification of the AdoMet-binding protein fold and these motifs. The current availability of several structures has allowed identification of a conserved structural motif in β6, as well as clarification of a sequence motif in β5 that had been previously identified (GxIxGxxE)20, both of which interact with the AdoMet adenine (Figure 4). The very low sequence homology between members of the Radical SAM superfamily prevents unambiguous naming of the motifs in a manner incorporating residue names. Therefore, the motif names do not always correspond to the sequence of each enzyme. The AdoMet binding motifs will be referred to in this discussion by the names they have already been given, and this section will attempt to clarify the function of each motif.
The CX3CXϕC motif of the Radical SAM superfamily functions mainly in ligation of the cluster and formation of the AdoMet adenine binding site (see above). The “GGE motif” or “glycine-rich region” observed at the end of strand β2 invariably binds the amino group of AdoMet, aiding in proper ligation of the cluster’s unique iron. This motif also forms the wall of this side of the active site and maintains the structure of the loop region after β2.
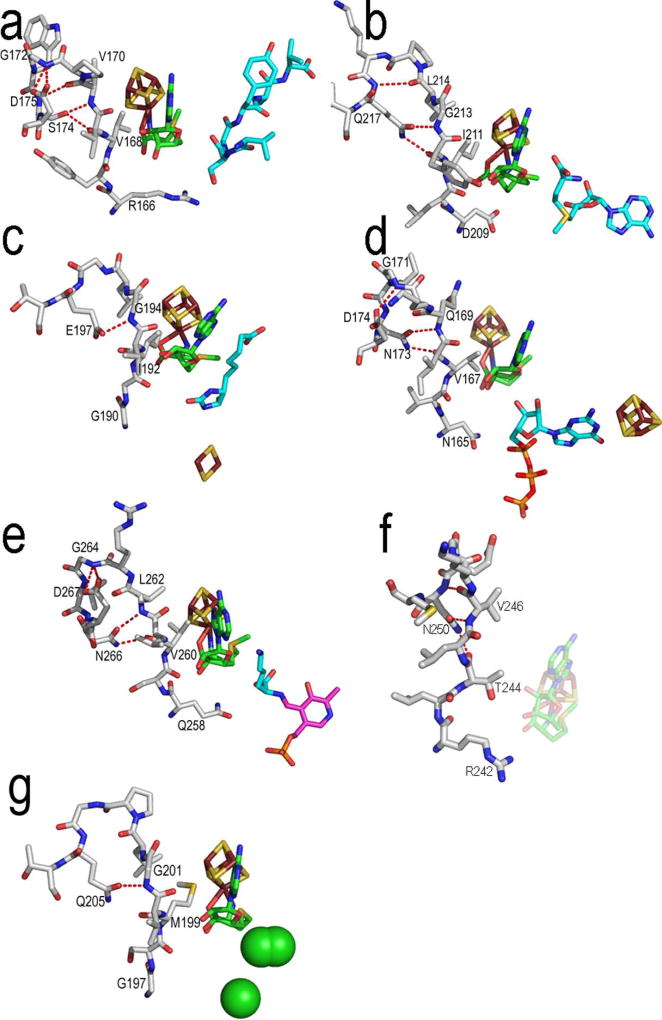
The GxIxGxxE motif (β5) and the β6 structural motif of the Radical SAM superfamily form most of the AdoMet adenine binding site. The GxIxGxxE motif, named in reference to the BioB sequence, provides hydrophobic interactions to the adenine portion of the binding site at its second conserved position (Figures 4, 6, 7, S8 and S9). Also, through one or more conserved sidechain-to-backbone interactions, this motif helps to preserve the structure of the AdoMet binding site as well (Figures 7 and S9). For example, the interaction observed in BioB from E197 (position 5) to the backbone nitrogen of G194 (position 3) is found in each of these seven structures, though the residue at position 5 in the other structures (with the exception of HydE) also interacts with the carboxyl backbone atom of the residue at position 2 (Figures 4 and 7). In addition, PFL-AE, MoaA and LAM share a conserved sidechain-to-backbone interaction from position 6 (PFL-AE D175; MoaA D174; LAM D267) to the backbone amino group of the residue in position 4. Sequence alignments suggest that this particular interaction may be conserved in several other Radical SAM enzymes, such as PylB, NifB, BssD and possibly ThiH (data not shown). Finally, position 1 of the GxIxGxxE motif is more difficult to characterize, as it is highly conserved but its identity differs in the individual Radical SAM subfamilies. As discussed above, the positioning of this residue near the AdoMet ribose and/or substrate in most of the known structures (Figure 7), along with its high conservation within each subfamily, implicates it as an important residue.
Figure 7. The GxIxGxxE motif.
AdoMet and the 4Fe-4S cluster and the protein residues making up this motif are shown in sticks, colored as in Figure 6. Shown are (a) PFL-AE (b) HemN (c) BioB (d) MoaA (e) LAM (f) phTYW1 with the 4Fe-4S cluster and AdoMet of the superimposed MoaA structure (shown as transparent sticks) and (g) HydE. Note that, with the exception of BioB and HydE, the sidechain of the first residue of the motif is positioned similarly with respect to the AdoMet ribose and atom C5'. Each protein also has a similar sidechain-to-backbone hydrogen bond in the loop following β5 (shown as red dashed lines). Three bound chloride ions observed in the HydE structure are displayed as green spheres. See text for more details and Figure S9 for these images in stereoview.
The β6 structural motif, which does not have a consensus sequence, provides hydrophobic interactions to the adenine from the residue in position 1 (with the exception of BioB, which uses this residue to interact with substrate, and HydE). The residue in the second position of this motif hydrogen bonds to the AdoMet adenine N1 and N6 atoms via its backbone amino nitrogen and carboxyl oxygen atoms (Figures 4 and 6).
6.5. Deviations and variations in AdoMet binding between the subfamilies
The major differences in AdoMet binding between these enzymes are readily apparent from Figures 4 and 6. The modes of interaction adopted by these enzymes to bind the AdoMet ribose and carboxylate groups specifically vary widely. As mentioned above, AdoMet carboxylate binding by these enzymes governs the position of the methionyl moiety, resulting in the elongated or compact conformations observed (Figure 5). For example, R173 of BioB is located at a distance farther away from the binding site, resulting in a more elongated methionyl conformation, while the location of LAM H230 in the center of the binding site causes the methionyl group to bend inwards (Figures 5 and 6). Comparison of the ribose binding interactions are less straightforward, with water mediating the contacts in MoaA and LAM, involvement of backbone atoms in LAM, and the use of both charged and polar residues in HemN and BioB. Beyond use of residues from the same strands, and the presence of a Glu or Asp nearby (with the exception of LAM), the Radical SAM enzymes share little similarity in this region of the AdoMet binding site.
Several minor differences are observed between these structures in their mode of interaction with the adenine of AdoMet, including the number and identity of the contacting hydrophobic residues. The details of the hydrogen bonding to N6 and N7 of the adenine ring also differ slightly. These differences are likely a result of experimental differences such as presence or absence of substrate, the resolution at which the structure was refined, or effects of the crystallization conditions. Overall, the AdoMet adenine binding sites are more or less the same.
