Abstract
Obesity is a risk factor for osteoarthritis (OA), the greatest cause of disability in the US. The impact of obesity on OA is driven by systemic inflammation, and increased systemic inflammation is now understood to be caused by gut microbiome dysbiosis. Oligofructose, a nondigestible prebiotic fiber, can restore a lean gut microbial community profile in the context of obesity, suggesting a potentially novel approach to treat the OA of obesity. Here, we report that — compared with the lean murine gut — obesity is associated with loss of beneficial Bifidobacteria, while key proinflammatory species gain in abundance. A downstream systemic inflammatory signature culminates with macrophage migration to the synovium and accelerated knee OA. Oligofructose supplementation restores the lean gut microbiome in obese mice, in part, by supporting key commensal microflora, particularly Bifidobacterium pseudolongum. This is associated with reduced inflammation in the colon, circulation, and knee and protection from OA. This observation of a gut microbiome–OA connection sets the stage for discovery of potentially new OA therapeutics involving strategic manipulation of specific microbial species inhabiting the intestinal space.
Keywords: Inflammation, Microbiology
Keywords: Obesity, Osteoarthritis
Correcting obesity-related gut microbiome dysbiosis using the prebiotic agent oligofructose may provide a therapeutic strategy to treat the osteoarthritis of obesity.
Introduction
Osteoarthritis (OA), a degenerative disease primarily affecting diarthrodial joints (1), afflicts 31 million people in the US (2), with a global prevalence of disease recently estimated to exceed 250 million (3). Overall, diarthrodial/spinal OA is the most prevalent cause of disability in the US, surpassing the next 4 medical disorders combined (4), with annual medical expenditures exceeding $185 billion (4, 5). Unfortunately, the only clinically accepted treatment strategies are palliative, with no disease-modifying OA drug approved for use in humans (6). Thus, an effective treatment strategy for OA is a critical unmet need.
Obesity, a key risk factor for the development of OA, has become a global epidemic due to decreased physical activity and the consumption of the typical Western diet (7, 8). Obesity is linked to increased morbidity and mortality, with an annual financial burden projected to exceed $344 billion in the US by 2018 (9). Concurrently, the obesity epidemic drives an increased incidence of OA in both weight-bearing and nonweight-bearing joints (10, 11), with 66% of all adults diagnosed with OA being either overweight, obese, or obese/type 2 diabetic (12). While it has been generally held that joint overloading is a central cause of accelerated OA in obesity, clinical and animal findings suggest that the association is more likely linked to obesity-related increases in systemic and local inflammation (13). This is driven by migration of activated macrophages and other inflammatory cells to adipose tissue, which release Tnf and other proinflammatory cytokines into the circulation (14–16). Since global ablation of Tnf protects against progression of joint degeneration in the OA of obesity (17), one potential therapeutic approach is to reduce the systemic inflammation in this disease.
It is now established that activation of inflammation in obesity is also caused by shifts in the gut microbiome (18, 19). The composition of the gut microbiome is dependent on both intrinsic and extrinsic factors — primarily diet (20) — and plays a large role in digestion and shaping the immune system (21). Alterations in the normal microbial flora in the gut have been implicated in various pathologies, including allergic asthma (22), obesity (20, 23), type 2 diabetes (24), Parkinson’s disease (25), rheumatoid arthritis (26), and osteoporosis (27). The potential role of the gut microbiome in OA has been speculated (28, 29) but not directly investigated.
Cause-and-effect links between the gut microbiome and systemic inflammation, metabolic dysregulation, and host disease have been established using prebiotic dietary supplements to manipulate gut microbial populations (30). For example, one beneficial prebiotic, the indigestible fiber oligofructose, supports expansion of Bifidobacteria in the intestine and consequently reduces inflammation (31) and improves insulin sensitivity (32). While a recent report suggests otherwise (33), the broad literature to date indicates that the health-promoting effects of oligofructose are not caused by a direct biological or metabolic action playing out in the host (reviewed in ref. 34). Specifically, unlike digestible polysaccharides including starch, the β-2,1 covalent bonds linking fructose monomers in oligofructose are not digestible by human intestinal enzymes. Thus, oligofructose passes through the gastrointestinal tract without being metabolized or absorbed until it enters the colon, where it is fermented by various species of the resident microbial community (35–39). It follows that host effects of oligofructose and other similar prebiotics are dependent on biological actions playing out in the gut microbiome. Because of their ability to predictably alter gut microbial communities without having direct host effects, prebiotics have emerged to be important research reagents that have uncovered gut microbiome–host relationships whose targeting could have potential therapeutic efficacy in various disease conditions.
Since obesity initiates and accelerates OA by inducing chronic systemic inflammation in parallel with alteration of the gut microbiome, we propose the hypotheses that (i) a proinflammatory shift in the obese gut microbiome is causal in the OA of obesity, and (ii) reversal of this shift via oral supplementation with oligofructose will ameliorate systemic and joint inflammation, thereby mitigating the influence of obesity on the progression of OA. To address this hypothesis, a mouse model of high-fat diet–induced obesity was used in the context of trauma-induced knee OA to examine the impact of obesity and oligofructose on the gut microbiome, the colon transcriptome, circulating cytokines, joint macrophage infiltration and inflammation, and endpoint OA degeneration. Results provide the first direct evidence to our knowledge of a gut-joint connection in the OA degenerative process, and they suggest that oligofructose supplementation, via its impact on the gut microbiome, is protective against trauma-induced OA of obesity. Thus, manipulation of the gut microbiome may represent a viable disease-modifying therapeutic strategy for the most common form of OA, the OA of obesity.
Results
Oligofructose does not prevent obesity in mice consuming a high-fat diet.
The connection between obesity, the gut microbiome, and the OA of obesity was investigated in a murine model of diet-induced obesity. A 12-week high-fat diet regimen induced obesity and increased body fat percentage and insulin resistance (Supplemental Figure 1, A, B, D, E; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.95997DS1). After 12 weeks, these diets were supplemented with the undigestible prebiotic fiber oligofructose or cellulose (control fiber). While oligofructose did partially rescue glucose intolerance in obese mice (Supplemental Figure 1, D and E), it did not affect body mass or body composition (Supplemental Figure 1, A–C).
Oligofructose reverses the effect of obesity on the gut microbiome.
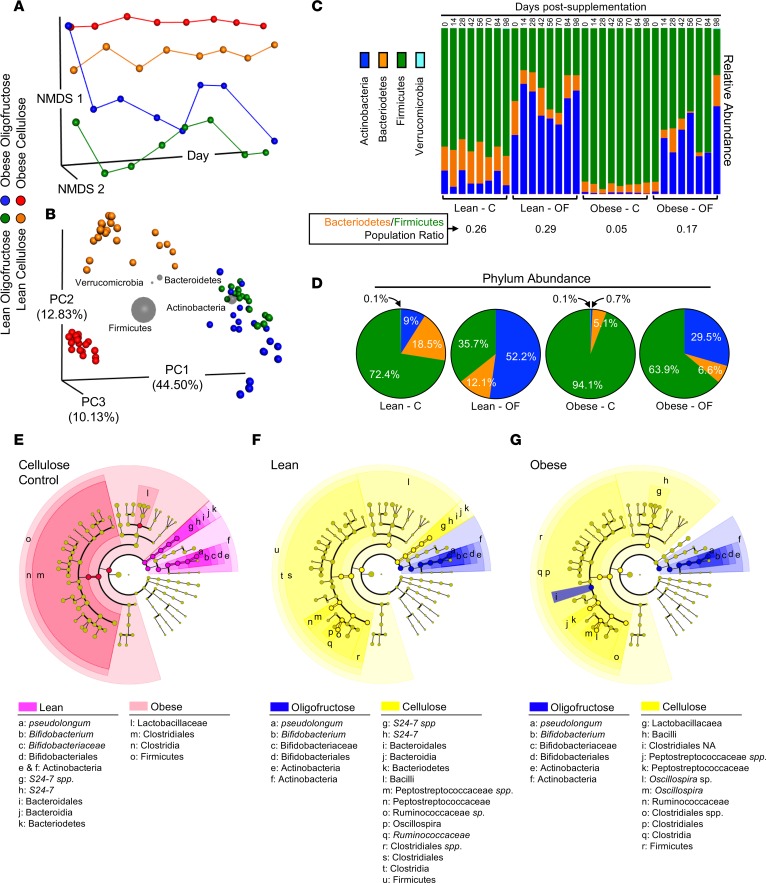
To characterize the gut microbiome in mice under various dietary and prebiotic conditions, microbiota were analyzed in fecal samples collected immediately prior to initiation of prebiotic treatment and at subsequent 2-week intervals for 98 days. Not only did obese and lean mice have distinct intestinal microbial populations, oligofructose tended to reverse key changes induced by obesity. Nonmetric multidimensional scaling (NMDS) revealed that the gut microbiome of obese and lean mice was stably distinct throughout the experiment, with oligofructose permanently shifting diversity of both groups within 2 weeks (Figure 1A). Principle coordinate analysis (PCoA) of microbial community dissimilarity identified 3 distinct profiles. Lean and obese groups were different from each other, with oligofructose altering both lean and obese microbial populations similarly, an effect that was primarily driven by the phylum Actinobacteria, as indicated by biplot analysis (Figure 1B). At the phylum level, obese mice had an increased abundance of Firmicutes with decreased Bacteriodetes and a nearly complete ablation of Actinobacteria (Figure 1C). The Bacteriodetes/Firmicutes ratio was reduced in obese compared with lean mice, a measure associated with obesity (40) and increased host inflammation (41) (Figure 1C). Notably, oligofructose increased the abundance of Actinobacteria, substantially rescuing its loss in obesity, and partially correcting the Bacteriodetes/Firmicutes ratio (Figure 1C). Since the dominant microbial phyla were stable once the prebiotic was introduced (Figure 1, A and C), absolute abundance values were averaged within each group and were represented as percentage of the entire microbial community (Figure 1D). Cladograms demonstrated that the intestinal microbiota of obese mice were dominated by Firmicutes, specifically from the order Clostridiales (Figure 1E). Conversely, lean mice were dominated by Bacteriodetes and Actinobacteria, with pronounced abundance of Bifidobacterium pseudolongum (B. pseudolongum). Oligofructose expanded Actinobacteria in both lean (Figure 1F) and obese (Figure 1G) mice, consistent with convergence of the lean and obese diversity profile in the PCoA analysis (Figure 1A). This effect was primarily due to expansion of B. pseudolongum by oligofructose in both lean and obese mice (Figure 1, F and G).
Figure 1. Oligofructose converts the obese gut microbiome to a lean diversity profile.
