Abstract
Exosomes are extracellular vesicles produced by all cells with a remarkable ability to efficiently transfer genetic material, including exogenously loaded siRNA, to cancer cells. Here, we report on a bioreactor-based, large-scale production of clinical-grade exosomes employing good manufacturing practice (GMP) standards. A standard operating procedure was established to generate engineered exosomes with the ability to target oncogenic Kras (iExosomes). The clinical-grade GMP iExosomes were tested in multiple in vitro and in vivo studies to confirm suppression of oncogenic Kras and an increase in the survival of several mouse models with pancreatic cancer. We perform studies to determine the shelf life, biodistribution, toxicology profile, and efficacy in combination with chemotherapy to inform future clinical testing of GMP iExosomes. Collectively, this report illustrates the process and feasibility of generating clinical-grade exosomes for various therapies of human diseases.
Keywords: Oncology
Keywords: Cancer gene therapy
Exosomes can be GMP manufactured to deliver siRNA for efficient therapeutic control of pancreatic cancer.
Introduction
Exosomes are nanosized vesicles released by cells, and they participate in intercellular exchange of DNA, RNA, proteins, and other cellular components. The capacity of exosomes has spurred a renewed interest in their utility as a delivery system for various therapeutics (1–10). Unlike liposomes and synthetic nanoparticles, the natural features of exosomes may offer unique advantages for the efficient delivery of therapeutic payloads into tumors. The regulatory machinery of exosomes that is associated with production and cellular uptake remains largely unknown. Nonetheless, the engineering of exosomes for therapeutic control of diseases, including cancer, indicated promising results (11–16).
We recently reported on the utility of engineered exosomes derived from fibroblasts for the delivery of siRNA targeting oncogenic Kras in the treatment of pancreas ductal adenocarcinoma (PDAC) in mice (16). Previous studies showed that oncogenic Kras-controlled macropinocytosis in pancreas cancer cells increases exosomes uptake (17, 18) and enhances the efficacy of exosomes in delivering their therapeutic payload to pancreatic tumors (16). Such results highlighted the potential utility of exosomes with the ability to target oncogenic Kras (iExosomes) for the treatment of patients with PDAC, an aggressive and lethal cancer with limited therapeutic options that is on the rise.
A major bottleneck in the advancement of iExosome-based therapy into the clinic is the development of high scale and efficient production of clinical-grade exosomes. This would require sterile generation of exosomes with therapeutic payloads, produced in sufficient amounts for clinical testing, without batch-to-batch variation leading to compromised efficacy. Therefore, we developed a process for production of good manufacturing practice–grade (GMP-grade) exosomes derived from bone marrow mesenchymal stem/stromal cells (MSCs). MSCs have been infused in patients with various disorders without any significant side effects (19). Additionally, bone marrow–derived MSCs have emerged as an attractive cellular source for the generation of clinical-grade exosomes for human therapies (20, 21). Here, we report on the production process and the potential of MSCs in the generation and engineering of iExosomes for human trials and tested the efficacy of GMP-grade MSC derived–iExosomes in several assays and mouse models of pancreatic cancer.
Results
iExosomes target oncogenic Kras to suppress patient-derived PDAC xenograft.
We performed global gene expression profiling of untreated/control and iExosome-treated pancreatic cancer cells with KrasG12D mutation (Panc-1 cells) or wild-type Kras (BxPC-3 cells). The expression profiles of untreated cells were compared with expression profiles of cells treated with control exosomes (no siRNA payload), exosomes with scrambled siRNA control (siScrbl Exo), or exosomes with KrasG12D siRNA (siKrasG12D iExo). Principal component analyses and hierarchical clustering analyses of gene expression revealed a specific segregation of the transcriptomic profiles of Panc-1 cells treated with siKrasG12D iExo, compared with all other control groups (Supplemental Figure 1, A–C; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.99263DS1). This was not observed when BxPC-3 cells were treated with siKrasG12D iExo, and the transcriptomic profile of siKrasG12D iExo–treated BxPC-3 cells did not segregate from that of BxPC-3 controls (Supplemental Figure 1A). Volcano plots of significantly deregulated genes depict a significant effect on the transcriptome of Panc-1 cells treated with siKrasG12D iExo compared with all other controls (Figure 1A and Supplemental Figure 1, B and C), whereas the transcriptome of BxPC-3 cells was largely unaffected (Figure 1A and Supplemental Figure 1, D and E). These results indicate that siKrasG12D iExo specifically affected cells with KrasG12D mutation in contrast with cells with wild-type Kras. Gene set enrichment analyses of the deregulated transcriptome of Panc-1 cells with siKrasG12D iExo compared with all other controls indicated a strong upregulation of genes associated with proteasome, lysosome, and phagosome pathways and others (Supplemental Figure 1F). Downregulated transcripts were associated with response to mineralocorticoid and nuclear chromosome segregation and positive regulation of Smad protein signal transduction, chemokine receptors and chemokines, and response to calcium (Supplemental Figure 1F). These analyses reflect previously reported transcriptomic changes associated with Kras signaling and oncogenic Kras targeting (22, 23) and offer additional insights into mechanisms associated with cell death by siKrasG12D iExo treatment.
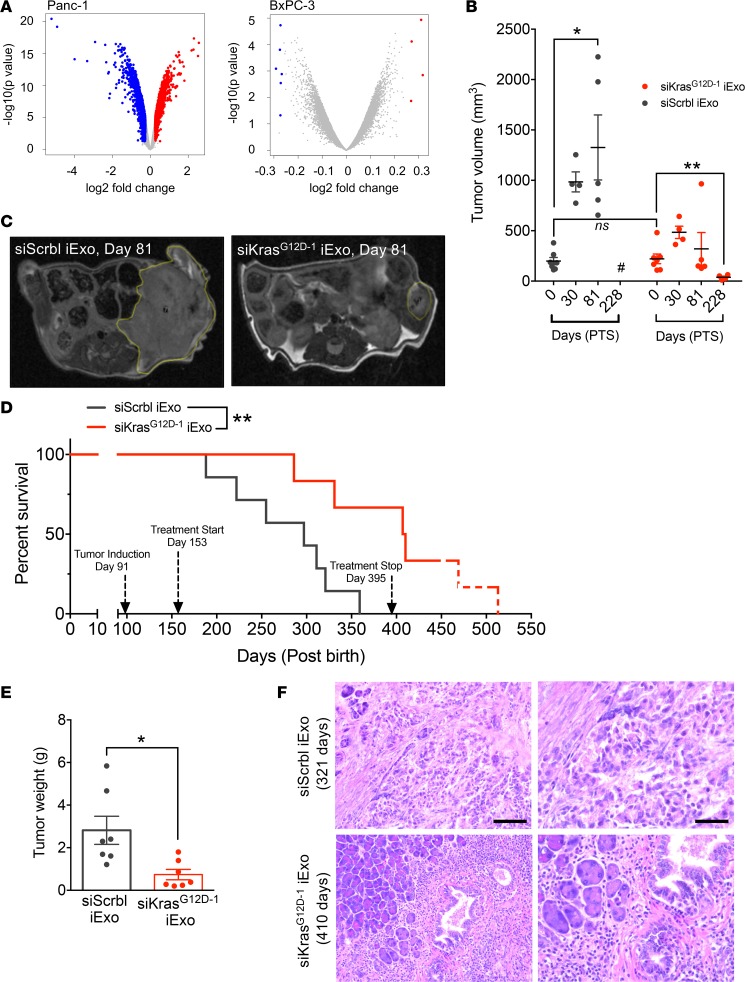
Figure 1. Specific targeting of iExosomes for human PDAC cells with KrasG12D.
(A) Volcano plots depicting log2 fold change (red, upregulated genes; blue, downregulated genes; gray, genes that were not significantly deregulated) and –log10 (P value) of differentially expressed genes between siKrasG12D–1 iExo–treated Panc-1 cells and all controls and BxPC-3 cells. (B) Tumor volume measured by MRI of siKrasG12D–1 iExo (n = 7) or siScrbl iExo (n = 7) at baseline (day 0 = day 62 after tumor induction) and after treatment (day 30, 81, and 228 posttreatment start [PTS]). #, no measurement available; mice died. (C) Representative MRI images of PDX pancreas tumors; a yellow dashed line encircles the tumors. (D) Kaplan-Meier curve indicating the survival of PDX mice in the listed treatment groups after birth (siKrasG12D–1 iExo [n = 7], siScrbl iExo [n = 7]; log-rank (Mantel-Cox) test). The approximated time for the natural life span of nude mice is indicated by the red dotted line. (E) Tumor weight at end point in studies with PDX mice (siKrasG12D iExo [n = 7], siScrbl iExo [n = 7]). (F) Representative H&E image of aggressively invasive pancreatic tumors in siScrbl iExo–treated mice, in comparison with predominant inflammation and markedly reduced tumors in pancreata of siKrasG12D–1 iExo–treated mice. Scale bars: 100 μm (left); 50 μm (right). The data are presented as the mean ± SEM. Unless otherwise stated, unpaired 2-tailed t test was used to determine statistical significance. *P < 0.05, ** P < 0.01. See Supplemental Source Data 1 and 2.
Next, we confirmed the efficacy of siKrasG12D iExo in mice orthotopically implanted with patient-derived xenograft (PDX) that harbors the oncogenic KrasG12D mutation (Supplemental Figure 2A). Mice with PDX were monitored for tumor burden by ultrasound imaging and MRI; both modalities reflected similar measurements in tumor volumes (Supplemental Figure 2B). MRI was chosen as the modality for follow-up imaging, and the results indicated that, while control mice treated with siScrbl Exo progressed with increased tumor volumes over time, mice treated with siKrasG12D iExo showed regression of tumor volume over time (Figure 1, B and C). The tumor burden control achieved with siKrasG12D iExo treatment was associated with a significant increase in survival of mice compared with control siScrbl Exo treatment (Figure 1D). siKrasG12D iExo treatment was stopped (Figure 1D) when all control mice succumbed to pancreatic cancer and presented with substantial tumor weight at endpoint (Figure 1E). Note that at the experimental endpoint, the pancreas weight of mice treated with siKrasG12D iExo was significantly reduced compared with that of controls (Figure 1E). Gross observations at necropsy and histopathological evaluation of tissues revealed that mice from the siKrasG12D iExo treatment group had markedly reduced tumor burden and less aggressive tumors in comparison with mice from siScrbl Exo treatment group (Figure 1F and Supplemental Figure 2, C and D). Some of the mice with siKrasG12D iExo treatment displayed a life span similar to the life span expected with untreated nude mice (Figure 1D). At the experimental endpoint, some of the old mice with siKrasG12D iExo treatment presented with minimal or mild inflammation of the bowel and kidneys (Supplemental Figure 2, C and D), which are common age-related lesions in nude mice, as they may present with increased susceptibility to infection.
MSC-derived iExosomes suppress PDAC in mice.
