Abstract
Background
Observational studies suggest greater height is associated with increased ovarian cancer risk, but cannot exclude bias and/or confounding as explanations for this. Mendelian randomisation (MR) can provide evidence which may be less prone to bias.
Methods
We pooled data from 39 Ovarian Cancer Association Consortium studies (16,395 cases; 23,003 controls). We applied two-stage predictor-substitution MR, using a weighted genetic risk score combining 609 single-nucleotide polymorphisms. Study-specific odds ratios (OR) and 95% confidence intervals (CI) for the association between genetically predicted height and risk were pooled using random-effects meta-analysis.
Results
Greater genetically predicted height was associated with increased ovarian cancer risk overall (pooled-OR (pOR) = 1.06; 95% CI: 1.01–1.11 per 5 cm increase in height), and separately for invasive (pOR = 1.06; 95% CI: 1.01–1.11) and borderline (pOR = 1.15; 95% CI: 1.02–1.29) tumours.
Conclusions
Women with a genetic propensity to being taller have increased risk of ovarian cancer. This suggests genes influencing height are involved in pathways promoting ovarian carcinogenesis.
Subject terms: Genetics research, Ovarian cancer, Cancer epidemiology, Risk factors
Introduction
Observational studies have reported a positive association between adult height and ovarian cancer risk.1–4 However, these studies were subject to the biases inherent in conventional observational studies, including selection bias, differential and non-differential reporting bias and confounding. The degree to which these factors could account for the observed association is uncertain. Mendelian randomisation (MR) uses genetic markers as proxies for environmental exposures and, due to the singular qualities of genotype data, can provide complementary evidence by overcoming many biases affecting conventional studies.5 We used MR to examine the relationship between height and ovarian cancer risk in the Ovarian Cancer Association Consortium (OCAC),6 aiming to provide more certainty about the relationship between height and ovarian cancer risk. We hypothesised that greater genetically predicted height would be associated with increased risk.
Materials and methods
Study population and outcomes
We pooled data from 16,395 genetically European women with primary ovarian/fallopian tube/peritoneal cancer and 23,003 controls from 39 OCAC studies (Table 1; Supplementary Table 1). The data set and methods have been described previously.7 Participants were genotyped via the Collaborative Oncological Gene-Environment Study.8 Twenty-two studies provided height data (16 provided parity, oral contraceptive (OC) use, education and age at menarche information) for >50% of their participants. We first considered all cases, then stratified by tumour behaviour. Secondary analyses stratified by histologic subtype/behaviour.
Table 1.
Characteristics of 39 OCAC studies and 39,398 participants of European ancestry included in the Mendelian randomisation analysis
| Study acronyma | Country | Diagnosis (years) | Median (range) age at diagnosis/interview | Invasive cases (N) | Borderline cases (N) | All cases (N)b | Controls (N) | Mean (SD) height (cm)c |
|---|---|---|---|---|---|---|---|---|
