Abstract
Bile acids are signaling molecules that critically control hepatocellular function. Disrupted bile acid homeostasis may be implicated in the pathogenesis of chronic liver diseases. Glutathione is an important antioxidant that protects the liver against oxidative injury. Various forms of liver disease share the common characteristics of reduced cellular glutathione and elevated oxidative stress. This study reports a potentially novel physiological function of bile acids in regulating hepatic sulfur amino acid and glutathione metabolism. We found that bile acids strongly inhibited the cysteine dioxygenase type-1–mediated (CDO1-mediated) cysteine catabolic pathway via a farnesoid X receptor–dependent mechanism. Attenuating this bile acid repressive effect depleted the free cysteine pool and reduced the glutathione concentration in mouse liver. Upon acetaminophen challenge, cholestyramine-fed mice showed impaired hepatic glutathione regeneration capacity and markedly worsened liver injury, which was fully prevented by N-acetylcysteine administration. These effects were recapitulated in CDO1-overexpressing hepatocytes. Findings from this study support the importance of maintaining bile acid homeostasis under physiological and pathophysiological conditions, as altered hepatic bile acid signaling may negatively impact the antioxidant defense mechanism and sensitivity to oxidative injury. Furthermore, this finding provides a possible explanation for the reported mild hepatotoxicity associated with the clinical use of bile acid sequestrants in human patients.
Keywords: Hepatology, Metabolism
Keywords: Amino acid metabolism, Toxicology, Transcription
Bile acids inhibit the cysteine dioxygenase type-1-mediated cysteine catabolic pathway via an FXR-dependent mechanism.
Introduction
Bile acids are synthesized from cholesterol in hepatocytes (1). Cholesterol 7α-hydroxylase (CYP7A1) catalyzes the rate-limiting step in the classic bile acid synthesis pathway. Bile acids circulating in the enterohepatic system activate nuclear farnesoid X receptor (FXR) to exert tight feedback inhibition on CYP7A1 gene expression and bile acid synthesis. Hepatocellular FXR induces the repressor small heterodimer partner (SHP) and intestinal FXR induces the endocrine hormone fibroblast growth factor 15 (FGF15) to transcriptionally repress the CYP7A1 gene in hepatocytes (2–4). Bile acids are efficiently conjugated to glycine or taurine to form N-acyl amidates in hepatocytes (5). The human bile acid pool contains about 2 to 3 times more glycine-conjugated bile acids than taurine-conjugated bile acids, while the mouse bile acid pool contains primarily taurine-conjugated bile acids (6, 7). Bile acid amidation increases bile acid aqueous solubility and enhances their digestive function in the gut (8, 9). In the enterohepatic system, bile acids play important roles in regulating metabolic homeostasis, immune response, and cell proliferation (1, 10–14).
The sulfur-containing amino acid cysteine supports various important cellular pathways including protein synthesis, glutathione (GSH) synthesis, taurine synthesis, and sulfate production (Figure 1A) (15). GSH plays important roles in cellular antioxidant defense and detoxification (16). The liver plays a central role in regulating whole-body GSH homeostasis and has the highest concentration of GSH among all organs (17). Cysteine availability is a major determinant of GSH synthesis (16). On the other hand, abnormally elevated cysteine can cause oxidative stress and cytotoxicity in certain cell types (18). Cysteine dioxygenase type-1 (CDO1) catalyzes the irreversible conversion of cysteine to cysteine sulfinic acid, which is the major cellular cysteine elimination mechanism (15). The liver takes up a significant amount of cysteine from the portal circulation, and CDO1 is expressed at markedly higher levels in hepatocytes than nonhepatic tissues, which makes the liver a major organ that regulates cysteine disposal (15, 19). Here, we report that CDO1-mediated hepatic cysteine catabolism is under the negative regulation of bile acids and FXR, and attenuating this bile acid repressive effect may deplete the hepatic free cysteine pool and impair GSH synthesis. This study therefore identified a potentially novel physiological function of bile acids in regulating hepatic sulfur amino acid metabolism and the GSH antioxidant defense mechanism, and further suggests that altering hepatic bile acid homeostasis and signaling may modulate the sensitivity of the liver to oxidative stress–induced injury.
Figure 1. Disrupting enterohepatic bile acid recycling decreased hepatic glutathione in mice.
(A) Illustration of cellular cysteine and glutathione (GSH) metabolism. Cysteine can be obtained from extracellular or intracellular sources including conversion from methionine via multistep reactions. In GSH synthesis, cysteine and glutamate are used by GCL to produce γ-glutamylcysteine, which is then linked to glycine to produce GSH. GSH can be transported out of the cells where extracellular membrane-bound GGT transfers the glutamyl moiety of GSH to amino acid acceptors to produce γ–glutamyl amino acids and cysteinylglycine, which is transported into the cells for GSH regeneration. The major cysteine degradation pathway is CDO1-mediated cysteine conversion to CSA, which can be further used by downstream enzymes to produce other metabolites such as taurine, sulfate, and pyruvate. Arrows indicate increased or decreased hepatic metabolites in cholestyramine-fed (ChTM-fed) mice. CDO1, cysteine dioxygenase-1; CSA, cysteine sulfinic acid; GCL, glutamate-cysteine ligase; CSAD, cysteine sulfinic acid decarboxylase; Cys-Gly, cysteinylglycine; GGT, γ-glutamyl transferase; γ–Glu-Cys, γ-glutamyl-cysteine. (B–E) Male C57BL/6J mice at 12 weeks of age were fed a chow diet or 2% ChTM-containing chow diet for 6 days. Livers were collected after approximately 6 hours of fasting. Liver metabolites were measured by metabolomics analysis. All results are expressed as mean ± SD. n = 5. *P < 0.05 versus chow-fed mice by 2-tailed Student’s t test. AUC, peak area under the curve.
Results
Disrupting enterohepatic bile acid recycling decreased hepatic cysteine and GSH in mice.
To better understand the endogenous bile acid signaling regulation of hepatic metabolism, we fed mice a cholestyramine-containing (ChTM-containing) chow diet for 6 days to decrease enterohepatic bile acid recycling and determined the hepatic metabolite profile. Interestingly, we found significantly decreased hepatic reduced GSH without increased GSH disulfide (GSSG) in the ChTM-fed mice (Figure 1, B and C). The γ-glutamyl cycle is a GSH-dependent amino acid transport system that plays an important role in supporting intracellular GSH regeneration (Figure 1A) (16). The outer plasma membrane–associated γ-glutamyl transferase (γ-GT) transfers the γ-glutamyl moiety of extracellular GSH to an amino acid to produce γ-glutamyl amino acids and cysteinylglycine, which are subsequently transported into the cells for GSH synthesis. Consistent with reduced GSH, liver cysteinylglycine and almost all detected γ-glutamyl amino acids were reduced by ChTM treatment (Figure 1, D and E). These results suggest that decreasing intestine bile acid recycling results in a hepatic GSH-deficient state. The hepatic cysteine level was significantly lower in the ChTM-fed mice (Figure 2A). In contrast, liver glutamate was not changed, and liver glycine was significantly higher in the ChTM-fed group (Figure 2, B and C). Consistent with decreased liver cysteine, the hepatic γ-glutamylcysteine concentration was also significantly lower in ChTM-fed mice (Figure 2D). Hepatic methionine, which can be converted to cysteine, was not altered in ChTM-fed mice (Figure 2E). Furthermore, ChTM feeding did not have a general effect on hepatic amino acid concentration (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.99676DS1). In summary, these metabolic profiling studies revealed the interesting finding that disrupting intestinal bile acid recycling selectively reduced the hepatic free cysteine pool and impaired hepatic GSH synthesis in mice.