7. Implications of AdoMet binding site architecture on function and reactivity
7.1. Tailoring the reaction to specific substrates
The parts of the AdoMet binding site that are conserved across the Radical SAM enzymes are limited to those interactions that are made to the adenine and methionyl amino group. This conservation results in a similar placement of those parts of AdoMet in all of the structures (Figure 5) and variations in the other groups, with the exception of the sulfur atom. The sulfur atom in each structure is located in roughly the same position, presumably to allow and optimize electron transfer to AdoMet from the cluster. The placement of the ribose and carboxyl portions of AdoMet differ more significantly in each enzyme than do the AdoMet adenine and methionyl amino groups, which likely allows each enzyme to tailor the reaction to its unique substrate. This theory is consistent with mutagenesis studies conducted on BioB, which showed that mutation of N153 or D155 (both of which interact directly with the AdoMet ribose) resulted in altered or abrogated activity108. Conformational flexibility of AdoMet presumably allows careful control over the position of the molecule to ensure proper hydrogen atom abstraction from substrate upon generation of 5'-dA•.
7.2. AdoMet usage as cofactor or cosubstrate
An interesting question concerning the AdoMet binding site of these enzymes is whether any specific structural features accompany use by some Radical SAM enzymes, LAM79 and SPL109 in particular, of AdoMet as a cofactor instead of as a cosubstrate. Comparison of AdoMet binding sites has not identified any obvious structural features that are related to this question. AdoMet does not appear to have more extensive interactions in LAM as compared to the other enzymes; indeed, the HemN and MoaA binding sites seem to provide more numerous contacts to AdoMet. R134 of LAM does appear capable of physically restricting movement of AdoMet out of the binding site by folding across the methionyl moiety of the cofactor to contact substrate (Figure 6 and S8) and spectroscopic studies have shown that Met remains bound to the cluster after cleavage of AdoMet during turnover33. However, R134 is not conserved in SPL, and the crystal structure of MoaA with 5'-GTP shows the AdoMet cleavage products 5'-dA and Met bound to the enzyme, demonstrating that the simple ability to retain the AdoMet cleavage products does not alone determine how the enzyme uses AdoMet. Other factors such as the requirements of the specific chemical reaction being catalyzed must govern this difference in AdoMet usage. For example, in the LAM reaction, there is no net change in oxidation state of the substrate, whereas in the PFL-AE reaction, a proton and electron are transferred irreversibly from substrate to 5'-dA•, and in the HemN reaction, two of the propionate side chains of coproporphyrinogen III are oxidized by two electrons each, thus requiring an additional electron acceptor46,64,110. Structural characterization of SPL or identification and characterization of another Radical SAM enzyme that uses AdoMet as a cofactor will be necessary to clarify the AdoMet binding site characteristics, if any exist, involved in this aspect of catalysis.
7.3. AdoMet reaction stoichiometry
While some Radical SAM enzymes, such as LAM12,13 and PFL-AE59, require one AdoMet to generate a single radical species, others - such as RlmN - appear to require two: one for radical generation and one for methyl transfer111. Although initially controversial112,113, still other enzymes such as BioB114–117, LipA118 and HemN63 appear to require two AdoMet molecules per single turnover, both for radical generation. Three theories have been proposed to explain the observed stoichiometry of AdoMet cleavage in BioB, LipA and HemN. First, as originally proposed for LipA, each individual protein monomer could carry out AdoMet cleavage once, releasing a stable intermediate and the AdoMet cleavage products118. A second possibility is that each enzyme binds and cleaves one molecule of AdoMet at a time, releasing the cleavage products and then binding a second AdoMet molecule, all the while retaining the substrate intermediate. This possibility is the most consistent with what we know of BioB. There does not appear to be room in the BioB active site for a second AdoMet molecule to bind52, and only minor readjustments of the BioB structure would be needed to allow release of the AdoMet cleavage products52. Further, the first radical generation yields an intermediate, 9-mercaptodethiobiotin, which is not released by the enzyme, requiring the binding of a second molecule of AdoMet to the same active site to complete the reaction117,119. Consistent with this aspect of the second theory, both sulfur atoms incorporated into LipA’s lipoate product are likely derived from the cluster of the same LipA polypeptide45, suggesting that one monomer binding a total of two AdoMet molecules is responsible for one full turnover. The final possibility, suggested by the observation that two AdoMet molecules bind within the active site in the HemN structure51, is that there are two distinct AdoMet binding sites on the enzyme. This second AdoMet molecule (termed SAM251) is either physiologically relevant or an artifact of the crystallization conditions, binding where substrate would normally bind. Layer et al. have proposed that SAM2 is involved in catalysis based on their modeling of the coproporphyrinogen III substrate in the active site, both with and without presence of SAM251. In their analysis, the propionate sidechains could be better accommodated in the presence of SAM251. Additionally, mutation of the residues contacting the structure’s SAM2 affects AdoMet cleavage63. However, the decreased AdoMet cleavage observed is also consistent with the theory that those specific residues form the substrate binding site (discussed in detail below). Although not fully resolved, growing sentiment in the field supports the second proposal discussed here, in which AdoMet is bound and cleaved sequentially, while the enzyme retains the substrate intermediate.
7.4. HemN’s second AdoMet (SAM2)
Several factors suggest an artifactual explanation of AdoMet binding to the SAM2 site. First, each of the four other structures with substrate present shows that substrate binds in a very specific location with respect to AdoMet, with the substrate atom from which hydrogen atom abstraction should occur approximately 4Å from the C5' atom of AdoMet (Figure 8). SAM2 of HemN is observed binding in this location. Second, the nature of the chemistry catalyzed requires close control of the orientation of each AdoMet as well as the substrate molecule involved in the reaction, and the structure of the SAM2 site (Figure S10) does not indicate the same high level of conformational control as exercised over the cluster-bound AdoMet. Although SAM2 is recognized reasonably well by HemN through six possible hydrogen bonds mainly to the ribose hydroxyls and through hydrophobic interactions with three HemN residues (Figure S10a), there are fewer direct contacts to SAM2 than to the first AdoMet, suggesting that recognition of SAM2 is less specific; indeed, the methionyl moiety of SAM2 is partially disordered51 (Figure S10b). If SAM2 binding were physiologically relevant, one would expect to see a tighter conformational control, as is wielded over the cluster-bound AdoMet (Figure 6) and the substrates in the other structures. Third, writing a mechanism for the reductive cleavage of the second AdoMet in the SAM2 site is challenging51,64, requiring either an electron transfer through the first AdoMet or an electron transfer from a substrate intermediate. Both of these modes of electron transfer are unprecedented, and due to the low reduction potential of a typical trialkylsulfonium ion84, seem unlikely. On the other hand, model systems have confirmed that iron-sulfur clusters can mediate electron transfer to and subsequent cleavage of AdoMet120,121. Further, studies on LAM indicate the value of the iron-sulfur cluster in lowering the barrier for reductive cleavage of AdoMet in an enzyme active site84. Since components of the HemN reaction such as the physiological electron acceptor have not been identified, a full mechanistic description for this enzyme awaits further biochemical and structural studies for clarification.
Figure 8. Stereoview of the substrate binding sites of the Radical SAM enzymes.