| AUS | Australia | 2002–2006 | 58 (19–80) | 859 | 1 | 860 | 977 | 163 (6.9) |
| BAV | Germany | 2002–2008 | 58 (24–83) | 96 | 5 | 102 | 143 | 164 (5.8) |
| BEL | Belgium | 2007–2010 | 46 (19–87) | 275 | 0 | 275 | 1347 | — |
| DOV | USA | 2002–2009 | 57 (35–74) | 904 | 327 | 1231 | 1487 | 166 (6.5) |
| GER | Germany | 1993–1998 | 57 (21–75) | 189 | 24 | 213 | 413 | 163 (6.0) |
| GRR | USA | 1981–2012 | 48 (21–83) | 125 | 0 | 125 | 0 | — |
| HAW | USA | 1993–2008 | 56 (27–87) | 60 | 20 | 80 | 157 | 163 (6.6) |
| HJO | Germany | 2007–2011 | 54 (18–88) | 261 | 13 | 290 | 273 | — |
| HMO | Belarus | 2006–2011 | 45 (22–76) | 142 | 0 | 143 | 138 | — |
| HOC | Finland | 1975–1999 | 46 (18–86) | 210 | 8 | 239 | 447 | — |
| HOP | USA | 2003–2009 | 58 (25–94) | 567 | 71 | 723 | 1464 | 163 (6.8) |
| HSK | Germany | 2000–2007 | 58 (18–81) | 147 | 9 | 156 | 0 | 165 (5.6) |
| LAX | USA | 1989–2008 | 58 (31–88) | 278 | 0 | 278 | 0 | — |
| MAL | Denmark | 1994–1999 | 57 (31–80) | 440 | 138 | 578 | 828 | 166 (6.1) |
| MAY | USA | 2000–2010 | 61 (20–93) | 699 | 79 | 778 | 743 | 165 (6.3) |
| MCC | Australia | 1990–2008 | 65 (45–79) | 66 | 0 | 66 | 66 | 159 (7.0) |
| MDA | USA | 1997–2009 | 62 (23–88) | 375 | 0 | 375 | 384 | — |
| MSK | USA | 1997–2010 | 57 (18–89) | 450 | 0 | 450 | 593 | — |
| NCO | USA | 1999–2008 | 57 (20–75) | 722 | 171 | 896 | 792 | 163 (6.4) |
| NEC | USA | 1992–2003 | 52 (21–78) | 654 | 232 | 904 | 1009 | 163 (6.7) |
| NJO | USA | 2002–2009 | 60 (25–88) | 169 | 0 | 169 | 181 | 163 (6.9) |
| NOR | Norway | 2001–2010 | 51 (18–86) | 236 | 12 | 248 | 371 | — |
| NTH | Netherlands | 1997–2008 | 55 (18–83) | 292 | 3 | 295 | 323 | 167 (6.0) |
| ORE | USA | 2007–2011 | 58 (22–86) | 55 | 9 | 65 | 0 | — |
| OVA | Canada | 2002–2009 | 58 (19–80) | 640 | 161 | 801 | 748 | — |
| POC | Poland | 1998–2008 | 55 (23–82) | 423 | 0 | 423 | 417 | — |
| POL | Poland | 2000–2004 | 56 (24–74) | 236 | 0 | 236 | 223 | 162 (5.6) |
| PVD | Denmark | 2004–2009 | 63 (30–88) | 168 | 0 | 168 | 0 | 165 (6.5) |
| RMH | UK | 1993–1996 | 52 (26–73) | 148 | 7 | 155 | 0 | — |
| SEA | UK | 1998–2011 | 57 (19–78) | 1447 | 76 | 1530 | 6004 | 162 (6.3) |
| SOC | UK | 1993–1998 | 62 (22–92) | 268 | 20 | 288 | 0 | — |
| SRO | UK | 1999–2001 | 59 (34–84) | 158 | 0 | 158 | 0 | — |
| STA | USA | 1997–2002 | 50 (20–64) | 251 | 10 | 261 | 313 | 165 (6.7) |
| TOR | Canada | 1995–2007 | 58 (26–85) | 603 | 0 | 605 | 440 | 163 (7.1) |
| UCI | USA | 1993–2005 | 56 (18–86) | 277 | 141 | 418 | 367 | 165 (6.6) |
| UKO | UK | 2006–2010 | 63 (19–89) | 718 | 0 | 718 | 1104 | 162 (6.7) |
| UKR | UK | 1991–2009 | 54 (24–77) | 47 | 0 | 47 | 0 | — |
| USC | USA | 1992–2010 | 57 (22–82) | 693 | 152 | 845 | 1047 | 165 (6.8) |
| WOC | Poland | 1997–2010 | 44 (20–81) | 201 | 2 | 203 | 204 | — |
All participants were of >90% European ancestry according to genetic markers of ancestry.
aOCAC is an international collaboration of largely case–control studies. See Supplementary Table 1 for study names and references. To maximise power, nine case-only studies were grouped for analysis with case–control studies from the same region: HSK combined with GER; GRR with HOP; PVD with MAL; RMH, SOC, SRO, UKR with SEA and UKO (‘UK group’); ORE with DOV; LAX with UCI.