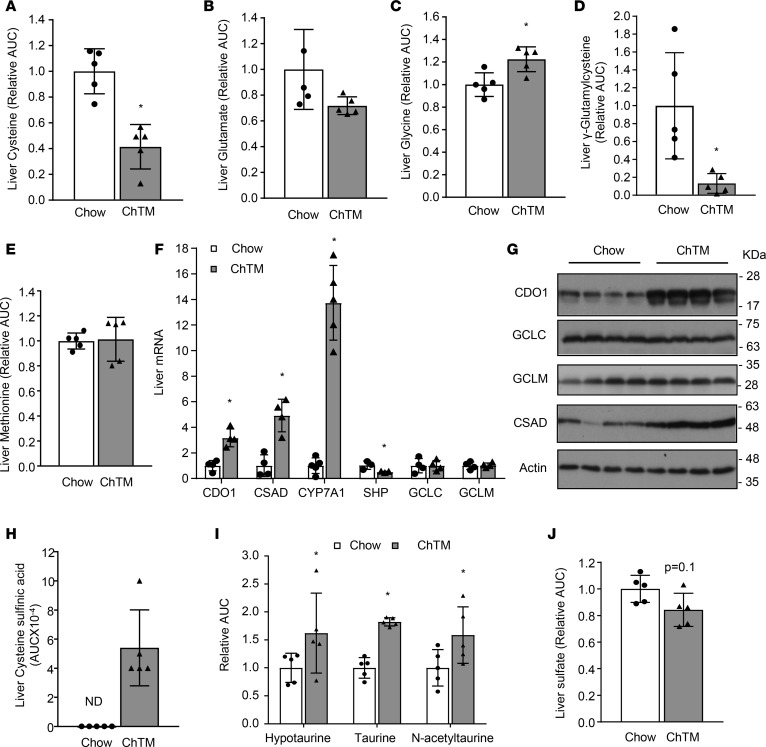
Figure 2. Cholestyramine induced CDO1-dependent cysteine catabolism.
Liver samples were the same as described in Figure 1. (A–E and H–J) Liver metabolites were measured by metabolomics analysis. Results are expressed as mean ± SD. n = 5. (F) Liver mRNA expression. Results are expressed as mean ± SD. n = 4. (G) Liver protein levels. AUC, peak area under the curve; CDO1, cysteine dioxygenase-1; CSAD, cysteine sulfinic acid decarboxylase; CYP7A1, cholesterol 7α-hydroxylase; SHP, small heterodimer partner; GCLC, glutamate-cysteine ligase catalytic unit; GCLM, glutamate-cysteine ligase modifier unit; ChTM, cholestyramine; ND, not detected. *P < 0.05 versus chow-fed mice by 2-tailed Student’s t test.
Attenuating the bile acid repression of hepatic CDO1 promoted hepatic cysteine catabolism in mice.
Hepatic gene expression analysis revealed that ChTM feeding did not alter the expression of glutamate-cysteine ligase (GCL) catalytic subunit (GCLC) or GCL modifier subunit (GCLM) (Figure 2, F and G), which further suggested that decreased cellular GSH concentration was a result of substrate deficiency but not decreased expression of the rate-limiting enzyme. Interestingly, ChTM feeding significantly increased liver CDO1 expression (Figure 2, F and G). The hepatic cysteine sulfinic acid concentration was also elevated in ChTM-fed mice (Figure 2H), which suggests increased CDO1-mediated cysteine catabolism. Consistent with a recent report (20), CSAD was induced in the ChTM-fed mice (Figure 2, F and G), which explains increased downstream hypotaurine, taurine, and N-acetyltaurine concentration (Figure 2I) but modestly decreased sulfate concentration (Figure 2J). ChTM feeding reduced hepatic SHP mRNA and induced hepatic CYP7A1 mRNA, indicating reduced FXR activation by bile acids (Figure 2F). In contrast, activation of FXR with GW4064 decreased CDO1 and CSAD in mouse livers (Figure 3, A and B). GW4064 did not alter hepatic GCLC and GCLM levels in mice (Figure 3B). Bile duct ligation (BDL) for 6 hours and 24 hours, which caused rapid intrahepatic bile acid accumulation and SHP induction (21), decreased hepatic CDO1 expression in mice (Figure 3, C and D). CSAD was also repressed 24 hours after BDL, while GCLC and GCLM were not altered after 6-hour or 24-hour BDL (Figure 3D). In cultured mouse hepatocytes, GW4064 or chenodeoxycholic acid (CDCA) induced SHP mRNA and decreased CDO1 expression (Figure 3E). Treating primary human hepatocytes with GW4064 or CDCA induced SHP and inhibited CYP7A1 and CDO1 (Figure 3, F and G). In summary, these results suggest that endogenous bile acids activate FXR to inhibit hepatic CDO1 and limit the cellular cysteine elimination rate. Disrupting gut bile acid recycling alleviates this repressive effect, which depletes the free cysteine pool and impairs GSH synthesis in the liver.
Figure 3. Bile acids and FXR agonist repressed hepatic CDO1 expression.
(A and B) Hepatic mRNA and protein expression in vehicle- or GW4064-treated mice (male, C57BL/6J, 12 weeks old). n = 4–5. Results are shown as mean ± SD. (C and D) Liver CDO1 mRNA and protein in male C57BL/6J mice that were subjected to 6-hour sham operation or common bile duct ligation (BDL) for 6 or 24 hours. Liver mRNA is shown as mean ± SD. n = 7–8. (E) Primary mouse hepatocytes were treated with vehicle, 1 μM GW4064, or 25 μM CDCA for 16 hours. Results of 3 independent hepatocyte preparations are expressed as mean ± SD. (F) Primary human hepatocytes were treated with vehicle, 1 μM GW4064, or 25 μM CDCA for 16 hours. Average mRNA expression of 4 batches of hepatocytes is shown as mean ± SD. (G) Primary human hepatocytes were treated as in F. A representative Western blot is shown. Relative band intensity of results from 4 batches is expressed as mean ± SD. *P < 0.05 versus vehicle or sham by 2-tailed Student’s t test in all data plots. CDO1, cysteine dioxygenase-1; CYP7A1, cholesterol 7α-hydroxylase; SHP, small heterodimer partner; CDCA, chenodeoxycholic acid; CSAD, cysteine sulfinic acid decarboxylase; GCLC, glutamate-cysteine ligase catalytic unit; GCLM, glutamate-cysteine ligase modifier unit.
FXR-induced SHP inhibits HNF4α transactivation of Cdo1 gene transcription.