In (a), five structures with substrate bound are superimposed based on their Radical SAM cores. Shown is the Radical SAM core, 4Fe-4S cluster and AdoMet of PFL-AE only, with AdoMet carbons colored teal. The five “substrates” of the enzymes are shown in sticks, colored as follows: teal, PFL-AE; magenta, HemN; orange, BioB; green, MoaA; purple, LAM. PLP of LAM and the BioB 2Fe-2S, MoaA 4Fe-4S and HydE 2Fe-2S clusters are also shown, displayed as in other figures. In (b), five enzyme structures are superimposed on their 4Fe-4S clusters and AdoMet only, to give a more accurate comparison of the relative positions of the substrates with respect to AdoMet. The core backbone, 4Fe-4S cluster and AdoMet of PFL-AE are shown, and colors are as described in (a). C5' of AdoMet is shown as a sphere. The atoms from which hydrogen abstraction is known to occur (i.e. Cα of G734 of the peptide, C6 and C8 of dethiobiotin, and Cβ of lysine) are also shown as spheres.
8. Substrate binding to Radical SAM enzymes
8.1. Positioning of the substrate
Tight conformational control over substrate position is also likely to be necessary to afford correct hydrogen atom abstraction. Structural superposition shows that substrates bind in a very similar site on all of the enzymes (Figure 8). The Radical SAM substrate binding site is located within the lateral opening of the core’s partial TIM barrel, which corresponds to the center of the full TIM barrel of BioB. As discussed above, the substrate binding site is formed between the core region and structural motifs located outside the core, mainly at the C-terminus (Figure 4). The combined interactions between protein, cofactors and substrate result in a similar position of the substrate abstraction point (Figure 8b). Overall, the orientations of the substrates observed in the structures of PFL-AE, BioB and LAM result in similar distances between the AdoMet C5' atom and the atom from which a hydrogen atom is abstracted (PFL-AE, Cα of peptide G734, 4.1 Å; BioB, C6 and C9 of the dethiobiotin, 4.1 Å and 3.9 Å respectively; LAM, Cβ of lysine, 3.8 Å).
Other than their position with respect to the Radical SAM core, there are virtually no similarities between the substrate binding sites, reflecting the diversity of the substrates and reactions of the Radical SAM enzymes (see Figures S10 – S15 for stereoviews of the individual substrate and cofactor binding sites of HemN, PFL-AE, BioB, MoaA, LAM and HydE). The enzymes have very different protein motifs outside of the Radical SAM core, and the size and protein sidechain makeup of the substrate binding sites are also dissimilar. There is also no conservation noted in the locations of additional bound cofactors (Figure 8, discussed below). These variations are presumably necessary in order to ensure that the various substrates bind properly to allow the correct hydrogen atom abstraction. From a structural point of view, the substrate conformation is certainly one of the most important aspects of catalysis.
8.2. Additional cofactors in some Radical SAM substrate binding sites
In addition to protein sidechain interactions, some of the enzymes use a separate cofactor to help anchor their substrate, such as the PLP of LAM and the second 4Fe-4S cluster of MoaA. However, the role of additional cofactors is not always for substrate binding. For example, the 2Fe-2S cluster of BioB serves as a source of sulfur for biotin production52,119. While the role of the HydE 2Fe-2S cluster is less clear, it has recently been purposed to also serve as a sulfur source in the formation of the H cluster on HydF102. The positions of the additional cofactors differ accordingly between the enzymes (Figure 8), reflecting the plasticity of the Radical SAM fold in its ability to accommodate different substrates.
8.3. Conformational changes associated with substrate binding in Radical SAM enzymes
As discussed above, the radical reactions catalyzed by Radical SAM enzymes would likely require sequestration of the active sites from solvent during catalysis, as in the active sites of BioB and LAM. This assumed requirement for sequestration implies that conformational changes will occur upon substrate binding in most of the Radical SAM enzymes, as has been observed for PFL-AE upon comparison of the substrate-free and substrate bound structures of the enzyme55. In PFL-AE, the loop preceding the core plays a major role in substrate binding, providing what appears to be a critical interaction between a protein sidechain (D16) and the amino group of the peptide glycine residue (which corresponds to PFL G734), as well as steric interactions with the peptide (Figure S11). This loop, which carries a motif specific to all of the Radical SAM activases, (D/N)GxGxR, undergoes a major conformational change, swinging up into the active site and becoming fully ordered to properly bind substrate55. The arginine of this motif interacts with neighboring β strands, preserving the structural integrity of the Radical SAM core during the loop’s conformational change. However, the HemN and MoaA active sites appear particularly solvent exposed51,54. In the case of MoaA, no major conformational change is observed between the substrate-free and substrate-bound forms, leaving the active site very solvent exposed47,54. However, an additional protein, MoaC, is required for activity, and its function during catalysis is as of yet unclear. It is possible that MoaC helps to bury the active site in some way, either directly or by enabling a conformational change. Obviously, further structural studies are required to identify conformational changes associated with substrate binding in the Radical SAM enzymes, as only PFL-AE and MoaA have more than one structure available for identification of changes.
9. Reductant binding in Radical SAM enzymes
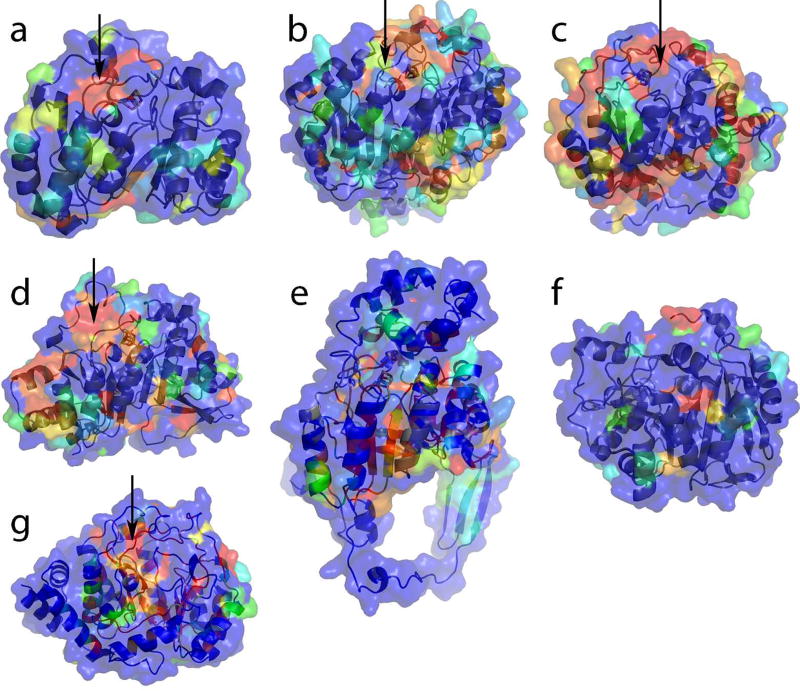
The physiological reductants for most of these enzymes are flavodoxin and ferredoxin, and they serve to reduce the 4Fe-4S cluster from the 2+ to the 1+ state. Since so many other aspects of the initial radical generation are conserved, the general location of the reductant binding site could be similar in each enzyme. Our laboratory52,55 and that of Dieter Jahn51 have proposed that the reductant binding site is the surface nearest the cluster-binding loop, formed mainly by this loop, with in some cases additional residues from loops following strands β2 and β4. Although this surface is conserved in most of the structures (Figures 9a–d, g and S16a–d,g), an extensive pattern of high conservation is not present on the surface of LAM or phTYW1 (Figures 9e, f and S16e, f). In the latter structure, disorder may prevent visualization of the full reductant binding site48. In LAM, the N-terminal helical domain sits atop the partial barrel above the 4Fe-4S cluster, obstructing the putative reductant-binding interface. It is not clear whether a conformational change could expose more of this surface, or a different surface on LAM interacts with the reductant. In fact, LAM may not require a reductant, as the [4Fe-4S]1+ cluster is regenerated during turnover9,79.
