Abstract
Eosinophilic esophagitis (EoE) is an allergic inflammatory esophageal disorder with a complex underlying genetic etiology often associated with other comorbidities. Using whole-exome sequencing (WES) of 63 patients with EoE and 60 unaffected family members and family-based trio analysis, we sought to uncover rare coding variants. WES analysis identified 5 rare, damaging variants in dehydrogenase E1 and transketolase domain–containing 1 (DHTKD1). Rare variant burden analysis revealed an overabundance of putative, potentially damaging DHTKD1 mutations in EoE (P = 0.01). Interestingly, we also identified 7 variants in the DHTKD1 homolog oxoglutarate dehydrogenase-like (OGDHL). Using shRNA-transduced esophageal epithelial cells and/or patient fibroblasts, we further showed that disruption of normal DHTKD1 or OGDHL expression blunts mitochondrial function. Finally, we demonstrated that the loss of DHTKD1 expression increased ROS production and induced the expression of viperin, a gene previously shown to be involved in production of Th2 cytokines in T cells. Viperin had increased expression in esophageal biopsies of EoE patients compared with control individuals and was upregulated by IL-13 in esophageal epithelial cells. These data identify a series of rare genetic variants implicating DHTKD1 and OGDHL in the genetic etiology of EoE and underscore a potential pathogenic role for mitochondrial dysfunction in EoE.
Keywords: Immunology, Inflammation
Keywords: Allergy, Cellular immune response, Mitochondria
Whole-exome sequencing reveals links between eosinophilic esophagitis, a Th2-driven allergic inflammatory disease, and mitochondrial dysfunction.
Introduction
Eosinophilic esophagitis (EoE) is a chronic allergic disorder that presents in infancy as vomiting and failure to thrive and as dysphagia and food impaction in adulthood (1, 2). Evidence is emerging that EoE involves the interplay of genetics and environment (3–5). Most genetic studies have focused on analysis of common genetic variants, and linkages at chromosomes 2p23 and 5q22 have been found (6, 7). Gene expression profiling and immunologic analyses have revealed that the disease is characterized by intense type 2 immune associated inflammatory processes (8, 9).
Although genetic predisposition has been demonstrated in EoE, the non-Mendelian inheritance pattern and polygenic nature of the disease have hindered the identification of disease-causal variants (2, 5, 6, 8). GWAS and candidate gene studies have identified select genes contributing to EoE risk, such as thymic stromal lymphopoietin (TSLP) and more recently calpain 14 (CAPN14); however, the causal variants remain controversial (6, 7, 10–13). There is now an increased recognition of the key role of environmental factors (exposures) (3).
Molecular analyses have identified IL-13 as a potent mediator of many pathologic features of the esophageal epithelium in EoE, including hyperplasia, barrier dysfunction, and release of proinflammatory chemokines and cytokines (9, 14–17). IL-13 is the chief stimulus for the production of eotaxin-3 (chemokine [C-C motif] ligand 26, encoded by CCL26), an eosinophil-selective chemoattractant and activating cytokine, as well as CAPN14 from esophageal epithelial cells. In fact, in vitro stimulation of primary esophageal epithelial cells with IL-13 alone can elicit a substantial portion of the disease-specific transcriptional responses (e.g., the EoE transcriptome) observed in inflamed esophageal tissues ex vivo (8, 13, 14, 18). Further evidence highlighting the critical role of IL-13 in EoE pathogenesis has come from a recent clinical trial showing a positive effect of anti–IL-13 treatment on esophageal eosinophilia and gene expression in patients (17).
In addition to atopic disorders such as atopic dermatitis and asthma (19, 20), EoE has been observed in the presence of other comorbidities, including a spectrum of inherited connective tissue disorders (CTD) characterized by hypermobility syndrome, suggesting common disease pathways (21). For instance, TGF-β–mediated fibrotic responses have been implicated in both esophageal remodeling and dysmotility in EoE and the CTDs Marfan syndrome and Loeys-Dietz syndrome (21–25). Acquired mitochondrial dysfunction, often associated with coenzyme Q deficiency, has been noted in a cohort of patients with EoE (26). Mitochondriopathies can affect virtually all organ systems with various phenotypic manifestations. During allergic inflammatory responses, mast cell degranulation — which has been demonstrated to occur in the esophagus of patients with EoE — is dependent on mitochondrial translocation to sites of exocytosis; conversely, mast cells and eosinophils can also secrete mitochondrial components that, in turn, can induce mast cell degranulation and histamine release (27–30). Disrupted synthesis of mitochondria by haploinsufficiency of the intracellular phosphatidylcholine transport protein steroidogenic acute regulatory protein–related (StAR-related) lipid transfer domain protein 7 (StARD7) in epithelial cells results in spontaneous allergic responses in mice, highlighting the potential of mitochondrial dysfunction to induce inflammatory responses (29, 31). In addition, activated eosinophils can also release mitochondrial DNA traps into the extracellular space as an antibacterial defense mechanism (30). Despite these findings, however, no direct link between mitochondrial dysfunction and EoE has been reported.