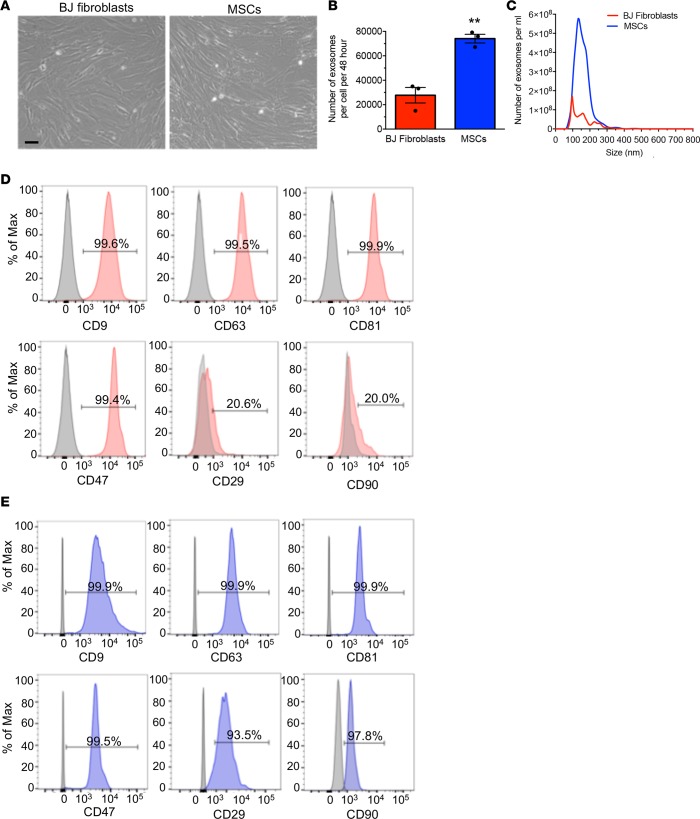
To translate the preclinical findings into the clinic, we opted for the use of GMP-grade compliant normal donor bone marrow–derived MSCs, approved for clinical use in patients, to produce exosomes. We evaluated exosome production of MSCs from 3 donors and chose MSCs from donor 3 for subsequent experiments and GMP production of iExosomes based on its superior exosome production rate (Supplemental Figure 3A). In some of our preclinical studies, we utilized MSCs from donor 2 (for one of the preclinical models, see Methods) and human foreskin fibroblasts (BJ fibroblasts) obtained from ATCC (ref. 16 and Figure 1, B–D). Both BJ fibroblasts and MSCs display similar morphology (Figure 2A) and similar surface marker expression (MSC markers and putative exosome markers), as evaluated by flow cytometry (Supplemental Figure 3, B and C). Both BJ fibroblasts and MSCs were positive for CD44, CD90, CD105, CD29, CD73, and HLA-ABC (Supplemental Figure 3, B and C). They were both negative for CD45, CD31, HLA-DR, and CD34 (Supplemental Figure 3, B and C). They both expressed exosome markers CD81, CD9, and CD63 (Supplemental Figure 3, B and C). BJ fibroblasts were also CD10+, in contrast with MSCs (Supplemental Figure 3, B and C). Both cells expressed high levels of CD47, previously characterized as important for impactful iExosome therapy (16) (Supplemental Figure 3, B and C). BJ fibroblasts and MSCs produced high levels of exosomes, with a mode and mean of 107 nm and 161 nm for BJ fibroblast–derived exosomes and 108 nm and 179 nm for MSC-derived exosomes, respectively, Figure 2, B and C). The MSC exosome production level was significantly higher when compared with that of BJ fibroblasts, with an overall higher count of exosomes per cell over time (Figure 2, B and C). The exosomes from both BJ fibroblasts and MSCs exhibited a similar presence of exosome markers (CD9, CD63, CD81) and CD47, although the mesenchymal markers (CD29, CD90) were predominantly noted on the MSC-derived exosomes (Figure 2, D and E).
Figure 2. Exosome production by MSCs.
(A) Representative bright-field images of BJ fibroblasts and MSCs. Scale bar: 100 μm. (B) Comparison of the number of exosomes quantified by NanoSight, produced by the same number of BJ fibroblasts and MSCs, and collected from the conditioned media over a period of 48 hours (n = 3 independent collections). (C) Particle size distribution analysis of BJ fibroblast exosomes and MSC exosomes by NanoSight. (D and E) Representative histogram of flow cytometry analysis of exosomal markers (CD9, CD63, CD81), CD47, and mesenchymal markers (CD29, CD90) on BJ fibroblasts (red, D) exosomes and MSC exosomes (blue, E). Numbers represent the percentage of positive beads (gray, isotype control). The data are presented as the mean ± SEM. Unpaired 2-tailed t test was used to determine statistical significance. **P < 0.01. See Supplemental Source Data 1 and 2.
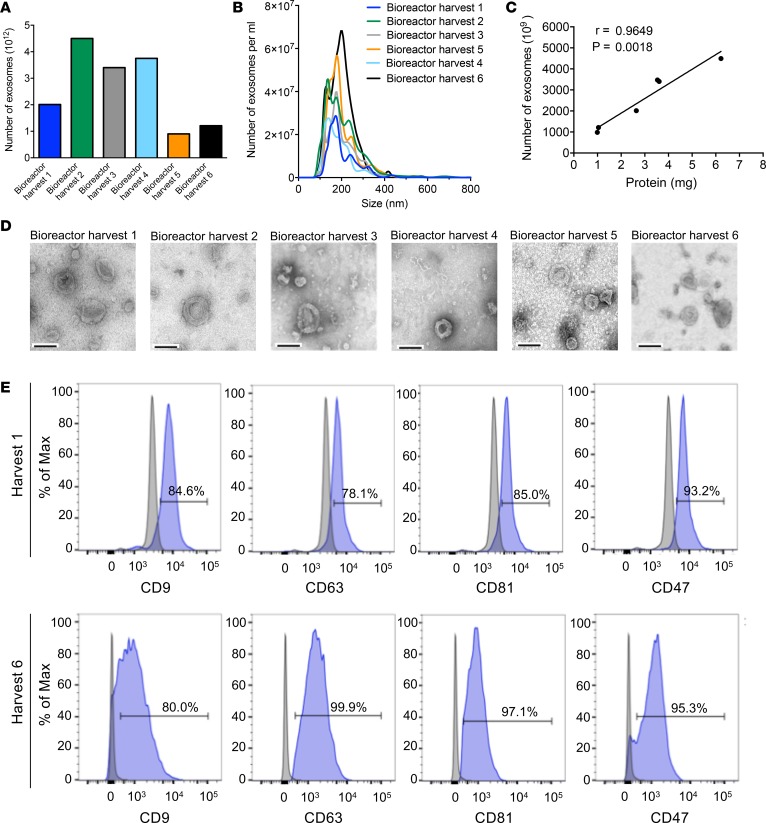
To generate large amounts of exosomes from MSCs, the bioreactor culture of bone marrow–derived MSCs was adapted to enable the harvest of 6 different 250-ml collections of conditioned media (Supplemental Figure 4A). Exosomes were enriched by filtration and ultracentrifugation (Supplemental Figure 4, B and C; see also Methods), and exosome numbers ranged between approximately 900 billion and approximately 4,500 billion per harvest (Figure 3A and Supplemental Figure 5A). Three distinct bioreactor experiments were run, and the sum number of exosomes generated per bioreactor run ranged between 9.8 and 15.6 trillion per bioreactor run (Supplemental Figure 5B). The average size of exosomes from harvests of a bioreactor run (run 1) showed a mode of 165.9 nm and a mean of 202.2 nm (Figure 3B). The measurement of total exosomal protein content correlated with the exosomes counts, as determined by NanoSight analyses (Figure 3C). The metabolic readouts in the bioreactor (glucose and lactose) during the course of the bioreactor harvest of conditioned media for the 6 different collections remained constant (Supplemental Figure 5C), suggesting that the MSCs remained equally viable during the entire process. Electron microscopy analyses and flow cytometry analyses of exosomes from each of the harvests revealed the presence of exosomes (Figure 3D), with consistent presentation of key exosomes biomarkers (CD9, CD63, CD81) and CD47 (Figure 3E and Supplemental Figure 5D). All 6 bioreactor exosome harvests were frozen after each collection. When the final collection was completed, the harvests were thawed and pooled for subsequent isolation and electroporation of siRNA against KrasG12D and generation of iExosomes for further testing (Supplemental Figure 4, B and C).
Figure 3. GMP-grade production of MSC-derived exosomes.
(A) The number of exosomes, quantified by NanoSight, produced by 6 consecutive 48-hour isolations (harvests) of MSC-conditioned media from the bioreactor. (B) Particle size distribution analysis using NanoSight. (C) Correlation between the number of exosomes and exosomal protein from the bioreactor harvests (Pearson correlation test). Data shown in A–C are representative of the same data, obtained from the same bioreactor experiment (Bioreactor run 1) (see also Supplemental Figure 5A). (D) Representative TEM of exosomes from each of the 6 consecutive bioreactor harvests. Scale bar: 100 nm. (E) Representative histogram of flow cytometry analysis of exosomal markers (CD9, CD63, CD81) and CD47 on exosomes from bioreactor harvests 1 and 6 (see also Supplemental Figure 5D). Numbers represent the percentage of positive beads (gray, isotype control). See Supplemental Source Data 1 and 2.
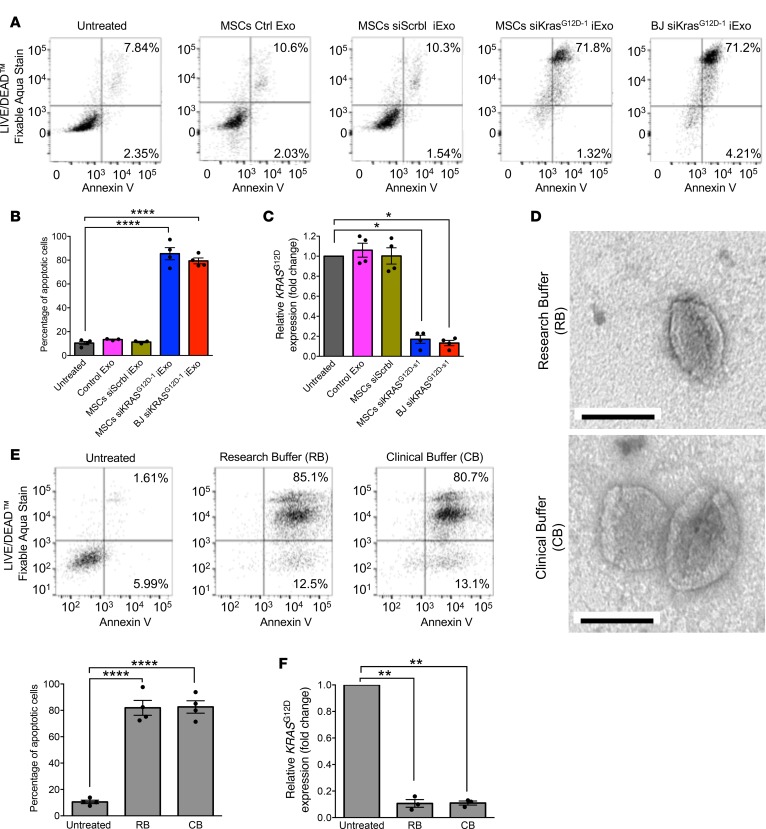
The efficacy of MSC-derived iExosomes to target oncogenic Kras and induce apoptosis of Panc-1 cells was similar to that of freshly prepared BJ fibroblast–derived iExosomes (Figure 4, A–C). For this experiment, MSC-derived exosomes were collected and frozen at –80°C for 2 weeks. After thawing, the MSC-derived exosomes and freshly prepared BJ fibroblast–derived exosomes were electroporated as previously described (16) with siRNAG12D (siRNAG12D source 1 [siRNAG12D–1]; see Methods). MSC-derived iExosomes induced robust apoptosis of Panc-1 cells (Figure 4, A and B) and significantly downregulated oncogenic Kras expression (Figure 4C) when compared with relevant controls (untreated cells, Panc-1 cells treated with control exosomes without siRNA cargo, and Panc-1 cells treated with siScrbl iExo). Previously, a defined electroporation buffer (research buffer [RB]) was used to introduce siRNA into exosomes, which necessitated a wash step of the iExosomes prior to treatment of cells or mice (16). The RB is not approved for human clinical testing. The wash step was associated with a loss of iExosomes (data not shown). To alleviate this hurdle, we employed a diluent known as Plasma-Lyte, referred to here as clinical buffer (CB), that enabled the successful electroporation of the siRNA into exosomes and direct use for administration to cells or mice without an additional wash step. Plasma-Lyte (CB), an FDA-approved diluent for human use, has been used to dilute MSCs for infusions into patients. Following electroporation of MSC-derived exosomes in CB, electron microscopy analyses confirmed the presence of intact exosomes (Figure 4D). Both RB and CB enabled the generation of iExosomes with similar efficacy, induced apoptosis of Panc-1 cells (Figure 4E), and suppressed the expression of oncogenic Kras (Figure 4F).