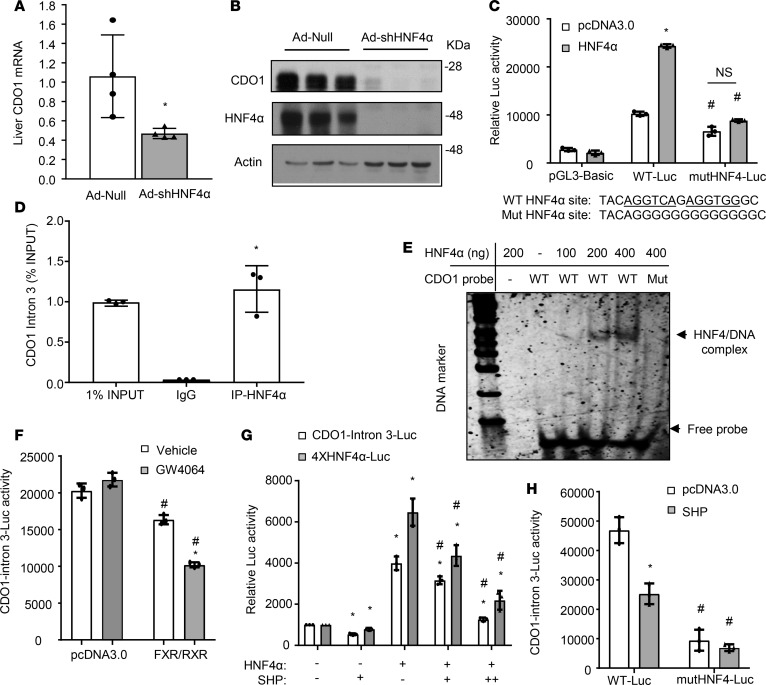
Hepatocyte nuclear factor 4α (HNF4α) is known to play an essential role in maintaining the expression of a large set of hepatically enriched genes (22, 23). Furthermore, it is well documented that FXR-induced SHP, via direct interaction with HNF4α as a corepressor, inhibits a number of HNF4α target genes including bile acid and glucose synthesis genes (23–25). Indeed, knockdown of liver HNF4α in mice markedly decreased hepatic CDO1 expression (Figure 4, A and B). Analysis of a published mouse liver HNF4α ChIP-seq data set revealed HNF4α binding peaks in the chromatin promoter, intron 3, and 3′-untranslated region (3′-UTR) of the Cdo1 gene (26) (Supplemental Figure 2A). Corresponding DNA fragments containing the putative HNF4α binding sites were subsequently cloned into a pGL3-basic luciferase reporter construct. In transient transfection assays, coexpression of HNF4α only induced the reporter activity driven by the intron 3 region (Figure 4C), but not by a 2,901-bp CDO1 promoter, a distal 3,009-bp upstream promoter region, or the 3′-UTR region (Supplemental Figure 2, B and C). Mutations introduced into this HNF4α intronic binding sequence largely abolished the stimulatory effect of HNF4α (Figure 4C). ChIP assay confirmed HNF4α occupancy in this Cdo1 intron 3 chromatin region in mouse livers (Figure 4D). Electrophoretic mobility shift assay (EMSA) further showed direct HNF4α binding to this CDO1 DNA probe, which was abolished when the HNF4α binding site was mutated (Figure 4E). Nuclear receptors can induce a gene via binding to non-promoter regions. For example, FXR strongly induces human FGF19 and mouse FGF15 via a binding motif located in the intron 2 region (4, 27). Our results suggest that HNF4α may stimulate CDO1 transcription via a functional intronic binding site. We further demonstrated that FXR/RXR cotransfection and GW6046 treatment inhibited the CDO1 intron 3 reporter activity (Figure 4F). Coexpression of SHP dose-dependently decreased basal and HNF4α-stimulated activity of the CDO1 intron 3 reporter containing the functional HNF4α binding site (Figure 4G). As positive controls, SHP also repressed HNF4α transactivation of an artificial promoter reporter containing 4 copies of a consensus HNF4α binding site (Figure 4G). Furthermore, SHP coexpression repressed the WT, but not the HNF4α binding site mutant CDO1 intron 3 reporter activity (Figure 4H). Taken together, these results suggest that SHP repressed the CDO1 reporter activity by inhibiting the HNF4α transactivating activity.
Figure 4. SHP repressed HNF4α transactivation of CDO1 reporter activity.
(A and B) C57BL/6J male mice at 12 weeks of age (n = 5) were injected with empty adenovirus (Ad-Null) or Ad-shHNF4α. After 7 days, mice were fasted for 6 hours and sacrificed. The mRNA results are expressed as mean ± SD. *P < 0.05 versus Ad-Null. (C) WT and HNF4α-mutant CDO1-Intron 3-Luc constructs and pGL3-basic construct (0.2 μg), β-gal construct (0.05 μg), and 0.1 μg pcDNA3.0 or HNF4α plasmid were cotransfected into AML12 cells. Luciferase and β-gal activities were measured 48 hours later. The putative HNF4α binding site and mutant sequences are shown below the bar graph. *P < 0.05 versus pcDNA3.0; #P < 0.05 versus corresponding WT-Luc. NS, not significant. (D) ChIP assay detection of HNF4α occupancy to the intron 3 of the Cdo1 chromatin in mouse livers. *P < 0.05 versus IgG. (E) EMSA detection of HNF4α binding to the WT but not the mutant HNF4α binding site in the CDO1 intron 3 DNA probe. Mut, CDO1 probe with mutations introduced into the HNF4α binding site as shown in C. (F) CDO1-Intron 3-Luc plasmids (0.2 μg), β-gal expression construct (0.05 μg), and 0.2 μg pcDNA3.0 or 0.1 μg FXR and 0.1 μg RXR were cotransfected into AML12 cells. After 24 hours, cells were treated with vehicle (DMSA) or GW4064 (1 μM) for 24 hours. *P < 0.05 versus vehicle; #P < 0.05 versus corresponding pcDNA3.0. (G) Luciferase reporter constructs (0.2 μg), β-gal expression construct (0.05 μg), and 0.1 μg (+) or 0.2 μg (++) HNF4α and/or SHP expression plasmids were cotransfected into AML12 cells. Luciferase and β-gal activities were measured 48 hours later. *P < 0.05 versus control (first bar); #P < 0.05 versus HNF4α (third bar). (H) WT and mutant CDO1-Intron 3-Luc plasmids (0.2 μg), β-gal expression construct (0.05 μg), and 0.1 μg pcDNA3.0 or HNF4α were cotransfected into AML12 cells. Luciferase and β-gal activities were measured 48 hours later. *P < 0.05 versus pcDNA3.0; #P < 0.05 versus corresponding WT-Luc. All reporter assay results are shown as mean of triplicates ± SD (technical repeats). Statistical significance determined by 2-tailed Student’s t test (A and D) or 2-way ANOVA with Tukey’s post hoc test (C, F–H). CDO1, cysteine dioxygenase 1; SHP, small heterodimer partner; FXR, farnesoid X receptor; RXR, retinoic X receptor; HNF4α, hepatocyte nuclear factor 4α.
Disrupting enterohepatic bile acid recycling impaired hepatic GSH regenerating capacity and sensitized mice to acetaminophen hepatotoxicity.
The ChTM-fed mice appeared to have normal liver histology, but showed approximately 3- to 4-fold higher plasma transaminases than chow-fed controls (Figure 5, A and B). This result is consistent with clinical findings that patients taking bile acid sequestrant therapies often showed elevated liver transaminases (28–31). Acetaminophen (APAP) overdose causes excess cytochrome p450 2E1–mediated (CYP2E1-mediated) production of toxic N-acetyl-p-benzoquinone imine (NAPQI), which results in hepatic GSH depletion, mitochondrial damage, and hepatocyte necrosis (32). APAP-induced liver injury is highly GSH dependent (32) and thus serves as a suitable experimental model to evaluate the impact of attenuated bile acid signaling on the hepatic sensitivity to oxidative injury. Interestingly, when challenged with a toxic dose of APAP (400 mg/kg), ChTM-fed mice showed markedly worsened liver injury, with higher plasma transaminases (Figure 5, B and C, and Supplemental Figure 3, A and B) and sustained JNK activation at 6 hours after APAP injection (Figure 5D) (32). ChTM did not alter hepatic expression of CYP2E1 (Figure 5D). In the control group, APAP overdose caused rapid hepatic GSH depletion, followed by partial GSH recovery at 6 hours after injection (Figure 5E). However, hepatic GSH levels remained markedly lower at 3 and 6 hours after APAP injection in ChTM-fed mice than chow-fed mice (Figure 5E). In this cohort of mice, hepatic GSSG concentration was also reduced in ChTM-fed mice (Figure 5F). This is in contrast to unaltered hepatic GSSG shown in Figure 1C, which could be due to slight differences in experimental conditions. Nevertheless, the lack of increased GSSG further confirmed that decreased liver GSH in ChTM-fed mice was not a result of increased conversion of GSH to GSSG. Hepatic GSH regeneration is an important factor that determines the severity of APAP-induced liver injury, which can be efficiently ameliorated by early administration of N-acetylcysteine to support GSH synthesis in humans and experimental models (32). Administration of N-acetylcysteine to mice 15 minutes after APAP injection largely prevented worsened liver injury caused by ChTM feeding (Figure 5, B and C, and Supplemental Figure 3B). Consistently, N-acetylcysteine administration significantly improved hepatic GSH regeneration after APAP injection in the ChTM-fed group (Figure 5E).
