Abstract
Objective
To determine whether the GNAQ R183Q mutation is present in the forme fruste cases of Sturge-Weber syndrome (SWS) to establish a definitive molecular diagnosis.
Methods
We used sensitive droplet digital PCR (ddPCR) to detect and quantify the GNAQ mutation in tissues from epilepsy surgery in 4 patients with leptomeningeal angiomatosis; none had ocular or cutaneous manifestations.
Results
Low levels of the GNAQ mutation were detected in the brain tissue of all 4 cases—ranging from 0.42% to 7.1% frequency—but not in blood-derived DNA. Molecular evaluation confirmed the diagnosis in 1 case in which the radiologic and pathologic data were equivocal.
Conclusions
We detected the mutation at low levels, consistent with mosaicism in the brain or skin (1.0%–18.1%) of classic cases. Our data confirm that the forme fruste is part of the spectrum of SWS, with the same molecular mechanism as the classic disease and that ddPCR is helpful where conventional diagnosis is uncertain.
Sturge-Weber syndrome (SWS) is a rare, sporadic neurocutaneous disorder that occurs in 1 in 20,000 newborns, typically characterized by brain pathology—leptomeningeal angiomatosis (LMA), cortical atrophy and calcification, and layer 1 fusion—port-wine stain, and vascular glaucoma.1 Clinical manifestations and severity are heterogeneous with drug-resistant epilepsy, hemiparesis and cognitive impairment the most common neurologic features, glaucoma the most frequent ocular presentation, and port-wine stain the predominant dermatological feature.1 Sometimes, the characteristic meningeal lesions of SWS are seen without skin or ocular features2,3—this is referred to as forme fruste of SWS, or sometimes type III SWS, and diagnosis can be challenging.
A somatic mosaic mutation (c.548G>A; p.R183Q) of the GNAQ gene that disrupts the activity of the encoded guanosine triphosphatase is present in classic SWS and also in patients who only have a port-wine stain.4 This mutation was found in studies from different populations to be present in the brain or skin of more than 80% of patients.4,5 Enrichment of this mutation in endothelial cells of both SWS skin and brain specimens,6,7 and SWS brain parenchyma not affected by LMA,6 has also recently been reported.
Droplet digital PCR (ddPCR) is an ultra-sensitive technique recently reported for detection of the SWS mutation.5,7 It uses microfluidics and surfactant chemistries to emulsify input DNA into thousands of uniformly sized droplets and then to amplify them with fluorescently labeled TaqMan probes before measuring fluorescence on a droplet reader, as we and others have previously described.8,9 Based on fluorescence intensity, the number of mutation-positive and wild-type templates is quantified to calculate the frequency of a mutant allele. Here, we used this approach to screen 4 patients with forme fruste SWS including 1 in which the diagnosis was equivocal.
Methods
Patients
We ascertained 4 patients with forme fruste SWS through our epilepsy surgery programs at Austin Health, Royal Children's Hospital, Melbourne, and the Lady Cilento Children's Hospital, Queensland, Australia. Genomic DNA was extracted from the brain using the DNA Genotek PrepIt 2CD Kit (Ontario, Canada) or Qiagen AllPrep DNA/RNA Kit and peripheral blood using the Macherey-Nagel NucleoBond CB 100 Kit (Duren, Germany) or Qiagen QIAamp DNA Maxi Kit (Hilden, Germany).
Standard protocol approvals, registrations, and patient consents
The Human Research Ethics Committees of The Royal Children's Hospital, Melbourne, Australia (project no. 29077F), and Austin Health, Melbourne, Australia (project no. H2007/02961), approved this study. Informed consent was obtained from the patients, or their parents in the case of minors, for participation in the study.