Figure 9. Surface conservation of Radical SAM enzymes suggests putative binding site for physiological reductant.
Shown are the cartoon representations and transparent surfaces of (a) PFL-AE, (b) HemN, (c) BioB, (d) MoaA, (e) LAM, (f) phTYW1 and (g) HydE, from the the opposite side of the partial barrel with respect to the lateral opening of the Radical SAM core, in roughly the same view as in Figure 3b. The transparent surface is colored as a rainbow according to the extent of sequence conservation with respect to the protein’s individual family, with red being 100% conserved and blue as 0% conserved. A BLAST search was conducted via the ExPASy proteomics server using the sequence of the enzyme that is structurally characterized as the query. The top matching sequences (up to 100 in number) were input for alignment by ClustalW. The alignment output by ClustalW was then used as input to ESPript, which calculated the extent of conservation of each residue. Arrows indicate proposed surface for interaction with reductant. See Figure S16 for stereoviews of each surface.
For most of these enzymes, the surface highlighted with an arrow in Figure 9 is an ideal candidate to interact with the physiological reductant. It is typically the closest surface to the cluster, within 7 to 10 Å in each of the structures, which is a reasonable distance for electron transfer122. Also, in order to prevent nonproductive cleavage of AdoMet, one would expect the reduction of the 4Fe-4S cluster to occur after substrate binds, when the active site is ready for catalysis. The location of this particular surface, on the opposite side of the enzyme from the lateral opening of the Radical SAM core and active site, allows for substrate to be bound to the enzyme at the same time as the reductant. Also the presence of a distant binding surface for reductant allows for multiple Radical SAM enzymes to use the same reductants while maintaining flexibility in their substrate preference.
10. Other known AdoMet-binding protein folds
The PDB (at the RCSB, http://www.rcsb.org/pdb) contains more than 290 total protein structures of known AdoMet-binding proteins, about half of which are unique (<95% identical by sequence), whose coordinates include AdoMet (88 total, 72 unique) or AdoHCys (207 total, 105 unique). The large majority of these proteins are methyltransferases (MTases). As discussed by Kozbial and Mushegian123, there are 15 distinct folds capable of binding AdoMet or AdoHCys, which can be divided into the following 4 categories: (1) Rossmann-like, (2) β barrel-like, (3) “Double-β” and (4) folds derived from other ligand-binding domains.
The most common fold utilized to bind AdoMet is the Rossmann-like domain. The typical MTase AdoMet-binding fold is a 7-stranded β sheet, adopting a mainly parallel orientation with one strand, typically β7, oriented antiparallel to the others (Figure S17). AdoMet binds straddling the top of the β sheet, at the C-terminal loops following strands β1, β2, β3 and β4. The Rossmann-like AdoMet-binding fold includes most of the MTases, such as catechol-O-MTase (COMTase)124 and the DNA:m5C MTases125, as well as some enzymes that bind AdoMet but are not MTases, like spermidine synthase126, the bacterial fluorinating enzyme127 and cyclopropane mycolic acid synthase128. Additionally, several classes of MTases employ a similar fold, such as the SPOUT MTases, thus named for the first identified enzymes of this class, SpoU and TrmD MTases129,130. The Rossmann-like domain is similar to the β barrel-like domain observed in the Radical SAM enzymes in that the loops near the C-terminal end of the β-sheet structure are responsible for binding AdoMet. Also, these fold types are both commonly found in the context of larger protein chains that contain both an AdoMet-binding region and a variable substrate-binding region. Low sequence conservation like that observed between members of the Radical SAM superfamily is also observed between the Rossmann-like AdoMet binding proteins.
The double-β fold can bind AdoMet between its two β sheets. Examples of the double-β fold include the SET domain (named because the first proteins identified with this conserved domain were the Su(var)3–9, E(Z) and Trithorax chromatin remodeling proteins123,131,132), such as that found in PrmA MTase133 (Figure S18), a class of MTases that transfer a methyl group to a nuclear protein lysine residue (Figure S18). Still other folds were generated by recruitment of broad-specificity ligand-binding folds to bind AdoMet, such as in the case of ACC synthase (AdoMet methylthioadenosine lyase, a member of the diverse PLP-dependant transferase family)134 and the MetJ repressor (Figure S18, a ribbon-helix-helix DNA-binding protein that uses AdoMet as a co-repressor)135. Finally, several enzymes, such as the methionine synthase (MetH) reactivation domain (Figure S18)136, do not fit easily into any of these categories. The usual “cup-like” shape of the MetH reactivation domain probably evolved to specifically recognize the MeCbl-binding domain of MetH. The Radical SAM enzymes clearly form their own unique class of AdoMet-binding proteins with little similarity to the other known AdoMet-binding folds.
11. Conclusions
Even with the exclusion of the structure of ThiC and RimO, structures of PFL-AE, HemN, BioB, MoaA, LAM, TYW1, and HydE provide an excellent cross-section of the superfamily allowing for a detailed description of the protein fold used for 5′dA• radical generation from AdoMet. These structures have allowed an analysis of the similarities and differences between the enzymes in terms of the core fold, AdoMet binding sites, overall AdoMet conformation, and tailoring of the protein chain to specific substrates. The Radical SAM core, with its observed (β/α)6 topology, is well-conserved and contains the canonical 4Fe-4S cluster binding motif and four low sequence identity AdoMet binding motifs. Though specific 4Fe-4S to AdoMet and protein to AdoMet interactions vary, the combined effects of these interactions appear to result in placement of the AdoMet sulfur atom in approximately the same position with respect to the 4Fe-4S cluster in each enzyme, which suggests a conserved mechanism for the radical-generating electron transfer step, consistent with recent calculations36. Examination of structures solved in the presence of substrate suggests that the substrate is bound by each enzyme such that the atom that undergoes hydrogen atom abstraction is located in approximately the same place with respect to C5' of AdoMet. Location of the second bound molecule of AdoMet at this particular place in the HemN structure suggests that this second AdoMet binding site is artifactual. Similar to the methyltransferases, the substrate binding sites of the Radical SAM enzymes are diverse and can involve various cofactors. Finally, a putative reductant binding site on the enzyme surfaces behind the 4Fe-4S cluster and opposite the substrate binding site is further suggested by analysis of the sequence conservation of surface residues. Our structure based sequence alignment, refined further by analysis of the additional structures, can now be used to predict more residues involved in AdoMet binding and catalysis in other superfamily members.
Although analysis of the available Radical SAM structures has yielded interesting insights into catalysis by the superfamily, important aspects such as the alteration of the 4Fe-4S and AdoMet reduction potentials to allow electron transfer, and the factors that govern AdoMet usage as a cofactor or cosubstrate, clearly need more study. Further, for the majority of these enzymes, catalysis beyond the initial radical generation step is only in the initial stages of characterization. The discovery of a new class of Radical SAM enzymes in the forms of ThiC and Dph2 is an important recent advance. The next few years will surely provide many new discoveries in terms of resolution of the remaining mechanistic questions and identification of diverse new Radical SAM enzyme functionalities.
Supplementary Material
Acknowledgments
The authors thank Yvain Nicolet and Joan B. Broderick for helpful discussions and JoAnne Stubbe, Stuart Licht, Anna Croft, and Rebekah Bjork for a critical reading of the text. This work was supported by the NSF (C.L.D.) and by a William Asbjornsen Albert Fellowship (J.L.V). C.L.D. is a Howard Hughes Medical Institution Investigator.