Figure 4. Validation of GMP-grade iExosome efficacy in vitro.
(A) Representative dot plot and (B) quantification of flow cytometry analysis of apoptosis in Panc-1 cells induced by MSC Ctrl Exo, MSC siScrbl iExo, MSC siKrasG12D–1 iExo, or BJ siKrasG12D–1 iExo after 48 hours, compared with untreated cells. Numbers represent the percentage of positive cells (n = 4 independent experiments, 1-way ANOVA compared with untreated). (C) KRASG12D transcript levels in Panc-1 cells treated with MSC Ctrl Exo, MSC siScrbl iExo, MSC siKrasG12D–1 iExo, or BJ G12D–1 iExo after 3 hours, compared with untreated cells (n = 4 independent experiments, 1-tailed unpaired t test). (D) Representative TEM of MSC exosomes, after electroporation, using either research buffer (RB) or clinical buffer (CB). Scale bar: 100 nm. (E) Representative dot plot of flow cytometry analyses and quantification of apoptosis in Panc-1 cells untreated or treated for 48 hours with MSC siKrasG12D–1 iExo electroporated using either RB or CB. Numbers represent the percentage of positive cells (n = 4 independent experiments, 1-way ANOVA compared with untreated). (F) KRASG12D transcript levels in Panc-1 cells (n = 3 independent experiments, 1-tailed unpaired t test). The mean ± SEM is depicted. *P < 0.05, **P < 0.01, ****P < 0.0001. See Supplemental Source Data 1 and 2.
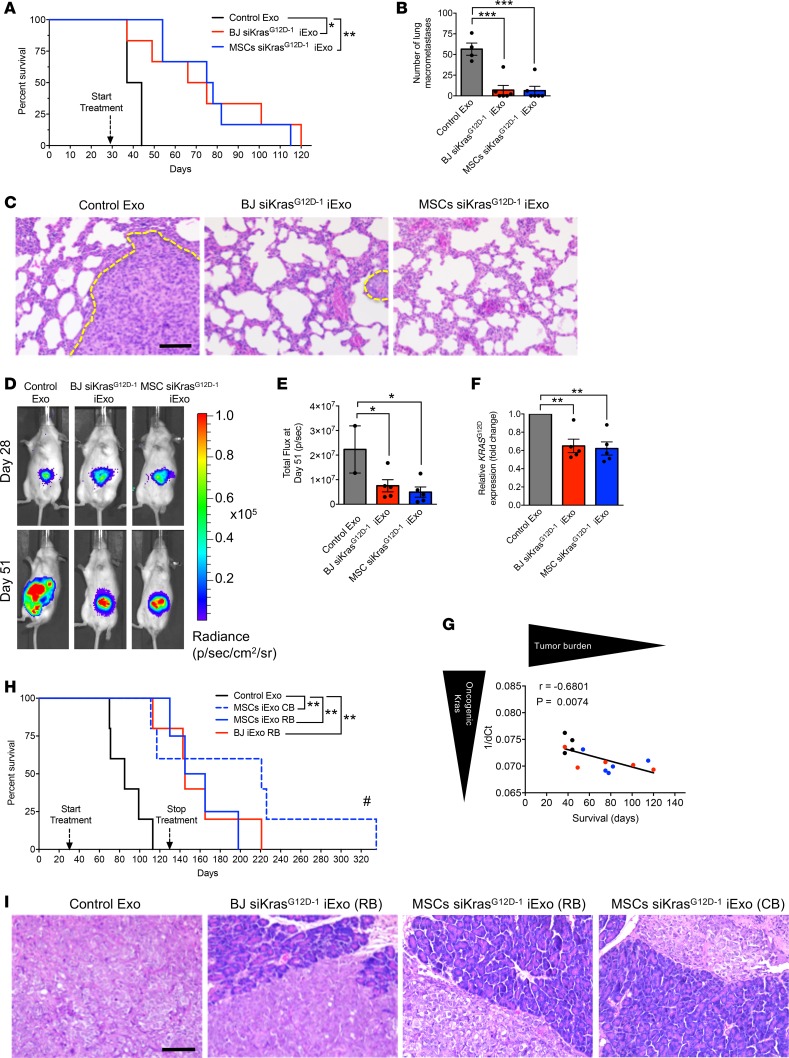
We next tested the efficacy of iExosomes from BJ fibroblasts and MSCs, produced using RB or CB, in PDAC models. iExosomes induced apoptosis of KPC689 cells (Supplemental Figure 6A). In the KPC-derived orthotopic model (KPC689) (16), we tested BJ fibroblast– (freshly isolated and subjected to electroporation) and MSC-derived exosomes (frozen for 45 days after isolation, thawed, and then subjected to electroporation) electroporated with siKrasG12D–1 using RB (Figure 5, A–G). Both BJ fibroblast– and MSC-derived iExosomes significantly enhanced the overall life span of mice with PDAC (Figure 5A) and showed a reduction in metastatic burden (Figure 5, B and C, and Supplemental Figure 6, B–D) when compared with control exosome-treated mice. While tumor burden and weight at experimental endpoint, when mice eventually succumbed to the disease, were similar (Supplemental Figure 6, E and F), IVIS imaging analyses during the treatment phase indicated a significant decrease in tumor burden 23 days after the start of treatment (day 51, Figure 5, D and E, and Supplemental Figure 6G). Both BJ fibroblast– and MSC-derived iExosomes suppressed oncogenic Kras expression in the KPC689 tumors (Figure 5F), and the level of oncogenic Kras expression inversely correlated with survival of mice (Figure 5G). In the Panc-1 orthotopic model, when treatment was initiated at an advance stage of disease, BJ fibroblast– and MSC-derived iExosomes (frozen for 2 months after isolation and then freshly electroporated in RB or CB after a thaw cycle) significantly enhanced the overall survival (Figure 5H and Supplemental Figure 7A). BJ fibroblast– or MSC-derived iExosome therapy, whether using RB or CB for electroporation, substantially improved the histopathology of pancreas (Figure 5I and Supplemental Figure 7B); reduced tumor burden, as evaluated by IVIS (Supplemental Figure 7, C and D); and exhibited a trending decrease in tumor weight and tumor burden at experimental endpoint (Supplemental Figure 7, E and F).
Figure 5. Validation of GMP-grade iExosome efficacy in vivo.
(A) Kaplan-Meier curve indicating survival after tumor induction of mice with KPC689 orthotopic tumors in the listed treatment groups (Control Exo [n = 4], BJ siKrasG12D–1 iExo [n = 6], MSCs siKrasG12D–1 iExo [n = 6]; log-rank [Mantel-Cox] test). (B) Surface lung nodules of KPC689 mice (Control Exo [n = 4], BJ siKrasG12D–1 iExo [n = 6], MSCs siKrasG12D–1 iExo [n = 6]). (C) Representative H&E-stained lung sections from KPC689 mice. Tumor metastasis is indicated by a dashed yellow line. Scale bar: 100 μm. (D) Representative images of luciferase activity of KPC689 tumors at day 28 and day 51 after tumor induction (Control Exo [n = 4], BJ siKrasG12D–1 iExo [n = 6], MSCs siKrasG12D–1 iExo [n = 6]). (E) KPC689 orthotopic tumor growth (bioluminescence) and total flux at day 51 after tumor induction (Control Exo [n = 4], BJ siKrasG12D–1 iExo [n = 6], MSCs siKrasG12D–1 iExo [n = 6]). (F) KrasG12D transcript levels in KPC689 tumors at endpoint in the listed experimental groups (Control Exo [n = 4], BJ siKrasG12D–1 iExo [n = 5], MSCs siKrasG12D–1 iExo [n = 5]; 1-tailed unpaired t test). (G) Correlation between survival and 1/dCT for KrasG12D transcript levels in KPC689 tumors (Pearson correlation test) (black dots, Control Exo [n = 4]; red dots, BJ siKrasG12D–1 iExo [n = 5]; blue dots, MSCs siKrasG12D–1 iExo [n = 5]). (H) Kaplan-Meier curve indicating the survival of Panc-1 tumor-bearing mice after tumor induction in the listed treatment groups (Control Exo [n = 5], MSCs siKrasG12D–1 iExo, CB [n = 5], MSCs siKrasG12D–1 iExo, RB [n = 4], BJ siKrasG12D–1 iExo, RB [n = 5]; log-rank (Mantel-Cox) test). #, pancreas was normal and mice were not moribund (see Supplemental Figure 7A for details). (I) Representative H&E-stained sections of tumors from Panc-1 tumor-bearing mice (Control Exo [n = 5], MSCs siKrasG12D–1 iExo, CB [n = 5], MSCs siKrasG12D–1 iExo, RB [n = 4], BJ siKrasG12D–1 iExo, RB [n = 5]). Scale bar: 100 μm. Data are also depicted in Supplemental Figure 7B. The mean ± SEM is depicted. Unless stated otherwise, 1-way ANOVA, comparing experimental groups to the control group, was used to determine statistical significance. Unless the P value is specified on the figure, *P < 0.05, **P < 0.01, ***P < 0.001. See Supplemental Source Data 1 and 2.
Generation of large-scale iExosomes for PDAC therapy studies.
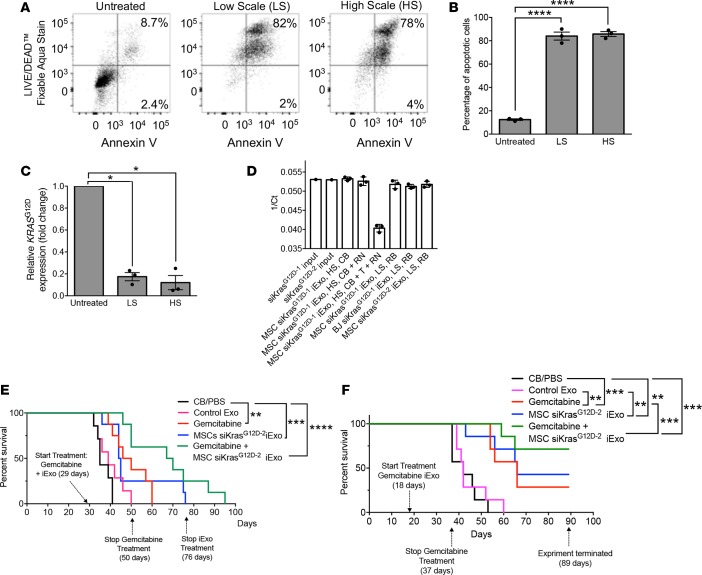
To enable large-scale production of iExosomes for potential clinical testing, we developed a scaled-up production of siRNA electroporation into exosomes. We empirically defined the electroporation parameters using the 4D Nucleofactor LV Unit (Lonza), hereafter referred to as HS for high-scale electroporation (in contrast with low-scale electroporation [LS] routinely used for laboratory studies, ref. 16). The HS would enable large volumes of MSC-derived exosomes to be electroporated in a closed system (Supplemental Figure 4, B and C). Employing Panc-1 apoptosis as a readout of iExosome efficacy, we defined the electroporation settings subsequently used for all experiments thereafter. The defined program (A-14) was chosen to electroporate siRNA into MSC-derived exosomes using CB as diluent/electroporation medium (Supplemental Figure 7G). Both the HS and LS procedures yielded MSC iExosomes that displayed similar efficacy in induction of apoptosis in Panc-1 cells (Figure 6, A and B) and suppressed expression of oncogenic Kras (Figure 6C). We also evaluated the incorporation of the siRNA using RB or CB, employing HS and LS equipment and two distinct sources of siRNA (siKrasG12D–1 and siKrasG12D–2). Under the defined experimental parameters (see Methods), the results indicated a highly efficient loading of the siRNAs into exosomes, regardless of diluent or equipment used (Figure 6D). In these experiments, we confirmed our previous findings (16), which showed that exosomes shielded the incorporated siRNA from RNAse-mediated degradation, which was compromised only when iExosomes were treated with Triton X-100 (Figure 6D).