Droplet digital PCR
We used a commercially available ddPCR Mutation Detection Assay (ID: 10049047; Bio-Rad, Hercules, CA) to detect the GNAQ c.548G>A (p.R183Q) mutation and wild-type allele. Briefly, the ddPCR reaction mixture was assembled from a 2× ddPCR Supermix for Probes (No dUTP; Bio-Rad), 20× ddPCR Mutation Detection Assay, and 10 ng of DNA sample to a final volume of 23 μL. Twenty microliters of each reaction mixture was then loaded into the sample well of an 8-channel droplet generator cartridge (Bio-Rad), and droplets were generated with 70 μL of droplet generation oil (Bio-Rad) using the manual QX200 Droplet Generator. Following droplet generation, samples were manually transferred to a 96-well PCR plate, heat-sealed, and amplified on a C1000 Touch thermal cycler using the following cycling conditions: 95°C for 10 minutes for 1 cycle, followed by 40 cycles at 94°C for 30 seconds and 55°C for 60 seconds, 1 cycle at 98°C for 10 minutes and 12°C for infinite. Post-PCR products were read on the QX200 droplet reader (Bio-Rad) and analyzed using QuantaSoft software. We established the detection limit of the ddPCR assay by serially diluting mutant samples with wild-type DNA to obtain different mutant/(mutant + wild-type) ratios: 5%, 1%, 0.5%, 0.25%, and 0.1%. These mixed DNA samples were subjected to ddPCR as described above.
Results
Clinical report
Four patients presented during childhood with forme fruste or SWS type III with drug-resistant epilepsy (table 1) and LMA on MRI and histopathology (figures 1, A–C and 2, A–C, figure e-1, links.lww.com/NXG/A48, table 1), without port-wine stains. Fresh-frozen (cases 1, 2, and 4) or formalin-fixed paraffin-embedded (case 3) brain tissue was available following epilepsy surgery. The diagnoses of SWS type III for cases 1, 2, and 4 were definitive based on imaging and pathologic data (figure 1, A–C, figure e-1, links.lww.com/NXG/A48, table 1). In case 3, the diagnosis was less certain, as CT and MRI showed calcification in the left occipital region posteroinferiorly without convincing focal atrophy (figure 2, A and B, table 1). Pathologically, in the subarachnoid plane, a small vascular malformation was seen with some arterial features, coupled with underlying parenchymal calcification and cortical dyslamination (figure 2C, table 1).
Table 1.
Clinical characteristics of forme fruste cases of Sturge-Weber syndrome
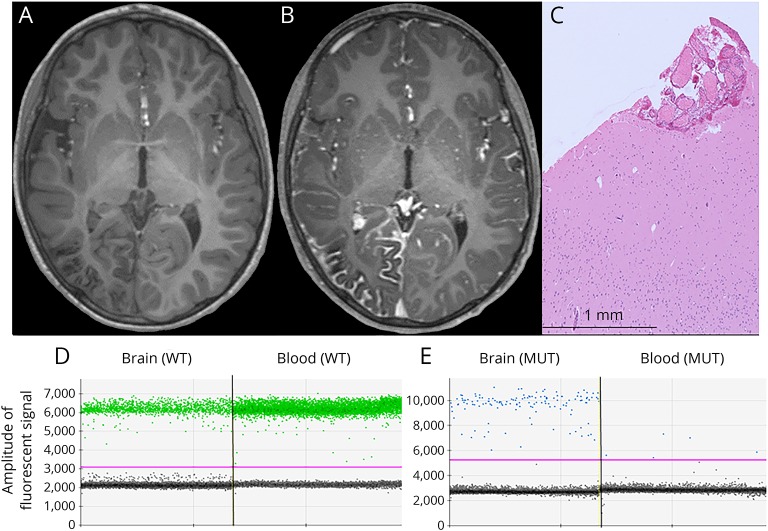
Figure 1. Imaging, histopathology, and molecular evaluation of case 1 with definite leptomeningeal angiomatosis.
(A) Precontrast T1-weighted axial MRI scan showing right temporal and occipital atrophy and right occipital cortical calcification. (B) Postcontrast T1-weighted axial MRI scan showing leptomeningeal enhancement. (C) Hematoxylin and eosin–stained image of the neocortex showing a small area of densely clustered leptomeningeal vessels. (D) Identification of the wild-type GNAQ allele in green (present in the brain and blood) by digital PCR. (E) Identification of the mutant GNAQ R183Q allele (in blue) in the brain-derived but not blood-derived DNA—rare blue dots in blood are signal from droplets containing multiple DNA templates (supplemental data, links.lww.com/NXG/A48). Droplets without DNA templates are gray. Y-axis, amplitude of fluorescent signal. WT = wild-type GNAQ probe; MUT = mutant GNAQ R183Q probe.