(A) Nonmetric multidimensional scaling (NMDS) was applied to the pairwise distance matrix between representative microbial rDNA analyses of fecal samples from lean and obese mice supplemented with cellulose (C) or oligofructose (OF). NMDS 1 and 2 portray similar samples as closer together and dissimilar samples as farther apart. The x axis represents time starting immediately prior to initiation of supplements and progressing to termination of the experiment. (B) Principal coordinate analysis was performed on all samples from each experimental group, excluding the time point prior to initiation of treatment. Each symbol represents 1 sample, with the distance between samples in 3-dimensional space corresponding to the dissimilarity between them. For each of the 4 main phyla identified, biplot analysis was performed. Gray spheres were positioned based on the weighted average of the coordinates of the samples that contained that phylum and were sized based on the weighted average of the phylum’s relative abundance in each sample. (C) The average relative abundance of phyla within in each experimental group (n = 3 at each time point) is depicted in the bar chart. The ratio of the relative abundance of Bacteriodetes to Firmicutes for each group was calculated and averaged over all nonzero time points, as was the percent abundance of the 4 most prevalent phyla (D). Cladograms were constructed showing taxa that are significantly enriched in lean versus obese comparison (E), the lean-C versus lean-OF comparison (F), and the obese-C versus obese-OF comparison (G).
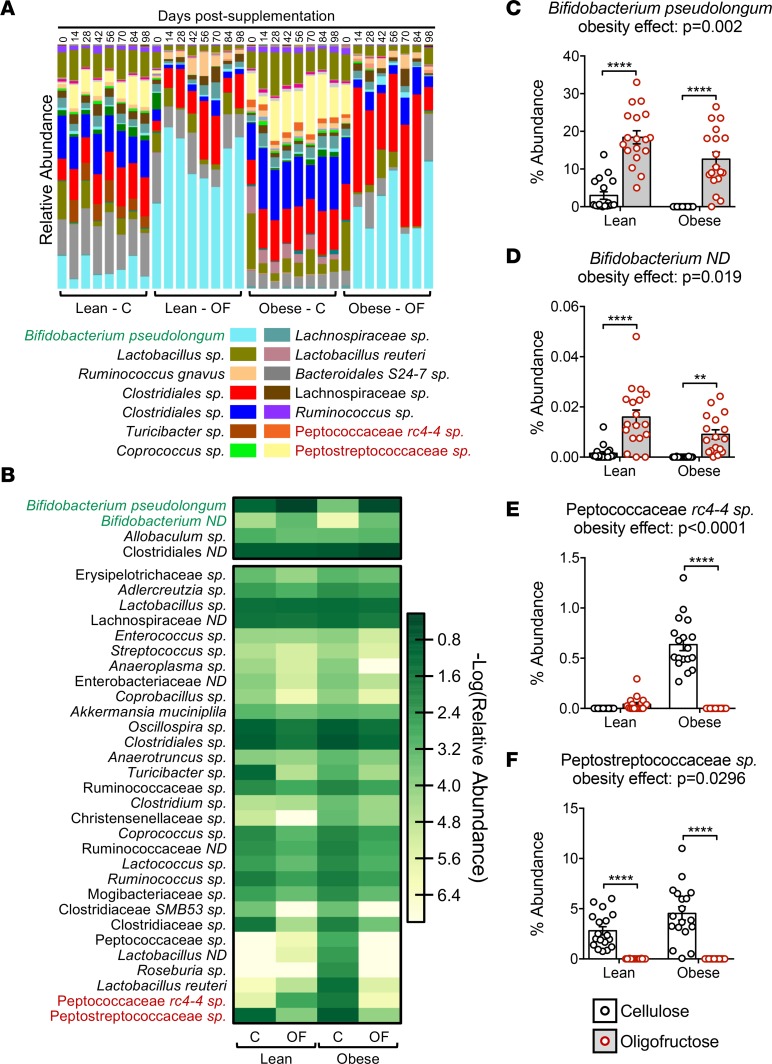
Deeper analysis of species-level abundance and diversity confirmed the findings depicted in the cladograms. Particularly, changes in Actinobacteria abundance (Figure 1C) were almost entirely driven by changes in B. pseudolongum (Figure 2, A–C). Compared with abundance of B. pseudolongum in lean mice, obesity was associated with complete loss of this species (Figure 2, A–C). Oligofructose enhanced this population in lean animals and rescued the obesity-related loss, raising abundance levels more than 1,000-fold (Figure 2C). An uncharacterized Bifidobacterium species (Bifidobacterium ND) showed a similar pattern (Figure 2, B and D). Since Bifidobacteria are associated with positive health effects, including increased gut barrier integrity and reduced inflammation (42), these shifts suggest protection against obesity-induced intestinal and systemic effects. The heatmap also revealed increased abundance of numerous proinflammatory bacteria in obese mice (Figure 2B). Of particular interest is an unidentified species in the family Peptostreptococcaceae, and 2 members of the Peptococcaceae family (rc4-4 sp and another unidentified species). All 3 are associated with obesity (43) and intestinal inflammation (44). Remarkably, all of these microbes were reduced with oligofructose (Figure 2B), particularly Peptococcaceae rc4-4 sp (Figure 2E) and Peptostreptococcaceae sp (Figure 2F).
Figure 2. B.pseudolongum is a key microbe lost in obesity and restored following OF supplementation.
(A) Average species abundance was determined in fecal samples collected across the various time points (n = 3/time point), with relative average abundance for detected species depicted in the bar chart. (B) A heatmap was constructed comparing abundance of species that were either decreased in obesity and significantly restored following oligofructose supplementation (top segment) or increased in obesity and significantly suppressed following oligofructose supplementation (bottom segment). Species highlighted in green or red in A and B are presented quantitatively, with summary statistics calculated for Bifidobacterium pseudolongum (C), an uncharacterized species in the genus Bifidobacterium (D), an unidentifiable species in the family Peptococcaceae (E), and an unidentifiable species in the family Peptostreptococcaceae (F). Since microbial diversity did not significantly change temporally after initiation of prebiotic supplementation (Figure 1A), abundance data for all time points from 14 days and after were averaged for each experimental group for each of these 4 species in order to perform statistical analysis. Significant differences between groups were identified via 2-way ANOVA with a Tukey multiple comparison post-test (P values for the obesity effect are reported; **P < 0.01, ****P < 0.0001).
Obesity effects on the colon transcriptome are modulated by oligofructose.
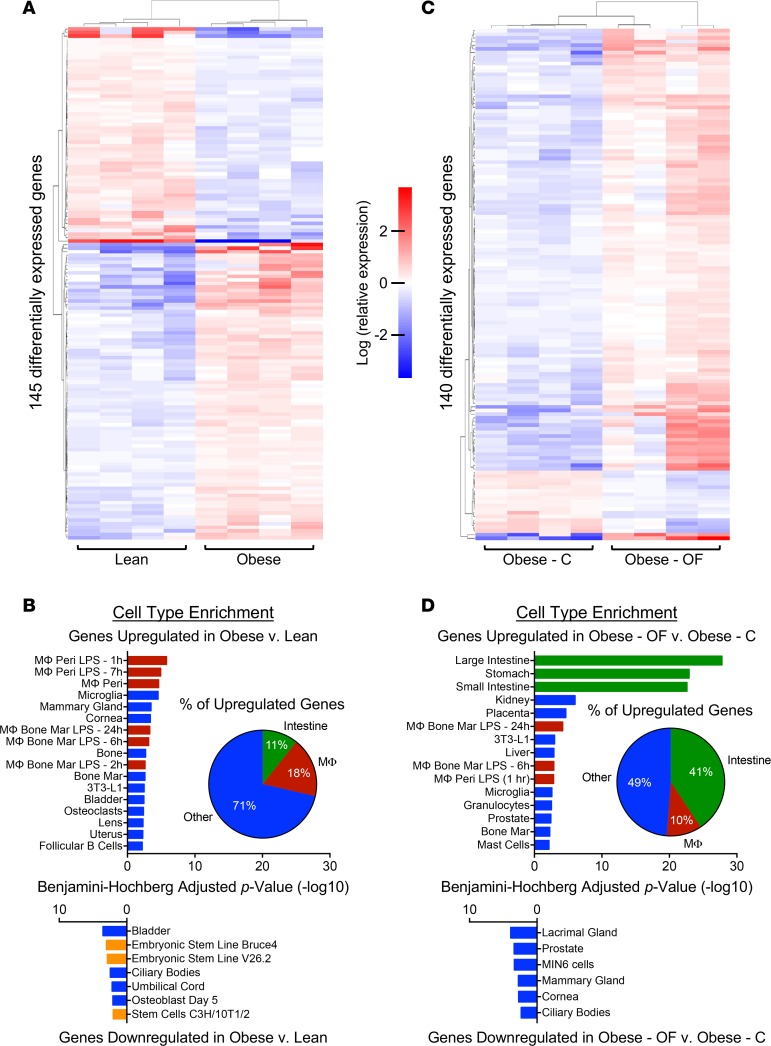
RNA sequencing (RNAseq) was performed on colonic tissues harvested from all treatment groups. Differential expression analysis between lean and obese mice identified 145 differentially expressed genes (Figure 3A and Supplemental Table 1). Cell-type enrichment analysis revealed that 18% of genes upregulated in obesity have a role in the macrophage lineage, suggesting increased macrophage activity in the obese state (Figure 3B). Conversely, genes downregulated in the signature annotate to stem-like cell types in the annotation (Figure 3B). Interestingly, while oligofructose had a significant impact on the obese colon transcriptome, it did not directly reverse the signature depicted in Figure 3, A and B. Overall, there were 140 differentially expressed genes in obese mice supplemented with oligofructose versus cellulose, with 123 genes upregulated and 17 genes downregulated (Figure 3C and Supplemental Table 2). While 10% of the upregulated genes in the oligofructose group continued to suggest macrophage enrichment, nearly half (41%) identified a significant increase in a digestive tract cell type signature (Figure 3D). Examples of genes annotating to this signature include Grp, a stimulator of intestinal epithelium proliferation (45), and Aqp4, a member of a family of genes involved in water reabsorption and protection against enteritis and inflammatory bowel disease (46); they were both increased in the oligofructose-supplemented group (Supplemental Table 2 and Supplemental Figure 2). Reg3g, a bactericidal lectin targeting pathogenic gram-positive bacteria at the intestinal epithelial surface (47), was also upregulated (Supplemental Table 2 and Supplemental Figure 2), as was Cdx2, a transcription factor that targets cell adhesion genes and whose activities are important for maintaining a normal intestinal epithelial barrier (48) (Supplemental Table 2 and Supplemental Figure 2). This latter effect is consistent with the observation that oligofructose-supplemented lean and obese mice had significantly reduced serum endotoxin levels (Supplemental Figure 3D). In aggregate, these findings suggest that oligofructose supplementation supports various intestinal epithelial functions, including gut barrier integrity.
Figure 3. Colon RNAseq reveals a macrophage signature in the obese gut, with mitigation of these effects by oligofructose.