Figure 6. Efficacy of large-scale produced-GMP iExosomes in combination with gemcitabine.
(A) Representative dot plot and (B) quantification of flow cytometry analyses of apoptosis in Panc-1 cells induced by MSCs siKrasG12D–2 iExo, comparing low scale (LS) or high scale (HS) electroporation of MSC exosomes. Numbers represent the percentage of positive cells (n = 3 independent experiments, 1-way ANOVA compared with untreated). (C) KRASG12D transcript levels in Panc-1 cells incubated 3 hours with MSCs siKrasG12D–2, comparing LS or HS electroporation of MSC exosomes (n = 3 independent experiments, 1-tailed unpaired t test). (D) qPCR of siRNA for KrasG12D (same siRNA sequence from 2 purchasing sources, siKrasG12D–1 and siKrasG12D–2 for source 1 and 2, respectively) in the indicated samples (n = 3 distinct samples treated on the same day; input siRNA: n = 1). The data are presented as 1/Ct and mean ± SD. CB, clinical buffer; RB, research buffer; T, Triton X-100; RN, RNase A. (E) Kaplan-Meier curve indicating the survival of KPC689 mice after tumor induction in the listed treatment groups (CB/PBS [n = 7], Control Exo [n = 7], gemcitabine [n = 8], MSC siKrasG12D–2 iExo [n = 8], gemcitabine + MSCs siKrasG12D–2 iExo [n = 8]; log-rank [Mantel-Cox] test). (F) Kaplan-Meier curve indicating the survival of KPC689 mice after tumor induction in the listed treatment groups (n = 7 mice in each of the listed groups; log-rank [Mantel-Cox] test). Unless otherwise specified, mean ± SEM is depicted. Unless stated otherwise, 1-way ANOVA, comparing experimental groups to control groups, was used to determine statistical significance. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. See Supplemental Source Data 1 and 2.
MSC-derived iExosomes using the CB and HS electroporation procedure were tested using an end-stage aggressive KPC689 orthotopic model, with a highly advanced disease burden, as a single agent and in combination with gemcitabine, a standard of care chemotherapeutic often used in the treatment of PDAC (Supplemental Figure 8A). In contrast with the first KPC689 experiment shown in Figure 5, A–G, IVIS imaging analyses confirmed that mice in this second experiment presented with more extensive tumor burden at the beginning of the treatment (Supplemental Figure 8B). Control mice received diluent (CB and PBS) or control exosomes (no siRNA payload). Experimental groups included mice given iExosomes alone, gemcitabine alone, and gemcitabine and iExosomes. Gemcitabine was administered for 3 weeks and suspended, while the iExosome treatment was continued (Figure 6E and Supplemental Figure 8A). iExosome therapy was also eventually suspended when mice were still alive at day 76 after tumor induction (Figure 6E and Supplemental Figure 8A). In this highly proliferative PDAC model, gemcitabine offered an extension in the overall survival, while iExosome therapy alone showed a reasonable with trend toward longer overall survival, albeit not statistically significant, at this highly advanced stage of disease (Figure 6E and Supplemental Figure 8, A, C, and D). However, the combination of gemcitabine and iExosomes yielded the best outcome, with a much longer overall survival benefit (Figure 6E and Supplemental Figure 8, A, C, and D). Next, we repeated this experiment with a change: the treatment of KPC689 tumor-bearing mice was initiated at an earlier time point in disease progression (Figure 6F and Supplemental Figure 9A). Similar, yet more impressive results were obtained, wherein combination therapy of iExosomes with gemcitabine yielded the most significant improvement in overall survival (Figure 6F and Supplemental Figure 9B). The rapid proliferation of cancer cells in this particular PDAC model (KPC689) likely enhances the response to gemcitabine, and chemotherapy-mediated debulking of large tumors potentially enhanced the efficacy of iExosomes.
Toxicity and immune response associated with iExosomes.
We evaluated the potential toxicity associated with long-term administration of exosomes in mice. The exosomes of human origin (BJ fibroblasts) were administered every 48 hours i.p. into immunocompetent mice for 4 months. Control mice were administered PBS. Hematologic and chemistry analyses of the blood and in-depth histopathological evaluation of several different tissues did not reveal abnormalities upon treatment with exosomes, other than natural age-related tissue changes that were also observed in PBS-administered mice (Supplemental Figure 10A and Supplemental Table 1). Albumin, alkaline phosphatase, ALT, AST, BUN, globulin, and total protein as well as white and red blood cell counts were comparable in both groups (Supplemental Figure 10A). Histopathological findings revealed minimal or mild lesions of inflammation in the liver, kidneys, lung, brain, mesentery, and spleen in both of the groups (Supplemental Table 1 and Supplemental Figure 10B). Minimal to mild acute inflammation of liver and gallbladder was commonly noted in exosome-treated mice and only occasionally in PBS-treated mice (Supplemental Table1 and Supplemental Figure 10B).
Immunocompetent mice were also administered CB, liposomes, BJ exosomes, MSC exosomes, and MSC iExosomes (electroporated to contain the siRNA to KrasG12D; see Methods) every 48 hours i.p. for 3 weeks. Immunotyping analyses of the spleen, bone marrow, and thymus revealed no significant changes in lymphocytes (CD3+, CD4+, CD8+, and CD19+) and myeloid cells (CD11b+, F4/80+) in exosome-administered mice, regardless of cell source for exosome production and whether the exosomes contained siRNA, when compared with CB (Supplemental Figure 11A). Liposome treatment was noted to depress thymic CD3+ levels (Supplemental Figure 11A). Circulating IL-6 and IFN-α levels were not elevated in any of the groups (Supplemental Figure 11B; below detection levels for IFN-α, data not shown), and they remained well below levels associated with immunostimulatory effects (24).
Biodistribution of administered exosomes in mice.
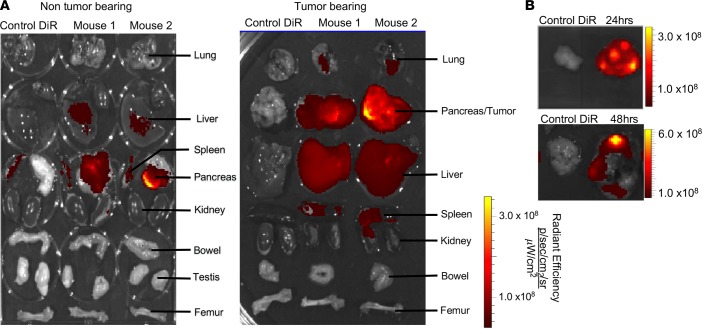
We explored the relative biodistribution of exosomes administered i.p. into tumor-bearing and non-tumor-bearing mice and noted, as previously reported using other readouts (16), a specific accumulation signal associated with exosomes in the normal pancreas and within the tumors in the pancreas, which was higher than the signal detected in the liver, spleen, and lung (Figure 7A). Furthermore, intravenous administration of exosomes into tumor-bearing mice also showed specific accumulation of signal associated with exosomes in the tumors, albeit to a lesser extent than when exosomes were delivered i.p. (Supplemental Figure 12, A and B). Distinct accumulation of signal associated with exosomes at 24 and 48 hours following i.p. administration in pancreatic tumors, compared with the liver, suggested a specific retention of exosomes in the tumors (Figure 7B and Supplemental Figure 12C).
Figure 7. Exosome biodistribution in mice.
(A) Imaging of the indicated organs for detection of DiR-labeled MSC exosomes, 6 hours after i.p. injection of non-tumor-bearing and tumor-bearing (KPC689) mice (DiR control only [n = 1], DiR-labeled MSC exosomes [n = 2]). (B) Imaging of the KPC689 tumors for detection of DiR-labeled MSC exosomes 24 and 48 hours after i.p. injection. Additional images are shown in Supplemental Figure 12C.
Structural and biological stability of iExosomes.
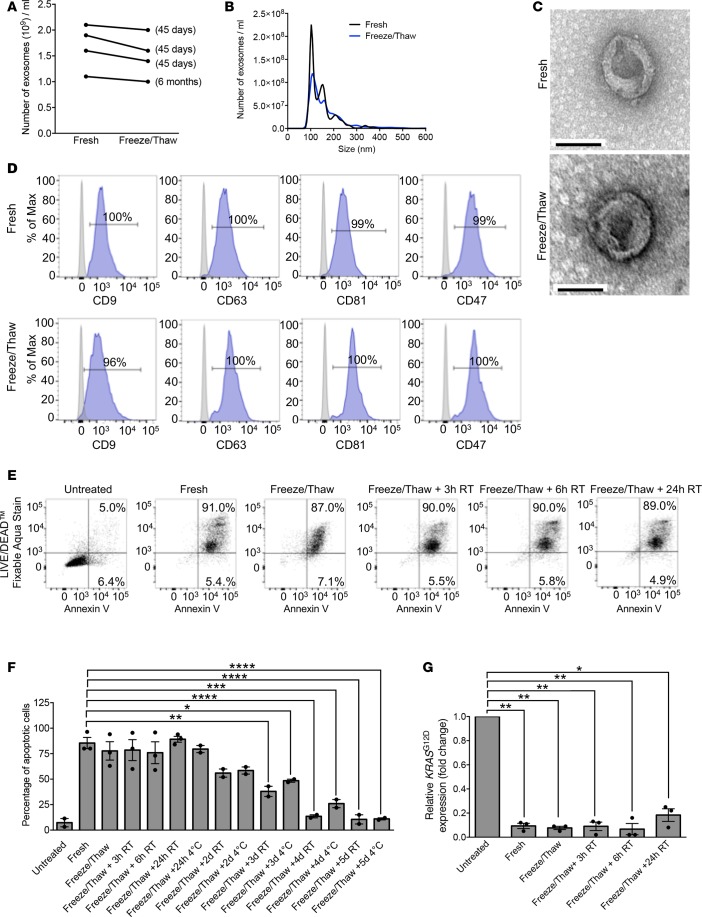
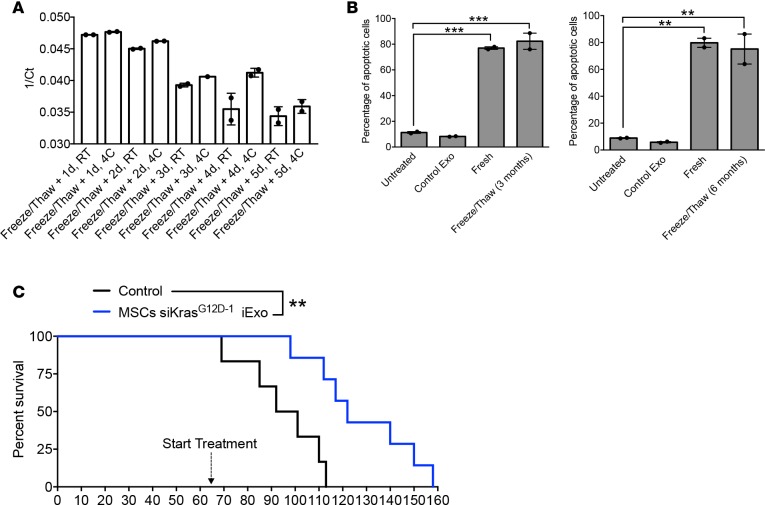
Keeping in mind the potential usage of iExosomes in clinics at different sites and banking of iExosome dosages for future therapy, we tested the structural and biological stability of the iExosomes stored at –80°C for extended periods of time. Compared with freshly prepared exosomes, freezing (for 45 days or 6 months) and thawing of exosomes did not affect their number and size distribution, as evaluated by NanoSight analyses (Figure 8, A and B) and electron microscopy (Figure 8C). Flow cytometry analyses for exosome markers and CD47 were also unaffected following a cycle of freezing (45 days) and thawing of exosomes (Figure 8D). The efficacy of iExosomes in inducing Panc-1 apoptosis and suppressing oncogenic Kras expression was also unaltered following a freezing (45 days) and thawing cycle (Figure 8, E–G). However, if the exosomes were left, after thawing, at room temperature or at 4°C for 2 or more days, a loss in biological activity was observed (Figure 8, F and G). This may possibly be due to the eventual degradation of the siRNA over time at higher temperatures, as determined by qPCR (Figure 9A).