As the number of genome-wide susceptibility loci associated with complex diseases continues to grow, an emphasis has now been placed on identifying causal variants that alter the expression or protein function of the associated genes, particularly those functions mediated by rare variants. Here, we report the genetic analysis of protein-coding variants in EoE through whole-exome sequencing (WES) of 37 unrelated families containing 63 patients with EoE and 60 unaffected family members. We identified 10,746 rare mutations predicted to be damaging, including 9 damaging de novo mutations and 477 damaging compound heterozygous variants in 181 genes, many of which are involved in innate immune system function. Interestingly, WES analysis revealed enrichment in rare, damaging variants in dehydrogenase E1 and transketolase domain–containing 1 (DHTKD1), a nuclear gene involved in mitochondrial lysine metabolism and ATP production (32, 33). In particular, we identified a potentially novel nonsense (arginine [Arg] 834*) of a 2-generation family with EoE and phenotypes suggestive of mitochondriopathy and a potentially novel splicing mutation (c.1897-1 guanine [G] > adenine [A], transcript NM_018706.6). Notably, DHTKD1 Arg834* resulted in haploinsufficiency in the affected family members through nonsense-mediated decay (NMD). Both the loss of DHTKD1 (through shRNA transduction of esophageal epithelial cell lines) and the c.1897-1G>A mutation (in patient esophageal fibroblasts) impaired mitochondrial function. We also identified 7 potentially damaging variants in the DHTKD1 homolog oxoglutarate dehydrogenase-like (OGDHL), and patient fibroblasts harboring a missense mutation (c.406 cytosine [C] > thymine [T], p.proline [Pro] 136 serine [Ser], transcript NM_001347819.1) in OGDHL also revealed mitochondrial dysfunction, including generation of ROS. Lastly, we show that DHTKD1 depletion in esophageal epithelial cells in vitro increased transcripts of radical S-adenosyl methionine domain–containing 2 (RSAD2) (also known as viperin), a Th2 promoting gene in T cells, both at baseline and with IL-13 treatment. Taken together, these data suggest genetic insult of DHTKD1 and OGDHL as predisposing factors in driving mitochondrial dysfunction and molecular pathways linked to the progression of allergic inflammation.
Results
WES analysis of EoE.
Analysis of WES data from the study cohort, including the filtering methods used to identify variants and the analyses performed, are depicted in Figure 1. WES was performed on 63 EoE patients from 25 unrelated trios and 12 multiplex families composed of 2 or more related patients with EoE. We focused on identifying rare variants with a minor allele frequency (MAF) < 0.01 that were predicted to be damaging to protein function (including de novo mutation events). In addition, we sought to identify compound heterozygous variants, using an initial MAF threshold of <0.05 for 1 variant and a MAF <0.01 for a second variant in the same gene; notably, less than 10% of the 442,356 coding variants had a MAF < 0.05 (n = 40,333). Using these filtering parameters, we identified a total of 10,725 rare, putatively damaging mutations in 63 patients with EoE; of these mutations, Sanger sequencing was used to validate 9 de novo variants, each of which occurred in only 1 patient with EoE (Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.99922DS1). In particular, we identified a de novo variant in dedicator of cytokinesis 8 (DOCK8), whose encoded protein is critical for lymphocyte survival and function; notably, DOCK8 loss-of-function mutations result in combined immunodeficiency characterized by severe atopy, including EoE (34–36). We also identified an Arg264 glutamine (Gln) de novo variant identified in myeloid differentiation primary response 88 (MYD88); interestingly, this mutation is proximal to a leucine (Leu) 265 Pro gain-of-function mutation in MYD88 that is associated with B cell lymphoma and Waldenstrom macroglobulinemia (37, 38).
Figure 1. EoE WES study design.
Schematic depicting the workflow for whole-exome sequencing (WES) filtering of rare, damaging variants, de novo variants, and compound heterozygous variants in 63 patients with eosinophilic esophagitis (EoE) and 60 unaffected family members. Filtering details can be found in Methods. Alt, alternate; GQ, genotype quality score; DP, read depth; MAF, minor allele frequency; KBAC, kernel-based adaptive cluster. Asterisk indicates one of the families (trio) included an unaffected sibling and a grandparent.
We also identified 477 compound heterozygous variants in 181 genes and used Sanger sequencing to validate 14 variants in 7 genes (Supplemental Table 2). Notably, 5 of 7 genes have a previous genetic association (7) and/or potential functional implications in EoE pathophysiology (8, 39–41). Arachidonate 15-lipoxygenase (ALOX15), one of the most highly induced genes in the esophageal mucosa of patients with active EoE, was found to contain 2 compound heterozygous variants (8, 42). Compound heterozygous variants were also found in IL5RA, the gene encoding the α subunit of the receptor for IL-5, an essential cytokine driving eosinophil maturation and survival (39). Laminin subunit α 5 (LAMA5), encoding a key cell adhesion molecule necessary for gastrointestinal development, also had 2 compound heterozygous variants (43). In addition, 2 compound heterozygous variants, including a nonsense mutation at Gln370, were identified in TLR10, whose protein product has been shown to act as an antiinflammatory-pattern recognition receptor that antagonizes TLR2 signaling (44). Lastly, 2 compound heterozygous variants in WD repeat-containing protein 36 (WDR36), which resides in the EoE susceptibility locus on chromosome 5q22 (6, 7), were found.
DHTKD1 mutation in multiplex family with EoE.