Figure 5. Disrupting gut bile acid recycling impaired hepatic glutathione regenerating capacity and sensitized mice to acetaminophen (APAP) hepatotoxicity.
C57BL/6J male mice (12 weeks old) were fed a chow or a 2% cholestyramine-containing (ChTM-containing) chow diet for 6 days. Mice were fasted for 6 hours and then injected with 400 mg/kg APAP for 3 hours or 6 hours. The no-APAP control groups were injected with vehicle and sacrificed at the 3-hour time point. In some mice, 500 mg/kg N-acetylcysteine (NAC) was administered 15 minutes after APAP injection. (A) Plasma transaminases in chow or ChTM-fed mice. AST, aspartate aminotransferase; ALT, alanine aminotransferase. n = 5. (B) Representative H&E staining of liver sections (×10 magnification). Scale bar: 200 μm. (C) Plasma ALT. n = 5–7. (D) Liver phosphorylated JNK, total JNK, and cytochrome p450 2E1 (CYP2E1) protein levels. (E and F) Liver GSH and GSSG concentrations. n = 5–8. All results are expressed as mean ± SD. *P < 0.05 versus chow at the same time point and experimental condition; #P < 0.05 versus no-APAP-injected controls on the same diet; §P < 0.05 versus 6-hour APAP-injected mice on the same diet. Statistical significance was determined by 2-tailed Student’s t test (A) or 2-way ANOVA with Tukey’s post hoc test (C, E, and F).
Overexpression of CDO1 in primary mouse hepatocytes decreased cellular GSH concentration and increased APAP-induced cell death.
Although hepatic CDO1 was strongly induced in ChTM-fed mice, decreasing endogenous bile acids in the enterohepatic system and subsequent alteration of FXR activity are expected to affect various cellular pathways. Therefore, to seek direct evidence that ChTM-induced hepatic CDO1 was sufficient to sensitize hepatocytes to oxidative injury, we overexpressed CDO1 in cultured mouse hepatocytes in vitro. CDO1 overexpression did not affect the expression of CYP2E1 (Figure 6A). Propidium iodide (PI), which stains DNA in cells with damaged plasma membranes, was used to evaluate APAP-induced cell injury. When cells were cultured in medium containing 100 μM cysteine to mimic the physiological concentration of portal plasma cysteine (15), CDO1 overexpression slightly increased PI-positive cells in non–APAP-treated cells (Figure 6, B and C). Upon APAP treatment, CDO1 overexpression significantly increased the number of PI-positive cells compared with empty adenovirus–infected (Ad-Null–infected) cells (Figure 6, B and C). Furthermore, when cells were cultured in cysteine-free medium to remove the extracellular cysteine supply, the CDO1-overexpression effect on APAP-induced cell injury was further amplified at the 3- and 6-hour time points (Figure 6, B and C). Under this condition, APAP treatment resulted in close to 100% PI-positive cells at the 12-hour time point in both groups (Figure 6C). APAP-induced alanine aminotransferase (ALT) release into the culture medium was not elevated in control cells before the 6-hour time point, but was already significantly elevated in CDO1-overexpressing cells at the 6-hour time point (Supplemental Figure 4), which provided further evidence that increased hepatic CDO1 expression accelerated APAP-induced cell injury. Consistently, CDO1 overexpression significantly decreased cellular GSH levels by approximately 40% in non–APAP-treated cells (Figure 6D). APAP treatment time-dependently reduced GSH levels, while CDO1-overexpressing cells showed lower GSH than controls at each time point (Figure 6D), which correlated with the degree of cell injury.
Figure 6. Overexpression of CDO1 in primary mouse hepatocytes decreased cellular glutathione concentration and increased acetaminophen-induced cytotoxicity.
(A) Western blotting. Primary mouse hepatocytes were infected with empty adenovirus (Ad-Null) or Ad-CDO1 for 16 hours. (B–D) On the same day of isolation, mouse hepatocytes were infected with Ad-Null or Ad-CDO1 (MOI = 10) in regular DMEM medium containing 100 μM cysteine. After 16 hours, cells were switched to DMEM without cysteine/cystine or with 100 μM cysteine and treated with 2.5 mM acetaminophen (APAP) for 3, 6, and 12 hours. (B and C) Primary mouse hepatocytes were stained with 1 μg/ml propidium iodide (PI, pink), and then fixed with 4% paraformaldehyde and stained with DAPI (blue). Scale bar: 200 μm. The percentage of PI-positive cells was determined with ImageJ software and expressed as mean ± SD. Ten to 40 images from 3 hepatocyte preparations were analyzed. (D) Measurement of cellular glutathione. Results represent mean ± SD of 6 assays from 2 hepatocyte preparations. *P < 0.05 versus Ad-Null + cysteine at the same time point; #P < 0.05 versus Ad-Null – cysteine at the same time point. Two-way ANOVA and Tukey’s post hoc test were used for all statistical analysis. CDO1, cysteine dioxygenase 1; CYP2E1, cytochrome p450 2E1.
Discussion
Hepatic sulfur amino acid metabolism is critically involved in various essential cellular pathways, and disrupted sulfur amino acid homeostasis is implicated in the pathogenesis of many human diseases (15, 16). However, how bile acid signaling regulates hepatic sulfur amino acid metabolism is incompletely understood. In this study, we report the findings that the major hepatic cysteine catabolic pathway is under the negative control of bile acids and FXR. As a result, disrupting enterohepatic bile acid recycling may reduce the free cysteine pool, impair GSH synthesis, and sensitize the liver to oxidative injury. γ-GCL is the rate-limiting enzyme in de novo GSH synthesis. However, cellular cysteine availability is often considered a major determinant of GSH synthesis because of the relatively high Km of γ-GCL (15). In the ChTM-fed mice, hepatic γ-GCL expression was unaltered, but its product γ-glutamylcysteine was decreased by approximately 90%. Such markedly reduced γ-glutamylcysteine concentration could underlie the significantly elevated glycine due to decreased glycine utilization in the GSH synthesis pathway. Furthermore, we showed that most of the intermediate metabolites in the γ-glutamyl cycle were significantly lower in the liver of ChTM-fed mice. The γ-glutamyl cycle plays an important role in maintaining cellular GSH homeostasis and redox status, and decreased γ-glutamyl cycle activity could further predispose the liver to oxidative stress–induced injury. Indeed, our proof-of-concept study in an APAP-induced liver injury model demonstrated that ChTM-fed mice were unable to rapidly regenerate GSH upon APAP challenge and developed worsened liver injury. It should be noted that decreasing circulating bile acids and subsequent alteration at the FXR level are expected to affect various cellular pathways, especially given that bile acid/FXR signaling has been reported to induce phase II and phase III detoxification genes (33) and promote liver-regenerative processes after partial hepatectomy or drug-induced liver injury (34–36). However, we showed that worsened liver injury in ChTM-fed mice could be prevented by N-acetylcysteine supplementation, supporting hepatic cysteine depletion as the major underlying cause. In addition, we demonstrated that direct CDO1 overexpression in cultured hepatocytes, which by-passed other FXR-regulated pathways, was sufficient to decrease cellular GSH and sensitize hepatocytes to APAP toxicity in vitro. These results establish a causative relationship between loss of bile acid and FXR repression of hepatic CDO1 and increased hepatic susceptibility to oxidative stress.
