Abstract
Angiogenesis, new blood vessel formation from preexisting vessels, is critical for solid tumor growth. As such, there have been efforts to inhibit angiogenesis as a means to obstruct tumor growth. However, antiangiogenic therapy faces major challenges to the selective targeting of tumor-associated-vessels, as current antiangiogenic targets also disrupt steady-state vessels. Here, we demonstrate that the developmentally critical transcription factor Etv2 is selectively upregulated in both human and mouse tumor-associated endothelial cells (TAECs) and is required for tumor angiogenesis. Two-photon imaging revealed that Etv2-deficient tumor-associated vasculature remained similar to that of steady-state vessels. Etv2-deficient TAECs displayed decreased Flk1 (also known as Vegfr2) expression, FLK1 activation, and proliferation. Endothelial tube formation, proliferation, and sprouting response to VEGF, but not to FGF2, was reduced in Etv2-deficient ECs. ROS activated Etv2 expression in ECs, and ROS blockade inhibited Etv2 expression in TAECs in vivo. Systemic administration of Etv2 siRNA nanoparticles potently inhibited tumor growth and angiogenesis without cardiovascular side effects. These studies highlight a link among vascular oxidative stress, Etv2 expression, and VEGF response that is critical for tumor angiogenesis. Targeting the ETV2 pathway might offer a unique opportunity for more selective antiangiogenic therapies.
Keywords: Angiogenesis
Keywords: Cancer gene therapy, endothelial cells, hypoxia
ETS transcription factor ETV2, critical for developmental angiogenesis, is reactivated in tumor endothelial cells and required for tumor angiogenesis.
Introduction
Angiogenesis is an important process for successful embryogenesis, postinjury tissue repair and regeneration, and progression of many pathologic conditions, including cancer. Thus, while promotion of angiogenesis is critical to tissue repair and regeneration, inhibition of angiogenesis is important to obstruct pathologic conditions, such as for tumor growth. Currently, it remains a challenge to selectively target tumor angiogenesis, as many of the factors and pathways presently targeted in therapies are shared between normal and tumor vessels (1–3). For example, anti-VEGF therapies, although considered to be most prevalent in inhibiting tumor angiogenesis, have severe shortcomings in specifically targeting tumor-associated vessels undergoing neoangiogenesis (4, 5), as VEGF is also required for physiological steady-state vessels (6, 7). Considering these limitations, it would be crucial to identify molecular marker(s) and signaling pathways uniquely functioning in tumor-associated vessels that can be exclusively targeted.
ETS transcription factors have emerged as critical regulators of angiogenesis (8–10). A winged helix-turn-helix motif formed by the ETS domain can bind a consensus sequence (GGAA/T) to regulate target gene expression (11). Many ETS factors are redundantly expressed in hematopoietic and endothelial cells (ECs) in the developing embryo (12, 13). Consistently, mice or zebrafish deficient in any of these Ets factors display varying degrees of hematopoietic and vascular defects (14), probably reflecting the redundant expression pattern of these Ets factors in hematopoietic cells and ECs. Distinct from these ETS factors, Etv2 (also known as Er71 and Etsrp) is transiently expressed in the primitive streak, yolk sac blood islands, and large vessels of the dorsal aorta during embryogenesis (15–18). Etv2-deficient mice display a complete block in blood and blood vessel formation, demonstrating that ETV2 performs a nonredundant and indispensable function in hematopoietic and vessel development. We recently established that ETV2 activates other Ets transcription factors in hematopoietic cell and EC lineage development. Moreover, the hematopoietic and EC program induced by ETV2 is maintained by other ETS factors through an ETS switching mechanism in the face of transient Etv2 expression (19). Importantly, ETV2 function is evolutionary conserved, as mouse or human Etv2 can induce hematopoietic and vascular development in zebrafish (20). Notably, ECs can be efficiently generated from mouse and human pluripotent stem cells by enforced Etv2 expression (17, 21, 22). Moreover, Etv2 can efficiently reprogram somatic cells to ECs (23–26). These studies have established that Etv2 is necessary and sufficient for EC generation and function.
While Etv2 function in the developmental context has been extensively investigated (27, 28), studies on this gene in adults are greatly lacking. Previous studies have established that Etv2 expression becomes silent once hematopoietic cells and ECs have been generated (17, 29–31). However, it becomes reactivated upon injury and is required for hematopoietic and EC regeneration (32, 33). Since developmental programs are often reactivated in pathological conditions and regulate disease progression, we investigated whether the developmentally critical factor Etv2 is also reactivated in tumor endothelium and is required for tumor angiogenesis. Our data demonstrated that ETV2 is expressed in human and mouse tumor-associated ECs (TAECs). Endothelial Etv2 is required for tumor angiogenesis. While Etv2-deficient TAECs failed to elevate Flk1 expression and had a defective response to VEGF, they displayed a normal response to FGF. We identified ROS, which were readily elevated in TAECs, as upstream activators of Etv2 expression. Systemic Etv2 siRNA nanoparticle delivery effectively inhibited tumor growth and angiogenesis without cardiovascular side effects. Our results indicate that Etv2 is a marker for TAECs and that targeting Etv2 maybe a valid choice for blocking tumor angiogenesis.
Results
Etv2 is upregulated in both human and mouse TAECs.
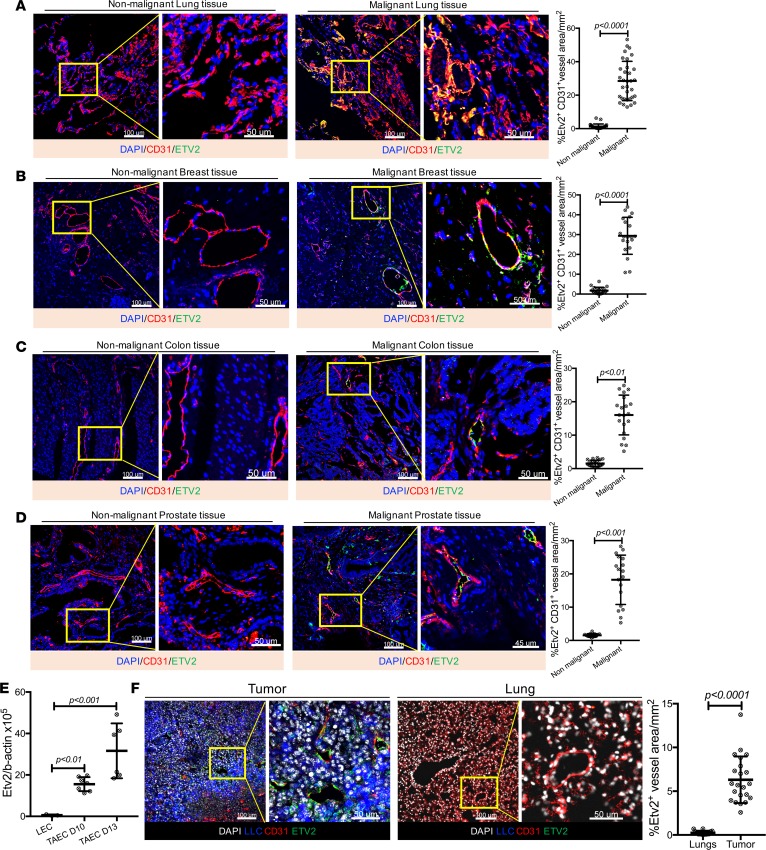
As Etv2 was required for ECs undergoing active angiogenesis (32), we assessed whether Etv2 also plays a role in tumor angiogenesis. We first examined human malignant and matched nonmalignant specimens from lung, breast, prostate, and colon cancer patients and found that ETV2 was expressed in the tumor vessels but not in the nonmalignant tissues (Figure 1, A–D). We next determined the kinetics of Etv2 expression in a mouse tumor transplantation model using Lewis lung carcinoma–GFP (LLC-GFP) tumor cells. In this tumor transplantation model, palpable tumor mass is detected by days 4–5 after tumor cell transplantation (ptt), followed by robust tumor growth. Etv2 expression was detected in the tumor mass from day 4 ptt, with its expression highly upregulated from day 10 ptt (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.97349DS1). We analyzed the FACS-sorted ECs from the tumor by quantitative real-time reverse-transcription PCR (qRT-PCR) and found that TAECs expressed high levels of Etv2 (Figure 1E). Immunofluorescence of the mouse tumor sections also revealed that ETV2 was present primarily in TAECs (Figure 1F). Intriguingly, ETV2 was also detected in some, not all, hematopoietic cells (Supplemental Figure 1B). It is notable that ETV2 was present in both nuclei and perinuclear spaces (Supplemental Figure 1, C and D). Since tumor cells and ECs in nonmalignant vessels do not express Etv2 (Figure 1, A–D, and Supplemental Figure 1A), we conclude that Etv2 expression is induced in TAECs.
Figure 1. Etv2 is upregulated in tumor endothelial cells.
(A–D), Representative images for ETV2 (green) and CD31 (red) immunofluorescence of malignant and matched nonmalignant specimens from (A) lung, (B) breast, (C) colon, and (D) prostate cancer patients. DAPI (blue) is used to counterstain nuclei. Associated graphs show the quantification of ETV2-expressing CD31+ vessels in malignant versus nonmalignant tissues. Every cancer type had malignant and nonmalignant tissue specimen pairs collected from at least 3 patients; at least 3 sections from each patient were subjected to immunofluorescence. (E) qRT-PCR analysis of Etv2 expression in CD31+CD45– endothelial cells obtained from lung (LEC) and tumor (TAEC), 10 and 13 days after tumor cell transplantation (n = 3 LEC, 7 TAEC day 10, and 6 TAEC day 13). (F) Representative images for ETV2 (green) and CD31 (red) immunofluorescence of mouse tumor and lung sections, processed after 20 days of tumor transplantation. LLC-GFP cells (blue) and nuclei counterstained with DAPI (gray) are shown (n = 15 or more/group). Scale bars: 100 μm; 50 μm (higher-magnification images). Data are presented as mean with standard deviation for all measurements. Statistical significance was analyzed by either a 2-tailed Student’s t test (A–D and F) or 1-way ANOVA with Dunnett’s multiple-comparison test (E).
Endothelial Etv2 is required for optimal tumor growth.