Biographies

CATHERINE L. DRENNAN is a professor of chemistry and biology at the Massachusetts Institute of Technology, and a professor and investigator with the Howard Hughes Medical Institute. She received an A.B. in chemistry from Vassar College and a Ph.D. in biological chemistry from the University of Michigan, working the laboratory of the late Professor Martha L. Ludwig. She was an NIH postdoctoral fellow with Professor Douglas C. Rees at the California Institute of Technology. She taught high school for several years prior to her graduate studies. In 1999, she joined the Massachusetts Institute of Technology. Her primary research interest is the use of X-ray crystallography to study the structure and mechanism of metalloproteins.

JESSICA L. VEY received a B.Sc. in chemistry from Temple University and a Ph.D. in biological chemistry from the Massachusetts Institute of Technology, where she worked in Professor Catherine Drennan’s laboratory on structural studies of iron-sulfur cluster assembly and their participation in radical generation. She is now a postdoctoral researcher at Vanderbilt University Medical Center, working in the laboratory of Professor Tina M. Iverson. Her research interests include X-ray crystallographic structural studies and analysis of conserved enzymatic folds.
Footnotes
Supporting Information Available. (1) Detailed description of the overall fold of each Radical SAM enzyme. (2) Figures S1 – S18. (3) Supporting Information References. This information is available free of charge via the Internet at http://pubs.acs.org/.
References
- 1.Sofia HJ, Chen G, Hetzler BG, Reyes-Spindola JF, Miller NE. Nucleic Acids Res. 2001;29:1097. doi: 10.1093/nar/29.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frey PA, Magnusson OT. Chem. Rev. 2003;103:2129. doi: 10.1021/cr020422m. [DOI] [PubMed] [Google Scholar]
- 3.Frey PA. FASEB J. 1993;7:662. doi: 10.1096/fasebj.7.8.8500691. [DOI] [PubMed] [Google Scholar]
- 4.Marquet A, Tse Sum Bui B, Smith AG, Warren MJ. Nat. Prod. Rep. 2007;24:1027. doi: 10.1039/b703109m. [DOI] [PubMed] [Google Scholar]
- 5.Wang SC, Frey PA. Trends Biochem. Sci. 2007;32:101. doi: 10.1016/j.tibs.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 6.Frey PA, Hegeman AD, Ruzicka F. Crit. Rev. Biochem. Mol. Biol. 2008;43:63. doi: 10.1080/10409230701829169. [DOI] [PubMed] [Google Scholar]
- 7.Atta M, Mulliez E, Arragain S, Forouhar F, Hunt JF, Fontecave M. Curr. Opin. Struct. Biol. 2010;20:684. doi: 10.1016/j.sbi.2010.09.009. [DOI] [PubMed] [Google Scholar]
- 8.Magnusson OT, Reed GH, Frey PA. J. Am. Chem. Soc. 1999;121:9764–9765. [Google Scholar]
- 9.Magnusson OT, Reed GH, Frey PA. Biochemistry. 2001;40:7773. doi: 10.1021/bi0104569. [DOI] [PubMed] [Google Scholar]
- 10.Magnusson OT, Frey PA. Biochemistry. 2002;41:1695. doi: 10.1021/bi011947w. [DOI] [PubMed] [Google Scholar]
- 11.Mansoorabadi SO, Magnusson OT, Poyner RR, Frey PA, Reed GH. Biochemistry. 2006;45:14362. doi: 10.1021/bi061586q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moss M, Frey PA. J. Biol. Chem. 1987;262:14859. [PubMed] [Google Scholar]
- 13.Baraniak J, Moss ML, Frey PA. J. Biol. Chem. 1989;264:1357. [PubMed] [Google Scholar]
- 14.Frey M, Rothe M, Wagner AF, Knappe J. J. Biol. Chem. 1994;269:12432. [PubMed] [Google Scholar]
- 15.Wagner AF, Demand J, Schilling G, Pils T, Knappe J. Biochem. Biophys. Res. Commun. 1999;254:306. doi: 10.1006/bbrc.1998.9931. [DOI] [PubMed] [Google Scholar]
- 16.Fontecave M, Mulliez E, Ollagnier-de-Choudens S. Curr. Opin. Chem. Biol. 2001;5:506. doi: 10.1016/s1367-5931(00)00237-4. [DOI] [PubMed] [Google Scholar]
- 17.Frey PA, Ballinger MD, Reed GH. Biochem. Soc. Trans. 1998;26:304. doi: 10.1042/bst0260304. [DOI] [PubMed] [Google Scholar]
- 18.Holliday GL, Thornton JM, Marquet A, Smith AG, Rebeille F, Mendel R, Schubert HL, Lawrence AD, Warren MJ. Nat. Prod. Rep. 2007;24:972. doi: 10.1039/b703107f. [DOI] [PubMed] [Google Scholar]
- 19.Eschenmoser A. Angew. Chem. Int. Ed. 1988;27:5. [Google Scholar]
- 20.Nicolet Y, Drennan CL. Nucleic Acids Res. 2004;32:4015. doi: 10.1093/nar/gkh728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duschene KS, Veneziano SE, Silver SC, Broderick JB. Curr. Opin. Chem. Biol. 2009;13:74. doi: 10.1016/j.cbpa.2009.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marsh ENG, Patterson DP, Li L. Chembiochem. 2010;11:604. doi: 10.1002/cbic.200900777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Petrovich RM, Ruzicka FJ, Reed GH, Frey PA. Biochemistry. 1992;31:10774. doi: 10.1021/bi00159a019. [DOI] [PubMed] [Google Scholar]
- 24.Lieder KW, Booker S, Ruzicka FJ, Beinert H, Reed GH, Frey PA. Biochemistry. 1998;37:2578. doi: 10.1021/bi972417w. [DOI] [PubMed] [Google Scholar]
- 25.Broderick JB, Henshaw TF, Cheek J, Wojtuszewski K, Smith SR, Trojan MR, McGhan RM, Kopf A, Kibbey M, Broderick WE. Biochem. Biophys. Res. Commun. 2000;269:451. doi: 10.1006/bbrc.2000.2313. [DOI] [PubMed] [Google Scholar]
- 26.Krebs C, Broderick WE, Henshaw TF, Broderick JB, Huynh BH. J. Am. Chem. Soc. 2002;124:912. doi: 10.1021/ja017562i. [DOI] [PubMed] [Google Scholar]
- 27.Walsby CJ, Hong W, Broderick WE, Cheek J, Ortillo D, Broderick JB, Hoffman BM. J. Am. Chem. Soc. 2002;124:3143. doi: 10.1021/ja012034s. [DOI] [PubMed] [Google Scholar]
- 28.Walsby CJ, Ortillo D, Broderick WE, Broderick JB, Hoffman BM. J. Am. Chem. Soc. 2002;124:11270. doi: 10.1021/ja027078v. [DOI] [PubMed] [Google Scholar]
- 29.Cosper MM, Jameson GN, Davydov R, Eidsness MK, Hoffman BM, Huynh BH, Johnson MK. J. Am. Chem. Soc. 2002;124:14006. doi: 10.1021/ja0283044. [DOI] [PubMed] [Google Scholar]