Transcriptome analysis of RNA extracted from colons of lean and obese mice with 4-weeks supplementation with cellulose (C) or oligofructose (OF). (A) The heatmap depicts clustering of 145 differentially expressed genes in obese compared with lean mice. Each column represents the expression profile of an individual mouse in each experimental group. (B) Cell type enrichment analysis was performed, depicting the most represented cell type signatures in the up- and downregulated gene sets. The percent of genes upregulated in obese colon associated with the intestinal, macrophage (MΦ), or other cell clusters is depicted in the inset pie chart. (C) The heatmap depicts clustering of 140 differentially expressed genes in obese-OF compared with obese-C. Each column represents the expression profile of an individual mouse in each experimental group. (D) Cell type enrichment analysis was performed, identifying the most represented cell type signatures in the up- and downregulated gene sets. The percent of genes upregulated in obese colon associated with the intestinal, MΦ, or other cell clusters is depicted in the inset pie chart. In B and D, the enrichment signature for intestinal cell types is denoted with green bars, macrophages with red bars, and stem cells with orange bars.
While various circulating short-chain fatty acids were not significantly altered under any experimental conditions (Supplemental Figure 3, A–C), this latter effect is consistent with the observation that oligofructose-supplemented lean and obese mice had significantly reduced serum endotoxin levels (Supplemental Figure 3D).
Oligofructose-supplemented obese mice have reduced systemic inflammation.
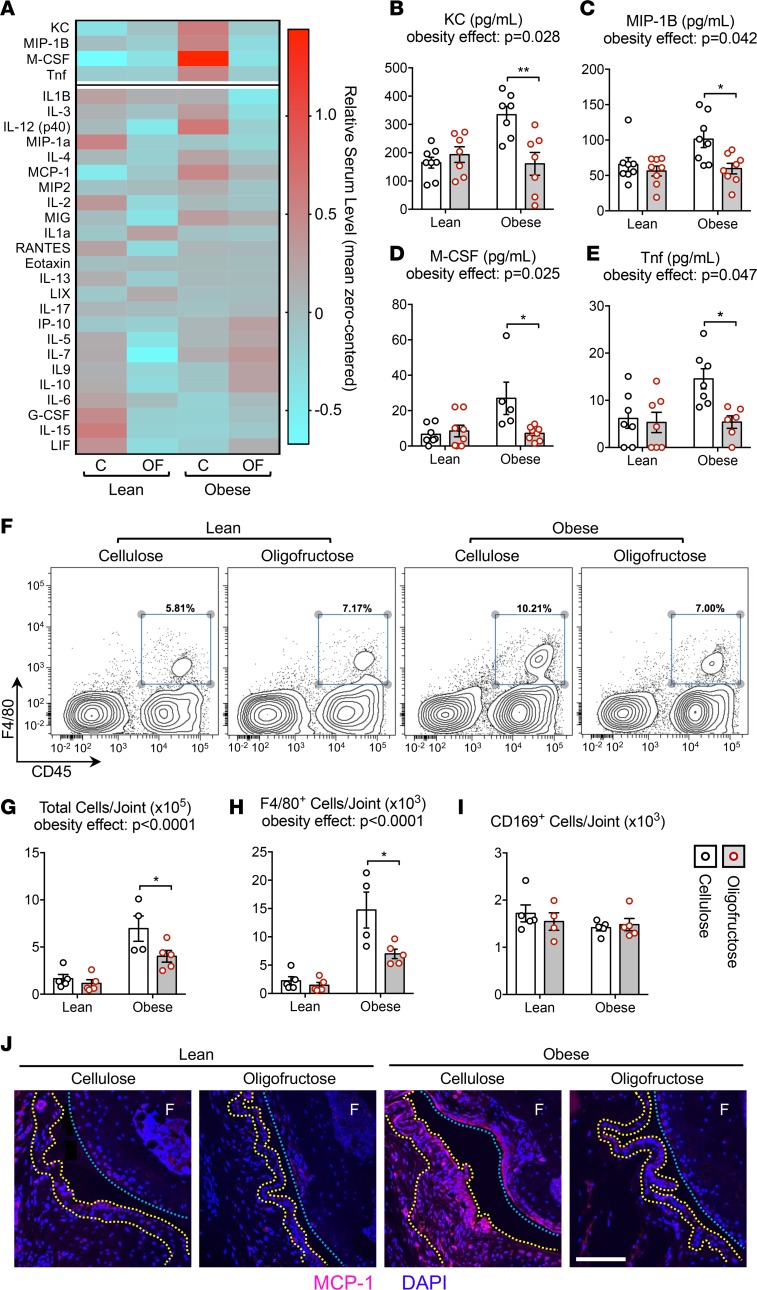
To determine if the microbiome and colon effects seen in mice supplemented with oligofructose were associated with changes in systemic inflammation in the context of obesity, we analyzed serum cytokine levels using a multiplex assay (Figure 4A). Four cytokines were significantly increased in obese mice that were also significantly reduced in the oligofructose-supplemented group, including KC (mouse homolog of IL-8), MIP-1B, M-CSF, and Tnf (Figure 4, B–E). Each has been implicated in rodent models of obesity (49). The proinflammatory mediators IL-12 and MCP-1 trended lower with oligofructose in obese mice, and the anti-inflammatory cytokine IL-10 trended upward. It warrants mentioning that IL-6, a proinflammatory cytokine involved in both obesity and OA, was not elevated in obese mice compared with lean controls. This is due to a large variance in the IL-6 datasets from each experimental group, with several individual outliers greater than 3 SDs away from the mean for each group (data not shown). Overall, these results suggest that oligofructose mitigates the systemic inflammation caused by high-fat diet–induced obesity, with several key cytokines significantly reduced following prebiotic supplementation.
Figure 4. Oligofructose mitigates systemic and synovial inflammation in obese mice.
(A) Serum harvested from all conditions were analyzed by multiplex ELISA-based assay, with results plotted in the heatmap. Columns depicts the average serum cytokine level (n = 6–8 mice) for each experimental condition. Color scaling was mean zero centered. The most significantly increased cytokines in obese compared with lean mice were further analyzed, with group effects for KC (B), MIP-1B (C), M-CSF (D), and Tnf (E) depicted in bar graphs. (F) PAST cells isolated from 4–5 individual knees, 4-weeks after lean or high-fat diet initiation, were subjected to flow cytometry. Representative histograms are presented, with F4/80 and CD45 double-positive populations gated (blue box) for quantification. Summary analysis of the results are shown in bar graphs depicting group effects on total number of cells recovered per joint (G), number of F4/80+ activated macrophages per joint (H), and number of CD169+ tissue resident macrophages per joint (I). (J) Sagittal tissue sections from contralateral knees were subjected to immunofluorescence detection of MCP-1, with representative staining presented. Nuclei are counterstained with DAPI, yellow dotted lines outline the synovial membrane, and blue dotted lines demarcate the articular cartilage surface on the femur (F). Scale bar: 50 μm. Significant differences between groups in (B–E) and (G–I) were identified via 2-way ANOVA with a Tukey multiple comparison post-test (P values for the obesity effect are reported; *P < 0.05, **P < 0.01).
Obesity-associated macrophage migration to the synovium is reduced in oligofructose-supplemented mice.
To analyze joint-specific inflammation, flow cytometry was performed on periarticular soft tissue (PAST) cells isolated from knee joints (Figure 4F). Confirming that obesity is associated with increased synovial hyperplasia and inflammation in mice (17, 50) and humans (17), obese mice displayed increased cell number (Figure 4G) and a 5-fold increase in the number of F4/80+/CD45+ infiltrating macrophages (Figure 4H). Remarkably, when supplemented with oligofructose, obese mice were significantly rescued from these proinflammatory changes (Figure 4, G and H). CD169+/CD45+ resident tissue macrophage numbers were not impacted by obesity or prebiotic (Figure 4I), suggesting migration of monocytes and/or macrophages to the knee in obesity. Consistent with this, MCP-1, a chemokine that recruits circulating inflammatory cells to the joint and has been implicated in posttraumatic OA (51), trended upward in serum from obese mice with apparent correction by oligofructose (Figure 4A). Mirroring this systemic profile, knee joint–specific MCP-1 levels were increased in the synovium of obese mice and suppressed by oligofructose (Figure 4J). Overall, these data suggest that the proinflammatory impact of obesity is substantially mitigated by oligofructose.
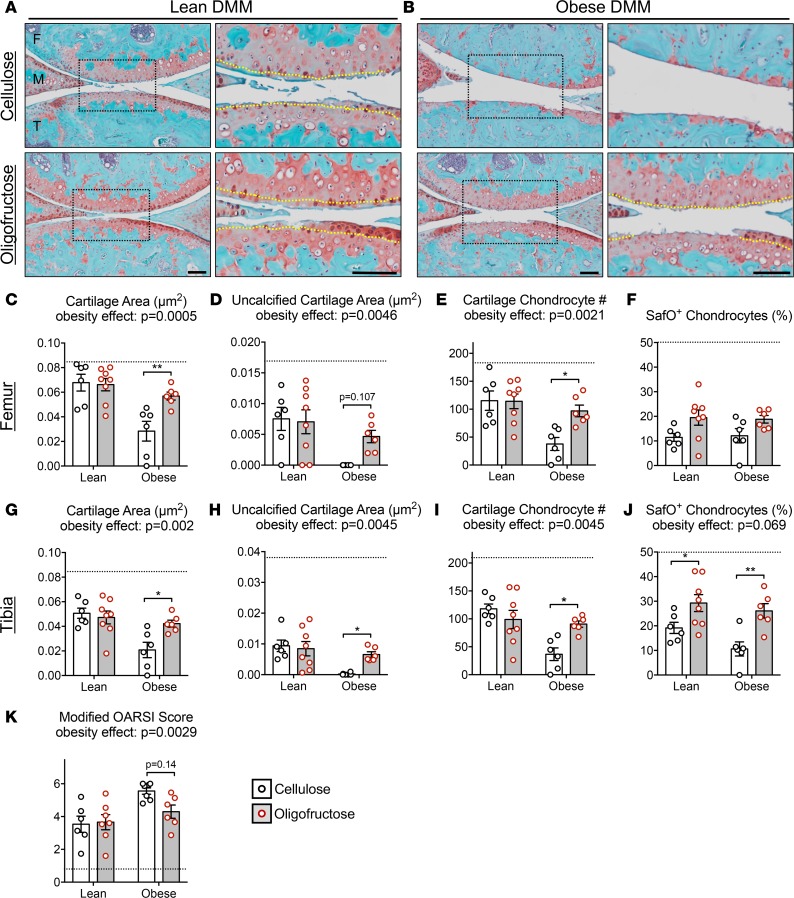
Oligofructose prevents cartilage loss in the OA of obesity.
To model the pathological process of the OA of obesity in humans (13), we employed the standard destabilization of the medial meniscus (DMM) (52) mouse injury to initiate OA, and we examined the impact of obesity on the progression of joint degeneration in this context. In obese mice supplemented with cellulose, the OA was more progressed than in lean mice, with most joints terminally degenerated (Figure 5, A and B). Cartilage and chondrocyte loss was more significant in every metric examined (Figure 5, C–K). Oligofructose did not impact the degenerative process in lean mice, except for percent of Safranin O+ chondrocytes (Figure 5J). Remarkably, oligofructose-supplemented obese mice were completely rescued from the deleterious effect of obesity (Figure 5B). With the exception of percent Safranin O+ chondrocytes on the femoral condyle (Figure 5F), every histomorphometric outcome was significantly improved to levels similar to those seen in lean controls (Figure 5, C–E, G–J), with Osteoarthritis Research Society International (OARSI) scores also trending toward improvement (Figure 5K). Note that, without injury, early joint degeneration was only discernible in a small percentage of obese mice with no significant change in any metric collected (Supplemental Figure 4). Based on these results, oligofructose provides nearly complete protection against the trauma-induced OA of obesity, suggesting its potential as a disease modifier in this context.