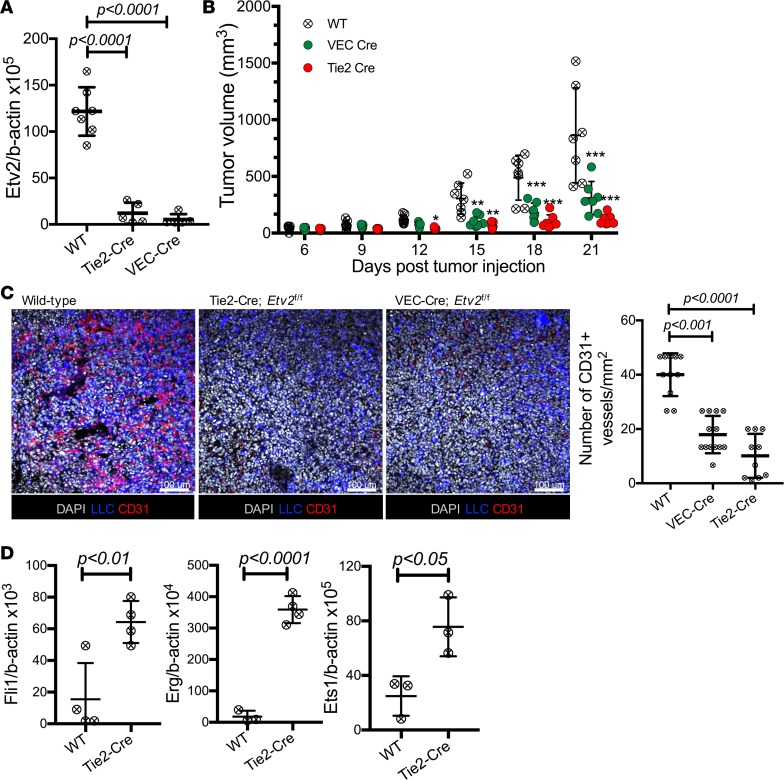
To determine whether the reactivation of Etv2 expression in TAECs reflected its functional requirement in tumor angiogenesis, we transplanted LLC-GFP cells into Tie2-Cre;Etv2f/f (Tie2-Cre;Etv2 conditional KO [CKO], lacks Etv2 in ECs and hematopoietic cells) and VECadherin-Cre;Etv2f/f (VEC-Cre;Etv2 CKO, lacks Etv2 in ECs) mice (32, 33). Efficient deletion of Etv2 was confirmed in ECs of both models (Supplemental Figure 2A). Etv2 expression in TAECs and tumor growth were significantly reduced in the Etv2 CKO mice (Tie2-Cre or VEC-Cre) compared with littermate control wild-type mice (Figure 2, A and B, and Supplemental Figure 2B). Similar tumor growth defects were observed with another well-studied B16 melanoma tumor model (Supplemental Figure 2C). Capillary densities within tumors were also significantly decreased (Figure 2C), suggesting that endothelial Etv2 is required for efficient tumor angiogenesis and growth. Ets transcription factors are redundantly expressed in ECs (12, 13). Notably, Etv2-deficient TAECs had elevated expression of Fli1, Erg, and Ets1 (Figure 2D), suggesting that there might be possible compensatory roles played by the other Ets factors in the absence of the Etv2, which could have contributed to the residual tumor growth in the Etv2-CKO mice. Hematopoietic Etv2 also appears to contribute to tumor progression, as Tie2-Cre;Etv2-CKO mice displayed more severe reduction in tumor growth than the VEC-Cre;Etv2-CKO mice. Indeed, Vav-Cre;Etv2f/f (Vav-Cre;Etv2 CKO, Etv2 deletion in hematopoietic cells) mice showed a mild, but significant, reduction in tumor growth and angiogenesis compared with littermate control mice (Supplemental Figure 2, D and E). Importantly, tumor-associated hematopoietic cells in the Vav-Cre;Etv2 mice had diminished expressions of angiogenic growth factors Vegf-a, Vegf-b, Fgf-2, Igf-1, and Igf-2 (Supplemental Figure 2F), which could explain why mice with deleted hematopoietic Etv2 displayed defective tumor angiogenesis.
Figure 2. Endothelial Etv2 is required for tumor growth and angiogenesis.
(A) qRT-PCR analysis of Etv2 expression in TAECs of wild-type control (n = 5), Tie2-Cre;Etv2 CKO (n = 7), and VEC-Cre;Etv2 CKO (n = 7) mice on day 13 after tumor cell transplantation (ptt). (B) Tumor growth in control, VEC-Cre;Etv2, and Tie2-Cre;Etv2-CKO mice. Tumor volume was measured on days 6, 9, 12, 15, 18, and 21 ptt (n = 7/group) (*P < 0.05, **P < 0.01, ***P < 0.001). (C) Representative images for CD31 (red) immunofluorescence and quantification of CD31+ vessel density of the tumor sections (n = at least 10/group). LLC-GFP cells (blue) and nuclei counterstained with DAPI (gray) are shown. Scale bars: 100 μm. Tumors were harvested from littermate control (wild-type), Tie2-Cre;Etv2 CKO, and VEC-Cre;Etv2-CKO mice on day 21 ptt. (D) qRT-PCR analysis of Fli1, Erg, and Ets1 expression in TAECs of littermate control (WT) and Tie2-Cre;Etv2-CKO mice on day 15 ptt (n = 4/group). Data are presented as mean with standard deviation for all measurements. Statistical significance was analyzed by either a 2-tailed Student’s t test (D), 1-way ANOVA with Dunnett’s multiple-comparison test (A and C), or 2-way repeated-measures ANOVA with Tukey’s multiple-comparison test (B).
2PM analysis reveals that Etv2-deficient tumor vessels are similar to steady-state vessels.
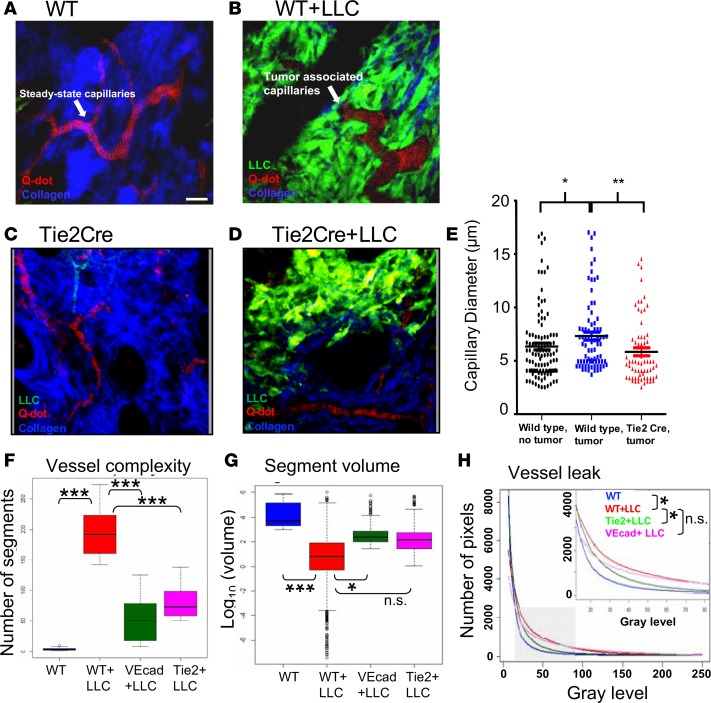
We examined the effect of Etv2 loss on the morphology and function of the tumor-associated vasculature in vivo using 2-photon microscopy (2PM). 2PM induces less photodamage than confocal microscopy and has higher spatiotemporal resolution (~30 fps and <0.5 μm/pixel) than magnetic resonance imaging, computed tomography, and positron emission tomography (34). To assess Etv2-deficient tumor vessels, we transplanted LLC-GFP cells into the ear pinna of Etv2-CKO mice, performed in vivo 2PM, and assessed vessel morphology. In wild-type mice, tumor-associated vessels, compared with steady-state vessels, had distinct tortuous morphology, larger diameter, and functional abnormalities, including sluggish blood flow and increased leakiness, as previously observed (35). However, tumor-associated vessels in Etv2-CKO mice resembled those of steady-state vessels (Figure 3, A–E, and Supplemental Video 1). It is important to note that the vessels of the control and Etv2-CKO mice were similar in steady states (Supplemental Figure 3A). Next, to better assess tumor-associated changes to the vascular network, we created 3D contour surfaces of all vessels in the images, including capillaries and larger venules and arterioles, to quantify the number of discontinuous vessel segments, as a measure of network complexity and the volume of each vessel segment. Tumors growing in wild-type mice showed increased vessel complexity and decreased average vessel segment volume, compared with steady-state vessels, suggesting the expansion of tumor capillaries via angiogenesis (Figure 3, F and G, and Supplemental Figure 3, B and C). However, tumors growing in Etv2-CKO mice had similar vessel complexity and average vessel segment volume compared with steady-state vessels (Figure 3, F and G, and Supplemental Figure 3, B and C), suggesting that new capillary formation requires Etv2. Finally, we evaluated vessel integrity in the tumor-bearing mice by plotting the average pixel intensity counts and identified extravascular leakage by an increase in intermediate pixel intensity counts in the red channel (10–80 gray levels, zoomed inset) and, when fitted to an exponential curve, a lower rate of decay. Tumor-associated vessels were significantly leakier than steady-state vessels in wild-type mice, whereas, the leakage was significantly reduced in Etv2-CKO mice (Figure 3H and Supplemental Figure 3D). We additionally performed a kymograph analysis on representative images to further validate vessel integrity of the Etv2-deficient tumor vessels. While tumor-associated vessels in wild-type mice showed noticeable broadening of the intensity profile and higher extravascular dextran signal, tumor-associated vessels in the Etv2-CKO mice had sharp well-delineated vessel edges and little extravascular dextran signal (Supplemental Figure 3E and Supplemental Video 2). These data suggested that Etv2-deficient tumor vessels, in the absence of new angiogenesis, remained similar to steady states.
Figure 3. In vivo assessment of tumor-associated vessel morphology in Etv2-CKO mice.
2PM was performed on anesthetized mice to assess vessel morphology in (A and B) wild-type (WT+LLC) and (C and D) Tie2-Cre;Etv2-CKO (Tie2Cre, Tie2Cre+LLC) tumor-bearing mice. Images show that LLC tumor (green) is associated with vascular remodeling and the growth of larger tortuous and irregular capillaries (red) and collagen fibers (second harmonic generation signal, blue). Scale bar: 10 μm. (E) Average capillary diameter in steady-state vessels (WT, black dots) and tumor-associated capillaries in littermate control (WT+LLC, blue dots) and CKO (Tie2-Cre+LLC, red dots) mice (n = 5/group; at least 10 capillaries were measured per mouse; *P < 0.05, **P < 0.01). (F) Vessel complexity was analyzed by generating 3D contour surfaces to identify vessels (Supplemental Video 1) and counting the number of discrete vessel segments in each image (n = at least 8/group; ***P < 0.001). (G) Vessel volume is plotted for each group, with the middle line in each bar representing the average volume of vessel segments in an image (n = 5/group; *P < 0.05, ***P < 0.001). (H) A plot of average pixel intensity counts for each group fitted to an exponential probability distribution function using maximum likelihood estimation methods to extract the rate of decay for each group (n = 4/group; *P < 0.05). Data are presented as mean with standard deviation for all measurements. Statistical significance was analyzed by 1-way ANOVA with Dunnett’s multiple-comparison test (E–G) or 2-way repeated-measures ANOVA with Tukey’s multiple-comparison test (H).
ETV2 regulates VEGF response in TAECs.