Identification of this potentially novel function of bile acids in the control of hepatic cysteine and GSH homeostasis not only emphasizes the importance of maintaining bile acid homeostasis, but also provides possible mechanistic insights into the reported hepatotoxicity associated with the clinical use of bile acid sequestrants (28–31). A nonabsorbable bile acid sequestrant has been clinically used to treat hypercholesterolemia, and was recently approved for treating hyperglycemia in diabetic patients (37). The clinical use of bile acid sequestrants is generally considered safe. However, multiple clinical trials reported frequently elevated plasma transaminases that resolved upon treatment discontinuation (28–31). The underlying causes of the reported hepatotoxicity are still unclear and somewhat puzzling because bile acid sequestrants are poorly absorbed into the systemic circulation and theoretically should possess little potential to cause direct hepatotoxicity. The serum alkaline phosphatase levels in these patients were usually normal and thus did not indicate biliary injury (28–31). On the other hand, a recent report showed that serum ALT elevation in ChTM-treated patients was accompanied by over 20-fold elevation of serum microRNA-122 (29), suggesting the presence of hepatocellular injury. Based on the major finding from this study, it is reasonable to speculate that the negative impact of bile acid sequestrants on hepatic GSH synthesis, in the presence of increased oxidative stress derived from environmental, pathological, or genetic factors, may potentially cause hepatotoxicity in some susceptible individuals.
The liver is considered the major organ for taurine synthesis (15, 38). From an evolutionary point of view, many mammalian species such as mice, rats, and dogs primarily use taurine for bile acid conjugation, and the use of glycine for bile acid conjugation in humans and other nonhuman primates is considered to be a more recent event (39, 40). Amidation increases bile acid aqueous solubility and enhances their digestive function in the gut. Defective bile acid amidation, such as in pediatric patients lacking functional bile acid-CoA synthase (BACS) or bile acid–CoA:amino acid N-acetyltransferase (BAAT), caused fat-soluble vitamin deficiency, growth retardation, and cholangiopathy (8, 9, 41). Therefore, coordinated regulation of bile acid synthesis and taurine synthesis by bile acids may ensure that sufficient taurine is available when de novo bile acid synthesis is induced. Given that taurine is not exclusively required for bile acid conjugation in humans, the tight coupling of bile acid synthesis and taurine synthesis may be less critical in humans. Therefore, it is possible that the degree of stimulated cysteine conversion to taurine in humans taking bile acid sequestrants is less than that in mice, which may explain the generally mild hepatotoxicity associated with the use of bile acid sequestrant therapies in humans (28–31). Furthermore, hepatic cysteine and GSH reduction alone may not be sufficient to cause liver injury, while its interaction with other genetic and environmental factors may contribute to the generally mild hepatotoxicity in a subgroup of patients taking bile acid sequestrants. The ratio of glycine and taurine conjugates can be controlled by the species-dependent substrate specificity of the BAAT enzyme and subcellular taurine concentration in the peroxisomes (42–44). However, the evolutionary forces driving the selection of a particular amino acid for bile acid conjugation is not fully understood. Findings from this study suggest that in species that primarily use taurine for bile acid conjugation, stimulated de novo bile acid synthesis can be met with upregulation of taurine production but potentially at the expense of cysteine depletion. Therefore, the use of glycine for bile acid amidation, and thus eliminating the need to tightly couple taurine production to de novo bile acid synthesis, may provide an advantage in maintaining bile acid homeostasis and at the same time preserving cellular cysteine for GSH synthesis.
Our results suggest that abnormally elevated hepatic CDO1 expression over baseline levels may be an unwanted effect in pathophysiological conditions or drug therapies. However, the effects of CDO1 inhibition are less clear and likely tissue dependent. It was recently found that CDO1 expression is frequently silenced in many types of cancer cells due to the CDO1 gene hypermethylation or missense mutations (45, 46). It is hypothesized that reduced CDO1 expression may provide an advantage to cancer cells to survive under an oxidative stress environment. However, a causative relationship has not been established so far, and the opposite effect of CDO1 expression in cancer cell growth has also been reported recently (47). A few studies conducted in genetic Cdo1-knockout mice revealed that whole-body complete CDO1 ablation caused more complex metabolic changes and subsequent pathological effects in mice. Global-Cdo1-knockout mice showed severe growth deficit, postnatal lethality, and toxicity in various organs include the liver, pancreas, and lung (48, 49). Mice lacking CDO1 had decreased taurine production, and taurine supplement only improved survival but did not correct other adverse phenotypes. Although the underlying causes are likely complex and remain to be elucidated, it was reported that global CDO1 ablation resulted in higher tissue cysteine and its conversion to hydrogen sulfide. Intracellular hydrogen sulfide accumulation may impair mitochondrial function and cause cytotoxicity in certain sensitive organs (48, 49). It has been suggested that several human diseases may also be potentially linked to decreased CDO1 activities (50–52). In contrast, liver-specific Cdo1-knockout mice were phenotypically normal, did not show taurine deficiency, and appeared to have an increased hepatic free cysteine pool and increased GSH levels (53). It is likely that complete loss or partial loss of cellular CDO1 function may have distinct and also tissue-specific effects. Currently, diseases associated with pathological changes of hepatic CDO1 expression are largely unknown. Future studies need to further investigate how pharmacological or genetic modulation of hepatic CDO1 impacts hepatic organelle function and metabolic homeostasis in various pathophysiological conditions.
The transcriptional regulation of the Cdo1 gene has not been well characterized so far. In this study, we show that the Cdo1 gene is a target gene of HNF4α, which can be targeted by the FXR/SHP pathway. These results provide a molecular mechanism by which bile acids regulate hepatic CDO1 expression. However, our study does not rule out the possibility that bile acids, FXR, and SHP can also target other transcriptional factors that positively regulate Cdo1 gene expression. This is because we also found that the CDO1 promoter reporter activity was repressed by GW4064 and FXR/RXR, and by cotransfection of SHP in AML12 cells (Supplemental Figure 2, D and E). These results suggest that although HNFα does not stimulate CDO1 promoter activity, FXR-induced SHP may repress CDO1 promoter activity. One possibility may be that SHP inhibits the transactivating activity of other transcriptional factors that bind the CDO1 promoter. Nevertheless, since HNF4α knockdown significantly decreased liver CDO1 expression in vivo, SHP repression of HNF4α likely plays an important role in mediating FXR repression of CDO1. Additional studies are required to further characterize the transcriptional regulation of CDO1 by transcriptional factors and bile acid signaling. Although our study provided in vivo and in vitro evidence showing that bile acids and FXR regulated hepatic CDO1, we noted that induction of hepatic CDO1 in ChTM-fed mice was not identified in a previous study (20), and the cause of such a discrepancy is currently unclear. It is also known that increased cellular cysteine can induce CDO1 mRNA expression and decrease CDO1 protein degradation via unknown mechanisms (54, 55). However, ChTM treatment decreases hepatic cysteine, which cannot account for increased hepatic CDO1 in these mice. Further studies are necessary to gain better understanding of the regulation of CDO1 under physiological and pathological conditions, and by pharmacological approaches.