Figure 5. Oligofructose protects against accelerated posttraumatic osteoarthritis (PTOA) in obesity.
Two weeks after lean and obese mice were initiated on cellulose or oligofructose, sham or DMM surgery was performed and knee joints were collected 12 weeks later. Representative Safranin O/Fast Green stained sections in lean (A) and obese (B) mice are presented, with black dotted boxes denoting areas that are shown at higher magnification to the immediate right (scale bars: 100 μm) and yellow dotted lines demarcating the tide marks. Sections like those shown in A and B were used to perform histomorphometry on femur (C–F) and tibia (G–J) articular cartilage, as well as OARSI scoring (K). Black dotted lines in the bar graphs depict the value for the respective Sham-lean-cellulose negative control, bars represent the group mean (± SEM), and individual symbols depict the average of measurements made in 3 histologic levels from 6–8 individual joints. Significant differences between groups in C–K were identified via 2-way ANOVA with a Tukey multiple comparison post-test (P values for the obesity effect are reported when significance was achieved; *P < 0.05, **P < 0.01). Similar histology and histomorphometry performed in Sham-operated joints is presented in Supplemental Figure 4.
Oligofructose suppresses obesity-induced molecular, cellular, and joint structure change in the context of injury.
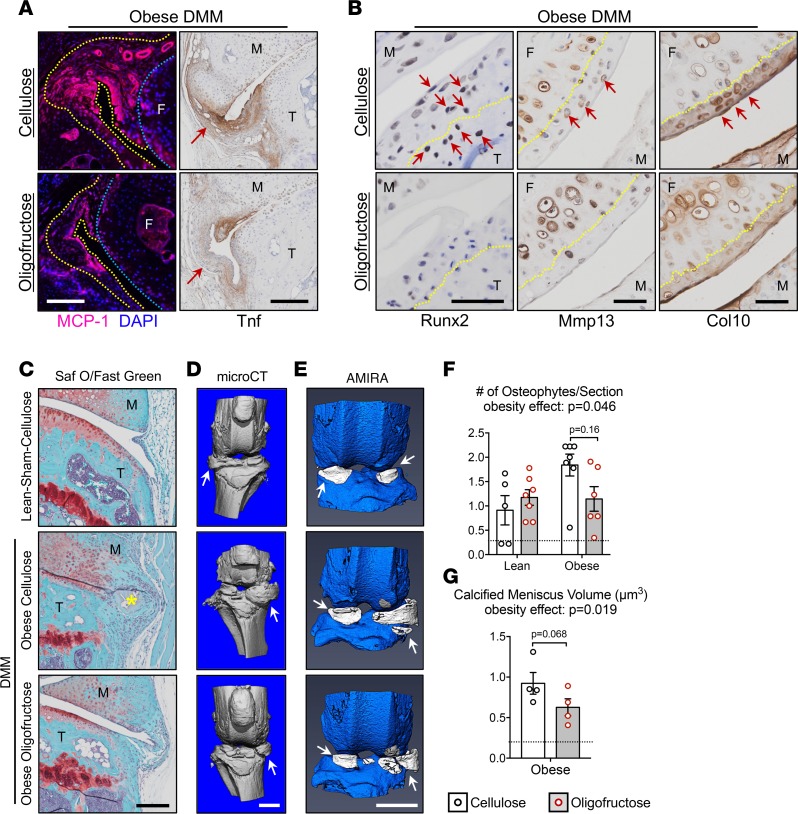
To extend findings related to cartilage architecture reported in Figure 5, joint inflammation in obese mice that were administered DMM injury was further examined. The oligofructose-associated reduction of MCP-1 that was identified in synovium of obese mice (Figure 4J) was also observed in injured joints (Figure 6A and Supplemental Figure 5B). These effects parallel the reduction in macrophage homing to the obese joint documented in Figure 4H. Consistent with the role of MCP-1 as a macrophage chemoattractant, synovial Tnf in obese DMM mice was increased (Figure 6A) relative to lean controls (Supplemental Figure 5A), with oligofructose reversing this inflammatory response (Figure 6A). Even without injury, obese mice showed increased circulating Tnf (Figure 4A) and increased synovial Tnf (Supplemental Figure 5B), as we previously observed in obese mice and humans (17), with effective suppression of these effects observed in mice supplemented with oligofructose (Figure 4, A, H–J, and Supplemental Figure 5B). These findings establish that the joint protective effects of oligofructose in the OA of obesity are aligned with suppression of a MCP-1/Tnf inflammatory profile and early joint changes.
Figure 6. Oligofructose mitigates obesity-induced synovial inflammation, chondrocyte hypertrophy, and meniscal mineralization.
(A) To assess the inflammatory signature of joints of obese mice supplemented with oligofructose or cellulose, MCP-1 and Tnf expression was examined. MCP-1 was detected via immunofluorescence, with DAPI counterstaining of nuclei. The blue dotted lines outline articular cartilage on the femoral condyle (F, femur), the yellow dotted lines demarcate the margins of the synovial membrane, and the white scale bar represents 50 µm. Tnf expression was assessed via IHC, with synovial staining identified with red arrows and black scale bars representing 200 µm (M, meniscus; T, tibia). (B) To evaluate articular chondrocyte hypertrophic differentiation and cartilage matrix changes, Runx2, Mmp13, and Col10 expression was examined via IHC, with yellow dotted lines demarcating the tidemarks, red arrows pointing to chondrocytes or areas with positive staining, and black scale bars representing 50 µm (M, meniscus; T, tibia; F, femur). (C) Representative Safranin O/Fast Green–stained sections depict posterior medial joint margins with an osteophyte identified by a yellow asterisk in the obese-DMM-cellulose group and the black scale bar representing 200 µm (M, meniscus; T, tibia). (D) MicroCT imaging reveals mineralized structures in experimental joints, with mineralized menisci identified with white arrows and the white scale bar representing 1 mm. (E) Amira-based reconstruction of the raw dicoms from the microCT imaging was performed to enable quantitative segmentation of calcified menisci colored white with the white scale bar representing 1 mm. (F) Histology sections similar to those shown in C were used to quantify the number of osteophytes per histologic section in each experimental group. Data represent the group mean (± SEM), each symbol represents the average number of osteophytes per section from 3 levels within a joint (n = 5–7 joints), and the black dotted line represents the average in the lean-sham-cellulose negative control group. (G) Amira reconstructions similar to those depicted in E were used to quantify the volume of calcified meniscus in various experimental groups. Data represent the group mean (± SEM), each symbol represents the average calcified meniscus volume from each joint (n = 4 joints), and the black dotted line represents the average in the lean-sham-cellulose negative control group. Significant differences between groups in F and G were identified via 2-way ANOVA with a Tukey multiple comparison post-test (P values for the obesity effect are reported).
It is established that OA involves inappropriate hypertrophic differentiation of articular chondrocytes (1). Consistent with this, Runx2 — a key transcription factor driver of chondrocyte hypertrophy that is upregulated in OA chondrocytes (53) — was detected in chondrocytes residing in both the uncalcified and calcified zones of the articular cartilage of obese mice with (Figure 6B) and without injury (Supplemental Figure 6A). Comparatively, Runx2 was not detected in the articular chondrocytes of obese mice supplemented with oligofructose in both uninjured (Supplemental Figure 6A) and DMM (Figure 6B) groups. Given the apparent increase in expression of this master regulator of chondrocyte differentiation in injury and obesity, it follows that the uncalcified cartilage of lean DMM mice showed increased Mmp13 positivity (Supplemental Figure 6B), with a trend toward a greater increase observed in both injured (Figure 6B) and uninjured (Supplemental Figure 6B) obese mice. Aligned with the chondroprotection reported in Figure 5, Mmp13 appeared to be reduced in oligofructose-supplemented groups regardless of obesity or injury status (Figure 6B and Supplemental Figure 6B). Similarly, a trend toward increased Col10 staining was observed in uncalcified cartilage matrix in lean and obese DMM mice, as well is in uninjured (sham) obese mice (Figure 6B and Supplemental Figure 6C), and was reduced with oligofructose supplementation in obese mice — again regardless of obesity or injury status (Figure 6B and Supplemental Figure 6B). In summary, these findings establish that the chondroprotective effects of oligofructose in the OA of obesity are paralleled by a reduction in the expression of the hallmarks of chondrocyte hypertrophy and cartilage degeneration, Runx2, Mmp13, and Col10.
Consistent with our previous reports (54, 55), knee joints from obese mice have an increased prevalence of osteophytes (Figure 6C) and expanded mineralization of menisci (Figure 6D). While obesity increased osteophyte prevalence, oligofructose tended to decrease the number of osteophytes detected per joint (Figure 6, C and F). Moreover, microCT reconstructions indicated that menisci in obese DMM mice supplemented with oligofructose were less mineralized than obese controls (Figure 6, D and G). An Amira-based imaging technique to segment mineralized menisci (Figure 6E) and quantify their volume revealed a strong trend toward decreased calcified meniscus volume in oligofructose-supplemented DMM joints (Figure 6G). This is further evidence that obesity-induced degeneration of the joint can be suppressed by oligofructose.
Discussion
The growing global prevalence of OA makes the current lack of a disease modifying therapy a critical unmet need. Noteworthy is the absence of clinical trials to address the OA of obesity, a disease process distinct from other forms of OA (56, 57) that accounts for more than half of all OA burden (12). The major driver of the OA of obesity is increased joint inflammation (13, 50, 58) downstream of systemic inflammation that is a central mediator of obesity’s comorbidities (49, 59–61), and we have shown that reducing Tnf-associated inflammation in obesity is profoundly protective against OA in mice (17). Thus, development of strategies that mitigate obesity-related inflammation could be an effective disease-modifying approach to treat OA of obesity.
It is now recognized that the gut microbiome can have a profound influence on systemic inflammation and chronic disease (62). Roles of key microbial species in inducing inflammation or protecting from it have already been established (63), and antagonizing proinflammatory species has been proposed as a disease-modifying strategy in pathologic conditions including obesity (19, 20, 23, 64) and several skeletal disorders including steroid-related bone loss (27) and rheumatoid arthritis (65). While the gut microbiome has been superficially analyzed in a diet-induced obesity model of OA (28) and has been speculated as a possible player in OA pathogenesis (29), a causal link between gut flora and OA has not been established.