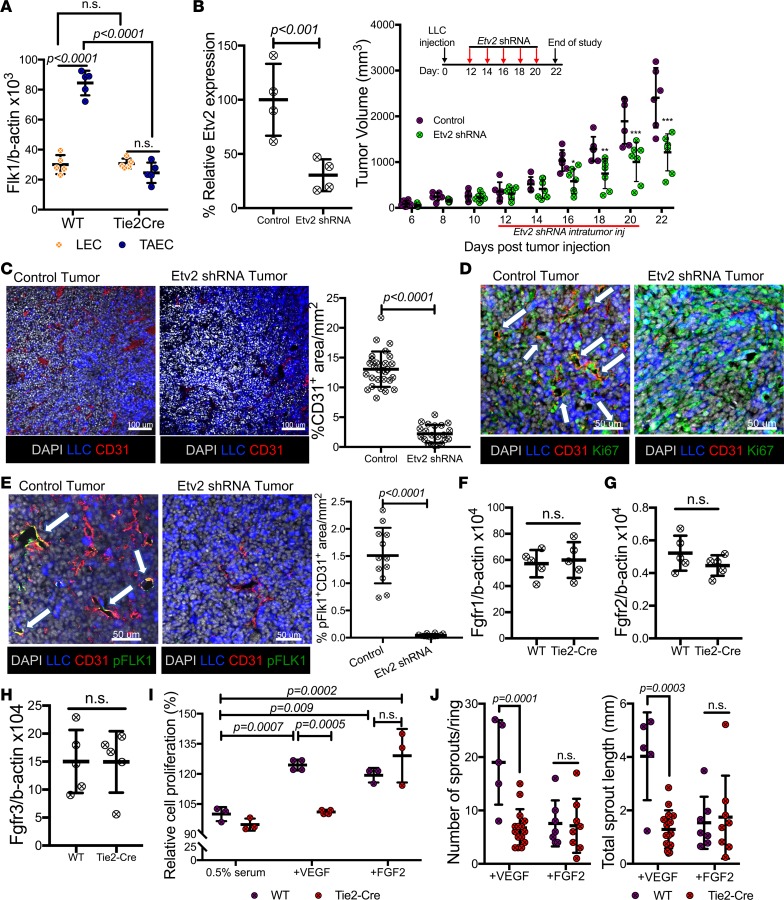
VEGF is crucial for tumor angiogenesis. Previous studies have established positive feedback regulation between Etv2 and Flk1 expression in development (19, 36). We therefore determined if Etv2 deficiency might lead to defects in VEGF responsiveness in TAECs. Flk1 expression was similar in steady-state ECs from both wild-type and Etv2-CKO mice. However, while Flk1 expression was readily upregulated in TAECs in the wild-type mice, Flk1 expression in the Etv2-deficient TAECs remained similar to that of steady-state ECs (Figure 4A). As expected, intratumoral lentiviral Etv2 shRNA injection significantly inhibited Etv2 expression in the TAECs, reduced TAEC proliferation, and restricted tumor growth and angiogenesis (Figure 4, B–D, and Supplemental Figure 4A). Importantly, tumor-associated vessels, but not the nontumor vessels, had reduced activating phosphorylation of FLK1 (37) after Etv2 shRNA injection, suggesting that Etv2 regulates Flk1 expression and activation in a TAEC-specific manner (Figure 4E and Supplemental Figure 4C). Notably, Etv2 deficiency does not appear to disturb FGF signaling, as expression of Fgfr1, Fgfr2, and Fgfr3 in both TAECs and nontumor ECs was similar in wild-type and Etv2-CKO mice (Figure 4, F–H, and Supplemental Figure 4C). Notably, Etv2-deficient lung ECs showed reduced proliferation to VEGF stimulation, but not FGF2, compared with controls (Figure 4I). Additionally, Etv2-deficient aortas displayed diminished VEGF-induced, but not FGF2-induced, sprouting in aortic ring assay (38) (Figure 4J and Supplemental Figure 4D). Moreover, lenti-Etv2 shRNA–infected yolk sac–derived endothelial (YSE) cells displayed defective tube formation in response to VEGF, but not FGF2, compared with wild-type YSE cells (Supplemental Figure 4E). Taken together, these results indicate that Etv2 regulates VEGF singling in TAECs.
Figure 4. Etv2 regulates tumor endothelial cell–specific Flk1.
(A) qRT-PCR analysis of Flk1 expression in lung ECs (LECs) and tumor ECs (TAECs) obtained from littermate control (WT) and Tie2-Cre;Etv2-CKO mice on day 15 ptt (n = 5/group). (B) qRT-PCR analysis of Etv2 expression in CD31+CD45– ECs obtained from tumors of Gfp shRNA– (control) and Etv2 shRNA–treated mice on day 15 ptt. Data are shown as percentage relative expression of control shRNA group (n = 4/group). Tumor growth in Gfp shRNA– (control) and Etv2 shRNA–treated mice (n = 6 control, 7 Etv2 shRNA; *P < 0.05, **P < 0.01,***P < 0.001). (C–E) Representative images and quantification for (C) CD31 (red) (n = 25/group), (D) Ki67 (green), and (E) pFLK1 (Y951) (green) (n = 12/group) immunofluorescence of tumor sections. LLC-GFP cells (blue) and nuclei counterstained with DAPI (gray) are shown. White arrows indicate the expression of Ki67 (D) and pFLK1 (E) in tumor vessels. Scale bars: 100 μm (C); 50 μm (D and E). (F–H) Fgfr1, Fgfr2, and Fgfr3 expression in TAECs of littermate control (WT) and Tie2-Cre;Etv2-CKO mice on day 15 ptt (n = 5/group). (I) Lung ECs (LEC) of littermate control (WT) or Tie2-Cre;Etv2-CKO mice were sorted and subjected to a cell proliferation assay. Data are shown as a percentage of control (0.5% serum) (n = 4/group). (J) Aortas from the controls (WT) and Tie2-Cre;Etv2-CKO mice were subjected to an angiogenic sprouting assay, and the mean sprout number and length were measured 8 days later (n = 5 or more/group). Data are presented as mean with standard deviation for all measurements. Statistical significance was analyzed by either a 2-tailed Student’s t test (B, left, C, and E–H), or 2-way ANOVA with Sidak’s (A, I, and J) or 2-way repeated-measures ANOVA with Tukey’s (B, right) multiple-comparison test.
ROS activate Etv2 expression.
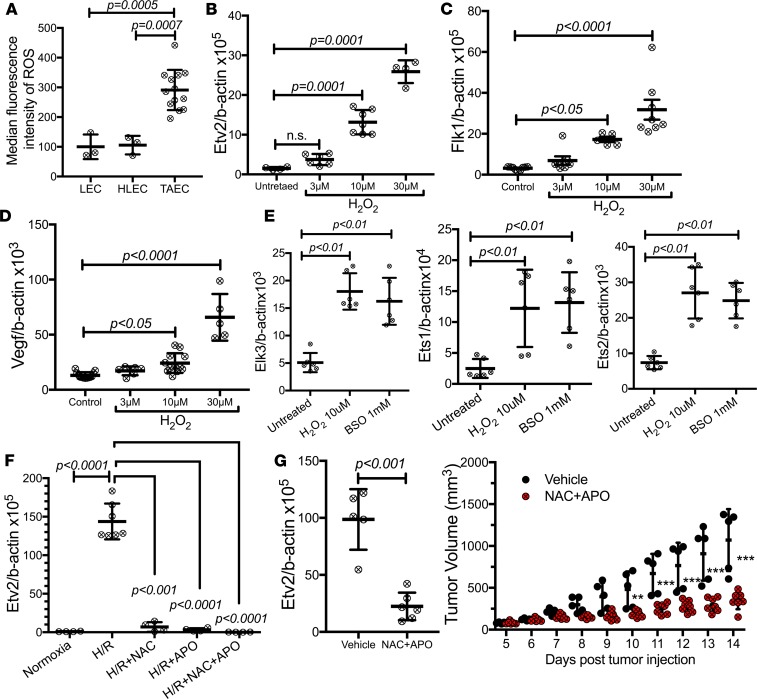
Developmental angiogenesis is controlled by changes in O2 tension (39, 40). Moreover, oxidative stress underlies many of the characteristics of cancer (41). Particularly, ROS, increased in response to ischemia, hypoxia, inflammation, or growth factors, such as VEGF and angiopoietin-1, play a critical role in angiogenesis (42). Thus, we determined if ROS could trigger Etv2 expression in TAECs. Higher ROS levels were detected in TAECs compared with steady-state lung or hind limb ECs (Figure 5A). Remarkably, Etv2, Flk1, or Vegf expression was greatly upregulated by H202 or L-buthionine-S,R-sulfoximine (BSO) in YSE cells (Figure 5, B–D, and Supplemental Figure 5, A–C). ROS also increased the expression of other Ets factors, including Ets1, Ets2, and Elk3 in YSE cells (Figure 5E). Additionally, 24-hour hypoxia followed by 24-hour reoxygenation, which mimics the pathophysiological ischemia/reperfusion condition (43, 44), or even 24-hour hypoxic exposure alone increased ROS and upregulated Etv2 expression in YSE cells; ROS scavengers inhibited this Etv2 expression (Figure 5F and Supplemental Figure 5, D and E). Remarkably, systemic ROS scavenger treatment reduced Etv2 expression in the TAECs and reduced tumor growth in vivo (Figure 5G).
Figure 5. ROS trigger Etv2 expression.
(A) ROS levels in ECs obtained from lung (LEC), hind limb tissue (HLEC), or tumor (TAEC) (n = 3 or more/group). (B–E) qRT-PCR analysis of expression of (B) Etv2, (C) Flk1, (D) Vegf, and (E) Ets factors Elk3, Ets1, and Ets2 in H2O2-treated yolk sac–derived endothelial (YSE) cells (n = 4 or more/group). (F) Etv2 expression in YSE cells underwent with 24-hour hypoxia (<1% oxygen), followed by 24-hour normoxia (H/R) with/without ROS scavengers N-Acetyl cysteine (NAC; 5 mM) and apocynin (APO; 100 mM) (n = 4 or more/group). (G) Etv2 expression in TAECs isolated from untreated and NAC+APO-treated mice on day 14 ptt. Tumor growth in untreated and NAC+APO-treated mice. Mice were treated with daily i.p. injections of vehicle or a combination of NAC (200 mg/kg) and APO (50 mg/kg) for 10 days, from day 4 to day 13 ptt (n = 5 vehicle, 7 NAC+APO) (**P < 0.01, ***P < 0.001). Data are presented as mean with standard deviation for all measurements. Statistical significance was analyzed by either a 2-tailed Student’s t test (G, left), 1-way ANOVA with Dunnett’s (B–F) or Bonferroni’s multiple-comparison test (A), or 2-way repeated-measures ANOVA with Sidak’s multiple comparison test (G, right).
Etv2 siRNA nanoparticles effectively inhibit tumor angiogenesis and tumor growth.