bCases had primary ovarian (n = 15,636), fallopian tube (n = 180) or peritoneal (n = 552) cancer or ovarian/tubal/peritoneal tumours of undetermined site (n = 27).
cUsual adult height. Height is summarised for 22 studies (20 case–control studies) where >50% participants had data available (AUS, BAV, DOV, GER, HAW, HOP, HSK, MAL, MAY, MCC, NCO, NEC, NJO, NTH, POL, PVD, SEA, STA, TOR, UCI, UKO, USC). Sixteen of these 22 studies were also used in conventional height analyses, as they provided data on potential confounders (age, parity, use of oral contraceptives, education, and age at menarche) for >50% of participants (AUS, DOV, GER, HAW, HOP, MAL, NCO, NEC, NJO, NTH, POL, STA, TOR, UCI, UKO, USC).
OCAC Ovarian Cancer Association Consortium, SD standard deviation
Genetic risk score
The Genetic Investigation of ANthropometric Traits (GIANT) Consortium had previously identified 697 single-nucleotide polymorphisms (SNPs) significantly associated with height.9 In our sample, 92 of these SNPs had been genotyped and the remainder were imputed using 1000 Genome Project data.8, 10 After excluding poorly-imputed SNPs (quality r2 < 0.6), 609 remained (92 genotyped/517 imputed) (Supplementary Table 2). In controls, minor allele frequencies (MAFs) were >5% (except for 16 SNPs, MAFs 1.7–4.9%).
We constructed a weighted genetic risk score (GRS) for height by summing height-increasing alleles across the 609 SNPs (‘GRS-609’/‘the GRS’), weighting alleles by β-coefficients for their association with height reported by GIANT. The score represents predicted additional height conferred by these variants, compared to having no height-increasing alleles. We report results for 5 cm increments.
Statistical analysis
Statistical methods have been described previously.7 Briefly, we used individual-level OCAC data for two-stage predictor-substitution MR 11, 12: first, we predicted height from the weighted GRS for all participants using coefficients from linear regression in 17,649 controls with height data; second, within each study, we used logistic regression to model disease status on GRS-predicted height. Models adjusted for age and five principal components for population structure.8 We combined study-specific estimates using meta-analysis,13 generating pooled odds ratios (pOR) and 95% confidence intervals (CI) for the trend in risk per 5 cm increase in predicted height. We had 97% power to detect an OR of 1.10 (mRnd tool).14
Sensitivity analyses included removing 16 SNPs with MAFs <5%, and restricting to SNPs with imputation r2 ≥ 0.9 (‘GRS-363’), SNPs representing distinct loci9 (‘GRS-377’), and directly-genotyped SNPs (‘GRS-92’). We examined whether potential confounders of the association in observational studies were associated with the GRS. To assess robustness to pleiotropy (where SNPs may influence risk via pathways not mediated through height), we conducted MR-Egger regression15 and assessed smaller GRSs excluding SNPs with the highest probability of acting via other pathways from GRS to outcome (SNPs associated with ovarian/other hormonal cancers (breast, prostate), hormone levels and in/near tumour initiation/growth genes). We identified these potentially pleiotropic, pathway-specific SNPs via the NHGRI GWAS Catalog,16 the UCSC Genome/Table Browsers17, 18 and from lists of SNPs nominated for iCOGS genotyping by ovarian, breast and prostate cancer researchers (to capture SNPs of interest unpublished at the time of analysis).
Secondary analyses defined cases by histologic subtype/behaviour. Among 16 studies with height/confounder data, we conducted conventional analysis (adjusted for parity, OC use, education, menarche age; stratified by study, 5-year age group) and compared results with MR-estimates among the same women.
Analyses were performed using SAS 9.2 (SAS Institute Inc., Cary, NC) and Stata 13.0 (StataCorp LP, College Station, TX). This work and each contributing study was approved by the appropriate institutional review board/ethics committee. All participants provided informed consent.