- 30.Chen D, Walsby C, Hoffman BM, Frey PA. J. Am. Chem. Soc. 2003;125:11788. doi: 10.1021/ja036120z. [DOI] [PubMed] [Google Scholar]
- 31.Henshaw TF, Cheek J, Broderick JB. J. Am. Chem. Soc. 2000;122:8331. [Google Scholar]
- 32.Padovani D, Thomas F, Trautwein AX, Mulliez E, Fontecave M. Biochemistry. 2001;40:6713. doi: 10.1021/bi002936q. [DOI] [PubMed] [Google Scholar]
- 33.Cosper NJ, Booker SJ, Ruzicka F, Frey PA, Scott RA. Biochemistry. 2000;39:15668. doi: 10.1021/bi0022184. [DOI] [PubMed] [Google Scholar]
- 34.Cosper MM, Cosper NJ, Hong W, Shokes JE, Broderick WE, Broderick JB, Johnson MK, Scott RA. Protein Sci. 2003;12:1573. doi: 10.1110/ps.0302203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kampmeier JA. Biochemistry. 2010 doi: 10.1021/bi101509u. Article ASAP. [DOI] [PubMed] [Google Scholar]
- 36.Nicolet Y, Amara P, Mouesca JM, Fontecilla-Camps JC. Proc. Natl. Acad. Sci. U. S. A. 2009;106:14867. doi: 10.1073/pnas.0904385106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dougherty MJ, Downs DM. Microbiology. 2006;152:2345. doi: 10.1099/mic.0.28926-0. [DOI] [PubMed] [Google Scholar]
- 38.Martinez-Gomez NC, Downs DM. Biochemistry. 2008;47:9054. doi: 10.1021/bi8010253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chatterjee A, Li Y, Zhang Y, Grove TL, Lee M, Krebs C, Booker SJ, Begley TP, Ealick SE. Nat. Chem. Biol. 2008;4:758. doi: 10.1038/nchembio.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martinez-Gomez NC, Poyner RR, Mansoorabadi SO, Reed GH, Downs DM. Biochemistry. 2009;48:217. doi: 10.1021/bi802154j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chatterjee A, Hazra AB, Abdelwahed S, Hilmey DG, Begley TP. Angew. Chem. Int. Ed. 2010;49:8653. doi: 10.1002/anie.201003419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu S, Milne GT, Kuremsky JG, Fink GR, Leppla SH. Mol. Cell. Biol. 2004;24:9487. doi: 10.1128/MCB.24.21.9487-9497.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Y, Zhu X, Torelli AT, Lee M, Dzikovski B, Koralewski RM, Wang E, Freed J, Krebs C, Ealick SE, Lin H. Nature. 2010;465:891. doi: 10.1038/nature09138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu X, Dzikovski B, Su X, Torelli AT, Zhang Y, Ealick SE, Freed JH, Lin H. Mol Biosyst. 2010;7:74. doi: 10.1039/c0mb00076k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cicchillo RM, Booker SJ. J. Am. Chem. Soc. 2005;127:2860. doi: 10.1021/ja042428u. [DOI] [PubMed] [Google Scholar]
- 46.Layer G, Verfurth K, Mahlitz E, Jahn D. J. Biol. Chem. 2002;277:34136. doi: 10.1074/jbc.M205247200. [DOI] [PubMed] [Google Scholar]
- 47.Hanzelmann P, Schindelin H. Proc. Natl. Acad. Sci. U. S. A. 2004;101:12870. doi: 10.1073/pnas.0404624101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Goto-Ito S, Ishii R, Ito T, Shibata R, Fusatomi E, Sekine SI, Bessho Y, Yokoyama S. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2007;63:1059. doi: 10.1107/S0907444907040668. [DOI] [PubMed] [Google Scholar]
- 49.Suzuki Y, Noma A, Suzuki T, Senda M, Senda T, Ishitani R, Nureki O. J. Mol. Biol. 2007;372:1204. doi: 10.1016/j.jmb.2007.07.024. [DOI] [PubMed] [Google Scholar]
- 50.Farrar CE, Siu KKW, Howell PL, Jarrett JT. Biochemistry. 2010;49:9985. doi: 10.1021/bi101023c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Layer G, Moser J, Heinz DW, Jahn D, Schubert WD. EMBO J. 2003;22:6214. doi: 10.1093/emboj/cdg598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Berkovitch F, Nicolet Y, Wan JT, Jarrett JT, Drennan CL. Science. 2004;303:76. doi: 10.1126/science.1088493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lepore BW, Ruzicka FJ, Frey PA, Ringe D. Proc. Natl. Acad. Sci. U. S. A. 2005;102:13819. doi: 10.1073/pnas.0505726102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hanzelmann P, Schindelin H. Proc. Natl. Acad. Sci. U. S. A. 2006;103:6829. doi: 10.1073/pnas.0510711103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vey JL, Yang J, Li M, Broderick WE, Broderick JB, Drennan CL. Proc. Natl. Acad. Sci. U. S. A. 2008;105:16137. doi: 10.1073/pnas.0806640105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nicolet Y, Rubach JK, Posewitz MC, Amara P, Mathevon C, Atta M, Fontecave M, Fontecilla-Camps JC. J. Biol. Chem. 2008;283:18861. doi: 10.1074/jbc.M801161200. [DOI] [PubMed] [Google Scholar]
- 57.Arragain S, Garcia-Serres R, Blondin G, Douki T, Clemancey M, Latour JM, Forouhar F, Neely H, Montelione GT, Hunt JF, Mulliez E, Fontecave M, Atta M. J. Biol. Chem. 2010;285:5792. doi: 10.1074/jbc.M109.065516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Conradt H, Hohmann-Berger M, Hohmann HP, Blaschkowski HP, Knappe J. Arch. Biochem. Biophys. 1984;228:133. doi: 10.1016/0003-9861(84)90054-7. [DOI] [PubMed] [Google Scholar]
- 59.Knappe J, Neugebauer FA, Blaschkowski HP, Ganzler M. Proc. Natl. Acad. Sci. U. S. A. 1984;81:1332. doi: 10.1073/pnas.81.5.1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wagner AF, Frey M, Neugebauer FA, Schafer W, Knappe J. Proc. Natl. Acad. Sci. U. S. A. 1992;89:996. doi: 10.1073/pnas.89.3.996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Knappe J, editor. Anaerobic dissimilation of pyruvate. American Society for Microbiology; Washington, D.C.: 1987. [Google Scholar]
- 62.Rand K, Noll C, Schiebel HM, Kemken D, Dulcks T, Kalesse M, Heinz DW, Layer G. Biol. Chem. 2010;391:55. doi: 10.1515/BC.2010.006. [DOI] [PubMed] [Google Scholar]
- 63.Layer G, Grage K, Teschner T, Schunemann V, Breckau D, Masoumi A, Jahn M, Heathcote P, Trautwein AX, Jahn D. J. Biol. Chem. 2005;280:29038. doi: 10.1074/jbc.M501275200. [DOI] [PubMed] [Google Scholar]