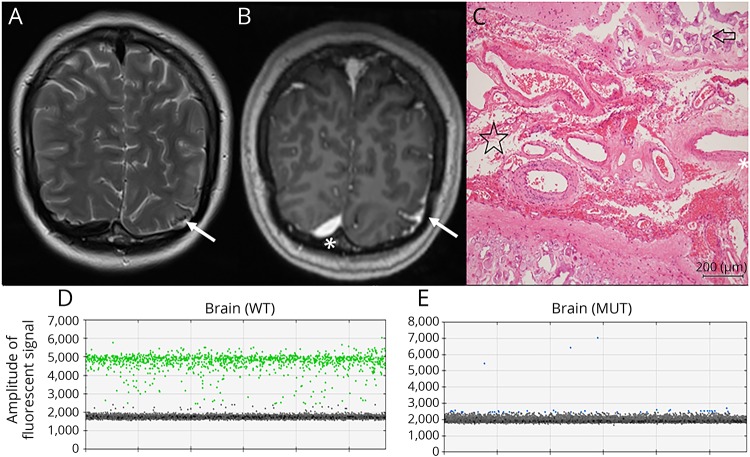
Figure 2. Imaging, histopathology, and molecular evaluation of case 3 with subtler MRI findings.
(A) Precontrast T2-weighted coronal MRI scan showing subtle signal change and calcification in the left occipital region (arrow) posteroinferiorly involving the occipital cortex or leptomeninges. Calcification was confirmed on CT (not shown). (B) Postcontrast T1-weighted coronal MRI scan showing leptomeningeal enhancement in the same region. Enhancement in the right occipital region (asterisk) is due to the normal transverse sinus. (C) Hematoxylin and eosin stained image showing subarachnoid angiomatosis (starred) between adjacent cerebral gyrae with cortical calcification (arrow). (D) Identification of the wild-type GNAQ allele (in green) in the brain by digital PCR. (E) Identification of the mutant GNAQ R183Q allele (in blue) in the brain. Droplets without genomic DNA templates are gray. Y-axis, amplitude of fluorescent signal. WT = wild-type GNAQ probe; MUT = mutant GNAQ R183Q probe.
Mutation detection in the brain-derived genomic DNA by ddPCR
We established the detection limit for the GNAQ mutation detection ddPCR by assaying serially mixed mutant and wild-type samples in triplicate. The mutant allele at a frequency ≥0.25% was consistently detected, while detection of the mutant allele at 0.1% was only achieved in 2 of the 3 wells (figure e-2, links.lww.com/NXG/A48, table e-1). Thus, the detection limit in our hands was 0.25% mutant allele frequency, comparable with a previously reported limit (0.1%) for a similar assay.5
Genomic DNA isolated from the resected brain tissue and that from the peripheral blood were analyzed using ddPCR. The GNAQ p.R183Q mutant allele was detected only in genomic DNA extracted from the brain tissue (7.1% frequency in case 1, 5.8% in case 2, 2.1% in case 3, and 0.42% in case 4) but not in genomic blood-derived DNA from 3 patients (figures 1, D, E and 2, D, E, figures e-1, e-3 to e-7, links.lww.com/NXG/A48). Blood-derived DNA was not available from case 3. It should be noted that although very low, the 0.42% mutant allele frequency of case 4 was above our established detection limit (figures e-2, e-3, and e-7, links.lww.com/NXG/A48, table e-1).
For case 1, fluorescent droplets were observed in the blood-derived genomic DNA below the expected amplitude, but these did not overlap with the true positive signal in the brain-derived genomic DNA when fluorescence intensity was viewed on 2D plots (figure e-3, links.lww.com/NXG/A48). Instead, this is fluorescent signal from droplets containing multiple genomic templates, a phenomenon not infrequently observed when running ddPCR assays.
Discussion
The important discovery of a recurrent, somatic GNAQ mutation provided the first insights into the molecular biology of SWS. Initial reports focused on classic SWS,4–6,10,11 and here, we extend these findings to forme fruste cases, a far more subtle, sometimes unrecognized, form of SWS. Our findings confirm that forme fruste cases are caused by the somatic GNAQ p.R183Q mutation present at low to very low levels in brain tissue due to mosaicism, consistent with a few reported cases.4,11 It is intriguing that the mutation was only present in the brain tissue of these forme fruste cases, and not in blood (of 3 cases), suggesting that the mutation may have arisen later during development than for classic cases, although we did not have other tissues available from our cases to confirm this. As MRI and even pathologic diagnosis can be equivocal for subtle LMA lesions, as for case 3 (figure 2, table 1), molecular evaluation may have specific diagnostic value. The relatively low level of the GNAQ mutation in the brain tissue of case 3 is consistent with the milder imaging and pathologic manifestations; however, case 4 had an even lower mutant load in terms of percentage mosaicism in the tissue tested, suggesting that there are other, as yet unidientified, influences on genotype-phenotype correlation.