Results
Population characteristics
We included 16,395 cases (14,549 invasive tumours, 1691 borderline, 155 of unknown behaviour) and 23,003 controls (Table 1). The median diagnosis year was 2003, with 74% diagnosed post-2000. Participants were aged 18–94 (median 56) years at diagnosis/interview. Mean height ranged from 159 to 167 cm across 22 studies with data, and was 163 (standard error (SE) = 0.05) cm for controls and 164 (SE = 0.06) cm for cases (p < 0.0001).
Genetic risk score characteristics
The GRS-609 was normally distributed in controls, ranging from 15.45 to 18.99 (median = 17.23; interquartile range = 16.92–17.54). It explained 13% of variance in height, 17% after adjusting for age and principal components (partial-R2 = 12%; first-stage regression partial-F-statistic = 2505.8 (df = 1), p < 0.001). A 1-unit GRS-609 increase was associated with 5.2 cm greater height. Average height was 6.2 cm greater in the highest vs. lowest GRS quartile.
Cochran’s I2 and p-values for heterogeneity19 showed no evidence of inter-study heterogeneity in the relationship between either the GRS-609 (I2 = 34%, p-heterogeneity = 0.07) or the simplified GRS-363 (I2 = 32%, p-heterogeneity = 0.08) and height among controls (Supplementary Figure 1a, b). The GRS-609 was not associated with most potential confounders of the height-ovarian cancer association in observational studies, including age, parity, OC use and education (Supplementary Table 3). The GRS was marginally positively associated with age at menarche (p = 0.03), consistent with known genetic overlap between these traits.20
Primary outcomes
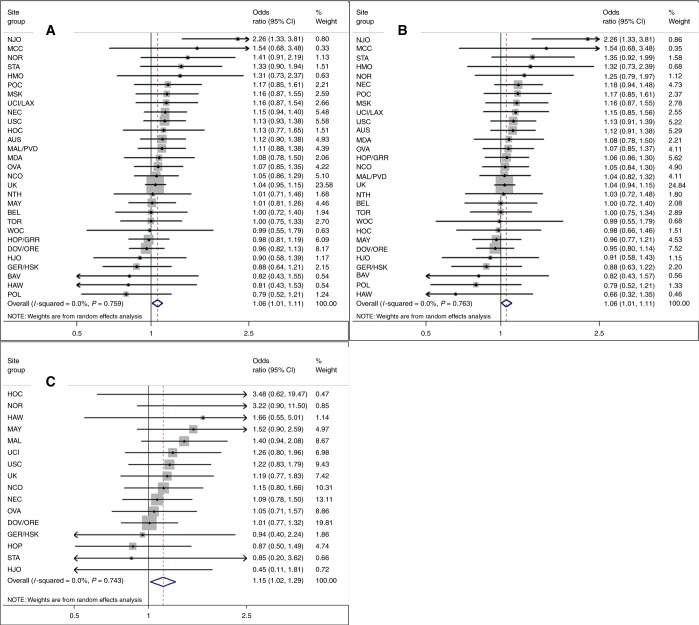
Women with greater genetically predicted height had a modestly increased risk of developing ovarian cancer (pOR = 1.06, 95% CI: 1.01–1.11 per 5 cm) (Fig. 1a; Table 2) with a greater magnitude of association for borderline (pOR = 1.15; 95% CI: 1.02–1.29) than invasive tumours (pOR = 1.06; 95% CI: 1.01–1.11; Fig. 1b, c; Table 2). No significant inter-study heterogeneity was noted (Fig. 1a–c). GRS-363 (pOR = 1.06, 95% CI: 1.00–1.11, all tumours) and GRS-377 (OR = 1.07; 95% CI: 1.01–1.12) results were similar to the GRS-609. The association was stronger when we restricted to 92 genotyped SNPs (pOR = 1.14; 95% CI: 1.04–1.25). Estimates from analyses excluding low-MAF SNPs, excluding case-only studies, or adjusting for age at menarche, were similar to primary analyses. When we sequentially excluded SNPs associated with ovarian or other hormonal cancers, hormone levels and tumour development, estimates were similar or stronger than GRS-609 results. MR-Egger suggested minimal bias from pleiotropy (p = 0.1; MR-Egger beta = 0.163 corresponded to an OR per 5 cm of 1.13 (95% CI: 1.02–1.25), confirming a significant positive association).