- 64.Layer G, Pierik AJ, Trost M, Rigby SE, Leech HK, Grage K, Breckau D, Astner I, Jansch L, Heathcote P, Warren MJ, Heinz DW, Jahn D. J. Biol. Chem. 2006;281:15727. doi: 10.1074/jbc.M512628200. [DOI] [PubMed] [Google Scholar]
- 65.Tse Sum Bui B, Florentin D, Marquet A, Benda R, Trautwein AX. FEBS Lett. 1999;459:411. doi: 10.1016/s0014-5793(99)01300-9. [DOI] [PubMed] [Google Scholar]
- 66.Ugulava NB, Gibney BR, Jarrett JT. Biochemistry. 2001;40:8343. doi: 10.1021/bi0104625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tse Sum Bui B, Benda R, Schunemann V, Florentin D, Trautwein AX, Marquet A. Biochemistry. 2003;42:8791. doi: 10.1021/bi034426c. [DOI] [PubMed] [Google Scholar]
- 68.Jameson GN, Cosper MM, Hernandez HL, Johnson MK, Huynh BH. Biochemistry. 2004;43:2022. doi: 10.1021/bi035666v. [DOI] [PubMed] [Google Scholar]
- 69.Choi-Rhee E, Cronan JE. Chem. Biol. 2005;12:461. doi: 10.1016/j.chembiol.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 70.Reyda MR, Fugate CJ, Jarrett JT. Biochemistry. 2009;48:10782. doi: 10.1021/bi901393t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wuebbens MM, Rajagopalan KV. J. Biol. Chem. 1995;270:1082. doi: 10.1074/jbc.270.3.1082. [DOI] [PubMed] [Google Scholar]
- 72.Rieder C, Eisenreich W, O'Brien J, Richter G, Gotze E, Boyle P, Blanchard S, Bacher A, Simon H. Eur. J. Biochem. 1998;255:24. doi: 10.1046/j.1432-1327.1998.2550024.x. [DOI] [PubMed] [Google Scholar]
- 73.Wuebbens MM, Liu MT, Rajagopalan K, Schindelin H. Structure. 2000;8:709. doi: 10.1016/s0969-2126(00)00157-x. [DOI] [PubMed] [Google Scholar]
- 74.Lees NS, Hanzelmann P, Hernandez HL, Subramanian S, Schindelin H, Johnson MK, Hoffman BM. J. Am. Chem. Soc. 2009;131:9184. doi: 10.1021/ja903978u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chirpich TP, Zappia V, Costilow RN, Barker HA. J. Biol. Chem. 1970;245:1778. [PubMed] [Google Scholar]
- 76.Ballinger MD, Reed GH, Frey PA. Biochemistry. 1992;31:949. doi: 10.1021/bi00119a001. [DOI] [PubMed] [Google Scholar]
- 77.Wu W, Lieder KW, Reed GH, Frey PA. Biochemistry. 1995;34:10532. doi: 10.1021/bi00033a027. [DOI] [PubMed] [Google Scholar]
- 78.Chang CH, Ballinger MD, Reed GH, Frey PA. Biochemistry. 1996;35:11081. doi: 10.1021/bi960850k. [DOI] [PubMed] [Google Scholar]
- 79.Wu W, Booker S, Lieder KW, Bandarian V, Reed GH, Frey PA. Biochemistry. 2000;39:9561. doi: 10.1021/bi000658p. [DOI] [PubMed] [Google Scholar]
- 80.Chen D, Frey PA. Biochemistry. 2001;40:596. doi: 10.1021/bi002265w. [DOI] [PubMed] [Google Scholar]
- 81.Frey PA, Chang CH, Ballinger MD, Reed GH. Methods Enzymol. 2002;354:426. doi: 10.1016/s0076-6879(02)54033-7. [DOI] [PubMed] [Google Scholar]
- 82.Hinckley GT, Ruzicka FJ, Thompson MJ, Blackburn GM, Frey PA. Arch. Biochem. Biophys. 2003;414:34. doi: 10.1016/s0003-9861(03)00160-7. [DOI] [PubMed] [Google Scholar]
- 83.Lees NS, Chen D, Walsby CJ, Behshad E, Frey PA, Hoffman BM. J. Am. Chem. Soc. 2006;128:10145. doi: 10.1021/ja061282r. [DOI] [PubMed] [Google Scholar]
- 84.Wang SC, Frey PA. Biochemistry. 2007;46:12889. doi: 10.1021/bi701745h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Noma A, Suzuki T. Nucleic Acids Symp. Ser. (Oxf); 2006. p. 65. [DOI] [PubMed] [Google Scholar]
- 86.Noma A, Kirino Y, Ikeuchi Y, Suzuki T. EMBO J. 2006;25:2142. doi: 10.1038/sj.emboj.7601105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Blobstein SH, Grunberger D, Weinstein IB, Nakanishi K. Biochemistry. 1973;12:188. doi: 10.1021/bi00726a002. [DOI] [PubMed] [Google Scholar]
- 88.Limbach PA, Crain PF, McCloskey JA. Nucleic Acids Res. 1994;22:2183. doi: 10.1093/nar/22.12.2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Urbonavicius J, Qian Q, Durand JM, Hagervall TG, Bjork GR. EMBO. J. 2001;20:4863. doi: 10.1093/emboj/20.17.4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li J, Esberg B, Curran JF, Bjork GR. J. Mol. Biol. 1997;271:209. doi: 10.1006/jmbi.1997.1176. [DOI] [PubMed] [Google Scholar]
- 91.Konevega AL, Soboleva NG, Makhno VI, Semenkov YP, Wintermeyer W, Rodnina MV, Katunin VI. RNA. 2004;10:90. doi: 10.1261/rna.5142404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Posewitz MC, King PW, Smolinski SL, Zhang L, Seibert M, Ghirardi ML. J. Biol. Chem. 2004;279:25711. doi: 10.1074/jbc.M403206200. [DOI] [PubMed] [Google Scholar]
- 93.King PW, Posewitz MC, Ghirardi ML, Seibert M. J. Bacteriol. 2006;188:2163. doi: 10.1128/JB.188.6.2163-2172.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.McGlynn SE, Ruebush SS, Naumov A, Nagy LE, Dubini A, King PW, Broderick JB, Posewitz MC, Peters JW. J. Biol. Inorg. Chem. 2007;12:443. doi: 10.1007/s00775-007-0224-z. [DOI] [PubMed] [Google Scholar]
- 95.Lill R, Muhlenhoff U. Annu. Rev. Cell. Dev. Biol. 2006;22:457. doi: 10.1146/annurev.cellbio.22.010305.104538. [DOI] [PubMed] [Google Scholar]
- 96.Ayala-Castro C, Saini A, Outten FW. Microbiol. Mol. Biol. Rev. 2008;72:110. doi: 10.1128/MMBR.00034-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bandyopadhyay S, Chandramouli K, Johnson MK. Biochem. Soc. Trans. 2008;36:1112. doi: 10.1042/BST0361112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mulder DW, Ortillo DO, Gardenghi DJ, Naumov AV, Ruebush SS, Szilagyi RK, Huynh BV, Broderick JB, Peters JW. Biochemistry. 2009;48:6240. doi: 10.1021/bi9000563. [DOI] [PubMed] [Google Scholar]
- 99.Pilet E, Nicolet Y, Mathevon C, Douki T, Fontecilla-Camps JC, Fontecave M. FEBS Lett. 2009;583:506. doi: 10.1016/j.febslet.2009.01.004. [DOI] [PubMed] [Google Scholar]
- 100.Brazzolotto X, Rubach JK, Gaillard J, Gambarelli S, Atta M, Fontecave M. J. Biol. Chem. 2006;281:769. doi: 10.1074/jbc.M510310200. [DOI] [PubMed] [Google Scholar]