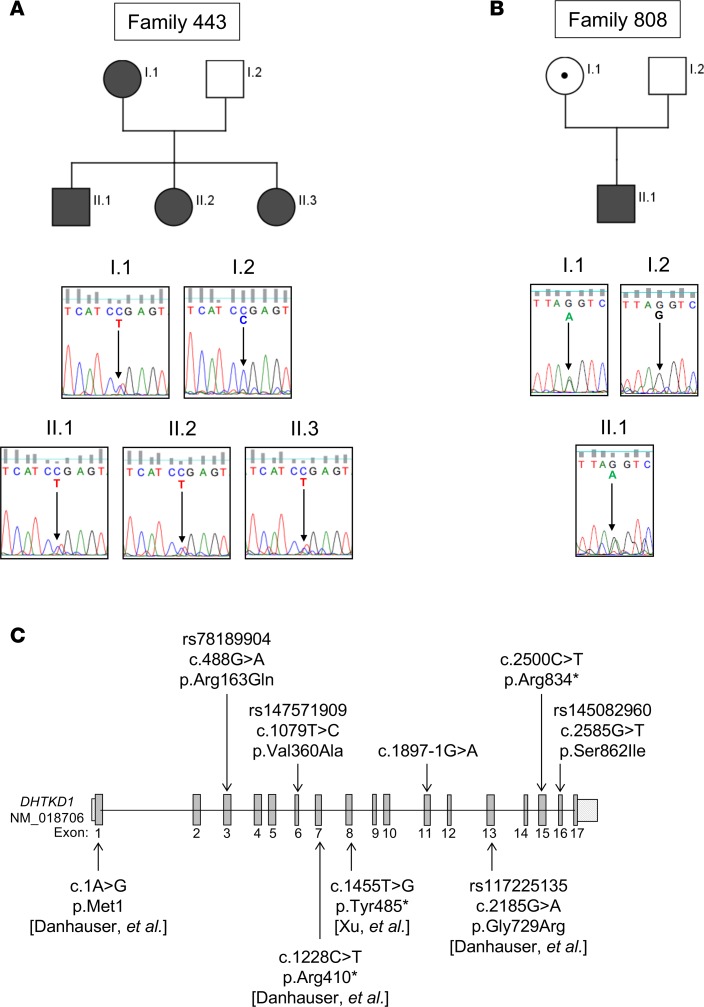
In an effort to identify causal variants, we focused our analysis on a multiplex family (family 443) exhibiting an autosomal dominant pattern of inheritance with extreme EoE phenotypes (Table 1 and Figure 2A). The affected mother (443, I.1) and all three offspring (443, II.1–3) have EoE diagnoses (including proton pump inhibitor [PPI] resistance) and multiple underlying atopic conditions. In addition, the offspring presented with phenotypic characteristics suggestive of mitochondriopathy, including gastrointestinal dysmotility, rhabodymyolysis (in 1 individual following anesthesia), kidney dysfunction, hypotonia, fatigue, and hyperopia. Notably, offspring II.3 also has the hypermobile form of Ehlers-Danlos syndrome, exhibiting skin elasticity and joint hypermobility with a Beighton score of 8; CTDs including Ehlers-Danlos syndrome have been identified as comorbidities of EoE (21, 45). No esophageal disease, atopy, or mitochondriopathy was reported in the unaffected father (I.2). Among the 90 rare (MAF > 0.01), maternally inherited (i.e., autosomal dominant) variants predicted to affect protein function, we identified and validated by Sanger sequencing a potentially novel nonsense Arg834* mutation in the gene encoding DHTKD1 (Figure 2A).
Table 1. Patient phenotypes of affected members of family 443 and patient 2.0.
Figure 2. Rare variants in DHTKD1 in EoE.
Pedigree of index family (family 443; affected individuals are noted by shaded symbols) and chromatograms from Sanger sequencing validating heterozygosity for c.2500 cytosine (C) > thymine (T), p.arginine (Arg) 834* in all 4 affected individuals from family 443 (A). Chromatograms from Sanger sequencing validating heterozygosity for c.1897-1 guanine (G) > adenine (A) mutation in a separate patient with eosinophilic esophagitis (EoE) (II.1) and his unaffected mother (I.1) in family 808 (B). Schematic of the DHTKD1 gene locus. The DHTKD1 variants noted above the gene locus schematic are those identified by whole-exome sequencing (WES) in this study, whereas those identified in other studies (with references in brackets) are indicated below the schematic (C).
DHTKD1 mutations are enriched in EoE.
Four other rare, damaging mutations in DHKTD1 were identified among 37 exomes of unrelated patients with EoE, including 3 nonsynonymous variants and 1 potentially novel splicing variant (Table 2 and Figure 2, B and C). We validated the c.1897-1G>A splicing mutation, which occurs immediately before exon 11 of DHTKD1, in family 808 by Sanger sequencing (Figure 2B). Despite the index patient (808, II.1) and his mother (808, I.1) being heterozygous for c.1897-1G>A, neither exhibited any phenotype suggestive of a potential mitochondriopathy, further suggesting that this variant is more likely to be associated with EoE. We next sought to determine whether DHTKD1 was enriched for rare variants in EoE. The Exome Aggregation Consortium (ExAC) database (http://exac.broadinstitute.org/) identifies 243 DHTKD1 amino acid–altering variants in the European (non-Finnish) population. From the MAF of these variants, it is expected that only 6.3% of the European population contains DHTKD1 amino acid–altering variants compared with the 18.75% (n = 7) identified in the 37 unrelated patients with EoE (P = 0.0015 χ2 with Yates correction). The 5 variants detected in the EoE cases were identified in 1.3% of the European population (P = 1.7 × 10–26) (Table 2).