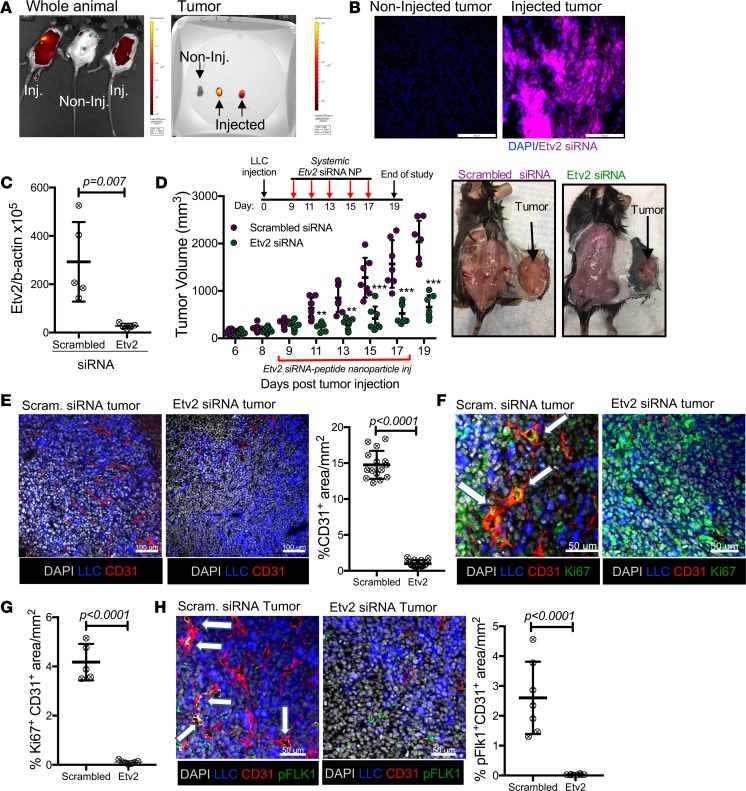
As Etv2 expression is silent in steady-state ECs, we speculated that a systemic anti-Etv2 strategy could effectively block Etv2 expression in TAECs, without causing any potential vascular adverse effects. To test this idea, we first determined if the Etv2 shRNA used was specific for inhibiting Etv2 but not other Ets factors. Particularly, YSE cells infected with Etv2 shRNA, upon H2O2 treatment, greatly suppressed Etv2 upregulation, whereas, upregulation of other Ets genes, including Fli1, Ets1, Ets2, and Elk3, was not affected, establishing the selectivity of Etv2 shRNA in inhibiting Etv2 (Supplemental Figure 6A). Next, we formulated nanoparticles composed of Etv2 siRNA (using the same sequence as the shRNA) and a self-assembling peptide carrier (“p5RHH”), which has been previously described (45) and used to deliver p65NF-kB siRNA to inhibit experimental arthritis (46, 47). The p5RHH-siRNA nanoparticles (~55-nm diameter) protect the siRNA from serum deactivation, while avoiding reticuloendothelial system uptake and deliver functional oligonucleotides to inflammatory targets (45, 47, 48). We injected Etv2 siRNA peptide nanoparticles through tail vein into the tumor-bearing mice, from days 9–17 ptt, 5 times, every other day. The nanoparticles reached the tumor effectively within 24 hours of the injection (Figure 6, A and B). Etv2 expression in TAECs and tumor growth and tumor angiogenesis were dramatically reduced, starting from 2 days after the first nanoparticle injection (Figure 6, C–E). An extended treatment scheme (days 5–23 ptt, 10 times, every other day) continuously attenuated the tumor progression until the end of the study on day 29 ptt (Supplemental Figure 6B). Importantly, proliferation and FLK1 activation were inhibited in the TAECs of Etv2 siRNA peptide nanoparticle–treated mice (Figure 6, F–H).
Figure 6. Systemic treatment with Etv2 siRNA peptide nanoparticle restricts tumor growth and tumor angiogenesis.
(A) IVIS imaging of whole-body and isolated tumor at 24 hours after the injection of Quasar705-tagged Etv2 siRNA peptide nanoparticles through the tail vein. (B) Representative images from a fluorescence microscope for Quasar705-tagged Etv2 siRNA nanoparticles (purple) in the tumor sections at 24 hours after the injection. (C) qRT-PCR analysis of Etv2 expression in TAECs isolated from scrambled and Etv2 siRNA nanoparticle–treated mice on day 15 ptt (n = 5/group). (D) Tumor growth in the scrambled and Etv2 siRNA nanoparticle–treated mice. Scrambled and Etv2 siRNA nanoparticles were injected through the tail veins of the mice on days 9, 11, 13, 15, and 17 ptt (n = 8 scrambled, 10 Etv2 siRNA; **P < 0.01, ***P < 0.001). Representative images of mice with tumor from the treatment and control groups. (E–H) Representative images and quantification for (E) CD31 (red), (F and G) Ki67 (green), and (H) pFLK1 (Y951) (green) immunofluorescence of the tumor sections (n = 7 or more/group). LLC-GFP cells (blue) and nuclei counterstained with DAPI (gray) are shown. White arrows indicate the expression of (F) Ki67 and (H) pFLK1 in tumor vessels. Scale bars: 200 μm (B); 100 μm (E); 50 μm (F and H). Data are presented as mean with standard deviation for all measurements. Statistical significance was analyzed by either a 2-tailed Student’s t test (C, E, G, and H) or 2-way repeated-measures ANOVA with Sidak’s multiple-comparison test (D).
Etv2 siRNA nanoparticles do not elicit cardiovascular side effects.
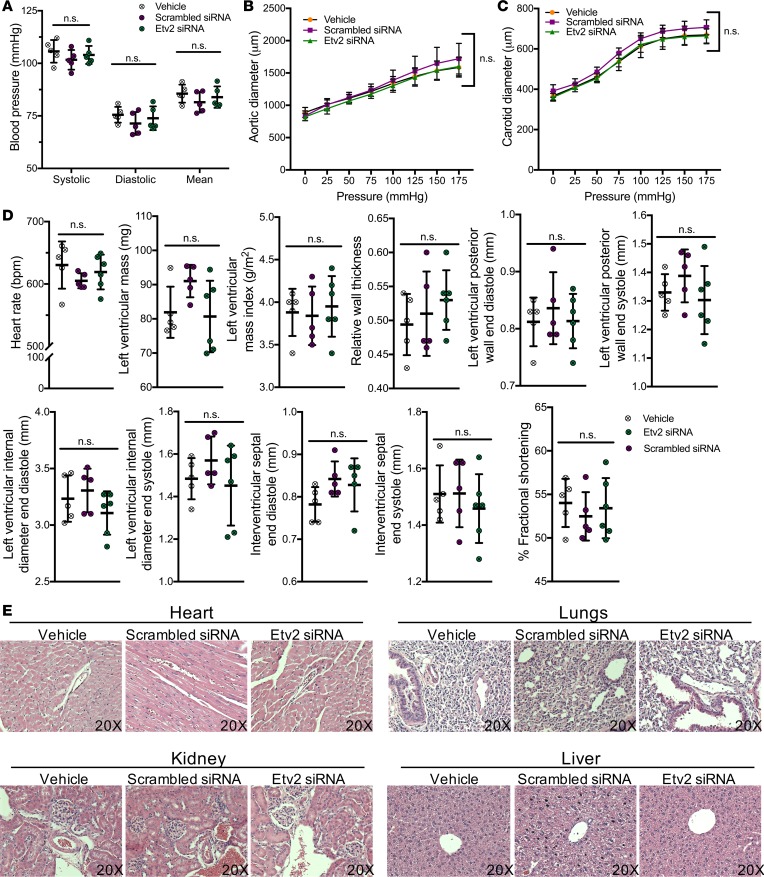
Antiangiogenic therapies, such as anti-VEGF treatments, often have systemic and vascular side effects that complicate and, at times, diminish the outcome of the treatment (4), as commonly targeted VEGF is also required for vascular homeostasis (6, 7). Prior work has shown that the p5RHH-based nanoparticles exert no untoward immune stimulation, complement activation, and appreciable toxicity toward blood components or major organs after multiple systemic doses (46). To assess the safety profile of our employed Etv2 siRNA peptide nanoparticle treatment in cardiovascular systems, we injected the Etv2 siRNA nanoparticles into wild-type mice (5 times, every other day). Following the treatment, systolic, diastolic, and mean blood pressures (49) were similar in the treated and control groups (Figure 7A). Pressure-diameter measurements, commonly used to evaluate vessel mechanics (50), showed that the compliances of the ascending aorta and carotid artery were equivalent for the treated and control groups (Figure 7, B and C). Clinically relevant cardiac functions, including left ventricular interior and posterior wall diameters following systole and diastole, relative wall thickness, mass index, and fractional shortening, were normal and equivalent across treated and control groups in echocardiogram analysis (Figure 7D). Additionally, histological inspection of the H&E-stained samples of major vascular organs revealed no pathological abnormalities (Figure 7E). To better understand the pharmacokinetics of the Etv2 siRNA nanoparticles, we injected a separate group of mice with a single dose of the siRNA nanoparticles and performed high-performance liquid chromatography (HPLC) on the blood samples. After the i.v. administration of 0.5 nmol of the siRNA nanoparticles to each wild-type mouse, the siRNA nanoparticle plasma concentrations were still detectable at 3 hours (Supplemental Figure 7). The mean elimination half-life of siRNA nanoparticles was calculated as 43 ± 27 minutes (Supplemental Figure 7). Together, these data suggest that Etv2 siRNA nanoparticle-based drugs can be considered in the future to inhibit tumor angiogenesis without any obvious cardiovascular side effects.
Figure 7. Etv2 siRNA peptide nanoparticle treatment do not adversely affect cardiovascular system and function.
Wild-type mice were intravenously treated with vehicle, scrambled siRNA, or Etv2 siRNA nanoparticles for 5 times, every other day. (A) Systolic, diastolic, and mean blood pressure measurements in control (vehicle and scrambled siRNA nanoparticles) or Etv2 siRNA nanoparticle–treated mice (n = 5/group). (B and C) Pressure diameter measurements of the (B) ascending aorta and (C) carotid artery over a range of pressures from 0 to 175 mmHg (n = 5/group). (D) Heart rate, left ventricular mass, left ventricular mas index, relative wall thickness, left ventricular posterior wall end diastole, left ventricular posterior wall end systole, left ventricular internal diameter end diastole, left ventricular internal diameter end systole, intraventricular septal end diastole, intraventricular septal end systole, and percentage fractional shortening in control (vehicle and scrambled siRNA nanoparticles) or Etv2 siRNA nanoparticle–treated mice, measured by echocardiogram analysis (n = 5/group). (E) Representative images of H&E-stained sections of heart, lungs, kidney, and liver harvested from control (vehicle and scrambled siRNA nanoparticles) or Etv2 siRNA nanoparticle–treated mice (original magnification, ×20). Data are presented as mean with standard deviation for all measurements. Statistical significance was analyzed by either 1-way ANOVA with Bonferroni’s multiple-comparison test (A and D) or 2-way repeated-measures ANOVA with Tukey’s multiple-comparison test (B and C).
Discussion
ETS factors have been found to play a role in tumorigenesis (51). However, most studies are restricted to tumor cell–intrinsic effects. Thus, despite their prominent roles in developmental angiogenesis (13, 27, 28), there have been limited studies on ETS factors in tumor angiogenesis (52, 53). Herein, we show that while Etv2 expression is silent in adult steady-state ECs, it is reactivated in TAECs in both humans and mice. Importantly, Etv2-deficient mice supported reduced tumor growth and angiogenesis. Etv2-deficient tumor vessels remained similar to physiological steady-state vessels. Previous studies have established that tumor angiogenesis is sensitive to oxygen tension. Particularly, the tumor environment is known to be hypoxic, and hypoxia induces angiogenesis through activating HIF1α and VEGF production (40, 42). Importantly, developmental angiogenesis is also regulated by oxygen tension (39, 54). Our data suggest that Etv2 is a redox-sensitive transcription factor, which is induced by ROS in TAECs. We show that ROS levels were elevated in TAECs. ROS scavenging inhibited Etv2 upregulation in TAECs. These studies enforce the notion that the genetic programs regulating developmental processes are recapitulated in the pathologic disease processes.