Regardless of etiology, many forms of liver diseases share the common characteristics of elevated oxidative stress and reduced cellular GSH (16). Findings from this study emphasize the importance of maintaining bile acid homeostasis and signaling under physiological and pathophysiological conditions. In addition, findings from this study suggest a possible link between altered GSH metabolism and the reported hepatotoxicity associated with the clinical use of bile acid sequestrants. This study added potentially new insights into the pathophysiological function of bile acid signaling, which is needed for future development of effective and safe bile acid–based therapies for human disease treatment.
Methods
Reagents.
Anti-CDO1 (catalog ab53436), anti-CSAD (catalog ab91016), and anti-CYP2E1 (catalog ab28146) antibodies were purchased from Abcam. Anti-HNF4α antibody (PP-H1415) was purchased from R&D Systems. Anti-actin antibody, ChTM, GW4064, APAP, and N-acetylcysteine were purchased from Sigma-Aldrich. Anti–phospho-JNK (catalog 4668) and anti-JNK (catalog 3708) antibodies were purchased from Cell Signaling Technology. Antibodies against GCLC and GCLM were provided by Terry Kavanagh (University of Washington, Seattle, Washington, USA). Aspartate aminotransferase and ALT assay kits were purchased from Pointe Scientific. GSH assay kit was purchased from Enzo Life Sciences.
Mice.
Male C57BL/6J mice (The Jackson Lab) at 10–12 weeks old were used for the study. ChTM (2% w/w) was mixed with food. GW4064 was prepared in 10% 2-hydroxypropyl-β-cyclodextrin and administered by oral gavage (30 mg/kg, twice/day) for 7 days. BDL in male C57BL/6J mice was performed as previously described (21). APAP was dissolved in warm sterile saline. Mice were fasted for approximately 6 hours and given APAP (400 mg/kg) via intraperitoneal injection. Mice were fasted for an additional 3 or 6 hours and sacrificed. N-acetylcysteine (500 mg/kg, dissolved in sterile saline) was given via intraperitoneal injection 15 minutes after APAP injection.
Cell culture.
Primary human hepatocytes and mouse hepatocytes were obtained from the Cell Isolation Core at the University of Kansas Medical Center. Male 10- to 12-week-old C57BL/6J mice were used for hepatocyte isolation. Primary hepatocytes were plated in collagen-coated plates and treatments were initiated within 24 hours after cells were plated. During the treatments, cells were cultured in serum-free DMEM supplemented with 1% penicillin-streptomycin. AML12 cells were a gift from Yanqiao Zhang (Northeast Ohio Medical University, Rootstown, Ohio, USA).
Metabolomics analysis.
Male C57BL/6J mice were fed a chow diet or a chow diet containing 2% ChTM for 6 days. Mice were fasted for 6 hours and liver tissues were collected. Metabolomics analysis was performed by Metabolon Inc. as previously described (56). Liver extract was analyzed on a Waters ACQUITY UPLC and a Thermo Fisher Scientific Q-Exactive high resolution/accurate mass spectrometer interfaced with a heated electrospray ionization (HESI-II) source and Orbitrap mass analyzer operated at 35,000 mass resolution. Metabolites were identified using Metabolon’s reference library and software. No standard curve was used, and results were expressed as relative peak area under the curve (AUC).
EMSA.
This assay was performed using an EMSA Kit with SYBR green detection (Thermo Fisher Scientific) following the manufacturer’s instructions. Recombinant human HNF4α protein (TP316588) was purchased from Origene. DNA probes were chemically synthesized. The sequences of DNA probes used in EMSA are shown in Supplemental Figure 2B. Images were acquired with a LI-COR Odyssey Imaging System.
Western blotting.
Cells or livers were homogenized in 1× RIPA buffer containing 1% SDS and protease inhibitors, incubated for 1 hour on ice, and sonicated briefly. Supernatant after centrifugation was used for SDS-PAGE and immunoblotting.
Real-time PCR.
Total RNA was isolated with TRI reagent (Sigma-Aldrich). Reverse transcription was performed with Superscript III reverse transcriptase (Thermo Fisher Scientific). SYBR primers were used in real-time PCR. Amplification of 18S was used for normalization. Relative mRNA expression was calculated using the comparative cycle threshold (Ct) method and expressed as 2−ΔΔCt. Real-time PCR was performed with a Bio-Rad CFX384 real-time PCR detection system. The sequences of real-time PCR primers are listed in Supplemental Table 1.
Recombinant adenovirus.
Ad-Null, Ad-CDO1, and Ad-shCDO1 were purchased from Vector Biolabs. Ad-shHNF4α was a gift from Yanqiao Zhang (Northeast Ohio Medical University). Adenovirus was purified from HEK293A cells by CsCl centrifugation. Adenovirus titer was determined with an Adeno-X rapid titer kit from Clontech. Mice were injected 1 × 109 pfu/mouse adenovirus via tail vein.
Luciferase reporter assay.
DNA fragments were generated either by standard PCR or by chemical synthesis and subcloned into the pGL3-basic vector (Promega). Mutations were introduced using a QuickChange Site-directed mutagenesis kit (Agilent Technologies Inc.). Expression plasmids used in this study were described previously (57, 58). Luciferase reporter constructs and expression plasmids were transfected into AML12 cells using Lipofectamine 3000 reagent (Thermo Fisher Scientific). Luciferase activity and β-galactosidase activity were measured using a Bright-Glo luciferase assay system and β-galactosidase enzyme assay system, respectively (Promega). Luciferase activity was normalized to β-galactosidase activity and expressed as relative luciferase activity. Results of triplicate assays were expressed as mean ± SD. A representative assay of 3 independent experiments is shown.
ChIP assay.
Freshly isolated nuclei from mouse livers were cross-linked in formaldehyde. ChIP assay was performed as described previously (59) with a ChIP assay kit (Millipore) following the manufacturer’s instructions. Anti-HNF4α antibody (PP-H1415, R&D Systems) was used in immunoprecipitation. One percent total lysate was used as input, and IgG was used as negative control. Real-time PCR was used to detect the HNF4α-bound chromatin region.
Liver histology.
Liver tissue sections were used for H&E staining. Liver pathological changes were evaluated by a clinical pathologist in a blinded fashion.
Statistics.
Results were expressed as mean ± SD. Statistical analysis was performed by either ANOVA and Tukey’s post hoc test, or 2-tailed Student’s t test. P less than 0.05 was considered statistically significant.
Study approval.
All animal protocols were approved by the Institutional Animal Care and Use Committee of the University of Kansas Medical Center.
Author contributions
YW, JL, and DM performed the experiments and analyzed the results. KD performed histological analysis. YZ, WXD, and TL were involved in experimental design and provided key reagents. TL supervised the study and wrote the manuscript.
Supplementary Material
Acknowledgments
This work was supported in part by an American Diabetes Association Junior Faculty Award (to T. Li), NIH grant 1R01DK102487-01 (to T. Li), the National Center for Research Resources (5P20RR021940-07), and the National Institute of General Medical Sciences (8 P20 GM103549-07) of the NIH.
Version 1. 04/19/2018
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: JCI Insight. 2018;3(8):e99676. https://doi.org/10.1172/jci.insight.99676.
Contributor Information
Yifeng Wang, Email: ywang5@kumc.edu.
Jibiao Li, Email: jli58@gsu.edu.
David Matye, Email: dmatye@kumc.edu.
Yuxia Zhang, Email: lzhang5@kumc.edu.
Katie Dennis, Email: KDENNIS2@kumc.edu.
Wen-Xing Ding, Email: wxding@kumc.edu.
Tiangang Li, Email: tli@kumc.edu.