Table 2. Rare, damaging DHTKD1 variants identified by WES in patients with EoE.
Among the 113 DHTKD1 variants detected in the European American population in the National Heart, Lung, and Blood Institute (NHLBI) GO Exome Sequencing Project (ESP) data, these variants were undetected or detected in less than 0.7% (MAF = 2.0 × 10–4 to 6.1 × 10–3) (data not shown). In order to determine whether DHTKD1 was enriched for rare, damaging variants in EoE, rare variant association testing was performed on 37 unrelated patients with EoE and 379 European ancestry controls from the 1000 Genomes Project (http://www.internationalgenome.org/) (Supplemental Table 3). Indeed, a significant association (P = 0.01) between DHTKD1 rare, damaging variants and EoE was observed, suggesting that DHTKD1 variants are linked to EoE, independently of mitochondriopathy.
Esophageal DHTKD1 expression in multiplex family with EoE.
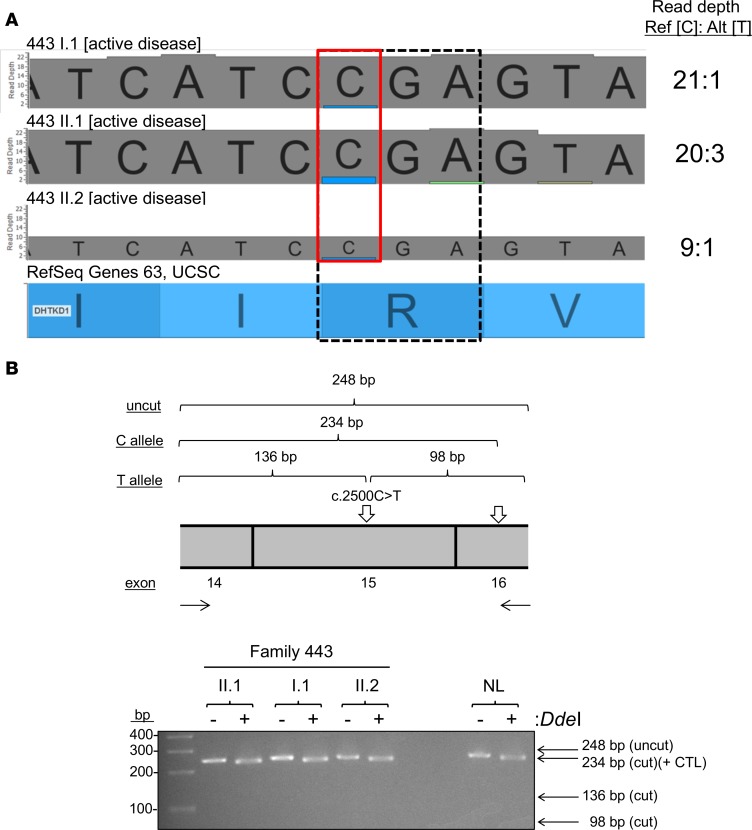
Gene expression profiling of esophageal biopsies by RNA sequencing (RNA-seq) demonstrated that esophageal expression of DHTKD1 was moderately increased (P = 6.7 × 10–3, fold change 1.67) in an unrelated cohort of patients with active EoE (n = 8) compared with normal controls (NL; n = 6) (Figure 3, A and B). Regardless of their disease activity at the time of biopsy, members of family 443 with EoE had significantly lower DHTKD1 expression than the EoE cohort (P = 0.029); interestingly, disease remission (443, II.2) had lower levels than disease flare (443, II.1 and I.1) within family 443 (Figure 3, A and B). A closer look at the RNA-seq read depths from the affected family 443 members revealed a disproportionate percentage of reads (87.0%–95.4%) detecting the reference C allele relative to the alternate T allele, suggesting NMD of the transcript with the T allele (Figure 4A). We validated the effect of c.2500C>T, p.Arg834*, which creates a unique DdeI recognition sequence in exon 15, on DHTKD1 transcript levels using RFLP mapping. PCR from esophageal cDNA samples from NL individuals and individuals with EoE from family 443 was performed across exons 14–16 of DHTKD1 (Figure 4B); the resulting amplicon also contains a natural DdeI restriction site 14 bp from the 3′ end as a positive control. Cleavage with DdeI demonstrated a similar fragment pattern between the heterozygous carriers from family 443 and a homozygous NL individual, indicating a lack of amplification from transcripts containing the alternate A allele in the affected family members (Figure 4B). IHC staining detected DHTKD1 protein throughout the esophageal epithelium in patients with EoE and lower expression, primarily in the basal region, in NL individuals (Figure 5). The staining intensities were reflective of the RNA-seq data: a more intense staining in patients with active EoE relative to NL individuals. Similar to the trends noted for family 443 and disease activity in the RNA-seq data (Figure 3B), DHTKD1 expression was reduced in the individual II.2 regardless of disease state (Figure 5).