Recent studies have established that the interplay among ETV2, VEGF, and its receptor VEGFR2 (also known as FLK1) is essential for hematopoietic and vascular development (19, 36). We have recently demonstrated that the VEGF/FLK1 pathway is also required downstream of ETV2 in EC regeneration (32). As in development and EC regeneration, the VEGF/FLK1 pathway seems to be the target of ETV2 in tumor angiogenesis. In particular, while Flk1 expression levels were elevated in wild-type TAECs, compared with steady-state vessels, Flk1 expression levels in Etv2-deficient TAECs were similar to steady-state vessels. Moreover, Etv2-deficient ECs showed decreased endothelial proliferation, tube formation, and sprouting response to VEGF, not FGF, compared with wild-type ECs. Thus, an important implication of our study is that VEGF targeting in TAECs maybe selectively achieved through Etv2 inhibition. Intriguingly, other Ets genes were greatly upregulated in Etv2-deficient TAECs. However, upregulation of other Ets genes was not apparently sufficient to overcome Etv2 deficiency in tumor angiogenesis. Potential compensatory mechanisms involving FGF signaling and other Ets factors in the residual tumor growth in Etv2-deficient mice need to be investigated in the future.
Currently, many RNAi-based drugs, including Vegf, Epha2, and Vegfr1, are in clinical trials, phase I or II (55–57), to inhibit tumor angiogenesis. In the present work, we sought to define the utility of a siRNA delivery approach using nanoparticles targeted to the Etv2. The nanoparticle construct is composed of a self-assembling peptide (p5RHH) siRNA complex that serves to protect the siRNA from serum deactivation, while avoiding reticuloendothelial system uptake and delivering functional oligonucleotides to inflammatory targets (45–47). Key features of the complex that promote sequentially coordinated endosomal uptake, endosomal lysis, and siRNA release depend on specific molecular features of the carrier peptide, which is derived through modification of the bee venom peptide melittin (48). Selected amino acid truncations and substitutions mitigate the undesirable pore-forming capacity of peptide, yet retain its ability to condense siRNA and facilitate endosomal escape, as previously reported (58). A simple mixing procedure of only 10 minutes yields a complex that is small enough (~55 nm) to passively diffuse rapidly into inflamed tissues, where it is retained, yet it avoids hepatic sequestration (48). Using a mouse tumor transplantation model, we demonstrated that systemic Etv2 siRNA-p5RHH nanoparticle delivery can effectively inhibit tumor angiogenesis, without eliciting any cardiovascular side effects. Future investigations on p5RHH-siRNA nanoparticles as a safe and effective clinical tool instead of viral vectors for delivering siRNA are warranted. Optimizing anti-Etv2 strategy and delineating Ets factor redundancy and potential compensatory pathways in Etv2-deficient TAECs would be critical for improved and selective clinical antiangiogenic interventions. Potentially, combined chemotherapy and/or immunotherapy with Etv2 blockade, leading to vessel normalization, could be beneficial for future treatment of solid tumors. Moreover, the role of Etv2 in the proangiogenic function of hematopoietic cells in the tumor microenvironment should also be addressed in the future studies. In summary, we have demonstrated that Etv2 is, to our knowledge, a novel and critical molecular marker specific for activated ECs and that targeting Etv2 is a valid approach for inhibiting tumor angiogenesis.
Methods
Mice.
Tie2-Cre;Etv2-CKO, VEC-Cre;Etv2-CKO, and Vav-Cre;Etv2-CKO mice were generated as described previously (32, 33). C57BL/6J mice were used as wild-type mice. Tie2-Cre;Etv2f/f mice were used as Tie2-Cre;Etv2-CKO mice. VECadherin-Cre;Etv2f/f mice were used as VECadherin-Cre;Etv2-CKO mice. Vav-Cre;Etv2f/f mice were used as Vav-Cre;Etv2-CKO mice. Littermate subjects were used as controls with the CKO mice. Both male and female mice were used in an equal quantity in any given experiment. The ages of the experimental animals were between 10 and 12 weeks. Animals were randomly distributed into groups when different treatments, such as Etv2 shRNA or Gfp shRNA treatment, N-acetyl cysteine plus apocynin (NAC+APO) or vehicle treatment, and Etv2 siRNA or scrambled siRNA nanoparticle treatment, were employed. A group of technicians was in charge of distributing and codifying the experimental animals. The investigators were not aware of the group allocation until the treatment, data collection, and data analysis were done.
Cell lines.
LLC-GFP cells were obtained from Alexander S. Krupnick, Washington University in St. Louis (St. Louis, Missouri, USA). B16F10 melanoma cells were purchased from ATCC. YSE cells were obtained from Laurence A. Lasky (Genentech, South San Francisco, California, USA). All cell lines have been tested negative for mycoplasma contamination.
Human tumor samples.
Deidentified patient-derived, formalin-fixed and paraffin-embedded (FFPE), malignant and matched nonmalignant breast, lung, prostate, and colon tissue specimens were collected from Tissue Procurement Core (TPC) Siteman Cancer Center at Washington University School of Medicine. TPC performs pathological reviews to determine the malignant versus nonmalignant status of the tissues following standard procedures before disseminating the tissue for downstream applications. According to TPC definition, any section associated with equal to or more than 1 neoplastic cell was considered malignant. However, most of our samples had more than 50% neoplastic cells in any given section. A summary of the pathological review of the tissues that we received and the SOP for TPC Pathology Specimen Review is provided in the supplemental materials (Supplemental Table 2 and Supplemental File 1)
Tumor transplantation studies.
LLC-GFP cells were cultured in growth medium consisting of DMEM (Gibco) supplemented with 10% (v/v) FBS, 100 unit/ml penicillin, and 100 μg/ml streptomycin. For tumor transplantation studies, 1 ml of growth factor–reduced Matrigel (catalog 354248; Corning) was mixed with 1 ml tumor cell suspension (2 × 106/ml in PBS); 100 μl tumor cell–Matrigel mixture was injected into the back of mice subcutaneously. Tumor growth was measured by a digital slide calipers (VWR International) at indicated days after tumor cell injection. Tumor volume was calculated by the equation, volume = (largest diameter) × (smallest diameter)2 × 0.5.
Immunofluorescence.
FFPE human tissue sections were processed for antigen retrieval using DIVA Decloaker (catalog DV2004, Biocare Medical), blocked using freshly made blocking buffer (3% essentially IgG free BSA, catalog A9085, MilliporeSigma; 0.3% Triton X-100; and Fc blocker, catalog 422301, Biolegend) to prevent nonspecific binding, incubated overnight at 4°C with mouse anti-human CD31 (1:100) (catalog MA5-13188, ThermoFisher Scientific) and rabbit anti-human ETV2 (1:300) (catalog ab181847, Abcam) primary antibodies, and finally visualized with Alexa Flour 594–labeled goat anti-mouse (1:400) (catalog R37121, ThermoFisher Scientific) and Alexa Fluor 647–labeled goat anti-rabbit (1:500) (catalog A-21245, ThermoFisher Scientific) secondary antibodies for 2 hours at room temperature. Later, the sections were processed to counterstain nuclei with DAPI (catalog D1306, ThermoFisher Scientific), cured with ProLong Diamond Antifade mountant (catalog P36970, ThermoFisher Scientific) for 24 hours, and finally sealed with nail polish for preservation. No primary antibody control, isotype control (Mouse IgG1 Isotype Control, catalog ab91353, Abcam, and Rabbit IgG Isotype Control, catalog ab172730, Abcam), and absorption control (using ETV2 Antibody Blocking Peptide at a 5:1 ratio with employed ETV2 antibody, catalog LS-E910, LifeSpan Biosciences) were used as negative controls for immunofluorescence staining. The stained sections were examined using the Olympus Fluoview 1200 confocal microscope and minimally processed with Imaris (Bitplane) software to prepare the figures. At least 6 pictures from every section were processed using ImageJ software (NIH) to quantify CD31+ and ETV2+ areas.
In mouse tumor transplantation studies, before harvesting the tumors, the mice underwent transcardiac whole-body perfusion of 10% buffered formalin. Harvested tumors were thin sliced and fixed in 10% buffered formalin for 36 hours, immersed in 30% (w/v) sucrose solution for 48 hours to cryoprotect the tissue, frozen in NEG-50 frozen section medium (catalog 6502, ThermoFisher Scientific) using liquid nitrogen and 2-methylbutane, and sectioned (16-μm thick) using a Cryostat Cryocut Microtome (Leica, CM1850). Tissue sections were first blocked using freshly made blocking buffer (3% essentially IgG free BSA, catalog A9085, MilliporeSigma; 0.3% Triton X-100; and Fc blocker, catalog 101301, Biolegend) to prevent nonspecific binding; then incubated overnight at 4°C with hamster anti-mouse CD31 (1:400) (catalog MA3105, ThermoFisher Scientific), along with rabbit anti-mouse ETV2 (1:200) (catalog orb156791, Biorbyt) or rabbit anti-mouse pFLK1 (Y951) (1:200) (catalog 4991S, Cell Signaling Technology) or rabbit anti-mouse Ki67 (1:200) (catalog Ab15580, Abcam) primary antibodies; and finally visualized with Alexa Fluor 568–labeled goat anti-hamster (1:500) (catalog A-21112, ThermoFisher Scientific) and Alexa Fluor 647–labeled goat anti-rabbit (1:500) (catalog A-21245, ThermoFisher Scientific) secondary antibodies for 2 hours at room temperature. Later, the sections were processed to counterstain nuclei with DAPI (catalog D1306, ThermoFisher Scientific), cured with ProLong Diamond Antifade mountant (catalog P36970, ThermoFisher Scientific) for 24 hours, and finally sealed with nail polish for preservation. No primary antibody control, isotype control (Armenian Hamster IgG Isotype Control, catalog 14-4888-85, ThermoFisher Scientific, and Rabbit IgG Isotype Control, catalog ab172730, Abcam), and absorption control (using ETV2 Antibody Blocking Peptide in a 5:1 ratio with employed ETV2 antibody, catalog LS-E910, LifeSpan Biosciences) were used as negative controls for immunofluorescence staining. The stained sections were examined using the Olympus Fluoview 1200 confocal microscope system and minimally processed with Imaris (Bitplane) software to prepare the figures. At least 5 pictures from every section were processed using ImageJ software (NIH) to quantify CD31+, ETV2+, pFLK1+, and Ki67+ areas.
To further confirm the specificity of the ETV2 antibody staining, we employed the iEtv2 embryonic stem (ES) cell system that expresses ETV2-V5 in a doxycycline-inducible manner, following protocol described previously (17, 19). Briefly, doxycycline-treated iEtv2 ES cells (cultured on Matrigel-coated coverslips) were fixed with ice-cold methanol for 10 minutes at room temperature. After fixation, the ES cells were first blocked using freshly made blocking buffer (10% Normal Goat Serum, catalog 5425S, Cell Signaling Technology, in 1× TBS) to prevent nonspecific binding, then incubated overnight at 4°C with rat anti-V5 antibody (1:1,000) (catalog ab206571, Abcam) and rabbit anti-mouse ETV2 (1:1,000) (catalog orb156791, Biorbyt), and finally visualized with Alexa Fluor 488–labeled goat anti-rabbit (1:500) (catalog ab150077, Abcam) and Alexa Fluor 568–labeled goat anti-rat (1:500) (catalog ab11077, Abcam) secondary antibodies for 1 hour at room temperature. Later, the cells were processed to counterstain nuclei with DAPI (catalog D1306, ThermoFisher Scientific), cured with ProLong Diamond Antifade mountant (catalog P36970, ThermoFisher Scientific) for 12 hours, and finally sealed with nail polish for preservation. The stained cells were examined using the Olympus Fluoview 1200 confocal microscope system and minimally processed with Imaris (Bitplane) software to prepare the figures. The data show that both the ETV2 and V5 tag were present in nuclei and perinuclear spaces, confirming the specificity of the Etv2 staining (Supplemental Figure 1, C and D).