Figure 8. GMP-grade iExosomes stability.
(A) Comparison of the number of MSC exosomes, quantified by NanoSight, prior to freezing and after thawing of frozen exosomes (n = 4 distinct exosomes aliquots). The times listed refer to the times that exosomes were stored for at –80°C. (B) Particle size distribution analysis of fresh and frozen (45 days) and then thawed MSC exosomes by NanoSight. (C) TEM of MSC exosomes, prior to freezing (fresh) and after freezing (45 days) and thawing. Scale bar: 100 nm. (D) Representative histogram of flow cytometry analyses of exosomal markers (CD9, CD63, CD81, CD47) on fresh versus freeze (45 days) and thaw MSC exosomes. Numbers represent the percentage of positive beads (gray, isotype control). (E and F) Representative dot plot of flow cytometry analyses (E) and quantification (F) of apoptosis in Panc-1 cells induced by MSCs siKrasG12D iExo (48 hours following iExo treatment), comparing the efficacy of freeze (3 months) and thaw iExosomes that were allowed to incubate for 3, 6, and 24 hours and 2, 3, 4, and 5 days at room temperature (RT) or 4°C (n = 2–3 independent experiments). Numbers represent the percentage of positive cells. One-way ANOVA compared with fresh exosomes. (G) KRASG12D transcript levels in Panc-1 cells treated with MSCs siKrasG12D iExo after 3 hours, comparing the efficacy of freeze (3 months) and thaw of iExosomes, under the listed conditions (n = 3 independent experiments, 1-tailed unpaired t test). The mean ± SEM is depicted. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. See Supplemental Source Data 1 and 2.
Figure 9. Efficacy of GMP-manufactured iExosomes.
(A) qPCR of siRNA for KrasG12D in the indicated samples (n = 2 independent experiments). MSC siKrasG12D iExo was frozen for 6 months and then thawed and allowed to incubate for 1, 2, 3, 4, and 5 days at room temperature (RT) or 4°C. (B) Quantification of flow cytometry analysis of apoptosis in Panc-1 cells after 48 hours induced by MSC iExo that were frozen for 3 and 6 months. Numbers represent the percentage of positive cells (n = 2 independent experiments). (C) Kaplan-Meier curve indicating the survival of PKS mice (Control Exo [n = 6], MSCs siKrasG12D–2 iExo [n = 7]; log-rank [Mantel-Cox] test). MSC iExo were generated in the GMP facility, and the electroporated iExosomes were frozen for at least 5 months, thawed, and directly injected in mice. The mean ± SEM is depicted. Unless stated otherwise, 1-way ANOVA comparing experimental groups to control groups was used to determine statistical significance. **P < 0.01, ***P < 0.001. See Supplemental Source Data 1 and 2.
GMP-produced MSC-derived iExosomes that were electroporated with siRNA and subsequently stored at –80°C for 3 or 6 months also retained their biological efficacy in inducing Panc-1 cell apoptosis (Figure 9B and Supplemental Figure 13A). These exosomes were further tested in a genetically engineered mouse model for PDAC, driven by pancreas-specific expression of KrasG12D and homozygous loss of Smad4 (PKS PDAC mice). MSC-derived exosomes were GMP produced and frozen for 5 months and subsequently thawed and administered i.p. into PKS mice 3 times per week. The GMP MSC iExosome treatment, initiated at an advanced stage of disease, significantly enhanced the survival of PKS mice (Figure 9C and Supplemental Figure 13B).
Discussion
Here, we report on the large-scale generation of GMP-compliant exosomes from bone marrow–derived MSCs. The detailed procedure provided here overcomes several hurdles that existed in our ability to produce therapeutic exosomes for potential clinical use. Our study demonstrates the large-scale manufacturing of iExosomes using a GMP-compliant process and the definition of criteria for the release of the product for human use. These criteria include the size distribution of the exosome preparation, ascertained by NanoSight; flow cytometry analyses of defined markers on exosomes; and potency assays. The MSCs used for the GMP production were obtained from the University of Texas MD Anderson Cancer Center clinical MSC bank.
We developed a bioreactor-based culture system to produce large quantities of MSC-derived exosomes and a procedure to electroporate them with siRNA to target KrasG12D using a clinical-grade diluent (Plasma-Lyte). Although modifications of the siRNA may be tested in the future to further improve the stability of the siRNA itself, both sources of siRNA employed in our studies to generate iExosomes showed robust antitumor efficacy in PDAC models. Notably, our efforts focused on multiple in vivo models of PDAC, in which the antitumor response to iExosomes was consistently observed. The use of unmodified siRNA in this study did not activate immune responses, as also reported by others (24, 25). We also failed to note an adverse immune reaction in mice, despite repeated injections (every 48 hours) over the course of 4 months. Additionally, a 3-week exposure of mice to MSC-derived iExosomes did not elicit any detectable adverse immune responses compared with control mice, as assayed by immunotyping of tissues, histopathological analyses, and cytokine production. Our production pipeline relies on ultracentrifugation, which may include nonexosomal contaminants; however, our iExosome preparations did not yield any measurable side effects and showed consistent in vitro and in vivo efficacy. We further tested the stability of the iExosomes generated in a GMP facility and noted insignificant loss of antitumor efficacy when iExosomes were stored at –80°C for up to 5 months. We do not anticipate any issues with respect to shelf life of iExosomes, although further testing will be required to confirm this notion. Notably, once thawed and prior to injection, the iExosomes appear to maintain robust anti-KrasG12D activity and the associated pancreatic cancer cell apoptosis efficacy for up to 48 hours when stored at room temperature or 4°C. These findings may suggest that there may be protection from degradation of the siRNA, even at room temperature for 48 hours, when contained within iExosomes. Based on the reported GMP-grade production, scalability, and biological stability described here, we envision potential phase I clinical testing in the future.
This study identified a robust accumulation of exosomes in the normal and tumor-bearing pancreas when delivered i.p. The specific mechanism for this tropism remains unclear, nonetheless these results are in agreement with our previous findings, in which we reported a specific accumulation of iExosomes in the pancreas and pancreatic tumors, the latter possibly further aided by enhanced macropinocytosis in cancer cells expressing oncogenic Kras (16). We also subjected immunocompetent mice to human-derived exosomes every 48 hours for 120 days and noted no obvious side effects associated with repeated exosomes administration. Our findings also support that repeated administration of iExosomes did not elicit signs of toxicity or immunogenicity, supporting a previous report on exosomes administered to mice (26). MSC-derived exosomes presented with CD47, a feature that facilitates their biological activity in vivo (16, 27). Together with the lack of transcriptional changes observed in pancreatic cancer cells with wild-type Kras subjected to iExosomes, these findings support that iExosome therapy may present with minimal side effects when administered to patients with pancreatic cancer.
Collectively, the reported findings lay the foundation for a clinically feasible approach for iExosome therapy targeting oncogenic Kras in other cancer types, such as lung and colon cancer. We believe that the iExosomes therapy strategy can be extended to other oncogenes and cancer-driving genes.
Methods
Cell culture.
BJ fibroblasts were cultured in DMEM supplemented with 10% FBS and 1% penicillin-streptomycin. Panc-1, BxPC-3, and KPC689 cells were cultured in RPMI supplemented with 10% FBS and 1% penicillin-streptomycin. These cell lines were from ATCC, and additional validation using SNP ID profiling via Sequenom confirmed the identity of the cell lines. The cells were negative for mycoplasma. The KPC689 cancer cell line was established from the pancreatic tumors of Pdx1cre/+;LSL-KRasG12D/+;LSL-Trp53R172H/+ (KPC) mice (28). Panc-1 and KPC689 cells were engineered to stably express GFP and luciferase following infection with F-Luc-GFP lentivirus (Capital Biosciences). Bone marrow–derived MSCs, passage 3 times, obtained from the Cell Therapy Laboratory at the University of Texas MD Anderson Cancer Center, were cultured in α MEM supplemented with 1% L-glutamine, 5% human platelet lysate, and 1% penicillin-streptomycin (complete media). MSCs from 3 distinct donors were evaluated, and a single donor was chosen based on high exosome production yield (donor 3) for nearly all experiments, with the exception of few of the iExosomes doses given to PKS mice (donor 2 was used, as detailed below). For transfection, Panc-1 or KPC689 cells were seeded in 6-well plates overnight. Before exosome treatment (as detailed below), the monolayer was washed with 1 ml PBS twice and then treated with iExosomes (detailed dosages below) in 1 ml serum-free media (RPMI-supplemented 1% penicillin-streptomycin) for the indicated time points, as described for each assay.
Isolation and purification of exosomes — research grade.
Exosomes were purified by a defined centrifugation processes, as described previously (16). Supernatant was collected from BJ fibroblasts or MSCs cultured as monolayers in serum-free media for 48 hours and was subsequently subjected to sequential centrifugation steps for 800 g for 5 minutes and 2,000 g for 10 minutes. The supernatant was then filtered using 0.2-μm filter, and a pellet was recovered at 100,000 g using a SW 32 Ti rotor after 3 hours of ultracentrifugation (Beckman). The supernatant was then aspirated, and the resulting pellet was then resuspended in either PBS, or CB and used for experimental procedures. A PBS wash step was performed when exosomes were used for microBCA, transmission electron microscopy analyses, and 1 in vivo experiment (see details below). These exosomes were then subsequently used for flow cytometry analysis, in vitro iExosome treatment assays, and treatment of tumor-bearing mice, as described below. For RNase treatment, purified exosomes were incubated (37°C, 30 minutes) with 2 mg/ml protease-free RNase A (Thermo Scientific) followed by addition of RNase inhibitor (Ambion). Exosomes were also treated with 1% Triton X-100 prior to RNAse A treatment. Briefly, exosomes were subjected to treatment with 1% Triton X-100 for 30 minutes at 37°C, after which RNAse A was added (following the steps detailed above). After resuspension, exosomes were transferred to microcentrifuge tubes (from Eppendorf) or cryo-glass vials and stored at –80°C. These exosomes were then thawed on ice and used for subsequent experiments. The CB (Plasma-Lyte, pH 7.4) was composed of 0.09 M sodium chloride, 0.23 M sodium gluconate, 0.27 M sodium acetate trihydrate, 5 mM potassium chloride, and 3 mM magnesium chloride. It contains no antimicrobial agents.
Isolation and purification of exosomes — clinical grade.