Figure 3. DHTKD1 expression in EoE.
Heatmap from RNA sequencing of patient esophageal biopsies showing log2 fragments per kilobase of transcript per million mapped reads (FPKM) values for dehydrogenase E1 and transketolase domain–containing 1 (DHTKD1) expression in normal controls (NL, n = 6; open squares), unrelated patients with active eosinophilic esophagitis (EoE) (n = 8, shaded circles), and select, affected individuals from family 443 (boxed with dashed line): I.1 and II.1 (active disease, shaded triangle) and II.2 (inactive disease, open triangle) (A). Data are plotted by FPKM in a profile plot below in A and clustered in B; bars in B represent the mean ± SD. Statistical analysis was performed using one-way ANOVA (and nonparametric) with Dunn’s multiple comparisons test. *P < 0.05 and **P < 0.01 compared with NL unless otherwise noted.
Figure 4. DHTKD1 Arg834* induces nonsense-mediated decay.
RNA sequencing alignments for I.1, II.1, and II.2 at the variation position c.2500 cytosine (C) > thymine (T) (red box) and arginine (Arg) 834* (dashed box) are shown (A). Reads per reference (Ref) C allele and alternate (Alt) T allele are shown to the right. RFLP mapping schematic showing fragments generated from undigested or DdeI-digested amplicons from esophageal DHTKD1 transcripts generated from the C vs. T allele (B, upper panel). Gel electrophoresis after RFLP mapping performed on individuals I.1, II.1, and II.2 and normal control individual (NL) homozygous for the C allele (B, lower panel).+ CTL, positive control.
Figure 5. DHTKD1 protein expression in family 443.
IHC staining for dehydrogenase E1 and transketolase domain–containing 1 (DHTKD1) expression in esophageal biopsies from normal controls (NL), unrelated patients with active EoE, and individual II.2 from family 443 during active disease (active) and disease remission (inactive). Images from 2 patients are shown for both the NL and unrelated active EoE groups (representative of 6 patients per group). Scale bar: 50 μM.
DHTKD1 gene silencing reduces mitochondrial function.
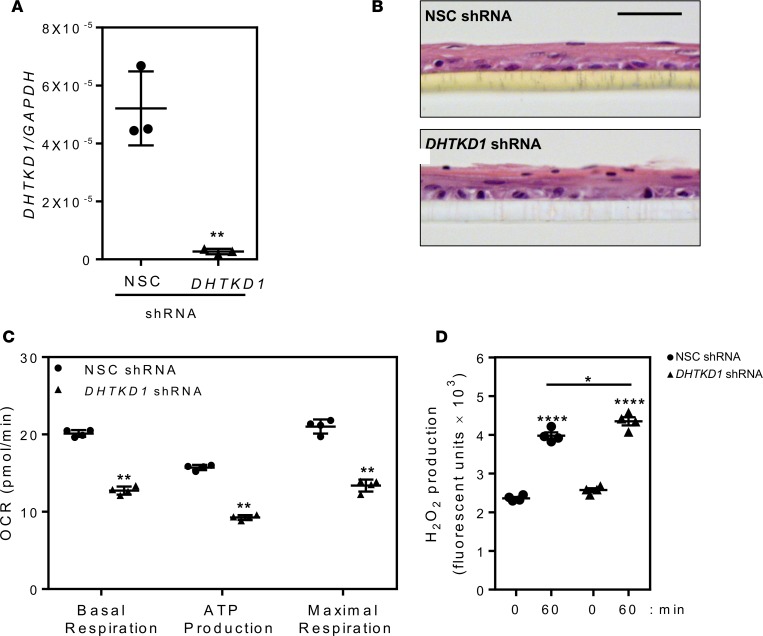
Given the haploinsufficiency in family 443, we aimed to determine the effects of reduced DHTKD1 expression on mitochondrial function in a cell type relevant to EoE. Therefore, we used shRNA to knock down DHTKD1 expression (compared with a nonsilencing control [NSC] shRNA) in an esophageal epithelial cell line (EPC2) (Figure 6A). DHTKD1 knockdown had no observable effect on cell differentiation under baseline conditions, as evident by H&E staining of air-liquid interface (ALI) cultures (Figure 6B). DHTKD1 knockdown resulted in reduced ATP production and blunted basal and maximal respiration rates in DHTKD1-deficient cells compared with control cells (Figure 6C). We also observed a significant increase in ROS (H2O2) production with DHTKD1 depletion (Figure 6D).
Figure 6. DHTKD1 deficiency impairs mitochondrial function in esophageal epithelial cells.