In formalin-fixed paraffin-embedded samples, low-level somatic mosaic mutations are challenging to detect because the DNA is of low quality and often has impurities. Despite these challenges, we were able to identify the somatic mutation in case 3 from a 3-year-old pathologic specimen. This and other sensitive mutation detection technologies are showing increasing utility in elucidating the role of somatic mosaicism in brain-specific neurologic disorders, as shown recently for tuberous sclerosis,12 in addition to SWS.
Acknowledgment
The authors thank the patients and their families for their participation in this study.
Glossary
- ddPCR
droplet digital PCR
- LMA
leptomeningeal angiomatosis
- SWS
Sturge-Weber syndrome
Author contributions
M.S.H., I.E.S., and S.F.B. initiated and directed the project. M.S.H., J.A.D., H.D., Z.Y., L.M., E.O., and G.G. performed molecular genetics experiments. A.S.H., S.M., W.M., B.N., M.W., K.P., R.J.L., I.E.S., and S.F.B. conducted clinical phenotyping. R.K. performed histopathologic analyses. M.S.H., N.C.J., P.J.L., A.D., and S.F.B. provided equipment and reagents. M.S.H., A.S.H., S.M., I.E.S., and S.F.B. wrote the paper. All authors discussed the results and commented on the manuscript.
Study funding
This study was supported by the National Health and Medical Research Council Program Grant (1091593) to I.E.S. and S.F.B., a Project Grant (1129054) to S.F.B., a Project Grant (1079058) to M.S.H., a Practitioner Fellowship (1006110) to I.E.S., and a R.D Wright Career Development Fellowship (1063799) to M.S.H. R.J.L. is supported by a Melbourne Children's Clinician Scientist Fellowship.
Disclosure
M.S. Hildebrand has received research support from the National Health and Medical Research Council. A.S. Harvey reports no disclosures. S. Malone has served on the scientific advisory boards of UCB Pharma and BioMarin; has received travel funding/speaker honoraria from UCB Pharma and BioMarin; and has received research support from UCB Pharma. J.A. Damiano reports no disclosures. H. Do has given lectures and educational presentations for Bio-Rad. Z. Ye and L. McQuillan report no disclosures. W. Maixner receives publishing royalties from Springer. R. Kalnins and B. Nolan report no disclosures. M. Wood has been a consultant to Medtronic. E. Ozturk and N.C. Jones report no disclosures. G. Gillies and K. Pope have received research support from the National Health and Medical Research Council. P.J. Lockhart has served on the editorial boards of Genetics Research International, Open Access (OA) Genetics and the Journal of Neurochemistry and has received research support from the National Health and Medical Research Council. A. Dobrovic has served on the editorial boards of BMC Cancer and Molecular Cancer; has been a consultant to Astra Zeneca; has given lectures and educational presentations for Bio-Rad; and has received research support from the National Health and Medical Research Council. R.J. Leventer has received research support from the National Health and Medical Research Council and Campbell Edwards Trust. I.E. Scheffer has received speaker honoraria from UCB Pharma, Eisai, GSK, Athena Diagnostics, and Transgenomics; has served on the scientific advisory boards of GSK, UCB Pharma, BioMarin, Ramaciotti Foundation, and Nutricia; received the 2014 Prime Ministers Prize For Science; has served on the editorial boards of Neurology, Epilepsy Currents, Annals of Neurology, Epileptic Disorders, Progress in Epileptic Disorders series, and Virtual Neuro Centre; holds patents/pending patents for SCN1A testing held by Bionomics Inc licensed to various diagnostic companies, Diagnostic and therapeutic methods for EFMR, A diagnostic method for epilepsy, A diagnostic method for epilepsy (also published as Methods for the diagnosis and treatment of epilepsy), Mutations in ion channels, Diagnostic and treatment methods relating to autosomal dominant nocturnal frontal lobe epilepsy, A gene and mutations thereof associated with