Clinical-grade exosomes were generated strictly from bone marrow–derived MSCs isolated and cultured in the GMP facility in the Cell Therapy Laboratory at the University of Texas MD Anderson Cancer Center. The Quantum bioreactor culture system (Terumo BCT) was primed (automatized process) with 1 l of 1× PBS and coated with 5 mg human fibronectin (BD Biosciences) diluted in 250 ml of 1× PBS for 24 hours. Then, the bioreactor was washed with 500 ml α MEM supplemented with 1% L-glutamine and 5% human platelet lysate (complete media) and loaded with 20 × 106 MSCs (passage 3) diluted in 25 ml complete media and expanded for 9 days using complete media. Fresh complete media were added continuously to cells and the inlet rate was adjusted as defined by the daily glucose and lactate measurements. After 9 days, when the cells reached approximately 80% confluence (as ascertained by glucose and lactose measurements), the cells were washed with 2 l of 1× PBS to replace the complete media with PLT-free media (α MEM supplemented with 1% L-glutamine). Bioreactor conditioned medium (250 ml) was then collected every 48 hours in a sealed bag (closed system) for a total of 6 collections (harvests). During these 12 days, the culture did not expand based upon the constant glucose levels measured daily. Exosomes were thus continuously harvested every 48 hours over 12 days. Harvests were stored at −80°C for further processing. Each harvest was tested for sterility (confirmed negative for anerobic and aerobic bacteria), endotoxin (<1 EU/ml), and mycoplasma (PCR, negative). The collections were then thawed overnight at 4°C, pooled, and centrifuged at 1,000 g for 15 minutes in a closed system using Cobe 2991 Cell Processor (Terumo BCT). After removing large-cell debris by centrifugation (1,000 g for 15 minutes at 4°C), the conditioned media were filtered in a closed system using a filtering bag with 0.2-μm filters (Terumo BCT). Then, 600 ml supernatant was transferred to 6 clear polycarbonate tubes (each tube has a 100-ml capacity) in a semiclosed system (Supplemental Figure 4C) using a syringe and a line connected directly with the polycarbonate tube (Beckman Coulter), sealed, and centrifuged for 3 hours at 100,000 g in a type 45 Ti rotor (Beckman Coulter). This process was repeated 3 times until all of the collections were spun (total of 1,500 ml). The supernatant was then aspirated using a 16-gauge syringe (BD Biosciences, catalog 14-826-18B) connected to a pump. The exosome pellet was resuspended manually using a 18-gauge syringe (BD, catalog 408360) in 4 ml (per tube) CB and transferred to sterile glass container (APP Pharma, capacity of 30 ml). This was maintained at 4°C for up to 72 hours until all centrifugating was completed. When all centrifugating was completed, the final pooled volume of resuspended exosomes was 72 ml. Pooled MSC exosomes were analyzed by NanoSight (0.5 ml) and flow cytometry (1 ml) and tested for endotoxin (using 0.5-ml pooled samples) and sterility (using 1 ml of pooled samples, as detailed above). The exosomes (69-ml final volume) were finally aliquoted in a cryo-glass vials, each containing 2 ml, and stored at −80°C. For the manufacture of future clinical product, the exosomes will be directly processed for large-scale electroporation (see below for details) and then aliquoted and stored at –80°C.
Measurement of particle size and concentration distribution with NTA.
Isolated exosome suspensions were analyzed using the NanoSight LM 10 instrument (NanoSight Ltd). The analysis settings were optimized and kept constant between samples.
Quantification of exosomes by microBCA assay.
MSC exosomes resuspended in CB were washed with 1× PBS and ultracentrifuged at 100,000 g for 3 hours in a type SW 41 Ti rotor (Beckman Coulter). The washed MSC exosomes were then measured again by NanoSight and analyzed for total protein content using the microBCA Protein Assay Reagent Kit (Thermo Scientific) following the manufacturer’s instructions.
LS of exosomes.
109 total exosomes (measured by NanoSight analysis) and 1 μg siRNA were mixed in either 400 μl RB (1.15 mM potassium phosphate, pH 7.2, 25 mM potassium chloride, 21% Optiprep, as previously described, ref. 16) or 400 μl CB. These exosomes were electroporated using a single 4-mm cuvette using a Gene Pulser Xcell Electroporation System (Bio-Rad, catalog 165-2081), as previously described (16). After electroporation, the exosomes were kept on ice and injected into the mice immediately, with a dose of approximately 109 exosomes resuspended in 100 μl PBS or CB, per mouse. This dosage represented approximately 0.8–1.2 μg exosome protein. Note that, in our previous studies (16), the electroporation was followed by a wash step, which led us to conservatively estimate a dose of 108 iExosomes per injection in mice, which we approximated to be equivalent to 0.15–0.2 μg exosome protein. As detailed in the results, the exosomes were either fresh or frozen and stored at –80°C and then thawed on ice prior to electroporation with siRNA. The siRNA sequence (sense 5′-GUUGGAGCUGAUGGCGUAGTT-3′, antisense 5′-CUACGCCAUCAGCUCCAACTT-3′) reflects a G to A nucleotide deviation from the wild-type Kras gene sequence (bold) to specifically target the glycine-to-aspartate amino acid substitution (KrasG12D) and include a TT nucleotide DNA overhang to promote silencing efficiency, as described previously (29–31). The siRNA was obtained from Qiagen (catalog1027424) (referred to as KrasG12D–1). AllStars Negative siRNA (scrambled siRNA) (catalog 1027287) was obtained from Qiagen. In addition, siRNA with the same sense and antisense sequence (referred to as KrasG12D–2 for “source 2”) was manufactured by Nitto Denko Avecia Inc.
HS of exosomes.
2 × 1012 total MSC-derived exosomes and 2 mg siRNA source 2 (Avecia) were mixed in 20 ml CB. These exosomes were electroporated using the 4D Nucleofactor LV unit (Lonza) in a closed system. The LV Nucleocuvette Cartridge is a new cuvette system that allows electroporation up to 20 ml. The Cartridge is connected to 2 reservoir bags (inlet and outlet) and a peristaltic pump that fills the cartridge with 1 ml per unit time. The outlet bag is maintained on ice during all of the procedure, and the procedure takes approximately 10 minutes to be completed. After electroporation, the exosomes were analyzed by NanoSight, tested for endotoxin and sterility (as detailed above), aliquoted in a cryovial, and stored at −80°C. These exosomes were then thawed on ice and used for subsequent in vitro and in vivo experiments. For in vitro experiments, the exosomes were diluted for downstream applications at detailed below. For in vivo experiments, 109 electroporated exosomes were diluted in 100 μl RB or CB.
Electron microscopy.
Electron microscopy was evaluated as previously described (16).
Flow cytometry analyses of exosomes.
Exosomes from BJ fibroblasts and MSCs were isolated as described above and resuspended in 200 μl PBS. Aldehyde/sulfate beads (10 μl, Life Technologies) were added to the solution, and beads and exosome mixture was allowed to mix using a benchtop rotator for 15 minutes at room temperature. PBS (600 μl) was then added to the solution and mixing was continued overnight at 4°C. 1 M glycine (400 μl) was added, and mixing was continued for 1 hour at room temperature. The mixture was then spun down at 8,000 g for 1 minute. The precipitate was then resuspended in 100 μl of 10% BSA in PBS and mixed for 45 minutes at room temperature. The mixture was spun down at 8,000 g for 1 minute, and the supernatant aspirated. The beads with the exosomes attached (pellet) were then resuspended in 20 μl of 2% BSA in PBS and immunolabeled for CD47, CD63, CD81, CD9, CD29, CD90, or an isotype control. The exosomes bound to beads were incubated with 1 μl anti-CD47 antibody (eBioscience, catalog 14-0479) or 1 μl anti-CD63 (BD Biosciences, catalog 556019) or 1 μl anti CD-81 antibody (BD Biosciences, catalog 555675) or 1 μl anti-CD9 antibody (MilliporeSigma, catalog SAB4700092) or 1 μl anti-CD29 antibody (Biolegend, catalog 303001) or 1 μl anti-CD90 antibody (Biolegend, catalog 328101), or 1 μl Mouse IgG1, κ isotype control antibody (BD Biosciences, catalog 555746) in 20-μl volume and mixed at room temperature for 30 minutes. The mixture was then centrifuged at 8,000 g for 1 minute, the supernatant was aspirated, and the pellet was resuspended in 20 μl of 2% BSA in PBS. Then, 1 μl secondary antibody (Invitrogen, catalog A21202) was added to the samples and isotype control. All samples were then mixed at room temperature for 1 hour. The samples were then centrifuged at 8,000 g for 1 minute, the supernatant was aspirated, and pellet was resuspended in 200 μl of 2% BSA in PBS. The exosomes bound to the beads were washed 3 times with 2% BSA in PBS. The expression of exosome markers (CD9, CD63, CD81, and CD47) and mesenchymal markers (CD29 and CD90) was analyzed using the LSR Fortessa X-20 cell analyzer. Data were analyzed using FlowJo software (TreeStar Inc.). The flow cytometry data were acquired side by side for both isotype control and samples for each experiment. The flow cytometry experiment was repeated 2 independent times using the same exosome preparation.
Flow cytometry analyses of cells.
MSC and BJ fibroblasts were detached with 0.25% trypsin and 0.02 % EDTA, suspended in ice-cold FACS buffer (106 cells per ml), and incubated for 30 minutes at 4°C with 1 μl monoclonal antibodies: CD44-PE (BD Biosciences, catalog 550989), CD90-Pe/Cy7 (Biolegend, catalog 328123), CD105-PerCP (Biolegend, catalog 323215), CD29-Alexa Fluor 488 (Biolegend, catalog 303015), CD73-APC (BD Biosciences, catalog 560847), HLA-ABC Brilliant Violet 421 (BD Biosciences, catalog 565332), CD81-FITC (BD Biosciences, catalog 551112), CD47 FITC (BD Biosciences, catalog 556045), CD63-PerCP (BD Biosciences, catalog 565426), CD10-Brilliant Violet 605 (Biolegend, catalog 312221), CD9-APC (ThermoFisher, catalog MA1-10307), CD45-Brilliant Violet 605 (Biolegend, catalog 304133), CD31-APCCy7 (Biolegend, catalog 303119), HLA-DR Brilliant Violet 421 (BD Biosciences, catalog 564244), and CD34-APC (Biolegend, catalog 343607) and their respective isotypes. Cells were stained with 1 μg/ml LIVE/DEAD Fixable Aqua (Thermo Fischer Scientific, catalog L34957) prior to flow cytometry analysis. The analysis was performed using the LSR Fortessa X-20 cell analyzer. All control samples were run side by side with experimental samples. Data were analyzed using FlowJo software (TreeStar Inc.). All control samples were run side by side with experimental samples. All the experiments were repeated 2 times.
Flow cytometry–based apoptosis assay.
For flow cytometry analysis of apoptosis in Panc-1 or KPC689 cells, approximately 250,000 cells per wells were seeded in a 6-well plate overnight. The next day, the cells were washed and treated with iExosomes (as detailed above), control nonelectroporated exosomes, or scramble iExosomes resuspended for 48 hours. Apoptosis was measured by Annexin V-FITC (BD 556547), according to the manufacturer’s instructions. Viability of the cells was measured by LIVE/DEAD Fixable Aqua (Thermo Fischer Scientific, catalog L34957). The samples were measured by using the LSR Fortessa X-20 cell analyzer, and data were analyzed using FlowJo software (TreeStar Inc.). All control samples were run side by side with experimental samples. Each sample was made in technical duplicate or triplicate. All the experiments were repeated 2 or 3 times, as indicated in the figure legends.
Real-time PCR analyses.
250,000 Panc-1 cells were seeded in a 6-well plate overnight, and then, the next day, they were treated with iExosomes, control (nonelectroporated) exosomes, or scramble siRNA containing iExosomes diluted in serum-free media. The cells were incubated with iExosomes for 3 hours and then trypsinized with 0.25% trypsin/0.02 % EDTA. The cells were spun and the cell pellet was processed for RNA extraction and cDNA synthesis as previously described (16). The transcripts of interest were normalized to 18S transcript levels. Primers for KrasG12D were previously described (32), and all primer sequences used are listed in our previous study (16). Each reaction included 3 technical replicates, which were averaged to define 1 biological replicate. The experiments were repeated 3 times on distinct days, and each experiment was defined a biological replicate. Statistical analyses were performed on ΔCt of biological replicates (3 independent experiments), and the results are expressed as relative fold change. The control group was arbitrarily set to 1. Several experimental conditions, depicted in individual graphs for clarity when carried out side by side with others, shared controls (e.g., untreated cells). These were identified in Supplemental Source Data 1 and 2.