Quantitative PCR showing efficiency of gene silencing by dehydrogenase E1 and transketolase domain–containing 1–specific (DHTKD1-specific ) shRNA compared with a nonsilencing control (NSC) shRNA in esophageal epithelial cell lines (EPC2) (A). Assessment of epithelial differentiation by H&E staining of EPC2 cells expressing NSC or DHTKD1 shRNA grown at the air-liquid interface (ALI) (B). Scale bar: 50 μM. Measurement of mitochondrial function (oxygen consumption rate [OCR]: basal respiration, ATP production, and maximal respiration) (C) and H2O2 production (D) in EPC2 cells expressing NSC (black circles) or DHTKD1 shRNA (black triangles). Data in A are a representative of 3 independent experiments performed in triplicates, images in B are representative image from 3 independent experiments performed in triplicates, and data in C are from 4 cycles per measurement and at least 6 wells for each cell line. Data in D are representative of 3 independent experiments performed in quadruplicate. Data in A and D are presented as the mean ± SEM compared with NSC or time 0 in D, and data in C are presented as mean ± SD. Statistical analysis done via 1-way ANOVA (and nonparametric) with Holm-Sidak multiple comparison test (A); unpaired t tests with Gaussian distribution and 95% confidence interval (C); 1-way ANOVA (and nonparametric) with Tukey multiple comparisons test (D). *P < 0.05; **P < 0.01; and ****P < 1 × 10–4.
In addition to esophageal epithelial cells, mitochondrial function testing was performed using primary esophageal fibroblasts from an NL patient and the patient with EoE with the heterozygous c.1897-1G>A splicing mutation (individual II.1 from family 808). The c.1897-1G allele and the flanking sequence at the 5′ boundary of exon 11 are highly conserved across multiple species (Supplemental Figure 2). In silico analysis using Human Splicing Finder (http://www.umd.be/HSF3/) (46) indicated that c.1897-1G>A ablates the canonical splice acceptor site at the boundary of exon 11, causing an alternative splice site 11 bases into exon 11 that produces a predicted frameshift and truncation of DHTKD1 protein (Figure 7A). Compared with the NL fibroblasts, the EoE fibroblasts from patient II.1 of family 808 had significantly attenuated mitochondrial function, as evidenced by decreased ATP production and reduced basal and maximal respiration rates (Figure 7B).
Figure 7. Effect of DHTKD1 splicing mutation on mitochondrial function in primary esophageal fibroblasts.
In silico analysis of c.1897-1G>A (in red and denoted by arrow), showing the aligned reference sequence (Ref, top strand) and variant sequence (Alt, bottom strand) with the predicted alternative splice site (in bold) and resulting truncated (*) DHTKD1 protein sequence (A). Mitochondrial function tests (oxygen consumption rate [OCR]: basal respiration, ATP production, and maximal respiration) from primary esophageal fibroblasts from a normal control (NL; unshaded squares) or individual II.1 from family 808 (eosinophilic esophagitis [EoE; shaded circles] c.1897-1G>A) (B). Data in B are presented as the mean ± SD from 4 cycles per measurement and at least 6 wells for each cell line. **P < 0.05 compared with NL, done via unpaired t tests with Gaussian distribution and 95% confidence interval. Ref, reference; Alt, alternate.
Genetic variants in the DHTKD1 homolog OGDHL.
Given the abundance of DHTKD1 variants across multiple patients with EoE, we hypothesized that perturbing DHTKD associated processes may be a widespread theme involved in disease pathology. Genetic variants in OGDHL, a nuclear gene that encodes a predicted functional homolog of DHTKD1 (47), was identified independently in a 21-year-old patient with EoE and clinical presentation of chronic fatigue and exercise intolerance who was being evaluated in the Mitochondrial Disorders Program at CCHMC. This patient (referred to as 2.0 in Table 1) had c.406C>T mutation in OGDHL that causes a Pro-to-Ser missense change at position 136 (p.Pro136Ser) (Figure 8, A and B), which is predicted to be damaging using SIFT prediction and Polymorphism Phenotyping version 2 (PolyPhen-2) (data not shown). The patient’s clinical background is described in Table 1. In brief, she experienced vomiting, stomach pain, and difficulty feeding as a child. A diagnosis of EoE was made at 12 years, and the patient was unsuccessfully treated with food trials but improved with prednisone. She eventually became dependent upon total parenteral nutrition, but her clinical symptoms progressed to include chronic fatigue, heat intolerance, and postural orthostatic tachycardia syndrome. She has an extensive work up for possible mitochondrial disease. She had complex IV deficiency as seen on muscle biopsy, but electron microscopy, mitochondrial DNA whole-genome sequencing, liver function, acylcarnitine level, organic acids, amino acid profiles, lactate/pyruvate levels, electromyogram, and brain MRI were all normal.
Figure 8. OGDHL mutation induces mitochondrial dysfunction.
Schematic of the oxoglutarate dehydrogenase-like (OGDHL) gene locus and location of variants identified in this study; the 2 OGDHL variants identified by whole-exome sequencing (WES) are denoted by dashed arrows (A). Chromatograms from Sanger sequencing validating the seven heterozygous OGDHL variants (B). Mitochondrial function tests (oxygen consumption rate [OCR]: basal respiration, ATP production, and maximal respiration) from primary fibroblasts from a normal control (NL; unshaded squares) or a patient with eosinophilic esophagitis (EoE; shaded circles) with an OGDHL mutation (EoE c.406 cytosine [C] > thymine [T]) (C). Metabolic pathway of dehydrogenase E1 and transketolase domain–containing 1 (DHTKD1) and OGDHL (D). Data in C are presented as the mean ± SD from 4 cycles per measurement and at least 6 wells for each cell line. *P < 0.05 and ***P < 0.01 compared with NL done by unpaired t tests with Gaussian Distribution and 95% confidence interval.