Fig. 1.
Association between increasing genetically predicted height and risks of all, invasive and borderline ovarian tumours. Increasing height per 5 cm predicted by weighted 609-locus genetic risk score among 39 studies. Risk of a all, b invasive and c borderline ovarian tumours. The UK grouping includes RMH, SOC, SRO, UKR, SEA and UKO for a and b, and RMH, SOC and SEA for c
Table 2.
Association between increasing height (per 5 cm)—predicted by a weighted 609-locus genetic risk score—and risk of ovarian cancer, stratified by study
| Histologic subtypea | N studies | N controls | N cases | Odds ratios (95% CI)b |
|---|---|---|---|---|
| Primary outcomes | ||||
| All ovarian cancers | 39 | 23,003 | 16,395 | 1.06 (1.01–1.11) |
| Invasive | 39 | 23,003 | 14,549 | 1.06 (1.01–1.11) |
| Borderlinec | 20 | 16,463 | 1680 | 1.15 (1.02–1.29) |
| Secondary outcomes, by histologic subtype and behaviour | ||||
| Serous | ||||
| High-graded | 39 | 23,003 | 7933 | 1.05 (0.99–1.11) |
| Invasive low-grade and borderline | 32 | 21,131 | 1408 | 1.15 (1.01–1.30) |
| Mucinous (invasive and borderline) | 38 | 22,410 | 1567 | 1.08 (0.96–1.21) |
| Endometrioid (invasive) | 39 | 23,003 | 2059 | 1.05 (0.95–1.16) |
| Clear cell (invasive) | 35 | 22,051 | 948 | 1.20 (1.04–1.38) |
Weights applied were β-coefficients for the relationship between each SNP and height as reported in the meta-analysis of genome-wide association studies conducted by the Genetic Investigation of ANthropometric Traits (GIANT) Consortium.9 On the basis of the additive SNP effects suggested by GIANT, the score summed alleles across the 609 SNPs. For the 92 genotyped SNPs, where values were missing (<2.5% per SNP), we used imputed probabilities.
aIncludes studies with >5 cases.
bPooled study-specific odds ratios are reported for primary outcomes; odds ratios stratified by study are reported for secondary outcomes (secondary analyses used single models stratified by study to maximise power).
cOf the 1691 borderline tumours included in the all-case analysis, 1680 were from 20 studies with >5 cases each.
dIncludes all invasive serous cancers except low-grade (G1) (n = 469) as well as invasive serous cancers of unknown grade (n = 1957) and primary peritoneal cancers of unknown behaviour (n = 44), because in both instances the majority would be high-grade serous.
CI confidence interval
In contrast, for women with height and confounder data (16 studies), the conventional analysis suggested no association (adjusted-OR = 1.01, 95% CI: 0.99–1.04 per 5 cm). Conducting MR within the same 16 studies yielded results similar to overall analyses (OR = 1.06, 95% CI: 1.00–1.13) (Supplementary Table 4).
Secondary outcomes
After stratifying by subtype/behaviour, the strongest associations were seen for clear cell (OR = 1.20, 95% CI: 1.04–1.38) and low-grade/borderline serous cancers (OR = 1.15, 95% CI: 1.01–1.30) (Table 2). However, CIs were wide and overlapping due to lower statistical power in these subgroup analyses. The estimate for clear cell cancers was also significantly elevated in our conventional analyses (Supplementary Table 4).