Lung EC isolation.
Lungs were harvested from the mice, dissociated into single-cell suspension by using a digestion buffer consisting of Collagenase II (catalog 17101-015, ThermoFisher Scientific) and Deoxyribonuclease 1 (catalog LS002139, Worthington) solution for 30 minutes, stained with rat anti-mouse CD31 (catalog 102501, Biolegend) conjugated magnetic sheep anti-rat IgG Dynabeads (catalog 11035, ThermoFisher Scientific), and CD31+ cells were sorted using MACS. Afterward, sorted CD31+ ECs were either cultured with M199 medium containing 20% (v/v) FBS and EC growth supplement (catalog E2759, MilliporeSigma) or directly processed for total RNA isolation by the RNeasy mini kit (catalog 74104, Qiagen), following the manufacturer’s instructions.
TAEC isolation.
Tumors were harvested from the mice, dissociated into single-cell suspension using a digestion buffer consisting of Collagenase i.v. (catalog LS004188, Worthington), Dispase (catalog 17105-041, ThermoFisher Scientific), and Deoxyribonuclease 1 (catalog LS002139, Worthington) for 90 minutes at 37oC and stained with PE-Cy7 anti-mouse CD31 (catalog102418, Biolegend) and APC anti-mouse CD45 (catalog103112, Biolegend) antibodies. GFP–CD45–CD31+ ECs were FACS sorted using BD FACSAria II (BD Bioscience). Sorted ECs were processed for total RNA isolation by the RNeasy mini kit (catalog 74104, Qiagen) following the manufacturer’s instructions. At least 5 independent tumors per group were used for gene expression analysis.
qRT-PCR.
RNA and cDNA were prepared according to the manufacturers’ protocols (Qiagen and Quantabio, respectively). Gene expression was measured by quantitative RT-PCR using primers detailed in the Supplemental Table 1.
Two-photon imaging and analysis of vessel morphology.
For 2-photon (2P) imaging experiments, tumor cells were implanted in the ear pinna (and in some cases the hind flank) by injecting 10 μl LLC-GFP and Matrigel mixture (2 × 106 cells/ml). Twenty-one days after tumor implantation, mice were anesthetized with isofluorane and placed in a custom chamber for imaging. 2PM was performed noninvasively through skin. Blood vessels were labeled with a retro-orbital injection of either 70-kDa Rhodamine Dextran (50 μl, ~1 μg; catalog D1824, ThermoFisher Scientific) or 655 nm Q-dots (15 μl; catalog Q21321MP, Invitrogen). Multiple images were collected near the tumor margin using a custom-built 2PM system equipped with a Chameleon Vision II Ti:Sapp laser and a 20× 1.0 NA Olympus water dipping objective. Each 3D image consisted of 31 Z-slices taken in 2.5-μm steps. Z-stacks were rendered in 3D, and morphometric analysis was performed to assess vessel diameter, the number of discontinuous segments in the tumor-associated vessel network, and the volume of each vessel segment. Imaris (Bitplane) was used to generate 3D contour surfaces to identify vessels (Supplemental Video 1). Surface quality was optimized using background subtraction and thresholding and common settings used across all images. Vessel complexity was determined by counting the number of discrete vessel segments in each image. The volume of each vessel segment in an image was measured and compared across groups by using a linear mixed-effects model where the fixed effects are the treatment groups themselves and the random effects are due to intraimage correlation. Normality of the outcome variable was achieved after a log10 transform, and all tests were performed on the transformed data. After the model was fitted, pairwise statistical contrasts were used to assess pairwise group differences. Vessel leak was variable and was often observed in discreet regions of an image. Therefore, to minimize selection bias in our analysis, we quantified leak by averaging histograms of maximum intensity projections for all images in each group and then plotting average pixel intensity counts for each group. These plots were analyzed by fitting them to an exponential probability distribution function using maximum likelihood estimation methods to extract the rate of decay for each group. When extravascular leak was present, it resulted in an increase in the intermediate pixel intensity counts (10–80 gray levels) and thus a lower rate of decay for the plot. Maximum likelihood estimates were obtained for each curve, and the estimates were compared by generating a null-hypothesis estimate and a likelihood ratio test to assess a goodness of fit. Vessel integrity was assessed using a standard kymograph analysis on representative images.
Lentiviral Etv2 shRNA production.
pLKpuro lentiviral Etv2 shRNA clones, TRCN0000084284 (NM_007959.1-93s1c1), TRCN0000084285 (NM_007959.1-52s1c1), TRCN0000084286 (NM_007959.1-887s1c1), and TRCN0000084287 (NM_007959.1-888s1c1), constructed and distributed by the RNAi Consortium at the Broad Institute (https://portals.broadinstitute.org/gpp/public/), were obtained from the McDonnell Genome Institute at Washington University School of Medicine and funded in part by the Children’s Discovery Institute of Washington University in St. Louis. 293T cells were transfected with pLKpuro Etv2 shRNA constructs, pCAG-HIVgp, and pCMV-VSV-G-RSV-Rev (4:3:1) by using the calcium phosphate method. Sixteen hours after transfection, media were changed and cells were then grown for additional 48 hours. Subsequently, the supernatant was harvested and concentrated by Lenti-X-Concentrator (Clontech). The virus titer was determined using the Lenti-X p24 Rapid Titer Kit (Clontech). For the intratumor injection, IFU was around 3 × 107/ml. In a preliminary screening with this panel of Etv2 shRNA lentiviruses, we identified lentiviral shRNA clone TRCN0000084284 (NM_007959.1-93s1c1) to be most effective in restricting tumor growth, and hence, it was used for all subsequent experiments (data not shown). Lentiviral Gfp shRNA was used as control treatment.
EC proliferation assay.
Lung ECs, previously sorted from wild-type and Tie2-Cre;Etv2-CKO mice, were cultured in growth medium consisting of M199 medium (Gibco), 10% (v/v) FBS, 100 unit/ml penicillin, and 100 μg/ml streptomycin. For proliferation assay, cells were plated in 96-well plates (3,000 cells/well). One day later, medium was changed to 0.5% serum-containing medium. After 24 hours, VEGF (50 ng/ml) or FGF2 (50 ng/ml) was added to the culture. After 3 days, 10 μl CCK-8 (catalog 96992, MilliporeSigma) solution was added to each well, and the plates were incubated for 3 hours at 37°C. Absorbance at 450 nm (A450) for each well was measured.
Aortic ring assay.
The aortic ring assay was performed as previously described (1, 38). Briefly, aortas were retrieved from control or Tie2-Cre;Etv2-CKO mice, cut into rings approximately 0.5 mm in width, and cultured for 24 hours in serum-free culture medium. Subsequently, aortic rings were embedded into Matrigel (catalog 356231, Corning), and culture medium containing 2.5% FBS and VEGF (50 ng/ml) or FGF2 (50 ng/ml) was added. Medium was changed on days 3, 5, and 7. Aortic rings were fixed with 4% PFA on day 8, and sprout number and sprout length were measured using ImageJ image analysis software (NIH).
Tube formation assay.
Wild-type and lentiviral Etv2 shRNA–infected YSE cells, cultured in growth medium consisting of M199 medium (Gibco), 10% (v/v) FBS, 100 unit/ml penicillin, and 100 μg/ml streptomycin, were kept in 24-hour hypoxia (<1% oxygen), followed by 24-hour normoxia (in which overnight serum starvation [0.5% serum] was used). Afterward, cells were plated on a Matrigel-coated (catalog 96992, Corning) 24-well plate with either VEGF (50 ng/ml) or FGF2 (50 ng/ml) and incubated at 37°C for 5 hours, before images were taken with a Leica DFC 310 FX microscope system.
ROS induction, measurement, and scavenger treatment.
For ROS induction, wild-type or Etv2 shRNA–infected YSE cells were plated in 6-well culture plate (1.5 × 105 cells/well). The following day, cells were treated with H2O2, BSO, or hypoxic condition (<1%O2) for 24 hours or hypoxia/reoxygenation (24 hours <1% O2, followed by 24 hours normoxia). For ROS measurement, YSE or tissue-derived ECs were incubated with PBS containing 2.5 μM DCFDA (catalog D399, ThermoFisher Scientific) for 10 minutes at 37°C. Tissue-derived cells were stained with PE-Cy7 anti-mouse CD31 (catalog102418, Biolegend) and APC anti-mouse CD45 (catalog 103112, Biolegend) antibodies. ROS levels in YSE cells or in CD45–CD31+ cells from the tissues were measured by BD LSRFortessa (BD Bioscience). For in vitro ROS scavenger treatment, NAC (dissolved in water; 5 mM) (catalog A9165, MilliporeSigma) and APO (dissolved in DMSO and later diluted with water; 100 mM) (catalog 178385EMD, MilliporeSigma) were added to the YSE cells at the beginning of reoxygenation period for next 24 hours. For the in vivo treatment, NAC (200 mg/kg, dissolved in water) and APO (50 mg/kg, dissolved in DMSO and later diluted with water) were intraperitoneally injected into the mice.
Preparation of Etv2 siRNA peptide nanoparticle.
For the siRNA nanoparticle treatment study, we designed the Etv2 siRNA against the sequence “GAGCGGAATTTGGTTTCTATT” [the sequence of the lentiviral shRNA clone TRCN0000084284(NM_007959.1-93s1c1) that was used for the shRNA studies] and obtained it from MilliporeSigma. MISSION siRNA Universal Negative Control #1 (catalog SIC001, MilliporeSigma) was used as scrambled siRNA control. For tracking the distribution of the siRNA, Quasar 705 tagged Etv2 siRNA was obtained from MilliporeSigma. 100 μM siRNA solution was prepared by dissolving in 1× siRNA buffer (catalog B-002000-UB-100, Dharmacon) and incubating on an orbital mixer (~500 rpm) at 37°C for 70–90 minutes; this stock siRNA was stored in 10 μl aliquot at –80°C until use. To prepare the nanoparticle complex, 10 μl of the siRNA suspension was mixed with 5 μl of 20 mM p5RHH peptide solution and 185 μl of 1× HBSS (Gibco) and incubated on ice for approximately 7 minutes for the siRNA and peptide to conjugate at a siRNA-to-peptide ratio of 1:100. The resultant 200 μl siRNA peptide nanoparticle was immediately injected into the mouse through the tail vein to deliver a siRNA dose of 1 nmol/mouse (2, 46).
IVIS imaging.
To track the siRNA distribution after nanoparticles injection, in vivo fluorescence images were acquired and analyzed with a Xenogen IVIS Spectrum imaging system (Caliper Life-Sciences). Mice were kept under isoflurane inhalation anesthesia during the whole-body image acquisition process. Tumor and other major organs were harvested immediately after that and processed for imaging as well. A fixed protocol (excitation, 640 nm; emission, 700 nm; exposure time, 2 s; binning factor, 8; f value, 2; field of view, 12.9) was used for image acquisitions at 24 hours after tail vein injection of nanoparticles containing Quasar705-labeled Etv2 siRNA into tumor-bearing wild-type mice. For the noninjected subjects, HBSS was injected through tail veins, and images were taken following the same protocol (2, 46).