To further test the hypothesis that mutations of OGDHL are associated with EoE, we mined the WES data for damaging mutations in the OGDHL genes and identified 2 rare, damaging mutations (Table 3). We next sought to identify additional OGDHL coding variants and performed Sanger sequencing of the 23 exons of OGDHL in a separate cohort of 49 patients with EoE. These direct sequencing efforts identified 5 additional OGDHL coding variants (Table 3 and Figure 8, A and B); similar sequencing efforts of the same cohort did not identify additional DHTKD1 variants (data not shown). Five of 7 OGDHL variants have not been reported in the NHLBI ESP6500 data, whereas the remaining 2 variants were detected with MAFs of 0.0028 and 0.0042 (Table 3). These results support that mutations in both DHTKD1 and its homolog OGDHL are associated with EoE. Notably, mitochondrial function testing on fibroblasts from this patient harboring the c.406C>T variant demonstrated attenuated mitochondrial function similar to the results of the patient’s muscle biopsy and the effects of the DHTKD1 c.1897-1G>A mutation (Figure 8C).
Table 3. Rare, damaging OGDHL variants identified in EoE patients by WES or Sanger sequencing in patients with EoE.
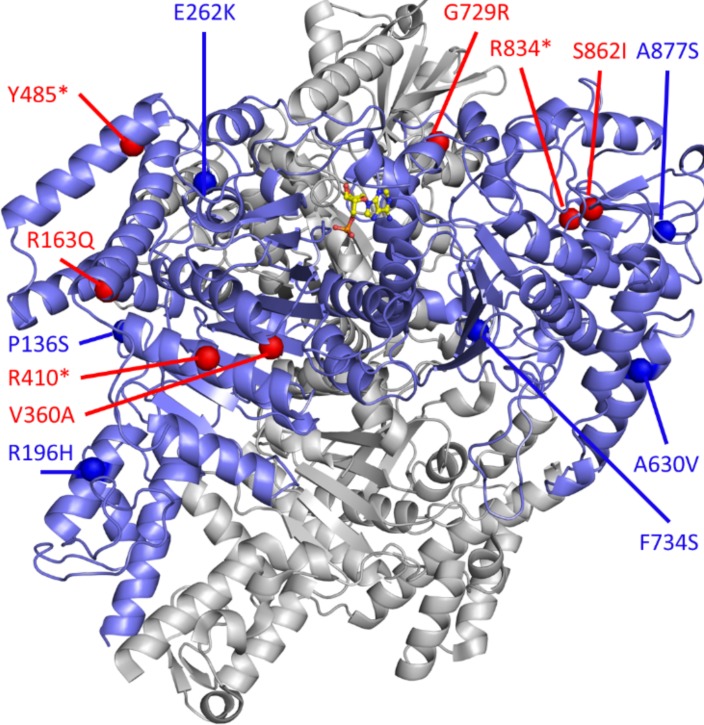
We ended up identifying 7 variants in OGDHL in patients with EoE. OGDHL exhibits high sequence homology to DHTKD1 and acts on a similar metabolic pathway (47) (Figure 8D and Supplemental Figure 1). To examine the possible mechanisms of how the variants possibly affect the protein function, we mapped all the variants found in OGDHL to the 3-D protein structure. As shown in Figure 9, we found that some of the missense variants in OGDHL (Pro136Ser, alanine [Ala] 630 valine [Val], glutamic acid [Glu] 262 lysine [Lys], phenylalanine [Phe] 734Ser, and Ala877Ser) were buried inside the protein and that other putative pathogenic variants (Arg196 histidine [His]) were at the protein surfaces. Surprisingly, we did not observe variants around the ligand-binding pocket. In general, variants buried inside the protein affect the stability of the protein, whereas variants on the surface are more likely to affect the protein-protein interaction (48). The variant Ala877Ser appears to be partially exposed and lies at the periphery of the structure, which could destabilize OGDHL.
Figure 9. 3-D protein structure of DHTKD1/OGDHL.
Dehydrogenase E1 and transketolase domain–containing 1 (DHTKD1)/oxoglutarate dehydrogenase-like (OGDHL) (blue) complexed with AMP (silver); the structure is built based on a homology model of E. coli sucA. The residue position of missense variants and starting point of frameshifts or truncations are indicated for DHTKD1 (red spheres) and OGDHL (blue spheres).
Inflammatory gene expression enriched in multiplex family with EoE and DHTKD1 mutation.