Discussion
We used a 609-SNP GRS to examine the relationship between height and ovarian cancer risk for women of European ancestry. Our data indicate a modest positive association between genetically predicted height and ovarian cancer risk, which may be stronger for borderline cancers. Height may be relevant to cancer risk as a marker for lifetime growth-factor levels (e.g. IGF-1) and/or early-life exposures (socio-economic/environmental/nutritional).3, 21, 22
Observational studies are subject to biases (reverse causality, selection bias, differential/non-differential reporting, confounding) which cannot be ruled out as possible explanations for observed associations. By using genotype, the MR technique can overcome some of these biases, given three assumptions. We confirmed the two verifiable assumptions: the GRS was associated with height, and not with most known confounders. The GRS-menarche age association is unlikely to explain the observed association, because age at menarche is only weakly associated with ovarian cancer, and women with later menarche have if anything lower ovarian cancer risk, so if this affected our results, we would expect the true effect to be at least as strong as the reported association. Also, removing hormone-related SNPs, or adjusting for menarche age, did not attenuate estimates. Owing to the limited current biological understanding of all 609 SNPs, we could not conclusively exclude the presence of alternate pathways from height genes to ovarian cancer (assumption three). However, MR-Egger and sensitivity analyses excluding pathway-specific SNPs provided some evidence for their absence, minimising the likelihood that our observed association is explained by pathways separate from height/growth. Although height data were not available for the entire population, this is unlikely to have affected our results as we used these data only to refine the height predictions from the GRS, and there is no reason to believe the GRS-height relationship would be different for women with and without height data. Further strengths of our analysis include the large number of SNPs and power to detect modest differences.
Despite potential limitations of conventional observational studies, our MR-estimate is almost identical to previously reported associations, suggesting previous estimates were not appreciably biased. The World Cancer Research Fund/American Institute for Cancer Research meta-analysis of 24 prospective studies, and a study pooling 47 prospective/case–control studies, both reported a significant 7–8% increase in risk (combining invasive/borderline cancers) per 5 cm height increase.3, 4 The lack of association seen in the OCAC conventional height analysis reflects the greater potential for bias in case–control studies and illustrates the value of MR in overcoming these biases. Few previous studies have examined borderline cancers separately, a strength of our analysis. Previous observational studies have not reported consistent patterns by histologic subtype2, 4, 23; our secondary analyses were under-powered to resolve this question.
Using MR, we have established that the previously observed association between height and ovarian cancer risk is unlikely to have been explained by bias, and that genetic factors influencing height play roles in ovarian cancer development. Height could therefore be used, with other risk factors, to identify women at elevated risk. Further research should continue to explore mechanisms underpinning this association.
Electronic supplementary material
Acknowledgements
We thank all the individuals who took part in this study and all the researchers, clinicians, and technical and administrative staff who have made possible the many studies contributing to this work. In particular, for their contribution to the design and conduct of the individual studies that contributed to the analysis, we thank: D. Bowtell, A. deFazio, D. Gertig, A. Green, P. Parsons, N. Hayward and D. Whiteman (AUS); G. Peuteman, T. Van Brussel and D. Smeets (BEL); U. Eilber (GER); S. Reckemeyer, A. Korotkaia and S. Reina-Campanon (HJO); J. Meyer, C. Hilker, S. Windebank, and J. Vollenweider (MAY); the Victorian Cancer Registry and the Australian Institute of Health and Welfare for ascertaining cases and vital status (MCC); I. Orlow, L. Paddock, and L. Rodriguez-Rodriguez (NJO); the SEARCH team, C. Luccarini, C. Baynes, and D. Conroy (SEA); the Scottish Gynaecological Clinical Trials group and SCOTROC1 investigators (SRO); I. Jacobs, M.Widschwendter, E. Wozniak, A. Ryan, J. Ford, N. Balogun and C. Karpinskyj (UKO); and C. Pye (UKR).
Competing interests
The authors declare no competing financial interests.