References
- 1.Li T, Chiang JY. Bile acid signaling in metabolic disease and drug therapy. Pharmacol Rev. 2014;66(4):948–983. doi: 10.1124/pr.113.008201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goodwin B, et al. A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1 represses bile acid biosynthesis. Mol Cell. 2000;6(3):517–526. doi: 10.1016/S1097-2765(00)00051-4. [DOI] [PubMed] [Google Scholar]
- 3.Lu TT, et al. Molecular basis for feedback regulation of bile acid synthesis by nuclear receptors. Mol Cell. 2000;6(3):507–515. doi: 10.1016/S1097-2765(00)00050-2. [DOI] [PubMed] [Google Scholar]
- 4.Inagaki T, et al. Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell Metab. 2005;2(4):217–225. doi: 10.1016/j.cmet.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 5.Shonsey EM, et al. Synthesis of bile acid coenzyme a thioesters in the amino acid conjugation of bile acids. Meth Enzymol. 2005;400:360–373. doi: 10.1016/S0076-6879(05)00021-2. [DOI] [PubMed] [Google Scholar]
- 6.Hardison WG, Grundy SM. Effect of bile acid conjugation pattern on bile acid metabolism in normal humans. Gastroenterology. 1983;84(3):617–620. [PubMed] [Google Scholar]
- 7.Hafkenscheid JC, Hectors MP. An enzymic method for the determination of the glycine/taurine ratio of conjugated bile acids in bile. Clin Chim Acta. 1975;65(1):67–74. doi: 10.1016/0009-8981(75)90335-6. [DOI] [PubMed] [Google Scholar]
- 8.Carlton VE, et al. Complex inheritance of familial hypercholanemia with associated mutations in TJP2 and BAAT. Nat Genet. 2003;34(1):91–96. doi: 10.1038/ng1147. [DOI] [PubMed] [Google Scholar]
- 9.Setchell KD, et al. Genetic defects in bile acid conjugation cause fat-soluble vitamin deficiency. Gastroenterology. 2013;144(5):945–955. doi: 10.1053/j.gastro.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Watanabe M, et al. Bile acids lower triglyceride levels via a pathway involving FXR, SHP, and SREBP-1c. J Clin Invest. 2004;113(10):1408–1418. doi: 10.1172/JCI21025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y, et al. Activation of the nuclear receptor FXR improves hyperglycemia and hyperlipidemia in diabetic mice. Proc Natl Acad Sci USA. 2006;103(4):1006–1011. doi: 10.1073/pnas.0506982103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Watanabe M, et al. Bile acids induce energy expenditure by promoting intracellular thyroid hormone activation. Nature. 2006;439(7075):484–489. doi: 10.1038/nature04330. [DOI] [PubMed] [Google Scholar]
- 13.Thomas C, et al. TGR5-mediated bile acid sensing controls glucose homeostasis. Cell Metab. 2009;10(3):167–177. doi: 10.1016/j.cmet.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Potthoff MJ, et al. FGF15/19 regulates hepatic glucose metabolism by inhibiting the CREB-PGC-1α pathway. Cell Metab. 2011;13(6):729–738. doi: 10.1016/j.cmet.2011.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stipanuk MH, Dominy JE, Lee JI, Coloso RM. Mammalian cysteine metabolism: new insights into regulation of cysteine metabolism. J Nutr. 2006;136(6 Suppl):1652S–1659S. doi: 10.1093/jn/136.6.1652S. [DOI] [PubMed] [Google Scholar]
- 16.Lu SC. Regulation of glutathione synthesis. Mol Aspects Med. 2009;30(1–2):42–59. doi: 10.1016/j.mam.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ookhtens M, Kaplowitz N. Role of the liver in interorgan homeostasis of glutathione and cyst(e)ine. Semin Liver Dis. 1998;18(4):313–329. doi: 10.1055/s-2007-1007167. [DOI] [PubMed] [Google Scholar]
- 18.Perry TL, et al. Hallervorden-Spatz disease: cysteine accumulation and cysteine dioxygenase deficiency in the globus pallidus. Ann Neurol. 1985;18(4):482–489. doi: 10.1002/ana.410180411. [DOI] [PubMed] [Google Scholar]
- 19.Stipanuk MH, Londono M, Lee JI, Hu M, Yu AF. Enzymes and metabolites of cysteine metabolism in nonhepatic tissues of rats show little response to changes in dietary protein or sulfur amino acid levels. J Nutr. 2002;132(11):3369–3378. doi: 10.1093/jn/132.11.3369. [DOI] [PubMed] [Google Scholar]
- 20.Kerr TA, et al. Cysteine sulfinic acid decarboxylase regulation: A role for farnesoid X receptor and small heterodimer partner in murine hepatic taurine metabolism. Hepatol Res. 2014;44(10):E218–E228. doi: 10.1111/hepr.12230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li J, et al. Sortilin 1 loss-of-function protects against cholestatic liver injury by attenuating hepatic bile acid accumulation in bile duct ligated mice. Toxicol Sci. 2018;161(1):34–47. doi: 10.1093/toxsci/kfx078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hayhurst GP, Lee YH, Lambert G, Ward JM, Gonzalez FJ. Hepatocyte nuclear factor 4alpha (nuclear receptor 2A1) is essential for maintenance of hepatic gene expression and lipid homeostasis. Mol Cell Biol. 2001;21(4):1393–1403. doi: 10.1128/MCB.21.4.1393-1403.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bolotin E, et al. Integrated approach for the identification of human hepatocyte nuclear factor 4alpha target genes using protein binding microarrays. Hepatology. 2010;51(2):642–653. doi: 10.1002/hep.23357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crestani M, Sadeghpour A, Stroup D, Galli G, Chiang JY. Transcriptional activation of the cholesterol 7alpha-hydroxylase gene (CYP7A) by nuclear hormone receptors. J Lipid Res. 1998;39(11):2192–2200. [PubMed] [Google Scholar]
- 25.Yamagata K, et al. Bile acids regulate gluconeogenic gene expression via small heterodimer partner-mediated repression of hepatocyte nuclear factor 4 and Foxo1. J Biol Chem. 2004;279(22):23158–23165. doi: 10.1074/jbc.M314322200. [DOI] [PubMed] [Google Scholar]
- 26.Alder O, et al. Hippo signaling influences HNF4A and FOXA2 enhancer switching during hepatocyte differentiation. Cell Rep. 2014;9(1):261–271. doi: 10.1016/j.celrep.2014.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holt JA, et al. Definition of a novel growth factor-dependent signal cascade for the suppression of bile acid biosynthesis. Genes Dev. 2003;17(13):1581–1591. doi: 10.1101/gad.1083503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.A multicenter comparison of lovastatin cholestyramine therapy for severe primary hypercholesterolemia The Lovastatin Study Group III. JAMA. 1988;260(3):359–366. [PubMed] [Google Scholar]
- 29.Singhal R, Harrill AH, Menguy-Vacheron F, Jayyosi Z, Benzerdjeb H, Watkins PB. Benign elevations in serum aminotransferases and biomarkers of hepatotoxicity in healthy volunteers treated with cholestyramine. BMC Pharmacol Toxicol. 2014;15:42. doi: 10.1186/2050-6511-15-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goldfine AB, Fonseca VA, Jones MR, Wang AC, Ford DM, Truitt KE. Long-term safety and tolerability of colesevelam HCl in subjects with type 2 diabetes. Horm Metab Res. 2010;42(1):23–30. doi: 10.1055/s-0029-1241195. [DOI] [PubMed] [Google Scholar]
- 31.Sirmans SM, Beck JK, Banh HL, Freeman DA. Colestipol-induced hepatotoxicity. Pharmacotherapy. 2001;21(4):513–516. doi: 10.1592/phco.21.5.513.34501. [DOI] [PubMed] [Google Scholar]