In this report, we show for the first time to our knowledge that correction of the obese gut microbiome can protect against the OA of obesity. Consistent with previous work (18, 19), induction of obesity via consumption of a high-fat diet led to a proinflammatory shift in the gut microbiome (Figures 1 and 2). Key changes included ablation of beneficial microbes from the genus Bifidobacterium (66) coupled with increased abundance of Peptostreptococcaceae sp and 2 members of the Peptococcaceae family, which are associated with obesity and intestinal inflammation (43, 44). This proinflammatory microbial shift coincided with an increased macrophage signature and reduced stem cell signature in the intestine (Figure 3, A and B), increases in key proinflammatory cytokines in the circulation (Figure 4, A–E), increased macrophage presence in the knee capsule (Figure 4, F–I), and upregulation of the monocyte chemokine MCP-1 in synovial tissue (Figure 4J). These processes were linked to accelerated cartilage degeneration, chondrocyte hypertrophy, formation of osteophytes, and mineralization of the menisci in the context of injury-accelerated disease (Figure 5 and 6). Prebiotic alteration of the gut microbiome via dietary supplementation with the indigestible fiber oligofructose mitigated this chain of events. Starting in the gut, oligofructose reestablished Bifidobacteria that were lost in obesity, particularly B. pseudolongum, and ablated Peptostreptococcaceae sp, Peptococcaceae rc4-4 sp, and Peptococcaceae sp (Figures 1 and 2). This was associated with support of intestinal cell types and key genes associated with epithelial proliferation, water reabsorption, and barrier function (Supplemental Figure 2), as well as reduced circulating endotoxin (Supplemental Figure 3D) and obesity-associated inflammatory cytokines (Figure 4, A–E) and protection against infiltration of macrophages into the joint capsule of obese mice (Figure 4, F–J). These effects were all upstream of an essentially complete rescue of the obesity effect on OA progression in DMM-injured mice from the perspective of cartilage architecture (Figure 5), joint inflammation (Figure 6A), chondrocyte hypertrophy (Figure 6B), osteophyte formation (Figure 6, C and F), and mineralization of the meniscus (Figure 6, D, E, and G). This protection was evident despite continued consumption of the high-fat diet and persistence of obesity (Supplemental Figure 1). While structural OA changes were not robust in obese mice that were uninjured, molecular evidence of obesity-induced joint inflammation and chondrocyte hypertrophy was mitigated following oligofructose supplementation in this context, as well (Supplemental Figure 5B and Supplemental Figure 6).
These results not only provide direct evidence that gut microbial shifts can influence inflammation and degeneration in diarthrodial joints, but they support prebiotic manipulation of the gut microbiome as the first disease modifying therapeutic strategy to our knowledge to address the OA of obesity. At every link in the chain of events connecting the influence of oligofructose on the microbial communities in the gut to protection against degeneration in the joint, there is a generalized and consistent reduction of the inflammatory signature induced by obesity. Given the broad literature indicating that effects of prebiotics like oligofructose on host biology and disease occur via alterations in the microbial community in the colon (34–39), we suggest that this is the primary mechanism underlying the therapeutic efficacy of oligofructose in the OA of obesity, which begins with expansion in the abundance of Bifidobacteria at the expense of numerous microbes implicated as proinflammatory. Deeper study of the give and take between microbial abundance in the context of obesity and under the influence of oligofructose is warranted. Particular focus on the possible influence of the metabolic and transcriptomic signatures of dominant species that could be impacting host immune system function and overall physiology in ways that have not been previously understood is an important future direction.
It warrants mention that oral supplements composed of cartilage and soft tissue matrix components, most notably glucosamine and chondroitin sulfate, are the only agents with clinical data supporting positive patient-reported functional improvement in OA (67–69). However, the validity of these studies is hotly debated (70, 71), largely due to lack of consensus on mechanism of action. Given results presented here, we speculate that positive results from glucosamine and chondroitin sulfate clinical trials in OA may be due to a never-before appreciated action of these agents as prebiotics that can affect the gut microbiome.
In summary, this report establishes that the OA of obesity is an inflammatory process driven by obesity-related dysbiosis of the gut microbiome that can be treated by restoring a healthy microbial community using the indigestible prebiotic fiber oligofructose. The oligofructose-induced changes in the abundance of key microbes in the obese gut are associated with colonic expression of genes associated with an intestinal cell-type annotation, while reducing the colonic macrophage cell signature, reducing systemic and knee joint inflammation, preserving articular cartilage, and protecting against OA in obese mice. In addition to suggesting a previously unappreciated connection between the gut microbiome and joints, findings reported here suggest that manipulation of the gut microbiome is a potentially novel candidate approach to address the OA of obesity, a clinical problem of massive global scope that is without an accepted disease-modifying therapy.
Methods
Animals.
C57BL/6J mice were purchased from The Jackson Laboratory and were housed in groups of 3–4 mice per micro-isolator cage in a room with a 12-hour light/dark schedule. At 5 weeks of age, mice were provided ad lib access to either a low-fat diet (Lean, 10% kcal from fat, Open Source Diets, D12450B) or a high-fat diet (Obese, 60% kcal from fat, Open Source Diets, D12492). At 17 weeks of age, mice were then offered ad lib access to Lean or Obese diet supplemented with either a control fiber (Cellulose, 10% w/w, JRS Pharma, VIVAPUR) or prebiotic (Oligofructose, 10% w/w, Beneo-Orafti, Orafti P95). After 2 weeks on supplemented diets, mice were administered DMM surgery to the right knee and sham surgery to the left knee as described (52). Briefly, mice were anesthetized via i.p. injection of 60 mg/kg ketamine (Ketalar, Par Pharmaceutical Inc.) and 4 mg/kg xylazine (AnaSed, Akorn Animal Health), and a 5-mm-long incision was made on the medial side of the knee. Under a dissecting microscope, an incision was made along the medial side of the patellar tendon, opening the joint space. Using a #11 scalpel, the medial meniscotibial ligament (MMTL) was transected, enabling the medial meniscus to move freely. A similar skin incision was made in the contralateral sham joint, but the MMTL was left intact. After surgery, 4-0 silk sutures were used to close the incision using an interrupted pattern. Mice were provided analgesia via i.p. injection of buprenorphine (0.5 mg/kg) every 12 hours for 72 hours, and sutures were removed after 7 days.
Glucose tolerance test (GTT).
Twelve weeks after DMM surgery, mice were fasted for 8 hours, and body weights were measured. Fasting glucose levels were measured in blood samples harvested from the tail vein using a commercially available glucometer (One Touch Ultra; Lifescan). A glucose bolus was delivered i.p. (200 mg/kg), and blood glucose levels were measured at 15, 30, 60, and 90 minutes after injection. To quantify the metabolic condition, an AUC was calculated using GraphPad Prism software (version 7), based on the curve of glucose levels on the GTT in each mouse.
Body composition.
Body fat percentage and lean mass were determined via Dual Energy X-ray Absorptiometry (DEXA) scanning (PIXImus2 Mouse Densitometer; GE Medical Systems). Twelve weeks after DMM surgery and prior to sacrifice for tissue harvest, mice were anesthetized via i.p. injection of ketamine (60 mg/kg) and xylazine (4 mg/kg), and scans were collected and analyzed using a previously described standard method (72).
16S rRNA bacterial sequencing.
Fecal pellets were freshly harvested from mice after scruffing and immediately frozen at –80°C. DNA was extracted using the ZR Fecal DNA Extraction Kit (Zymo Research) as directed by the manufacturer. 16S ribosomal DNA (rDNA) was amplified with Phusion High-Fidelity polymerase (Thermo Fisher Scientific) and dual indexed primers specific to the V3-V4 hypervariable regions (319F: 5′ ACTCCTACGGGAGGCAGCAG 3′; 806R: 3′ ACTCCTACGGGAGGCAGCAG 5′). Amplicons were pooled and paired-end sequenced on an Illumina MiSeq (Illumina) in the University of Rochester Genomics Research Center. Each sequencing run included: positive controls consisting of a 1:5 mixture of Staphylococcus aureus, Lactococcus lactis, Porphyromonas gingivalis, Streptococcus mutans, and Escherichia coli, and negative controls consisting of sterile saline. Raw data from the Illumina MiSeq was first converted into FASTQ format 2 × 300 paired-end sequence files using the bcl2fastq program, version 1.8.4, provided by Illumina. Format conversion was performed without demultiplexing, and the Extended Adaptive Multi-mode Search (EAMMS) algorithm was disabled. All other settings were default. Sequence processing and microbial composition analysis were performed with the Quantitative Insights into Microbial Ecology (QIIME) software package (73), version 1.9.1. Reads were multiplexed using a configuration described previously (74). Briefly, for both reads in a pair, the first 12 bases were a barcode, which was followed by a primer, then a heterogeneity spacer, and then the target 16S rRNA sequence. Using a custom Python script, the barcodes from each read pair were removed, concatenated together, and stored in a separate file. Read pairs were assembled using fastq-join from the ea-utils package, requiring at least 40 bases of overlap and allowing a maximum of 10% mismatched bases. Read pairs that could not be assembled were discarded. The concatenated barcode sequences were prepended to the corresponding assembled reads, and the resulting sequences were converted from FASTQ to FASTA and QUAL files for QIIME analysis. Barcodes, forward primer, spacer, and reverse primer sequences were removed during demultiplexing. Reads containing more than 4 mismatches to the known primer sequences or more than 3 mismatches to all barcode sequences were excluded from subsequent processing and analysis. Assembled reads were truncated at the beginning of the first 30-base window with a mean Phred quality score of less than 20 or at the first ambiguous base — whichever came first. Resulting sequences shorter than 300 bases or containing a homopolymer longer than 6 bases were discarded. Operational taxonomic units (OTU) were picked using the reference-based USEARCH (version 5.2) (75) pipeline in QIIME, using the May 2013 release of the GreenGenes 99% OTU database as a closed reference (76, 77). An indexed word length of 128 and otherwise default parameters were used with USEARCH. Chimera detection was performed de novo with UCHIME, using default parameters (75). OTU clusters with fewer than 4 sequences were removed, and representative sequences used to make taxonomic assignments for each cluster were selected on the basis of abundance. The RDP Naive Bayesian Classifier was used for taxonomic classification with the GreenGenes reference database, using a minimum confidence threshold of 0.85 and otherwise default parameters (78).
Colon RNAseq.
At sacrifice, colon tissue was harvested, residual fecal material was removed, and tissue was frozen at –80°C. Total RNA was extracted using the GeneJET RNA purification kit (Thermo Fisher Scientific) per manufacturer’s instructions, and its concentration was determined with the NanopDrop 1000 spectrophotometer and RNA quality assessed with the Agilent Bioanalyzer. The TruSeq Stranded mRNA Sample Preparation Kit (Illumina) was used for next-generation sequencing library construction per manufacturer’s protocols. Briefly, mRNA was purified from 200 ng total RNA with oligo-dT magnetic beads and fragmented. First-strand cDNA synthesis was performed with random hexamer priming followed by second-strand cDNA synthesis using dUTP incorporation for strand marking. End repair and 3′ adenylation was then performed on the double-stranded cDNA. Illumina adaptors were ligated to both ends of the cDNA, purified by gel electrophoresis and amplified with PCR primers specific to the adaptor sequences to generate cDNA amplicons of approximately 200–500 bp in size. The amplified libraries were hybridized to the Illumina single-end flow cell and amplified using the cBot (Illumina). Single-end reads of 100 nt were generated for each sample using Illumina’s HiSeq2500v4. The approach used for data processing and alignment can be found in Supplemental Material.
qPCR analysis.
cDNA used to perform colon RNAseq was also used to perform validation quantitative PCR (qPCR) for a small subset of genes of interest. The abundance of mouse Reg3g, Grp, Cdx2, Aqp4, and Gapdh was using SYBR Green Real Time PCR Master Mix (Qiagen). Reactions were carried out using a Rotor Gene 6000 PCR machine. Forward and reverse primer sequences used for each of these gene transcripts of interest are reported in Supplemental Table 3.