Funding
This work was supported by: the National Cancer Institute at the U.S. National Institutes of Health [K07-CA095666, K07-CA80668, K07-CA143047, K22-CA138563, N01-CN025403, N01-CN55424, N01-PC67001, N01-PC67010, P01-CA17054, P30-CA072720, P30-CA008748, P30-CA14089, P30-CA15083, P50-CA105009, P50-CA136393, P50-CA159981, R01-CA058860, R01 CA063678, R01 CA063682, R01-CA092044, R01-CA095023, R01-CA16056, R01-CA54419, R01-CA58598, R01-CA61107, R01-CA61132, R01-CA76016, R01-CA83918, R01-CA87538, R01-CA112523, R01-CA122443, R03-CA113148, R03-CA115195, U01-CA69417, U01-CA71966 and Intramural Research funds]; the European Commission's Seventh Framework Programme [agreement number 223175 HEALTH F2 2009-223175]; Cancer Research UK [C490/A16561, C536/A13086, C536/A6689, C1287/A10118, C1287/A 10710, C12292/A11174, C1281/A12014, C5047/A8384, C5047/A15007, C5047/A10692 and C8197/A16565]; a National Institutes of Health (CA128978), Cancer Post-GWAS Initiative [1U19 CA148537, 1U19 CA148065 and 1U19 CA148112—the Genetic Associations and Mechanisms in Oncology (GAME‐ON) initiative]; the U.S. Department of Defense [DAMD17-02-1-0669, W81XWH-07-0449, DAMD17-02-1-0666, W81XWH-10-1-0280 and W81XWH-10-1-0341]; the Canadian Institutes of Health Research (CIHR) [MOP-86727 and MSH-87734 to L.E.K.] and the CIHR Team in Familial Risks of Breast Cancer; the Komen Foundation for the Cure; the Breast Cancer Research Foundation; the Ovarian Cancer Research Fund (thanks to donations by the family and friends of Kathryn Sladek Smith); the U.S. Army Medical Research and Materiel Command [DAMD17-01-1-0729 and DAMD17-02-1-0669]; the National Health and Medical Research Council of Australia [199600, 209057, 251553, 400281, 504711, 1073898 and fellowships to G.C.-T. and P.M.W.]; Cancer Councils of Queensland, Victoria, New South Wales, South Australia and Tasmania and the Cancer Foundation of Western Australia [Multi-State Grant Applications 191, 211 and 182]; VicHealth; the ELAN Program of the University of Erlangen-Nuremberg; the Nationaal Kankerplan of Belgium; the German Federal Ministry of Education and Research of Germany Programme of Clinical Biomedical Research [01 GB 9401]; the German Cancer Research Center; the Roswell Park Cancer Institute Alliance Foundation [P30 CA016056]; the Rudolf-Bartling Foundation; the Helsinki University Central Hospital Research Fund; the National Institutes of Health/National Center for Research Resources/General Clinical Research Center [M01-RR000056]; an American Cancer Society Early Detection Professorship [SIOP-06-258-01-COUN to B.Y.K.]; the National Center for Advancing Translational Sciences (NCATS) [UL1TR000124 to B.Y.K.]; the Danish Cancer Society [94-222-52]; the Mermaid I project; the Mayo Foundation; the Minnesota Ovarian Cancer Alliance; the Fred C. and Katherine B. Andersen Foundation; the Cancer Institute of New Jersey; Helse Vest; the Norwegian Cancer Society; the Research Council of Norway; Radboud University Medical Centre; the Oregon Health and Science University (OHSU) Foundation; Pomeranian Medical University; the UK National Institute for Health Research Biomedical Research Centres at the University of Cambridge, University College London Hospital, and the Royal Marsden Hospital; the Imperial Experimental Cancer Research Centre [C1312/A15589]; the U.S. Public Health Service [PSA-042205]; the Lon V. Smith Foundation [LVS-39420]; The Eve Appeal; The Oak Foundation; the California Cancer Research Program [00-01389V-20170 and 2II0200]; the Polish Ministry of Science and Higher Education [4 PO5C 028 14 and 2 PO5A 068 27]; and the Maria Sklodowska-Curie Memorial Cancer Center and Institute of Oncology, Warsaw.
Footnotes
Deceased: Helga B. Salvesen, Kenneth D. Swenerton, Catherine Phelan
Electronic supplementary material
Supplementary information is available for this paper at 10.1038/s41416-018-0011-3.