Blood pressure measurement and compliance studies.
Arterial blood pressure and arterial compliance measurements were performed after 5 consecutive injections (every other day) of vehicle or Etv2 siRNA or scrambled siRNA nanoparticles into healthy wild-type mice as described previously (3, 49). Briefly, for blood pressure, mice were anesthetized with 1.5% isoflurane and kept on a heating pad to maintain normal body temperature and monitored via a rectal thermometer. A 2- to 3-mm midline incision was made in the neck; the lobes of the thymus were separated to expose the right common carotid artery. After clamping and making a small nick in the right common carotid artery, a Millar pressure transducer (model SPR-671) was introduced and advanced to the ascending aorta. Systolic blood pressure, diastolic blood pressure, and heart rate were recorded and analyzed using the PowerLab data acquisition system and LabChart 7 for Mac software, respectively (ADInstruments). For compliance studies, following the blood pressure measurement, mice were euthanized under isoflurane anesthesia, and the ascending aorta and the left common carotid artery were excised and placed in physiology saline (PSS) at 37°C. After cleaning the fatty tissues, vessels were placed on a pressure arteriograph (Danish Myo Technology), maintained in PSS at 37°C, and examined with an inverted microscope attached to a CCD camera and a computerized system that allows continuous recording of the vessel diameter measurement (4, 50).
Echocardiography.
The Mouse Cardiovascular Phenotyping Core at Washington University School of Medicine performed noninvasive cardiac ultrasound examination of healthy wild-type mice under light anesthesia, after 5 consecutive injections of vehicle or Etv2 siRNA or scrambled siRNA nanoparticle (every other day). VisualSonics Vevo-2100 cardiac echocardiography system (FUJIFILM, VisualSonics Inc.) was used to characterize the structure and function of the heart and great vessels.
Histological analysis.
Heart, lungs, kidneys, and liver were harvested after 5 injections (every other day) of vehicle or Etv2 siRNA or scrambled siRNA nanoparticles into healthy wild-type mice and fixed in 10% buffered formalin for 48 hours. The tissues were decalcified in EDTA solution, embedded in paraffin, and cut to 5-μm sections. Finally, the sections were stained with H&E. Digital images of 4 random areas per H&E-stained organ section were acquired at 20× magnification and checked by a double blinded histologist/pathologist.
Single-dose siRNA nanoparticle administration, blood sampling, HPLC, and pharmacokinetic analysis.
Five wild-type mice (C57BL/6J; ~17 weeks old) received a single i.v. dose of siRNA nanoparticle and underwent PK measurements. A carotid artery catheter was inserted before the siRNA nanoparticle dosing for blood sampling. Blood samples were taken prior to and at 10, 40, 90, and 180 minutes after the i.v. administration of p5RHH-Cy3 siRNA nanoparticle. Blood drawn was replaced with 80 μl heparinized saline (5 U/ml) after each collection to avoid volume depletion. Plasma was separated by centrifugation and then stored at –80oC until analyzed for siRNA.
Individual plasma samples (~60 μl) were added with 0.4 ml Clarity OTX lysis-loading buffer (Phenomenex Inc.). The mixture was vortex mixed for 30 seconds and then loaded onto the solid-phase extraction (SPE) cartridge. The siRNA nanoparticles were extracted from individual plasma samples using Clarity OTX SPE cartridges with polymeric sorbent (100 mg/3 ml; Phenomenex Inc.). Sorbent was conditioned with 1.0 ml methanol and equilibrated with 1 ml of 150 mM NaH2PO4 buffer (pH 5.5) containing 2 mM NaN3. For plasma samples, following sample loading, the cartridge was washed with 4.0 ml washing buffer (50 mM NaH2PO4 buffer [pH 5.5] containing 50% acetonitrile). The siRNA nanoparticles were eluted with 1 ml elution buffer (100 mM NH4HCO3 containing 10% tetrahydrofuran and 40% acetonitrile). The eluent was evaporated to near dryness using a Savant SC110 SpeedVac concentrator. Samples were reconstituted with 100 μl HPLC-grade water, and a 25-μl aliquot was injected into the HPLC column. The HPLC assay was carried out using an Agilent series 1200 HPLC system equipped with a binary pump, autosampler, and degasser coupled to an Agilent 1260 fluorescence detector. Separation was achieved on an Acquity UPLC oligonucleotide BEH C18 column (50 × 2.1 mm, 1.7-μm particle size; Waters Corporation). The analyte was eluted using acetonitrile/100 mM hexylammonium acetate (48:52, v/v) as the mobile phase with a flow rate of 0.1 ml/min. The column temperature was set at 60°C. The siRNA nanoparticle was detected by use of an excitation wavelength of 548 nm and an emission wavelength of 566 nm.
A 1-compartment open model with first-order elimination (C = C0 e–kt) was used to characterize drug disposition in plasma, where C0 represents siRNA concentration at time 0 and k denotes the first-order elimination rate constant. Elimination half-life was calculated as t1/2 = 0.693/k
Statistics.
GraphPad Prism 7 software was used for performing statistical analysis and generating graphs/plots. Data are presented as mean with standard deviation for all the measurements. All experimental data were reliably reproduced in 2 or more individual biological replicates. Comparison between 2 independent samples was performed using a 2-tailed Student’s t test. In case of comparing more than 2 groups, 1-way ANOVA with post hoc Dunnett’s or Tukey’s or Bonferroni’s multiple-comparison test or 2-way ANOVA with either post hoc Tukey’s or Sidak’s multiple-comparison test was used, whichever appropriate. Tumor volume data was analyzed using or 2-way repeated measures ANOVA with either post hoc Tukey’s or Sidak’s multiple-comparison test. A P < 0.05 was considered statistically significant.
Study approval.
Animal husbandry, generation, handling, and experimentation were performed in accordance with protocols approved by the Institutional Animal Care and Use Committee of Washington University School of Medicine in St. Louis.
Author contributions
AUK, TJL, and KC conceived and designed experiments and wrote the paper. AUK and TJL performed the experiments and analyzed the data. JCB, LY, SH, and MJM helped with 2P imaging, 2P data analysis, and preparation. HP and SAW helped with the siRNA design and nanoparticles preparation. QZ helped with the pharmacokinetics study of the siRNA nanoparticles. KK, JW, and HKK helped with characterizing CKO mice and performing the tube formation assay. FL helped with Ets factor expression analysis. RPM and KH helped to measure blood pressure and perform compliance studies. DJV helped in planning of tissue collection for histology and performed histologic inspection of H&E-stained tissue sections. KC provided overall supervision and coordinated all the experimental activities.
Supplementary Material
Acknowledgments
We thank members of the Choi lab for constructive criticism, encouragement, and discussion. We would like to thank Alexander S. Krupnick for providing LLC-GFP cells, Laurence A. Lasky for providing YSE cells, and Bernd H. Zinselmeyer for helping with immunofluorescence image processing. We also thank Stephen T. Oh for providing the access to a hypoxic (regulated oxygen) incubator. We thank The RNAi Consortium at Broad Institute, Children’s Discovery Institute, and McDonnell Genome Institute at Washington University for providing shRNA clones. We thank the Washington University Center for Cellular Imaging for providing the access to the confocal microscope and the Washington University Department of Medicine In vivo Imaging Core for providing access to 2P imaging. We thank Attila Kovacs and Carla J. Weinheimer of the Mouse Cardiovascular Phenotyping Core at Washington University School of Medicine for the echocardiogram analysis. We thank the Alvin J. Siteman Cancer Center at Washington University School of Medicine and Barnes-Jewish Hospital in St. Louis, Missouri, USA, for the use of the TPC, which provided the human tumor specimens. The Siteman Cancer Center is supported in part by a National Cancer Institute Cancer Center support grant (P30 CA91842). This work was supported by NIH grants R01HL63736 and R01HL55337 (to KC), R01AI077600 (to MM), R01HL105314 (to RM), and R01DK102691, R01AR067491, and R01HL073646 (to SW) and the Basic Science Research Program through the National Research Foundation of Korea funded by the Ministry of Education (2016R1A6A3A01012289, to TL).
Version 1. 04/19/2018
Electronic publication
Footnotes
Conflict of interest: SAW has equity in Trasir Therapeutics Inc.
License: This work is licensed under the Creative Commons Attribution 4.0 International License. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
Reference information: JCI Insight. 2018;3(8):e97349. https://doi.org/10.1172/jci.insight.97349.
Contributor Information
Ashraf Ul Kabir, Email: a.kabir@wustl.edu.
Tae-Jin Lee, Email: leetj@skku.edu.
Hua Pan, Email: huapan@health.usf.edu.
Karen Krchma, Email: kkrchma@wustl.edu.
Jun Wu, Email: 377545520wustl@gmail.com.
Fang Liu, Email: fliu0523@gmail.com.
Hee-Kyoung Kang, Email: pharmkhk@jejunu.ac.kr.
Kristina Hinman, Email: kristinadhinman@gmail.com.
Lihua Yang, Email: lihuayang@wustl.edu.
Samantha Hamilton, Email: samantha.hamilton@wustl.edu.
Qingyu Zhou, Email: qzhou1@health.usf.edu.
Kyunghee Choi, Email: kchoi@wustl.edu.