Serum cytokine analysis.
At the time of sacrifice, blood samples were collected by cardiac puncture, and serum was separated from plasma. Serum samples were analyzed by a multiplex assay for levels of various cytokines and inflammatory mediators. Eve technologies was contracted to perform the analysis using one of their routine cytokine arrays (Mouse Cytokine/Chemokine Array 31-plex). Analytes tested included Eotaxin, G-CSF, GM-CSF, IFN-γ, IL-1α, IL-1β, IL-2, IL-3, IL-4, IL-5, IL-6, IL-7, IL-9, IL-10, IL-12 (p40), IL-12 (p70), IL-13, IL-15, IL-17A, IP-10, KC, LIF, LIX, MCP-1, M-CSF, MIG, MIP-1α, MIP-1β, MIP-2, RANTES, Tnf, and VEGF.
Serum fatty acid analysis.
Serum collected for Multiplex assay of cytokine levels was also used to measure free fatty acids and endotoxin levels. Lipids were extracted from serum with heptane/isopropanol, and the solvent was evaporated and the free fatty acids were methylated using diazomethane. The Yale University Mouse Metabolic Phenotyping Center was contracted to quantify serum acetate, butyrate, and propionate levels via GC-MS (HP 5973MSD, Hewlett-Packard Instrument Corporation).
Endotoxin quantification.
Serum samples that were used for fatty acid analysis were also used to quantify endotoxin levels using the Pierce LAL Chromogenic Endotoxin Quantification Kit (Thermo Fisher Scientific) per the manufacturer’s instructions.
PAST cell isolation and flow cytometry.
Intact joints were harvested from mice and digested in collagenase for 2 hours to obtain periarticular soft tissue cells. Cell suspensions were achieved by filtering digested samples through a 70-µM nylon strainer (Corning Inc.) in ice cold PBS with 1% FBS. Cells were then pelleted and stained with fixable viability stain v450 (catalog 562247, BD Biosciences) for 40 minutes. Following washing, Fc block with rat anti–mouse CD16/CD32 (catalog 565502, BD Biosciences) was performed for 5 minutes prior to staining. All staining was performed for 40 minutes on ice at a concentration of 1:100. The following antibodies were used for phenotyping of cells: APC-Cy 7 rat anti–mouse CD19 (catalog 557655, BD Biosciences), BV510 rat anti–mouse Ly-6G and –Ly-6C (catalog 563040, BD Biosciences), Alexa Fluor 700 rat anti–mouse CD3 (catalog 561388, BD Biosciences), Per-CP Cy5.5 hamster anti–mouse CD69 (catalog 551113, BD Biosciences), BV711 rat anti–mouse F4/80 (catalog 565612, BD Biosciences), Alexa Fluor 700 rat anti–mouse CD4 (catalog 561025, BD Biosciences), BV605 rat anti–mouse CD45 (catalog 563053, BD Biosciences), PE-Cy7 anti–mouse CD11c (catalog 25-0114-82, eBioscience), PE anti–mouse CD11b (catalog 15-0112-82, eBioscience), and eFluor 660 anti–mouse CD169 (catalog 50-5755-82, eBioscience). All analyses were performed on a BD LSR II 18-color flow cytometer (BD Biosciences). Compensation was completed using anti-mouse or anti-rat/hamster compensation control beads (catalog 560497 or catalog 552845, BD Biosciences), and fluorescent minus 1 (FMO) controls were used to determine positive gating. All data was analyzed using Cytobank software.
Tissue fixation and histologic preparation.
At the time of sacrifice, mice were euthanized using an American Veterinary Medical Association–approved (AMVA-approved) method. Both knee joints were removed and fixed for 72 hours at room temperature in 10% neutral buffered formalin. Knee joints were then decalcified in EDTA for 14 days, and all tissues were processed using a microwave processor and were embedded in paraffin. Knee tissue blocks were sectioned in the midsagittal plane through the medial compartment of the joint. Sections (5-μm thick)were cut at 3 levels from each joint, with each level being 15 μm from the previous level. Knee tissue sections were stained with Safranin O/Fast Green for OARSI scoring and histomorphometric analysis. Unstained knee sections were employed for IHC staining.
Mouse OARSI scoring.
To quantify the amount of cartilage degeneration, a semiquantitative histopathologic grading system established by the OARSI histopathologic initiative was employed (79). Utilizing this system, Safranin O/Fast Green–stained knee joint sections were graded based on the following scale: 0, normal cartilage; 0.5, loss of proteoglycan stain without cartilage damage; 1, mild superficial fibrillation without cartilage loss; 2, fibrillation/clefting extending below the superficial zone coupled with partial loss of surface lamina; 3, vertical clefts/erosion of the cartilage to the calcified zone over <25% of the cartilage surface; 4, vertical clefts/erosion of the cartilage to the calcified zone over 25%–50% of the cartilage surface; 5, vertical clefts/erosion of the cartilage to the calcified zone over 50%–75% of the cartilage surface; and 6, vertical clefts/erosion extending to the calcified cartilage over >75% of the cartilage surface. Scoring was performed by 4 blinded observers (E.M. Schott, F. Kamal, R.A. Mooney, and M.J. Zuscik), and observer agreement for each score was evaluated in pairs via calculation of a weighted κ coefficient, using Fleiss-Cohen weights, as previously described (80).
Histomorphometric analysis of cartilage.
Tibial and femoral cartilage area, hypertrophic chondrocyte number, total chondrocyte number, and Safranin O+ chondrocytes were quantified by a blinded observer via histomorphometry as previously described (80). Briefly, using Safranin O/Fast Green–stained sections, articular cartilage area was measured using the OsteoMetrics system. The OsteoMetrics stylus pen was used to outline projected images obtained with an Olympus microscope (400× magnification) outfitted with a video camera. For each specimen, measurements were taken from one slide at each of the 3 levels (50 μm apart) on both the tibial plateau and the femoral condyle in a 200-μm wide area of interest centered on the joint. The OsteoMetrics stylus pen was then used to mark and count each chondrocyte, hypertrophic chondrocyte, and Safranin O+ chondrocyte. An average of all measurements was then obtained across the 3 levels for each given specimen.
IHC.
Mouse knee joint sections evaluated by IHC were deparaffinized in three changes of xylene for 5 minutes each, rehydrated in ethanols (2 changes of 100% ethanol, followed by 2 changes of 95% ethanol, followed by 1 change of 70% ethanol), and rinsed twice in deionized water. For Mmp13 and Col10, antigen retrieval was performed using 0.4% pepsin A (catalog P-7000, MilliporeSigma) made in 0.1 M HCl for 10 minutes at 37°C. Tnf did not require antigen retrieval. Endogenous peroxidases were quenched with BLOXALL (catalog SP-6000, Vector Laboratories) for 10 minutes at room temperature, and sections were rinsed in deionized water, followed by a rinse in phosphate-buffered saline containing 0.1% tween 20 (PBST, catalog P1397, MilliporeSigma). Next, slides were blocked with 5% normal goat serum (Mmp13) or 5% normal horse serum (Tnf and Col10) for 30 minutes at room temperature, followed by an overnight incubation at 4°C with rabbit anti–mouse Tnf polyclonal antibody (1:200 dilution, catalog ab6671, Abcam), rabbit anti–mouse Mmp13 polyclonal antibody (1:250 dilution, catalog ab75606, Abcam), mouse anti–mouse Col10 polyclonal antibody (1:500 dilution, catalog 1-CO097-05 Quartett), or a rabbit anti–mouse Runx2 polyclonal antibody (1:800 dilution, catalog HPA022040, MilliporeSigma). For Tnf and Col10 detection, slides were rinsed with PBST and incubated with mouse-on-mouse peroxidase polymer (catalog MPX-2402, Vector Laboratories) for 30 minutes at room temperature. For detection of Mmp13, slides were incubated with biotinylated goat anti–rabbit secondary antibody (Vectastain Elite ABC Kit, catalog PK-6100, Vector Laboratories) for 30 minutes at room temperature. Slides were once again rinsed in PBST 4 times for 5 minutes each, followed by 2 washes in deionized water for 5 minutes each. Antibody binding to Tnf and Col10 antigen was detected by a 3-minute incubation with ImmPACT DAB peroxidase substrate (catalog SK-4105, Vector Laboratories). ABC reagent from Vectastain Elite ABC Kit was applied to Mmp13 slides for 30 minutes, followed by a 3-minute application of ImmPACT DAB peroxidase substrate. Nuclei were then counterstained with Mayer’s hematoxylin solution (catalog MHS1, MilliporeSigma) for 15 seconds.
Immunofluorescence.
Mouse knee joint sections evaluated for MCP-1 protein by immunofluorescence were deparaffinized in 3 changes of xylene for 5 minutes each, rehydrated in ethanols (2 changes of 100% ethanol, followed by 2 changes of 95% ethanol, followed by 1 change of 70% ethanol), and rinsed twice in deionized water. Antigen retrieval was performed using 0.4% pepsin A made in 0.1 M HCl for 10 minutes at 37°C and was followed by permeabilization using 0.03% Triton X-100 (catalog T8787, MilliporeSigma) in TBS for 30 minutes at room temperature. Sections were then blocked in 10% normal goat serum made in 1× TBS for 1 hour at room temperature. Tissue sections were incubated at 4°C overnight with a rabbit anti–mouse MCP-1 polyclonal antibody (1:200 dilution, catalog ab25124, Abcam). Following two 5-minute washes in 1× TBS, slides were incubated with an Alexa Fluor 488 goat anti–rabbit secondary antibody (1:300 dilution, catalog A-11008, Invitrogen) for 1 hour at room temperature. Slides were then washed with 3 changes of 1× TBS for 5 minutes each, followed by 1 wash with 1× TBST, and a final 5-minute wash in 1× TBS. Nuclei were stained using ProLong Gold antifade reagent with DAPI (catalog 8961, Cell Signaling Technologies).
MicroCT and Amira analyses.
Prior to histologic processing, knee joints were evaluated via microCT using a Scanco vivaCT 40 scanner with 55-kVp source (Scanco). Joints were scanned at a resolution of 12 μm, with a slice increment of 10 μm. Images from each group were reconstructed at identical thresholds to allow 3-dimensional structural rendering of each joint. Analysis of mineralized meniscus volume was performed using an Amira-based algorithm that utilizes raw microCT Scanco DICOMS to threshold and renders volumetric information.
Statistics.