Quantification of loading efficiency within exosomes by RT-PCR.
Quantification of loading efficiency within exosomes by RT-PCR was carried out as previously described (16).
Microarray analysis of iExosome treatment in vitro.
The cells were treated for 3 hours, as detailed above, with BJ fibroblasts iExosomes containing siRNA to KrasG12D–1; controls included untreated cells and cells treated with control exosomes and scramble siRNA containing exosomes. In this experiment, the exosomes were washed to prior to treatment on the cells as previously described (16). The Illumina HumanHT-12 V4.0 expression beadchip was used to ascertain global transcriptomic changes, and data were deposited in the NCBI Gene Expression Omnibus database (GEO accession GSE97594). The Limma package (33) from R Bioconductor was used to do quantile normalization of expression arrays and analyze differentially expressed genes between any 2 sample groups (for example, Panc-1 sirnaExo vs. Panc-1 Ctrl) (P ≤ 0.05 and fold change ≥ 1.5). The principle components analysis was performed using the R function plotMDS (34), which uses multidimensional scaling to plot differences between different samples. Distances between samples on the plot represent leading log2 fold change, which is the log2 fold change between the samples for the genes that most distinguish those samples (34). Pathway analyses of differentially expressed genes between sample groups were performed using gene set enrichment analysis (35).
Mice.
Female athymic nu/nu mice (Experimental Radiation Oncology, University of Texas MD Anderson Cancer Center) between 8 and 10 weeks of age as well as C57BL/6 and albino C57BL/6 mice (Jackson Laboratory) were housed in individually ventilated cages on a 12-hour-light/dark cycle at 21°C–23°C and 40%–60% humidity. In some cases, male mice were used (biodistribution studies). Mice were allowed free access to an irradiated diet and sterilized water. The PDAC models studied include Panc-1 orthotopic tumors in nu/nu mice, KPC689 orthotopic tumors in C57BL/6 and albino C57BL/6 mice, the genetically engineered mouse model Ptf1aCre/+; LSL-KrasG12D/+; Smad4lox/lox (PKS, C57BL/6;129 mixed genetic background, both males and females were used), and PATX-60 PDXs in nude mice. Under general anesthesia, Panc-1 (106 cells in 10 μl PBS) or KPC689 cells (106 cells in 20 μl PBS) were injected into the tail of the pancreas using a 27-gauge syringe. For detection of luciferase expression, the mice were injected i.p. with 100 mg/kg of body weight of luciferin (200 μl of a 10 mg/ml solution of luciferin in PBS) 12–15 minutes before imaging, anesthetized with isoflurane, and imaged using IVIS (Xenogen Spectrum). For tumor burden analyses, Living Image version 4.4 (Caliper Life Sciences) was used to quantify all tumors. A circular region of interest around the pancreas and tumor was set within the same experimental groups. In addition, exposure conditions (time, aperture, stage position, binning) were kept identical for all measurements within each experiment. Tumor measurements were obtained under the same conditions for all experimental groups. Panc-1 and KPC689 tumor-bearing mice received 109 exosomes i.p. in a 10-μl volume of PBS/CB, and PKS mice received 109 exosomes i.p. in 100-μl volume of CB, 3 times a week (Monday-Wednesday-Friday schedule). Treatment of the PKS mice was initiated when they reached approximately 60 days of age (range from 58 to 75 days of age, see Supplemental Figure 13B for details). Some of the PKS mice received iExosomes from donor 2, all other mice received iExosomes from donor 3. For treatment of the PDX model, mice received conservatively 108 exosomes (due to the extra wash step as previously described, ref. 16) i.p. in a 100-μl volume of RB, 3 times a week (Monday-Wednesday-Friday schedule). Exosomes were electroporated with 1 μg siRNA (Qiagen or Avecia). For exosome biodistribution studies, adult C57BL/6 mice were injected i.p. with 8 × 109 exosomes labeled with XenoLight DiR (Perkin Elmer; additional details are listed below). This high dose of exosomes was necessary to enable a reliable capture of fluorescent signal in the dissected organs. DiR-labeled exosomes were strictly used for biodistribution studies and were not used for therapeutic or toxicity studies. For KPC689 tumor-bearing mice, treatment was initiated at 29 and 30 days after tumor cell injection; although we noted a more aggressive progression in the second experiment (see results for details, Supplemental Figure 8B), possibly accounted by a lower overall body weight of the mice at time of surgery for the second experiment. For gemcitabine studies, in the gemcitabine-only group, a 100 mg/kg dosage was administered 3 times a week (Monday-Wednesday-Friday schedule) for 3 weeks, after which treatment was suspended. For the combination of MSC iExosomes and gemcitabine, a 100 mg/kg dosage was administered similarly 3 times a week for 3 weeks, along with iExosomes, after which only the gemcitabine treatment was suspended and the iExosome treatment was continued further until the indicated time. Control mice were administered similar volumes of PBS (diluent for gemcitabine) and CB. The orthotopic human pancreatic cancer mouse model was developed using 6- to 8-week-old female CD1 nude mice (Charles River). Mice were anesthetized using 3% isoflurane gas via induction chamber and then secured in dorsal recumbency on the surgical table with a nose cone for maintenance of anesthesia at 1.5%–2%. A 5-mm abdominal insertion was made to expose the pancreas, 2 × 2 mm3 PATX-60 tumor fragments were surgically attached and sutured into pancreatic tail, and then the insertions were closed with 5-0 synthetic absorbable sutures (ETHICON Inc.). Tumor growth was monitored by ultrasound imaging once a week at early stage and once every 2 weeks at the late stage until termination due to tumor burden. The Visual Sonics Vevo 2100 and Vevo 770 Imaging Stations were used for all ultrasound imaging. Image acquisitions were performed using the enhanced abdominal measurement package in the B-mode and 3-D mode settings. Mice were imaged prior to tumor xenografting to establish baseline images and then imaged weekly or biweekly to monitor the size of the tumors. MRI was performed and analyzed as previously described (16). At time of necropsy or euthanasia, gross observation of the metastatic burden and measurement of primary tumor burden were performed in a blinded fashion: the experimentalist performing the tissue collection and recording disease burden and metastasis was blinded to the treatment groups. In all orthotopic mouse models (Panc-1, KPC689, and PDX), all control groups were treated side by side with the experimental groups. The percentage tumor burden was expressed as the percentage tumor weight relative to body weight at the experimental endpoint. Supplemental Table 2 lists the details of the exosome preparation used for each in vivo experiment.
Visualization of exosome biodistribution in vivo.
Biodistribution of MSC-derived exosomes was assayed using 8 × 109 prelabeled resuspended exosomes in 100 μl injected i.p. in 2 non-tumor-bearing and KPC689 tumor-bearing C57BL/6 mice. As controls, 1 nontumor and 1 KPC689 tumor-bearing C57BL/6 mouse and 1 tumor-bearing nude mouse were injected with 100 μl diluted DiR (see below for details). At the end of the experiment (6 hours after injection for Figure 7A and Supplemental 12A; 3 hours after injection for Supplemental Figure 12B; 24 and 48 hours after injection for Figure 7B and Supplemental Figure 12C), the mice were euthanized, and dissected tissues (brain, kidneys, spleen, liver, lungs, heart, pancreas, intestine, testis, and femur) were imaged immediately. Briefly, MSC exosomes were directly labeled with 1 μM 1,1′-dioctadecyltetramethyl indotricarbocyanine iodide (DiIC18, ref. 7, XenoLight DiR) (Perkin Elmer, catalog 125964), in a proportion of 5 × 109 exosomes per ml, by incubation for 5 minutes at 37°C and 15 minutes at 4°C and then washed overnight by ultracentrifugation in 10 ml of 1× PBS at 100,000 g in a type SW 41 Ti rotor (Beckman Coulter). The labeled exosomes (8 × 109) were resuspended in 100 μl of 1× PBS. For control, 1 μl DiR was diluted in 11 ml of 1× PBS and spun overnight by ultracentrifugation as carried out for the exosome-labeling procedure. Control samples (only DiR) were resuspended in 100 μl of 1× PBS. Fluorescence imaging was performed using the IVIS 200 small animal imaging system (PerkinElmer) using the Ex filter at 700 nm and the Em filter at 780 nm. Background fluorescence was measured and subtracted by setting up a background measurement at time of data acquisition.
Toxicology and necropsy analysis.
Adult C57BL/6 mice were injected with 108 BJ fibroblast exosomes conservatively or PBS, i.p. every other day, for a total of 120 days. These mice were then subjected to a comprehensive necropsy and toxicology analysis, performed by the Department of Veterinary Medicine and Surgery, University of Texas MD Anderson Cancer Center.
In addition, adult C57BL/6 immunocompetent mice were also administered CB, 109 liposomes, 109 BJ fibroblast exosomes, 109 MSC exosomes, and 109 MSC iExosomes (electroporated to contain the siRNA to KrasG12D–1) i.p. every other day for 3 weeks. Single-cell suspension was prepared from bone marrow and filtered through 100-μm cell strainer. Half of spleen and thymus were flushed through 100-μm cell strainer to generate a single-cell suspension. Cells were washed in PBS containing 2% FBS and stained with 100 μl surface antibody cocktail diluted (in PBS, 2%FBS, 20% Brilliant Stain Buffer [BD Bioscience], 50 μg/ml CD16/32 block, viability dye) for 30 minutes on ice. After 2 washes, the cells were fixed-permeabilized with the FoxP3 Staining Buffer Set (eBioscience, 00-5523-00) and stained with intracellular CD3 and Ki67 antibodies. Cells were washed twice, fixed with Cytofix Fixation buffer (BD Bioscience, 554655), and washed in PBS, 2% FBS. Data were acquired on Fortessa-X20 (BD Bioscience) and analyzed by FlowJo V10. Immune populations were gated on single live CD45+ cells. The antibodies used are listed in Supplemental Table 3. Circulating (serum) IL-6 and IFN-α levels were evaluated by ELISA (RayBiotech, ELM-IL6-1; eBioscience, BMS6027). Sensitivity for detection of IL-6 ELISA was 2 pg/ml. Note that there was no measurable serum IFN-α (sensitivity limit is 7.48 pg/ml, higher than anticipated levels in healthy C56BL/6 mice).
Histological analyses.
Tissues were fixed in formalin and processed for paraffin embedding. Tissue sections of 5-μm thickness were cut and stained for H&E.
Statistics.
Statistical analyses were performed using tests detailed in the figure legends. One-way ANOVA with Dunnett’s or Tukey’s multiple comparison test or 2-tailed Student’s t test was used to establish statistical significance using GraphPad Prism. For survival analyses, Kaplan-Meier plots were drawn and statistical differences were evaluated using the log-rank (Mantel-Cox) test. A P value of less than 0.05 was considered statistically significant.
Study approval.
All animal procedures were reviewed and approved by the Institute for Animal Care and Use Committee at the University of Texas MD Anderson Cancer Center.
Data availability.
Source data for all figures are provided as Supplemental Source Data 1 and 2, and reagents will be provided upon availability and reasonable request.