References
- 1.Emerging Risk Factors Collaboration. Adult height and the risk of cause-specific death and vascular morbidity in 1 million people: individual participant meta-analysis. Int. J. Epidemiol. 2012;41:1419–1433. doi: 10.1093/ije/dys086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schouten LJ, et al. Height, body mass index, and ovarian cancer: a pooled analysis of 12 cohort studies. Cancer Epidemiol. Biomark. Prev. 2008;17:902–912. doi: 10.1158/1055-9965.EPI-07-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.World Cancer Research Fund/American Institute for Cancer Research. Food, Nutrition, Physical Activity, and the Prevention of OvarianCancer 2014. Washington DC: AICR; 2014. [Google Scholar]
- 4.Collaborative Group on Epidemiological Studies of Ovarian Cancer. Ovarian cancer and body size: individual participant meta-analysis including 25,157 women with ovarian cancer from 47 epidemiological studies. PLoS Med. 2012;9:e1001200. doi: 10.1371/journal.pmed.1001200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davey Smith G, Ebrahim S. 'Mendelian randomization': can genetic epidemiology contribute to understanding environmental determinants of disease? Int. J. Epidemiol. 2003;32:1–22. doi: 10.1093/ije/dyg070. [DOI] [PubMed] [Google Scholar]
- 6.Gayther SA, et al. Tagging single-nucleotide polymorphisms in cell cycle control genes and susceptibility to invasive epithelial ovarian cancer. Cancer Res. 2007;67:3027–3035. doi: 10.1158/0008-5472.CAN-06-3261. [DOI] [PubMed] [Google Scholar]
- 7.Dixon SC, et al. Adult body mass index and risk of ovarian cancer by subtype: a Mendelian randomization study. Int. J. Epidemiol. 2016;45:884–895. doi: 10.1093/ije/dyw158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pharoah PDP, et al. GWAS meta-analysis and replication identifies three new susceptibility loci for ovarian cancer. Nat. Genet. 2013;45:362–370. doi: 10.1038/ng.2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wood AR, et al. Defining the role of common variation in the genomic and biological architecture of adult human height. Nat. Genet. 2014;46:1173–1186. doi: 10.1038/ng.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abecasis GR, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burgess S. Identifying the odds ratio estimated by a two-stage instrumental variable analysis with a logistic regression model. Stat. Med. 2013;32:4726–4747. doi: 10.1002/sim.5871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Didelez V, Meng S, Sheehan NA. Assumptions of IV methods for observational epidemiology. Stat. Sci. 2010;25:22–40. doi: 10.1214/09-STS316. [DOI] [Google Scholar]
- 13.Stukel TA, Demidenko E, Dykes J, Karagas MR. Two‐stage methods for the analysis of pooled data. Stat. Med. 2001;20:2115–2130. doi: 10.1002/sim.852. [DOI] [PubMed] [Google Scholar]
- 14.Brion MJ, Shakhbazov K, Visscher PM. Calculating statistical power in Mendelian randomization studies. Int. J. Epidemiol. 2013;42:1497–1. doi: 10.1093/ije/dyt179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int. J. Epidemiol. 2015;44:512–525. doi: 10.1093/ije/dyv080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Welter D, et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42:D1001–D1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karolchik D, et al. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 2004;32:D493–D496. doi: 10.1093/nar/gkh103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kent WJ, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–996. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Higgins JPT, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat. Med. 2002;21:1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 20.Bulik-Sullivan B, et al. An atlas of genetic correlations across human diseases and traits. Nat. Genet. 2015;47:1236–1241. doi: 10.1038/ng.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stefan N, Haring HU, Hu FB, Schulze MB. Divergent associations of height with cardiometabolic disease and cancer: epidemiology, pathophysiology, and global implications. Lancet Diabetes Endocrinol. 2016;4:457–467. doi: 10.1016/S2213-8587(15)00474-X. [DOI] [PubMed] [Google Scholar]
- 22.Clayton P, Banerjee I, Renehan AG, Murray PG, Clayton PE. Growth hormone, the insulin-like growth factor axis, insulin and cancer risk. Nat. Rev. Endocrinol. 2011;7:11–24. doi: 10.1038/nrendo.2010.171. [DOI] [PubMed] [Google Scholar]
- 23.Jordan SJ, Webb PM, Green AC. Height, age at menarche, and risk of epithelial ovarian cancer. Cancer Epidemiol. Biomark. Prev. 2005;14:2045–2048. doi: 10.1158/1055-9965.EPI-05-0085. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.