Statistical analyses for bacterial abundance studies and colon RNAseq is described above. For body mass and glucose tolerance parameters, abundance of single microbial species, individual serum cytokines or fatty acids/endotoxin, cell counts from flow cytometry, osteophyte counting, and mineralized meniscus volume, 2-way ANOVA with a Tukey’s multiple comparisons post-test was performed. This test was also used for all histomorphometry-based analysis of cartilage architecture, including cartilage area and chondrocyte population studies. After confirming blinded observer agreement via calculation of weighted κ coefficients, OARSI scoring studies were analyzed using a Kruskal-Wallis test with a Dunn’s multiple comparisons post-test. Statistical analysis of qPCR data involved comparisons between 2 experimental groups and was performed using an unpaired 2-tailed t test. All analyses and graphing of the data were carried out using Graphpad Prism software. Differences between groups were considered significant when P < 0.05 was achieved.
Study approval.
All handling and experimental procedures involving mice were reviewed and approved by University of Rochester IACUC (University Committee on Animal Resources). This committee sits at the University of Rochester Medical Center in Rochester, New York, USA.
Author contributions
EMS, CWF, RAM, and MJZ contributed to the conception and design of the study. EMS, CWF, SS, RDB, MLD, DAV, and FK contributed to data acquisition. EMS, CWF, AG, JAL, SS, GHD, RDB, MLD, DAV, HA, JPK, FK, CAB, JMA, SRG, RAM, and MJZ contributed to data analysis and interpretation. EMS, CWF, AG, JAL, HA, JPK, SRG, RAM, and MJZ contributed to drafting and revising the text. EMS, CWF, AG, JAL, SS, GHD, RDB, MLD, DAV, HA, JPK, FK, CAB, JMA, SRG, RAM, and MJZ read the final draft and provided final approval of the manuscript.
Supplementary Material
Acknowledgments
The authors wish to acknowledge the outstanding technical contributions of Sarah Mack and Kathy Maltby of the Histology, Biochemistry and Molecular Imaging Core in the Center for Musculoskeletal Research at the University of Rochester Medical Center. This work was supported by NIH NCATS UL1 TR000042 (CA-B, RAM, and MJZ), NIH NIAMS P50 AR054041-5471 (MJZ), a trainee slot on NIH NIAMS T32 AR053459 (CWF), and Core services supported by NIH NIAMS P30 AR061307 and P30 AR069665.
Version 1. 04/19/2018
Electronic publication
Version 2. 04/24/2018
Updated acknowledgments with correct NIH funding number.
Footnotes
License: This work is licensed under the Creative Commons Attribution 4.0 International License. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
Reference information: JCI Insight. 2018;3(8):e95997. https://doi.org/10.1172/jci.insight.95997.
Contributor Information
Alex Grier, Email: Alex_Grier@URMC.Rochester.edu.
Sarah Soniwala, Email: ssoniwal@u.rochester.edu.
Richard D. Bell, Email: Richard_Bell@URMC.Rochester.edu.
Hani Awad, Email: Hani_awad@urmc.rochester.edu.
Fadia Kamal, Email: Fadia_Kamal@URMC.Rochester.edu.
Cheryl Ackert-Bicknell, Email: Cheryl_AckertBicknell@URMC.Rochester.edu.
Robert A. Mooney, Email: robert_mooney@urmc.rochester.edu.
Michael J. Zuscik, Email: michael_zuscik@urmc.rochester.edu.
References
- 1.Martel-Pelletier J, et al. Osteoarthritis. Nat Rev Dis Primers. 2016;2:16072. doi: 10.1038/nrdp.2016.72. [DOI] [PubMed] [Google Scholar]
- 2.Cisternas MG, Murphy L, Sacks JJ, Solomon DH, Pasta DJ, Helmick CG. Alternative Methods for Defining Osteoarthritis and the Impact on Estimating Prevalence in a US Population-Based Survey. Arthritis Care Res (Hoboken) 2016;68(5):574–580. doi: 10.1002/acr.22721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Murray CJ, et al. UK health performance: findings of the Global Burden of Disease Study 2010. Lancet. 2013;381(9871):997–1020. doi: 10.1016/S0140-6736(13)60355-4. [DOI] [PubMed] [Google Scholar]
- 4.Centers for Disease Control Prevention (CDC) Prevalence and most common causes of disability among adults--United States, 2005. MMWR Morb Mortal Wkly Rep. 2009;58(16):421–426. [PubMed] [Google Scholar]
- 5.Kotlarz H, Gunnarsson CL, Fang H, Rizzo JA. Insurer and out-of-pocket costs of osteoarthritis in the US: evidence from national survey data. Arthritis Rheum. 2009;60(12):3546–3553. doi: 10.1002/art.24984. [DOI] [PubMed] [Google Scholar]
- 6.Yu SP, Hunter DJ. Emerging drugs for the treatment of knee osteoarthritis. Expert Opin Emerg Drugs. 2015;20(3):361–378. doi: 10.1517/14728214.2015.1037275. [DOI] [PubMed] [Google Scholar]
- 7.Zimmet P, Alberti KG, Shaw J. Global and societal implications of the diabetes epidemic. Nature. 2001;414(6865):782–787. doi: 10.1038/414782a. [DOI] [PubMed] [Google Scholar]
- 8.Ogden CL, Carroll MD, Curtin LR, Lamb MM, Flegal KM. Prevalence of high body mass index in US children and adolescents, 2007-2008. JAMA. 2010;303(3):242–249. doi: 10.1001/jama.2009.2012. [DOI] [PubMed] [Google Scholar]
- 9.Flegal KM, Carroll MD, Ogden CL, Curtin LR. Prevalence and trends in obesity among US adults, 1999-2008. JAMA. 2010;303(3):235–241. doi: 10.1001/jama.2009.2014. [DOI] [PubMed] [Google Scholar]
- 10.Centers for Disease Control Prevention (CDC) Arthritis as a potential barrier to physical activity among adults with diabetes--United States, 2005 and 2007. MMWR Morb Mortal Wkly Rep. 2008;57(18):486–489. [PubMed] [Google Scholar]
- 11.Dunbar MJ, Howard A, Bogoch ER, Parvizi J, Kreder HJ. Orthopaedics in 2020: predictors of musculoskeletal need. J Bone Joint Surg Am. 2009;91(9):2276–2286. doi: 10.2106/JBJS.H.01521. [DOI] [PubMed] [Google Scholar]
- 12.Shih M, Hootman JM, Kruger J, Helmick CG. Physical activity in men and women with arthritis National Health Interview Survey, 2002. Am J Prev Med. 2006;30(5):385–393. doi: 10.1016/j.amepre.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 13.Berenbaum F, Eymard F, Houard X. Osteoarthritis, inflammation and obesity. Curr Opin Rheumatol. 2013;25(1):114–118. doi: 10.1097/BOR.0b013e32835a9414. [DOI] [PubMed] [Google Scholar]
- 14.Xu H, et al. Chronic inflammation in fat plays a crucial role in the development of obesity-related insulin resistance. J Clin Invest. 2003;112(12):1821–1830. doi: 10.1172/JCI19451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weisberg SP, McCann D, Desai M, Rosenbaum M, Leibel RL, Ferrante AW. Obesity is associated with macrophage accumulation in adipose tissue. J Clin Invest. 2003;112(12):1796–1808. doi: 10.1172/JCI19246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lumeng CN, Bodzin JL, Saltiel AR. Obesity induces a phenotypic switch in adipose tissue macrophage polarization. J Clin Invest. 2007;117(1):175–184. doi: 10.1172/JCI29881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hamada D, et al. Suppressive Effects of Insulin on Tumor Necrosis Factor-Dependent Early Osteoarthritic Changes Associated With Obesity and Type 2 Diabetes Mellitus. Arthritis Rheumatol. 2016;68(6):1392–1402. doi: 10.1002/art.39561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Portune KJ, Benítez-Páez A, Del Pulgar EM, Cerrudo V, Sanz Y. Gut microbiota, diet, and obesity-related disorders-The good, the bad, and the future challenges. Mol Nutr Food Res. 2017;61(1) doi: 10.1002/mnfr.201600252. [DOI] [PubMed] [Google Scholar]
- 19.John GK, Mullin GE. The Gut Microbiome and Obesity. Curr Oncol Rep. 2016;18(7):45. doi: 10.1007/s11912-016-0528-7. [DOI] [PubMed] [Google Scholar]
- 20.Turnbaugh PJ, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457(7228):480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kau AL, Ahern PP, Griffin NW, Goodman AL, Gordon JI. Human nutrition, the gut microbiome and the immune system. Nature. 2011;474(7351):327–336. doi: 10.1038/nature10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trompette A, et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat Med. 2014;20(2):159–166. doi: 10.1038/nm.3444. [DOI] [PubMed] [Google Scholar]
- 23.Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med. 2009;1(6):6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cani PD, et al. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes. 2007;56(7):1761–1772. doi: 10.2337/db06-1491. [DOI] [PubMed] [Google Scholar]
- 25.Malkki H. Parkinson disease: Could gut microbiota influence severity of Parkinson disease? Nat Rev Neurol. 2017;13(2):66–67. doi: 10.1038/nrneurol.2016.195. [DOI] [PubMed] [Google Scholar]
- 26.Zhang X, et al. The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat Med. 2015;21(8):895–905. doi: 10.1038/nm.3914. [DOI] [PubMed] [Google Scholar]
- 27.Li JY, et al. Sex steroid deficiency-associated bone loss is microbiota dependent and prevented by probiotics. J Clin Invest. 2016;126(6):2049–2063. doi: 10.1172/JCI86062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Collins KH, Paul HA, Reimer RA, Seerattan RA, Hart DA, Herzog W. Relationship between inflammation, the gut microbiota, and metabolic osteoarthritis development: studies in a rat model. Osteoarthr Cartil. 2015;23(11):1989–1998. doi: 10.1016/j.joca.2015.03.014. [DOI] [PubMed] [Google Scholar]
- 29.Li Y, Luo W, Deng Z, Lei G. Diet-Intestinal Microbiota Axis in Osteoarthritis: A Possible Role. Mediators Inflamm. 2016;2016:3495173. doi: 10.1155/2016/3495173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoo JY, Kim SS. Probiotics and Prebiotics: Present Status and Future Perspectives on Metabolic Disorders. Nutrients. 2016;8(3):173. doi: 10.3390/nu8030173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kellow NJ, Coughlan MT, Savige GS, Reid CM. Effect of dietary prebiotic supplementation on advanced glycation, insulin resistance and inflammatory biomarkers in adults with pre-diabetes: a study protocol for a double-blind placebo-controlled randomised crossover clinical trial. BMC Endocr Disord. 2014;14:55. doi: 10.1186/1472-6823-14-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Everard A, et al. Responses of gut microbiota and glucose and lipid metabolism to prebiotics in genetic obese and diet-induced leptin-resistant mice. Diabetes. 2011;60(11):2775–2786. doi: 10.2337/db11-0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu RY, et al. Non-digestible oligosaccharides directly regulate host kinome to modulate host inflammatory responses without alterations in the gut microbiota. Microbiome. 2017;5(1):135. doi: 10.1186/s40168-017-0357-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Slavin J. Fiber and prebiotics: mechanisms and health benefits. Nutrients. 2013;5(4):1417–1435. doi: 10.3390/nu5041417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ziesenitz SC, Siebert G. In vitro assessment of nystose as a sugar substitute. J Nutr. 1987;117(5):846–851. doi: 10.1093/jn/117.5.846. [DOI] [PubMed] [Google Scholar]