References
- 1.Folkman J. Tumor angiogenesis: therapeutic implications. N Engl J Med. 1971;285(21):1182–1186. doi: 10.1056/NEJM197111182852108. [DOI] [PubMed] [Google Scholar]
- 2.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 3.Ye W. The complexity of translating anti-angiogenesis therapy from basic science to the clinic. Dev Cell. 2016;37(2):114–125. doi: 10.1016/j.devcel.2016.03.015. [DOI] [PubMed] [Google Scholar]
- 4.Ferrara N, Adamis AP. Ten years of anti-vascular endothelial growth factor therapy. Nat Rev Drug Discov. 2016;15(6):385–403. doi: 10.1038/nrd.2015.17. [DOI] [PubMed] [Google Scholar]
- 5.Kamba T, McDonald DM. Mechanisms of adverse effects of anti-VEGF therapy for cancer. Br J Cancer. 2007;96(12):1788–1795. doi: 10.1038/sj.bjc.6603813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee S, et al. Autocrine VEGF signaling is required for vascular homeostasis. Cell. 2007;130(4):691–703. doi: 10.1016/j.cell.2007.06.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kamba T, et al. VEGF-dependent plasticity of fenestrated capillaries in the normal adult microvasculature. Am J Physiol Heart Circ Physiol. 2006;290(2):H560–H576. doi: 10.1152/ajpheart.00133.2005. [DOI] [PubMed] [Google Scholar]
- 8.De Val S, et al. Combinatorial regulation of endothelial gene expression by ets and forkhead transcription factors. Cell. 2008;135(6):1053–1064. doi: 10.1016/j.cell.2008.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wythe JD, et al. ETS factors regulate Vegf-dependent arterial specification. Dev Cell. 2013;26(1):45–58. doi: 10.1016/j.devcel.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Randi AM, Sperone A, Dryden NH, Birdsey GM. Regulation of angiogenesis by ETS transcription factors. Biochem Soc Trans. 2009;37(Pt 6):1248–1253. doi: 10.1042/BST0371248. [DOI] [PubMed] [Google Scholar]
- 11.Wei GH, et al. Genome-wide analysis of ETS-family DNA-binding in vitro and in vivo. EMBO J. 2010;29(13):2147–2160. doi: 10.1038/emboj.2010.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pham VN, et al. Combinatorial function of ETS transcription factors in the developing vasculature. Dev Biol. 2007;303(2):772–783. doi: 10.1016/j.ydbio.2006.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meadows SM, Myers CT, Krieg PA. Regulation of endothelial cell development by ETS transcription factors. Semin Cell Dev Biol. 2011;22(9):976–984. doi: 10.1016/j.semcdb.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dejana E, Taddei A, Randi AM. Foxs and Ets in the transcriptional regulation of endothelial cell differentiation and angiogenesis. Biochim Biophys Acta. 2007;1775(2):298–312. doi: 10.1016/j.bbcan.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 15.Ferdous A, et al. Nkx2-5 transactivates the Ets-related protein 71 gene and specifies an endothelial/endocardial fate in the developing embryo. Proc Natl Acad Sci USA. 2009;106(3):814–819. doi: 10.1073/pnas.0807583106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kataoka H, et al. Etv2/ER71 induces vascular mesoderm from Flk1+PDGFRα+ primitive mesoderm. Blood. 2011;118(26):6975–6986. doi: 10.1182/blood-2011-05-352658. [DOI] [PubMed] [Google Scholar]
- 17.Lee D, et al. ER71 acts downstream of BMP, Notch, and Wnt signaling in blood and vessel progenitor specification. Cell Stem Cell. 2008;2(5):497–507. doi: 10.1016/j.stem.2008.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rasmussen TL, et al. VEGF/Flk1 signaling cascade transactivates Etv2 gene expression. PLoS One. 2012;7(11):e50103. doi: 10.1371/journal.pone.0050103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu F, et al. Induction of hematopoietic and endothelial cell program orchestrated by ETS transcription factor ER71/ETV2. EMBO Rep. 2015;16(5):654–669. doi: 10.15252/embr.201439939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sumanas S, Gomez G, Zhao Y, Park C, Choi K, Lin S. Interplay among Etsrp/ER71, Scl, and Alk8 signaling controls endothelial and myeloid cell formation. Blood. 2008;111(9):4500–4510. doi: 10.1182/blood-2007-09-110569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elcheva I, et al. Direct induction of haematoendothelial programs in human pluripotent stem cells by transcriptional regulators. Nat Commun. 2014;5:4372. doi: 10.1038/ncomms5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lindgren AG, Veldman MB, Lin S. ETV2 expression increases the efficiency of primitive endothelial cell derivation from human embryonic stem cells. Cell Regen (Lond) 2015;4(1):1. doi: 10.1186/s13619-014-0014-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ginsberg M, et al. Efficient direct reprogramming of mature amniotic cells into endothelial cells by ETS factors and TGFβ suppression. Cell. 2012;151(3):559–575. doi: 10.1016/j.cell.2012.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Han JK, et al. Direct conversion of adult skin fibroblasts to endothelial cells by defined factors. Circulation. 2014;130(14):1168–1178. doi: 10.1161/CIRCULATIONAHA.113.007727. [DOI] [PubMed] [Google Scholar]
- 25.Morita R, et al. ETS transcription factor ETV2 directly converts human fibroblasts into functional endothelial cells. Proc Natl Acad Sci USA. 2015;112(1):160–165. doi: 10.1073/pnas.1413234112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee S, et al. Direct reprogramming of human dermal fibroblasts into endothelial cells using ER71/ETV2. Circ Res. 2017;120(5):848–861. doi: 10.1161/CIRCRESAHA.116.309833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Val S, Black BL. Transcriptional control of endothelial cell development. Dev Cell. 2009;16(2):180–195. doi: 10.1016/j.devcel.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sumanas S, Choi K. ETS transcription factor ETV2/ER71/Etsrp in hematopoietic and vascular development. Curr Top Dev Biol. 2016;118:77–111. doi: 10.1016/bs.ctdb.2016.01.005. [DOI] [PubMed] [Google Scholar]
- 29.Brown TA, McKnight SL. Specificities of protein-protein and protein-DNA interaction of GABP alpha and two newly defined ets-related proteins. Genes Dev. 1992;6(12B):2502–2512. doi: 10.1101/gad.6.12b.2502. [DOI] [PubMed] [Google Scholar]
- 30.Rasmussen TL, et al. ER71 directs mesodermal fate decisions during embryogenesis. Development. 2011;138(21):4801–4812. doi: 10.1242/dev.070912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wareing S, Mazan A, Pearson S, Göttgens B, Lacaud G, Kouskoff V. The Flk1-Cre-mediated deletion of ETV2 defines its narrow temporal requirement during embryonic hematopoietic development. Stem Cells. 2012;30(7):1521–1531. doi: 10.1002/stem.1115. [DOI] [PubMed] [Google Scholar]
- 32.Park C, et al. Injury-mediated vascular regeneration requires endothelial ER71/ETV2. Arterioscler Thromb Vasc Biol. 2016;36(1):86–96. doi: 10.1161/ATVBAHA.115.306430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu CX, et al. ETV2/ER71 regulates hematopoietic regeneration by promoting hematopoietic stem cell proliferation. J Exp Med. 2017;214(6):1643–1653. doi: 10.1084/jem.20160923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cahalan MD, Parker I, Wei SH, Miller MJ. Two-photon tissue imaging: seeing the immune system in a fresh light. Nat Rev Immunol. 2002;2(11):872–880. doi: 10.1038/nri935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fukumura D, Duda DG, Munn LL, Jain RK. Tumor microvasculature and microenvironment: novel insights through intravital imaging in pre-clinical models. Microcirculation. 2010;17(3):206–225. doi: 10.1111/j.1549-8719.2010.00029.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao H, Choi K. A CRISPR screen identifies genes controlling Etv2 threshold expression in murine hemangiogenic fate commitment. Nat Commun. 2017;8(1):541. doi: 10.1038/s41467-017-00667-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matsumoto T, et al. VEGF receptor-2 Y951 signaling and a role for the adapter molecule TSAd in tumor angiogenesis. EMBO J. 2005;24(13):2342–2353. doi: 10.1038/sj.emboj.7600709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baker M, et al. Use of the mouse aortic ring assay to study angiogenesis. Nat Protoc. 2011;7(1):89–104. doi: 10.1038/nprot.2011.435. [DOI] [PubMed] [Google Scholar]
- 39.Fraisl P, Mazzone M, Schmidt T, Carmeliet P. Regulation of angiogenesis by oxygen and metabolism. Dev Cell. 2009;16(2):167–179. doi: 10.1016/j.devcel.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 40.Kim YW, Byzova TV. Oxidative stress in angiogenesis and vascular disease. Blood. 2014;123(5):625–631. doi: 10.1182/blood-2013-09-512749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Szatrowski TP, Nathan CF. Production of large amounts of hydrogen peroxide by human tumor cells. Cancer Res. 1991;51(3):794–798. [PubMed] [Google Scholar]
- 42.Xia C, Meng Q, Liu LZ, Rojanasakul Y, Wang XR, Jiang BH. Reactive oxygen species regulate angiogenesis and tumor growth through vascular endothelial growth factor. Cancer Res. 2007;67(22):10823–10830. doi: 10.1158/0008-5472.CAN-07-0783. [DOI] [PubMed] [Google Scholar]
- 43.Michiels C, Arnould T, Houbion A, Remacle J. Human umbilical vein endothelial cells submitted to hypoxia-reoxygenation in vitro: implication of free radicals, xanthine oxidase, and energy deficiency. J Cell Physiol. 1992;153(1):53–61. doi: 10.1002/jcp.1041530109. [DOI] [PubMed] [Google Scholar]
- 44.Zhu X, Zuo L, Cardounel AJ, Zweier JL, He G. Characterization of in vivo tissue redox status, oxygenation, and formation of reactive oxygen species in postischemic myocardium. Antioxid Redox Signal. 2007;9(4):447–455. doi: 10.1089/ars.2006.1389. [DOI] [PubMed] [Google Scholar]
- 45.Hou KK, Pan H, Lanza GM, Wickline SA. Melittin derived peptides for nanoparticle based siRNA transfection. Biomaterials. 2013;34(12):3110–3119. doi: 10.1016/j.biomaterials.2013.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou HF, et al. Peptide-siRNA nanocomplexes targeting NF-κB subunit p65 suppress nascent experimental arthritis. J Clin Invest. 2014;124(10):4363–4374. doi: 10.1172/JCI75673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yan H, et al. Suppression of NF-κB activity via nanoparticle-based siRNA delivery alters early cartilage responses to injury. Proc Natl Acad Sci USA. 2016;113(41):E6199–E6208. doi: 10.1073/pnas.1608245113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hou KK, Pan H, Ratner L, Schlesinger PH, Wickline SA. Mechanisms of nanoparticle-mediated siRNA transfection by melittin-derived peptides. ACS Nano. 2013;7(10):8605–8615. doi: 10.1021/nn403311c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Halabi CM, Broekelmann TJ, Lin M, Lee VS, Chu ML, Mecham RP. Fibulin-4 is essential for maintaining arterial wall integrity in conduit but not muscular arteries. Sci Adv. 2017;3(5):e1602532. doi: 10.1126/sciadv.1602532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Halabi CM, Broekelmann TJ, Knutsen RH, Ye L, Mecham RP, Kozel BA. Chronic antihypertensive treatment improves pulse pressure but not large artery mechanics in a mouse model of congenital vascular stiffness. Am J Physiol Heart Circ Physiol. 2015;309(5):H1008–H1016. doi: 10.1152/ajpheart.00288.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sizemore GM, Pitarresi JR, Balakrishnan S, Ostrowski MC. The ETS family of oncogenic transcription factors in solid tumours. Nat Rev Cancer. 2017;17(6):337–351. doi: 10.1038/nrc.2017.20. [DOI] [PubMed] [Google Scholar]
- 52.Baltrunaite K, et al. ETS transcription factors Etv2 and Fli1b are required for tumor angiogenesis. Angiogenesis. 2017;20(3):307–323. doi: 10.1007/s10456-017-9539-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Oettgen P. The role of ets factors in tumor angiogenesis. J Oncol. 2010;2010:767384. doi: 10.1155/2010/767384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chung AS, Ferrara N. Developmental and pathological angiogenesis. Annu Rev Cell Dev Biol. 2011;27:563–584. doi: 10.1146/annurev-cellbio-092910-154002. [DOI] [PubMed] [Google Scholar]
- 55.Yin H, Kanasty RL, Eltoukhy AA, Vegas AJ, Dorkin JR, Anderson DG. Non-viral vectors for gene-based therapy. Nat Rev Genet. 2014;15(8):541–555. doi: 10.1038/nrg3763. [DOI] [PubMed] [Google Scholar]
- 56.Burnett JC, Rossi JJ, Tiemann K. Current progress of siRNA/shRNA therapeutics in clinical trials. Biotechnol J. 2011;6(9):1130–1146. doi: 10.1002/biot.201100054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kanasty R, Dorkin JR, Vegas A, Anderson D. Delivery materials for siRNA therapeutics. Nat Mater. 2013;12(11):967–977. doi: 10.1038/nmat3765. [DOI] [PubMed] [Google Scholar]
- 58.Pan H, et al. Lipid membrane editing with peptide cargo linkers in cells and synthetic nanostructures. FASEB J. 2010;24(8):2928–2937. doi: 10.1096/fj.09-153130. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.