- 32.Jaeschke H, Bajt ML. Intracellular signaling mechanisms of acetaminophen-induced liver cell death. Toxicol Sci. 2006;89(1):31–41. doi: 10.1093/toxsci/kfi336. [DOI] [PubMed] [Google Scholar]
- 33.Lee FY, et al. Activation of the farnesoid X receptor provides protection against acetaminophen-induced hepatic toxicity. Mol Endocrinol. 2010;24(8):1626–1636. doi: 10.1210/me.2010-0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang W, et al. Nuclear receptor-dependent bile acid signaling is required for normal liver regeneration. Science. 2006;312(5771):233–236. doi: 10.1126/science.1121435. [DOI] [PubMed] [Google Scholar]
- 35.Bhushan B, et al. Role of bile acids in liver injury and regeneration following acetaminophen overdose. Am J Pathol. 2013;183(5):1518–1526. doi: 10.1016/j.ajpath.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang L, et al. Promotion of liver regeneration/repair by farnesoid X receptor in both liver and intestine in mice. Hepatology. 2012;56(6):2336–2343. doi: 10.1002/hep.25905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Staels B. A review of bile acid sequestrants: potential mechanism(s) for glucose-lowering effects in type 2 diabetes mellitus. Postgrad Med. 2009;121(3 Suppl 1):25–30. doi: 10.3810/pgm.2009.05.suppl53.290. [DOI] [PubMed] [Google Scholar]
- 38.Pasantes-Morales H, Chatagner F, Mandel P. Synthesis of taurine in rat liver and brain in vivo. Neurochem Res. 1980;5(4):441–451. doi: 10.1007/BF00964232. [DOI] [PubMed] [Google Scholar]
- 39.Hofmann AF, Hagey LR, Krasowski MD. Bile salts of vertebrates: structural variation and possible evolutionary significance. J Lipid Res. 2010;51(2):226–246. doi: 10.1194/jlr.R000042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moschetta A, et al. A phylogenetic survey of biliary lipids in vertebrates. J Lipid Res. 2005;46(10):2221–2232. doi: 10.1194/jlr.M500178-JLR200. [DOI] [PubMed] [Google Scholar]
- 41.Chong CP, et al. Bile acid-CoA ligase deficiency--a new inborn error of bile acid metabolism. J Inherit Metab Dis. 2012;35(3):521–530. doi: 10.1007/s10545-011-9416-3. [DOI] [PubMed] [Google Scholar]
- 42.Falany CN, Johnson MR, Barnes S, Diasio RB. Glycine and taurine conjugation of bile acids by a single enzyme. Molecular cloning and expression of human liver bile acid CoA:amino acid N-acyltransferase. J Biol Chem. 1994;269(30):19375–19379. [PubMed] [Google Scholar]
- 43.Falany CN, Fortinberry H, Leiter EH, Barnes S. Cloning, expression, and chromosomal localization of mouse liver bile acid CoA:amino acid N-acyltransferase. J Lipid Res. 1997;38(6):1139–1148. [PubMed] [Google Scholar]
- 44.Hardison WG. Hepatic taurine concentration and dietary taurine as regulators of bile acid conjugation with taurine. Gastroenterology. 1978;75(1):71–75. [PubMed] [Google Scholar]
- 45.Jeschke J, et al. Frequent inactivation of cysteine dioxygenase type 1 contributes to survival of breast cancer cells and resistance to anthracyclines. Clin Cancer Res. 2013;19(12):3201–3211. doi: 10.1158/1078-0432.CCR-12-3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brait M, et al. Cysteine dioxygenase 1 is a tumor suppressor gene silenced by promoter methylation in multiple human cancers. PLoS ONE. 2012;7(9):e44951. doi: 10.1371/journal.pone.0044951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Prabhu A, et al. Cysteine catabolism: a novel metabolic pathway contributing to glioblastoma growth. Cancer Res. 2014;74(3):787–796. doi: 10.1158/0008-5472.CAN-13-1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ueki I, et al. Knockout of the murine cysteine dioxygenase gene results in severe impairment in ability to synthesize taurine and an increased catabolism of cysteine to hydrogen sulfide. Am J Physiol Endocrinol Metab. 2011;301(4):E668–E684. doi: 10.1152/ajpendo.00151.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roman HB, Hirschberger LL, Krijt J, Valli A, Kožich V, Stipanuk MH. The cysteine dioxgenase knockout mouse: altered cysteine metabolism in nonhepatic tissues leads to excess H2S/HS(-) production and evidence of pancreatic and lung toxicity. Antioxid Redox Signal. 2013;19(12):1321–1336. doi: 10.1089/ars.2012.5010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bradley H, Gough A, Sokhi RS, Hassell A, Waring R, Emery P. Sulfate metabolism is abnormal in patients with rheumatoid arthritis. Confirmation by in vivo biochemical findings. J Rheumatol. 1994;21(7):1192–1196. [PubMed] [Google Scholar]
- 51.Davies MH, Ngong JM, Pean A, Vickers CR, Waring RH, Elias E. Sulphoxidation and sulphation capacity in patients with primary biliary cirrhosis. J Hepatol. 1995;22(5):551–560. doi: 10.1016/0168-8278(95)80450-1. [DOI] [PubMed] [Google Scholar]
- 52.Heafield MT, Fearn S, Steventon GB, Waring RH, Williams AC, Sturman SG. Plasma cysteine and sulphate levels in patients with motor neurone, Parkinson’s and Alzheimer’s disease. Neurosci Lett. 1990;110(1–2):216–220. doi: 10.1016/0304-3940(90)90814-p. [DOI] [PubMed] [Google Scholar]
- 53.Ueki I, Roman HB, Hirschberger LL, Junior C, Stipanuk MH. Extrahepatic tissues compensate for loss of hepatic taurine synthesis in mice with liver-specific knockout of cysteine dioxygenase. Am J Physiol Endocrinol Metab. 2012;302(10):E1292–E1299. doi: 10.1152/ajpendo.00589.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dominy JE, Hirschberger LL, Coloso RM, Stipanuk MH. Regulation of cysteine dioxygenase degradation is mediated by intracellular cysteine levels and the ubiquitin-26 S proteasome system in the living rat. Biochem J. 2006;394(Pt 1):267–273. doi: 10.1042/BJ20051510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kwon YH, Stipanuk MH. Cysteine regulates expression of cysteine dioxygenase and gamma-glutamylcysteine synthetase in cultured rat hepatocytes. Am J Physiol Endocrinol Metab. 2001;280(5):E804–E815. doi: 10.1152/ajpendo.2001.280.5.E804. [DOI] [PubMed] [Google Scholar]
- 56.Wang Y, et al. Targeting the enterohepatic bile acid signaling induces hepatic autophagy via a CYP7A1-AKT-mTOR axis in mice. Cell Mol Gastroenterol Hepatol. 2017;3(2):245–260. doi: 10.1016/j.jcmgh.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li T, Chiang JY. Rifampicin induction of CYP3A4 requires pregnane X receptor cross talk with hepatocyte nuclear factor 4alpha and coactivators, and suppression of small heterodimer partner gene expression. Drug Metab Dispos. 2006;34(5):756–764. doi: 10.1124/dmd.105.007575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Song KH, Li T, Chiang JY. A Prospero-related homeodomain protein is a novel co-regulator of hepatocyte nuclear factor 4alpha that regulates the cholesterol 7alpha-hydroxylase gene. J Biol Chem. 2006;281(15):10081–10088. doi: 10.1074/jbc.M513420200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li T, et al. Overexpression of cholesterol 7α-hydroxylase promotes hepatic bile acid synthesis and secretion and maintains cholesterol homeostasis. Hepatology. 2011;53(3):996–1006. doi: 10.1002/hep.24107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.