- 36.Nilsson U, Oste R, Jägerstad M, Birkhed D. Cereal fructans: in vitro and in vivo studies on availability in rats and humans. J Nutr. 1988;118(11):1325–1330. doi: 10.1093/jn/118.11.1325. [DOI] [PubMed] [Google Scholar]
- 37.Rumessen JJ, Bodé S, Hamberg O, Gudmand-Høyer E. Fructans of Jerusalem artichokes: intestinal transport, absorption, fermentation, and influence on blood glucose, insulin, and C-peptide responses in healthy subjects. Am J Clin Nutr. 1990;52(4):675–681. doi: 10.1093/ajcn/52.4.675. [DOI] [PubMed] [Google Scholar]
- 38.Bach Knudsen KE, Hessov I. Recovery of inulin from Jerusalem artichoke (Helianthus tuberosus L.) in the small intestine of man. Br J Nutr. 1995;74(1):101–113. doi: 10.1079/BJN19950110. [DOI] [PubMed] [Google Scholar]
- 39.Ellegård L, Andersson H, Bosaeus I. Inulin and oligofructose do not influence the absorption of cholesterol, or the excretion of cholesterol, Ca, Mg, Zn, Fe, or bile acids but increases energy excretion in ileostomy subjects. Eur J Clin Nutr. 1997;51(1):1–5. doi: 10.1038/sj.ejcn.1600320. [DOI] [PubMed] [Google Scholar]
- 40.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444(7122):1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 41.Verdam FJ, et al. Human intestinal microbiota composition is associated with local and systemic inflammation in obesity. Obesity (Silver Spring) 2013;21(12):E607–E615. doi: 10.1002/oby.20466. [DOI] [PubMed] [Google Scholar]
- 42.Arboleya S, Watkins C, Stanton C, Ross RP. Gut Bifidobacteria Populations in Human Health and Aging. Front Microbiol. 2016;7:1204. doi: 10.3389/fmicb.2016.01204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Clarke SF, et al. Targeting the microbiota to address diet-induced obesity: a time dependent challenge. PLoS ONE. 2013;8(6):e65790. doi: 10.1371/journal.pone.0065790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Monk JM, et al. Diets enriched with cranberry beans alter the microbiota and mitigate colitis severity and associated inflammation. J Nutr Biochem. 2016;28:129–139. doi: 10.1016/j.jnutbio.2015.10.014. [DOI] [PubMed] [Google Scholar]
- 45.Thomas RP, Hellmich MR, Townsend CM, Evers BM. Role of gastrointestinal hormones in the proliferation of normal and neoplastic tissues. Endocr Rev. 2003;24(5):571–599. doi: 10.1210/er.2002-0028. [DOI] [PubMed] [Google Scholar]
- 46.Zhu C, Chen Z, Jiang Z. Expression, Distribution and Role of Aquaporin Water Channels in Human and Animal Stomach and Intestines. Int J Mol Sci. 2016;17(9):1399. doi: 10.3390/ijms17091399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vaishnava S, et al. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science. 2011;334(6053):255–258. doi: 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Coskun M, Troelsen JT, Nielsen OH. The role of CDX2 in intestinal homeostasis and inflammation. Biochim Biophys Acta. 2011;1812(3):283–289. doi: 10.1016/j.bbadis.2010.11.008. [DOI] [PubMed] [Google Scholar]
- 49.Donath MY, Shoelson SE. Type 2 diabetes as an inflammatory disease. Nat Rev Immunol. 2011;11(2):98–107. doi: 10.1038/nri2925. [DOI] [PubMed] [Google Scholar]
- 50.Griffin TM, Huebner JL, Kraus VB, Yan Z, Guilak F. Induction of osteoarthritis and metabolic inflammation by a very high-fat diet in mice: effects of short-term exercise. Arthritis Rheum. 2012;64(2):443–453. doi: 10.1002/art.33332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Raghu H, et al. CCL2/CCR2, but not CCL5/CCR5, mediates monocyte recruitment, inflammation and cartilage destruction in osteoarthritis. Ann Rheum Dis. 2017;76(5):914–922. doi: 10.1136/annrheumdis-2016-210426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Glasson SS, Blanchet TJ, Morris EA. The surgical destabilization of the medial meniscus (DMM) model of osteoarthritis in the 129/SvEv mouse. Osteoarthr Cartil. 2007;15(9):1061–1069. doi: 10.1016/j.joca.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 53.Kamekura S, et al. Contribution of runt-related transcription factor 2 to the pathogenesis of osteoarthritis in mice after induction of knee joint instability. Arthritis Rheum. 2006;54(8):2462–2470. doi: 10.1002/art.22041. [DOI] [PubMed] [Google Scholar]
- 54.Bagi CM, Zakur DE, Berryman E, Andresen CJ, Wilkie D. Correlation between μCT imaging, histology and functional capacity of the osteoarthritic knee in the rat model of osteoarthritis. J Transl Med. 2015;13:276. doi: 10.1186/s12967-015-0641-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ruan MZ, Dawson B, Jiang MM, Gannon F, Heggeness M, Lee BH. Quantitative imaging of murine osteoarthritic cartilage by phase-contrast micro-computed tomography. Arthritis Rheum. 2013;65(2):388–396. doi: 10.1002/art.37766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lane NE, et al. OARSI-FDA initiative: defining the disease state of osteoarthritis. Osteoarthr Cartil. 2011;19(5):478–482. doi: 10.1016/j.joca.2010.09.013. [DOI] [PubMed] [Google Scholar]
- 57.Mobasheri A, Bay-Jensen AC, van Spil WE, Larkin J, Levesque MC. Osteoarthritis Year in Review 2016: biomarkers (biochemical markers) Osteoarthr Cartil. 2017;25(2):199–208. doi: 10.1016/j.joca.2016.12.016. [DOI] [PubMed] [Google Scholar]
- 58.Mooney RA, Sampson ER, Lerea J, Rosier RN, Zuscik MJ. High-fat diet accelerates progression of osteoarthritis after meniscal/ligamentous injury. Arthritis Res Ther. 2011;13(6):R198. doi: 10.1186/ar3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brostow DP, Hirsch AT, Collins TC, Kurzer MS. The role of nutrition and body composition in peripheral arterial disease. Nat Rev Cardiol. 2012;9(11):634–643. doi: 10.1038/nrcardio.2012.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Landsberg L, et al. Obesity-related hypertension: pathogenesis, cardiovascular risk, and treatment: a position paper of The Obesity Society and the American Society of Hypertension. J Clin Hypertens (Greenwich) 2013;15(1):14–33. doi: 10.1111/jch.12049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Borlaug BA. The pathophysiology of heart failure with preserved ejection fraction. Nat Rev Cardiol. 2014;11(9):507–515. doi: 10.1038/nrcardio.2014.83. [DOI] [PubMed] [Google Scholar]
- 62.Hand TW, Vujkovic-Cvijin I, Ridaura VK, Belkaid Y. Linking the Microbiota, Chronic Disease, and the Immune System. Trends Endocrinol Metab. 2016;27(12):831–843. doi: 10.1016/j.tem.2016.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Reinoso Webb C, Koboziev I, Furr KL, Grisham MB. Protective and pro-inflammatory roles of intestinal bacteria. Pathophysiology. 2016;23(2):67–80. doi: 10.1016/j.pathophys.2016.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bleau C, Karelis AD, St-Pierre DH, Lamontagne L. Crosstalk between intestinal microbiota, adipose tissue and skeletal muscle as an early event in systemic low-grade inflammation and the development of obesity and diabetes. Diabetes Metab Res Rev. 2015;31(6):545–561. doi: 10.1002/dmrr.2617. [DOI] [PubMed] [Google Scholar]
- 65.Scher JU, et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. Elife. 2013;2:e01202. doi: 10.7554/eLife.01202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Moya-Pérez A, Neef A, Sanz Y. Bifidobacterium pseudocatenulatum CECT 7765 Reduces Obesity-Associated Inflammation by Restoring the Lymphocyte-Macrophage Balance and Gut Microbiota Structure in High-Fat Diet-Fed Mice. PLoS One. 2015;10(7):e0126976. doi: 10.1371/journal.pone.0126976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bottegoni C, Muzzarelli RA, Giovannini F, Busilacchi A, Gigante A. Oral chondroprotection with nutraceuticals made of chondroitin sulphate plus glucosamine sulphate in osteoarthritis. Carbohydr Polym. 2014;109:126–138. doi: 10.1016/j.carbpol.2014.03.033. [DOI] [PubMed] [Google Scholar]
- 68.Henrotin Y, Lambert C, Richette P. Importance of synovitis in osteoarthritis: evidence for the use of glycosaminoglycans against synovial inflammation. Semin Arthritis Rheum. 2014;43(5):579–587. doi: 10.1016/j.semarthrit.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 69.Bruyere O, Reginster JY. Glucosamine and chondroitin sulfate as therapeutic agents for knee and hip osteoarthritis. Drugs Aging. 2007;24(7):573–580. doi: 10.2165/00002512-200724070-00005. [DOI] [PubMed] [Google Scholar]
- 70.Vangsness CT, Spiker W, Erickson J. A review of evidence-based medicine for glucosamine and chondroitin sulfate use in knee osteoarthritis. Arthroscopy. 2009;25(1):86–94. doi: 10.1016/j.arthro.2008.07.020. [DOI] [PubMed] [Google Scholar]
- 71.Bruyère O, Altman RD, Reginster JY. Efficacy and safety of glucosamine sulfate in the management of osteoarthritis: Evidence from real-life setting trials and surveys. Semin Arthritis Rheum. 2016;45(4 Suppl):S12–S17. doi: 10.1016/j.semarthrit.2015.11.011. [DOI] [PubMed] [Google Scholar]
- 72.Ackert-Bicknell CL, et al. Genetic variation in TRPS1 may regulate hip geometry as well as bone mineral density. Bone. 2012;50(5):1188–1195. doi: 10.1016/j.bone.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fadrosh DW, et al. An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform. Microbiome. 2014;2(1):6. doi: 10.1186/2049-2618-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27(16):2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.DeSantis TZ, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72(7):5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.McDonald D, et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012;6(3):610–618. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73(16):5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Glasson SS, Chambers MG, Van Den Berg WB, Little CB. The OARSI histopathology initiative - recommendations for histological assessments of osteoarthritis in the mouse. Osteoarthr Cartil. 2010;18 Suppl 3:S17–S23. doi: 10.1016/j.joca.2010.05.025. [DOI] [PubMed] [Google Scholar]
- 80.Sampson ER, et al. Establishment of an index with increased sensitivity for assessing murine arthritis. J Orthop Res. 2011;29(8):1145–1151. doi: 10.1002/jor.21368. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.