Author contributions
RK conceptually designed the strategy for this study, provided intellectual input, and contributed to the writing of the manuscript. VSL provided intellectual input, extracted the RNA, quantified the siRNA loading in exosomes by qPCR analyses, performed in vitro analyses and ELISA measurements, orthotopically injected cancer cells in mice, helped with the monitoring of in vivo experiments, designed the experimental strategy, analyzed the microarray data with CCW, prepared figures, and wrote the manuscript. MM and KMM prepared research-grade MSC exosomes and analyzed BJ fibroblasts and MSC exosomes (flow cytometry and NanoSight measurements) and BJ fibroblasts and MSCs (flow cytometry). MM designed the protocol and prepared and analyzed the MSC exosomes from the bioreactor (cultured cells in the bioreactor, harvested conditioned media, prepared by ultracentrifugation, performed electroporation, and performed flow cytometry analysis and NanoSight measurements). MM performed experiments, including treatment of mice with iExosomes, biodistribution studies, microBCA, apoptosis, and qPCR analyses; analyzed data; prepared figures; provided intellectual input; and helped to write the manuscript. SK prepared and treated cells with BJ iExosomes, extracted RNA for microarray analyses, prepared iExosomes for the treatment of PDX mice, and treated PDX mice and PKS mice with SY. SK monitored preclinical trials, analyzed the IVIS data and MRI for in vivo experiments, injected mice for cytotoxicity analyses, performed experiments, including qPCR and apoptosis assays, and treated mice with MM and SY. SK provided intellectual input, helped design the experimental strategy, and helped in the preparation of figures and writing of the manuscript. HS performed necropsy analyses (blinded). CCW analyzed the microarray data. MM performed the necropsy analyses, blood chemistry, and cytology for the toxicology study. SY treated PKS mice and carried out additional toxicology studies with HS and EVRB. EVRB performed the flow cytometry analyses of the bone marrow, spleens, and thymi. QP performed IVIS imaging (blinded), data acquisition, and cell culture and helped with ELISA. XM and JRM established the orthotopic PDX model and performed ultrasound imaging and analyses. AM offered intellectual input for the initial design of the PDX study. KR, ES, and CY offered intellectual input for the GMP production of exosomes. MG carried out the necropsy and histopathological and toxicity analyses of mice shown in Supplemental Table 1.
Supplementary Material
Acknowledgments
This work was primarily supported by the Cancer Prevention and Research Institute of Texas and Pancreas Cancer Moonshot funding from University of Texas MD Anderson Cancer Center. Research in RK’s laboratory is supported by National Cancer Institute grants CA213233, P01 CA117969, and CA195733. Research in VSL’s laboratory is supported by the University of Texas MD Anderson Cancer Center Khalifa Bin Zayed Al Nahya Foundation. Other support came from the High-Resolution Electron Microscopy Facility Institutional Core grant CA16672, the University of Texas MD Anderson Cancer Center Flow Cytometry core facility, the University of Texas MD Anderson Cancer Center Small Animal Imaging Facility (NIH grants P30-CA016672 and 5U24-CA126577). We thank Kenneth Dunner Jr. for the help with the electron microscopy analyses, PE Phillips for tissue processing, A. Haltom for sequencing analyses, J. Kim for biodistribution experiment analyses, S. Lovisa and PC de Sampaio for mycoplasma testing, C. Kingsley for MRI support, and N. Feng for support with the PDX experiment. We thank R. Shroff for helpful discussions regarding clinical trial design.
Version 1. 04/19/2018
Electronic publication
Footnotes
Conflict of interest: The University of Texas MD Anderson Cancer Center and RK are stock equity holders in Codiak Biosciences. RK receives research support from Codiak Biosciences and serves as a member of the board of directors. VSL served as a one-time paid consultant for Codiak Biosciences.
Reference information: JCI Insight. 2018;3(8):e99263. https://doi.org/10.1172/jci.insight.99263.
Contributor Information
Mayela Mendt, Email: MCMendt@mdanderson.org.
Sushrut Kamerkar, Email: skamerkarmda@gmail.com.
Hikaru Sugimoto, Email: Hsugimoto@mdanderson.org.
Chia-Chin Wu, Email: CWu9@mdanderson.org.
Mihai Gagea, Email: mgagea@mdanderson.org.
Sujuan Yang, Email: SYang3@mdanderson.org.
Qian Peng, Email: QPeng@mdanderson.org.
Xiaoyan Ma, Email: XMa1@mdanderson.org.
Anirban Maitra, Email: amaitra@mdanderson.org.
Cassian Yee, Email: cyee@mdanderson.org.
Elizabeth Shpall, Email: eshpall@mdanderson.org.
Raghu Kalluri, Email: rkalluri@mdanderson.org.
References
- 1.Kalluri R. The biology and function of exosomes in cancer. J Clin Invest. 2016;126(4):1208–1215. doi: 10.1172/JCI81135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kowal J, Tkach M, Théry C. Biogenesis and secretion of exosomes. Curr Opin Cell Biol. 2014;29:116–125. doi: 10.1016/j.ceb.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 3.Ha D, Yang N, Nadithe V. Exosomes as therapeutic drug carriers and delivery vehicles across biological membranes: current perspectives and future challenges. Acta Pharm Sin B. 2016;6(4):287–296. doi: 10.1016/j.apsb.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marcus ME, Leonard JN. FedExosomes: Engineering therapeutic biological nanoparticles that truly deliver. Pharmaceuticals (Basel) 2013;6(5):659–680. doi: 10.3390/ph6050659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van den Boorn JG, Dassler J, Coch C, Schlee M, Hartmann G. Exosomes as nucleic acid nanocarriers. Adv Drug Deliv Rev. 2013;65(3):331–335. doi: 10.1016/j.addr.2012.06.011. [DOI] [PubMed] [Google Scholar]
- 6.EL Andaloussi S, Mäger I, Breakefield XO, Wood MJ. Extracellular vesicles: biology and emerging therapeutic opportunities. Nat Rev Drug Discov. 2013;12(5):347–357. doi: 10.1038/nrd3978. [DOI] [PubMed] [Google Scholar]
- 7.El-Andaloussi S, et al. Exosome-mediated delivery of siRNA in vitro and in vivo. Nat Protoc. 2012;7(12):2112–2126. doi: 10.1038/nprot.2012.131. [DOI] [PubMed] [Google Scholar]
- 8.Gilligan KE, Dwyer RM. Engineering exosomes for cancer therapy. Int J Mol Sci. 2017;18(6):1122. doi: 10.3390/ijms18061122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rezaie J, et al. Exosomes and their application in biomedical field: Difficulties and advantages. Mol Neurobiol. 2018;55(4):3372–3393. doi: 10.1007/s12035-017-0582-7. [DOI] [PubMed] [Google Scholar]
- 10.Johnsen KB, Gudbergsson JM, Skov MN, Pilgaard L, Moos T, Duroux M. A comprehensive overview of exosomes as drug delivery vehicles - endogenous nanocarriers for targeted cancer therapy. Biochim Biophys Acta. 2014;1846(1):75–87. doi: 10.1016/j.bbcan.2014.04.005. [DOI] [PubMed] [Google Scholar]
- 11.Xitong D, Xiaorong Z. Targeted therapeutic delivery using engineered exosomes and its applications in cardiovascular diseases. Gene. 2016;575(2 Pt 2):377–384. doi: 10.1016/j.gene.2015.08.067. [DOI] [PubMed] [Google Scholar]
- 12.Cooper JM, et al. Systemic exosomal siRNA delivery reduced alpha-synuclein aggregates in brains of transgenic mice. Mov Disord. 2014;29(12):1476–1485. doi: 10.1002/mds.25978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kordelas L, et al. MSC-derived exosomes: a novel tool to treat therapy-refractory graft-versus-host disease. Leukemia. 2014;28(4):970–973. doi: 10.1038/leu.2014.41. [DOI] [PubMed] [Google Scholar]
- 14.Alvarez-Erviti L, Seow Y, Yin H, Betts C, Lakhal S, Wood MJ. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol. 2011;29(4):341–345. doi: 10.1038/nbt.1807. [DOI] [PubMed] [Google Scholar]
- 15.Morse MA, et al. A phase I study of dexosome immunotherapy in patients with advanced non-small cell lung cancer. J Transl Med. 2005;3(1):9. doi: 10.1186/1479-5876-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kamerkar S, et al. Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature. 2017;546(7659):498–503. doi: 10.1038/nature22341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Commisso C, et al. Macropinocytosis of protein is an amino acid supply route in Ras-transformed cells. Nature. 2013;497(7451):633–637. doi: 10.1038/nature12138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakase I, Kobayashi NB, Takatani-Nakase T, Yoshida T. Active macropinocytosis induction by stimulation of epidermal growth factor receptor and oncogenic Ras expression potentiates cellular uptake efficacy of exosomes. Sci Rep. 2015;5:10300. doi: 10.1038/srep10300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lalu MM, et al. Safety of cell therapy with mesenchymal stromal cells (SafeCell): a systematic review and meta-analysis of clinical trials. PLoS ONE. 2012;7(10):e47559. doi: 10.1371/journal.pone.0047559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gimona M, Pachler K, Laner-Plamberger S, Schallmoser K, Rohde E. Manufacturing of human extracellular vesicle-based therapeutics for clinical use. Int J Mol Sci. 2017;18(6):E1190. doi: 10.3390/ijms18061190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pachler K, et al. A good manufacturing practice-grade standard protocol for exclusively human mesenchymal stromal cell-derived extracellular vesicles. Cytotherapy. 2017;19(4):458–472. doi: 10.1016/j.jcyt.2017.01.001. [DOI] [PubMed] [Google Scholar]
- 22.Cullen PJ, Lockyer PJ. Integration of calcium and Ras signalling. Nat Rev Mol Cell Biol. 2002;3(5):339–348. doi: 10.1038/nrm808. [DOI] [PubMed] [Google Scholar]
- 23.Steckel M, et al. Determination of synthetic lethal interactions in KRAS oncogene-dependent cancer cells reveals novel therapeutic targeting strategies. Cell Res. 2012;22(8):1227–1245. doi: 10.1038/cr.2012.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xue W, et al. Small RNA combination therapy for lung cancer. Proc Natl Acad Sci USA. 2014;111(34):E3553–E3561. doi: 10.1073/pnas.1412686111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heidel JD, Hu S, Liu XF, Triche TJ, Davis ME. Lack of interferon response in animals to naked siRNAs. Nat Biotechnol. 2004;22(12):1579–1582. doi: 10.1038/nbt1038. [DOI] [PubMed] [Google Scholar]
- 26.Zhu X, et al. Comprehensive toxicity and immunogenicity studies reveal minimal effects in mice following sustained dosing of extracellular vesicles derived from HEK293T cells. J Extracell Vesicles. 2017;6(1):1324730. doi: 10.1080/20013078.2017.1324730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rodriguez PL, Harada T, Christian DA, Pantano DA, Tsai RK, Discher DE. Minimal “Self” peptides that inhibit phagocytic clearance and enhance delivery of nanoparticles. Science. 2013;339(6122):971–975. doi: 10.1126/science.1229568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zheng X, et al. Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature. 2015;527(7579):525–530. doi: 10.1038/nature16064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Réjiba S, Wack S, Aprahamian M, Hajri A. K-ras oncogene silencing strategy reduces tumor growth and enhances gemcitabine chemotherapy efficacy for pancreatic cancer treatment. Cancer Sci. 2007;98(7):1128–1136. doi: 10.1111/j.1349-7006.2007.00506.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma JB, Ye K, Patel DJ. Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. Nature. 2004;429(6989):318–322. doi: 10.1038/nature02519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Du Q, Thonberg H, Wang J, Wahlestedt C, Liang Z. A systematic analysis of the silencing effects of an active siRNA at all single-nucleotide mismatched target sites. Nucleic Acids Res. 2005;33(5):1671–1677. doi: 10.1093/nar/gki312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rachagani S, et al. Activated KrasG¹²D is associated with invasion and metastasis of pancreatic cancer cells through inhibition of E-cadherin. Br J Cancer. 2011;104(6):1038–1048. doi: 10.1038/bjc.2011.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Smyth GK. Limma: Linear Models for Microarray Data. In: Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W, eds. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. New York, NY: Springer; 2005:397–420. [Google Scholar]
- 34.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Source data for all figures are provided as Supplemental Source Data 1 and 2, and reagents will be provided upon availability and reasonable request.