seizure and movement disorders, and Diagnostic and therapeutic methods for EFMR; participated in the Epilepsy Drug Consortium as an investigator for GW Pharmaceuticals; has received research support from the National Health and Medical Research Foundation, National Institute of Health, University of Melbourne School of Health Sciences, March of Dimes Foundation, Queensland Emergency Medicine Research Foundation, Health Research Council New Zealand, Medical Research Future Fund, and Rebecca L Cooper Medical Research Foundation; and receives royalty payments for Diagnostic and Therapeutic Methods for EFMR (Epilepsy and Mental Retardation Limited to Females). S.F. Berkovic has served on the scientific advisory boards of UCB Pharma and Eisai Australia; has served on the editorial boards of Brain, Epileptic Disorders, and Lancet Neurology; has received speaker honoraria from UCB Pharma, Novartis Pharmaceuticals, Sanofi-Aventis, and Jansen Cilag; holds a patent for SCN1A testing held by Bionomics Inc and licensed to various diagnostic companies; is one of the inventors listed on a patent held by Bionomics Inc on Diagnostic testing of using the SCN1A gene; is one of the inventors on pending patent WO61/010176: Therapeutic compound that relates to discovery of PCDH19 gene as the cause of familial epilepsy with mental retardation limited to females; and has received research support from the National Health and Medical Research Council. Full disclosure form information provided by the authors is available with the full text of this article at Neurology.org/NG.
References
- 1.Comi AM. Sturge-Weber syndrome. Handb Clin Neurol 2015;132:157–168. [DOI] [PubMed] [Google Scholar]
- 2.Crosley CJ, Binet EF. Sturge-Weber syndrome: presentation as a focal seizure disorder without nevus flammeus. Clin Pediatr (Phila) 1978;17:606–609. [DOI] [PubMed] [Google Scholar]
- 3.Rasmussen T, Mathieson G, Le Blanc F. Surgical therapy of typical and a forme fruste variety of the Sturge-Weber syndrome. Schweiz Arch Neurol Neurochir Psychiatr 1972;111:393–409. [PubMed] [Google Scholar]
- 4.Shirley MD, Tang H, Gallione CJ, et al. Sturge-Weber syndrome and port-wine stains caused by somatic mutation in GNAQ. N Engl J Med 2013;368:1971–1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Uchiyama Y, Nakashima M, Watanabe S, et al. Ultra-sensitive droplet digital PCR for detecting a low-prevalence somatic GNAQ mutation in Sturge-Weber syndrome. Sci Rep 2016;6:22985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sundaram SK, Michelhaugh SK, Klinger NV, et al. GNAQ mutation in the venous vascular malformation and underlying brain tissue in Sturge-Weber syndrome. Neuropediatrics 2017;48:385–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang L, Couto JA, Pinto A, et al. Somatic GNAQ mutation is enriched in brain endothelial cells in Sturge-Weber syndrome. Pediatr Neurol 2017;67:59–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oxnard GR, Paweletz CP, Kuang Y, et al. Noninvasive detection of response and resistance in EGFR-mutant lung cancer using quantitative next-generation genotyping of cell-free plasma DNA. Clin Cancer Res 2014;20:1698–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsao SC, Weiss J, Hudson C, et al. Monitoring response to therapy in melanoma by quantifying circulating tumour DNA with droplet digital PCR for BRAF and NRAS mutations. Sci Rep 2015;5:11198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lian CG, Sholl LM, Zakka LR, et al. Novel genetic mutations in a sporadic port-wine stain. JAMA Dermatol 2014;150:1336–1340. [DOI] [PubMed] [Google Scholar]
- 11.Nakashima M, Miyajima M, Sugano H, et al. The somatic GNAQ mutation c.548G>A (p.R183Q) is consistently found in Sturge-Weber syndrome. J Hum Genet 2014;59:691–693. [DOI] [PubMed] [Google Scholar]
- 12.Lim JS, Gopalappa R, Kim SH, et al. Somatic mutations in TSC1 and TSC2 cause focal cortical dysplasia. Am J Hum Genet 2017;100:454–472. [DOI] [PMC free article] [PubMed] [Google Scholar]