- 101.McGlynn SE, Shepard EM, Winslow MA, Naumov AV, Duschene KS, Posewitz MC, Broderick WE, Broderick JB, Peters JW. FEBS Lett. 2008;582:2183. doi: 10.1016/j.febslet.2008.04.063. [DOI] [PubMed] [Google Scholar]
- 102.Shepard EM, McGlynn SE, Bueling AL, Grady-Smith CS, George SJ, Winslow MA, Cramer SP, Peters JW, Broderick JB. Proc. Natl. Acad. Sci. U. S. A. 2010;107:10448. doi: 10.1073/pnas.1001937107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Shepard EM, Duffus BR, George SJ, McGlynn SE, Challand MR, Swanson KD, Roach PL, Cramer SP, Peters JW, Broderick JB. J. Am. Chem. Soc. 2010;132:9247. doi: 10.1021/ja1012273. [DOI] [PubMed] [Google Scholar]
- 104.Driesener RC, Challand MR, McGlynn SE, Shepard EM, Boyd ES, Broderick JB, Peters JW, Roach PL. Angew. Chem. Int. Ed. 2010;49:1687. doi: 10.1002/anie.200907047. [DOI] [PubMed] [Google Scholar]
- 105.Layer G, Heinz DW, Jahn D, Schubert WD. Curr. Opin. Chem. Biol. 2004;8:468. doi: 10.1016/j.cbpa.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 106.Layer G, Kervio E, Morlock G, Heinz DW, Jahn D, Retey J, Schubert WD. Biol. Chem. 2005;386:971. doi: 10.1515/BC.2005.113. [DOI] [PubMed] [Google Scholar]
- 107.Markham GD, Norrby PO, Bock CW. Biochemistry. 2002;41:7636. doi: 10.1021/bi025682c. [DOI] [PubMed] [Google Scholar]
- 108.Farrar C, Jarrett J. Biochemistry. 2009;48:2448. doi: 10.1021/bi8022569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Cheek J, Broderick JB. J. Am. Chem. Soc. 2002;124:2860. doi: 10.1021/ja017784g. [DOI] [PubMed] [Google Scholar]
- 110.Seehra JS, Jordan PM, Akhtar M. Biochem. J. 1983;209:709. doi: 10.1042/bj2090709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yan F, LaMarre JM, Rohrich R, Wiesner J, Jomaa H, Mankin AS, Fujimori DG. J. Am. Chem. Soc. 2010;132:3953. doi: 10.1021/ja910850y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ollagnier-de-Choudens S, Mulliez E, Fontecave M. FEBS Lett. 2002;532:465. doi: 10.1016/s0014-5793(02)03733-x. [DOI] [PubMed] [Google Scholar]
- 113.Ollagnier-de Choudens S, Sanakis Y, Hewitson KS, Roach P, Munck E, Fontecave M. J. Biol. Chem. 2002;277:13449. doi: 10.1074/jbc.M111324200. [DOI] [PubMed] [Google Scholar]
- 114.Guianvarc'h D, Florentin D, Tse Sum Bui B, Nunzi F, Marquet A. Biochem. Biophys. Res. Commun. 1997;236:402. doi: 10.1006/bbrc.1997.6952. [DOI] [PubMed] [Google Scholar]
- 115.Escalettes F, Florentin D, Tse Sum Bui B, Lesage D, Marquet A. J. Am. Chem. Soc. 1999;121:3571. [Google Scholar]
- 116.Lotierzo M, Tse Sum Bui B, Florentin D, Escalettes F, Marquet A. Biochem. Soc. Trans. 2005;33:820. doi: 10.1042/BST0330820. [DOI] [PubMed] [Google Scholar]
- 117.Taylor AM, Farrar CE, Jarrett JT. Biochemistry. 2008;47:9309. doi: 10.1021/bi801035b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Cicchillo RM, Iwig DF, Jones AD, Nesbitt NM, Baleanu-Gogonea C, Souder MG, Tu L, Booker SJ. Biochemistry. 2004;43:6378. doi: 10.1021/bi049528x. [DOI] [PubMed] [Google Scholar]
- 119.Tse Sum Bui B, Lotierzo M, Escalettes F, Florentin D, Marquet A. Biochemistry. 2004;43:16432. doi: 10.1021/bi048040t. [DOI] [PubMed] [Google Scholar]
- 120.Daley CJ, Holm RH. Inorg. Chem. 2001;40:2785. doi: 10.1021/ic010039k. [DOI] [PubMed] [Google Scholar]
- 121.Daley CJ, Holm RH. J. Inorg. Biochem. 2003;97:287. doi: 10.1016/s0162-0134(03)00280-0. [DOI] [PubMed] [Google Scholar]
- 122.Page CC, Moser CC, Dutton PL. Curr. Opin. Chem. Biol. 2003;7:551. doi: 10.1016/j.cbpa.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 123.Kozbial PZ, Mushegian AR. BMC Struct. Biol. 2005;5:19. doi: 10.1186/1472-6807-5-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Vidgren J, Svensson LA, Liljas A. Nature. 1994;368:354. doi: 10.1038/368354a0. [DOI] [PubMed] [Google Scholar]
- 125.Cheng X, Kumar S, Posfai J, Pflugrath JW, Roberts RJ. Cell. 1993;74:299. doi: 10.1016/0092-8674(93)90421-l. [DOI] [PubMed] [Google Scholar]
- 126.Korolev S, Ikeguchi Y, Skarina T, Beasley S, Arrowsmith C, Edwards A, Joachimiak A, Pegg AE, Savchenko A. Nat. Struct. Mol. Biol. 2002;9:27. doi: 10.1038/nsb737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Dong C, Huang F, Deng H, Schaffrath C, Spencer JB, O'Hagan D, Naismith JH. Nature. 2004;427:561. doi: 10.1038/nature02280. [DOI] [PubMed] [Google Scholar]
- 128.Huang CC, Smith CV, Glickman MS, Jacobs WR, Jr, Sacchettini JC. J. Biol. Chem. 2002;277:11559. doi: 10.1074/jbc.M111698200. [DOI] [PubMed] [Google Scholar]
- 129.Anantharaman V, Koonin EV, Aravind L. J. Mol. Microbiol. Biotechnol. 2002;4:71. [PubMed] [Google Scholar]
- 130.Tkaczuk KL, Dunin-Horkawicz S, Purta E, Bujnicki JM. BMC Bioinformatics. 2007;8:73. doi: 10.1186/1471-2105-8-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Jenuwein T, Laible G, Dorn R, Reuter G. Cell. Mol. Life. Sci. 1998;54:80. doi: 10.1007/s000180050127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Alvarez-Venegas R, Avramova Z. Gene. 2002;285:25. doi: 10.1016/s0378-1119(02)00401-8. [DOI] [PubMed] [Google Scholar]
- 133.Demirci H, Gregory ST, Dahlberg AE, Jogl G. EMBO J. 2007;26:567. doi: 10.1038/sj.emboj.7601508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Capitani G, Tschopp M, Eliot AC, Kirsch JF, Grutter MG. FEBS Lett. 2005;579:2458. doi: 10.1016/j.febslet.2005.03.048. [DOI] [PubMed] [Google Scholar]
- 135.Rafferty JB, Somers WS, Saint-Girons I, Phillips SE. Nature. 1989;341:705. doi: 10.1038/341705a0. [DOI] [PubMed] [Google Scholar]
- 136.Dixon MM, Huang S, Matthews RG, Ludwig M. Structure. 1996;4:1263. doi: 10.1016/s0969-2126(96)00135-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.