We were interested in determining how, in the context of a proallergic inflammatory environment, the loss of DHTKD1 could contribute to EoE pathogenesis. We sought to determine family-specific changes within the 1,607 EoE transcriptome genes (8) in individuals I.1 and II.1 from family 443 when compared with other patients with active EoE. Thirty-six EoE transcriptome genes were found to be significantly dysregulated in the select, affected members of family 443 (I.1 and II.1) compared with unrelated patients with EoE (P < 0.05, fold change > 2.0) (Figure 10A and Supplemental Table 4). Moreover, the family 443–specific gene signature was significantly enriched in immune/inflammatory response genes — including CCL26, which was upregulated 2.3-fold in the affected members of family 443 compared with other patients with active EoE — and also enriched in the mast cell genes carboxypeptidase A3 (CPA3) and tryptases α/β1 and β2 (TPSAB1 and TPSB2, respectively) (Figure 10B and Supplemental Table 4). The increase in CCL26 was not due to differences in esophageal IL13 levels, which were not significantly different across the 2 cohorts (data not shown). Gene network analysis also showed enrichment of 3 genes encoding mitochondrial outer membrane proteins (RSAD2, acyl-CoA synthetase long chain family member 5 [ACSL5], and IFN-α inducible protein 27 [IFI27]) that were significantly dysregulated in individuals from family 443 (Figure 10B). Notably, 12 of the 36 family 443–specific genes, including RSAD2 and ACSL5, were differentially regulated after IL-13 treatment in an esophageal epithelial cell line grown in a 3-D ALI culture system (15) (P < 0.05, fold-change > 2.0) (Figure 10C).
Figure 10. Dysregulated EoE transcriptome genes in family 443.
Heatmap from RNA sequencing of patient esophageal biopsies showing log2 fragments per kilobase of transcript per million mapped reads (FPKM) values for EoE transcriptome genes (n = 36) and significant dysregulation in family 443 individuals I.1 and II.1 during active disease (dashed rectangle) compared with unrelated patients with active eosinophilic esophagitis (EoE) and normal controls (NL) (P < 0.05, fold change > 2.0) (A). Network analysis of family 443–specific EoE transcriptome genes (red hexagons) showing significantly enriched Gene Ontology (GO) terms (B). Heatmap from RNA sequencing of esophageal epithelial cell line (EPC2) cells grown at the air-liquid interface (ALI) showing log2 FPKM values of dysregulated genes in family 443 (from A) that are also regulated by IL-13 (+ IL-13; 100 ng/ml for 6 days) in vitro (n = 12 genes) (P < 0.05, fold change > 2.0 compared with untreated [untx] cells) (C). FPKM plot for RSAD2 from normal control (NL, n = 6; unshaded squares) and EoE patients (EoE, n = 10; shaded circles) (D).Quantitative PCR for radical S-adenosyl methionine domain containing 2 (RSAD2) from nonsilencing control (NSC; black circles) and dehydrogenase E1 and transketolase domain–containing 1 (DHTKD1) shRNA–transduced (black triangles) EPC2 cells untreated or treated with IL-13 (100 ng/mL for 24 h) in the ALI system (E). Data in D are presented as mean ± SD, and data in E are presented as the mean ± SEM from 3 independent experiments performed in triplicate and are normalized to untreated nonsilencing control (NSC) cells. Statistical analyses used were unpaired t test with Welch’s correction (D) and 1-way ANOVA with Sidak multiple comparison test (E). NL, normal control. *P < 0.05, **P < 0.01.
In an effort to mechanistically connect the DHTKD1 haploinsufficiency with exacerbated Th2 responses, we focused on the increased expression of RSAD2 that was observed in family 443 and in IL-13–treated cells. RNA-sequencing of esophageal biopsies from EoE and normal patients indeed showed that RSAD2 was elevated in diseased individuals (Figure 10D). RSAD2 encodes a protein known as viperin, an intracellular IFN-inducible antiviral protein (49) that has been shown to promote Th2 cytokine production in T cells. Using our in vitro system, DHTKD1 gene silencing led to a significant increase in RSAD2 expression at baseline compared with control shRNA (nonsilencing control; NSC) cells (Figure 10E). Interestingly, 24-hour treatment of DHTKD1-deficient cells with IL-13 increased RSAD2 expression relative to IL-13–treated NSC cells.
Discussion
The data presented herein provide the first genetic and functional evidence to our knowledge supporting an association of DHTKD1 and OGDHL variants and mitochondrial dysfunction with EoE. We have identified 2 DHTKD1 mutations in patients with EoE, Arg834* in family 443 and c.1897-1G>A in family 808, which alter endogenous DHTKD1 expression and/or impair mitochondrial function. A recent study demonstrated reduced mitochondrial respiratory chain activities and coenzyme Q10 (CoQ10) levels in muscle tissue from patients with EoE (26). Indeed, individual II.3 from family 443 had a history of low plasma CoQ10 levels while on a formula-supplemented diet devoid of CoQ10. Given the muscle pathology in family 443 (e.g., muscle weakness, constipation), future studies to fully assess mitochondrial function in the muscle tissue of these patients are warranted. However, in addition to the DHTKD1 mutations in families 443 and 808, we observed enrichment in rare, potentially damaging DHTKD1 mutations in a larger cohort of patients with EoE when compared with individuals without EoE of similar ancestry from the 1000 Genomes Project. We also identified rare mutations in the DHTKD1 homolog OGDHL, adding credence to the potential importance of DHTKD1/OGDHL in the 2-oxoglutarate/2-oxoadipate pathway as an underlying genetic predisposition to EoE and to the idea that this may, in part, contribute to the associated mitochondrial phenotype in a subset of patients.
Interestingly, DHTKD1 and OGDHL are structurally and functionally related and have similarity with 2-oxoglutarate dehydrogenase (OGDH), although they function in distinct steps in 2-oxogulutarate/2-oxoadipate metabolism. The OGDH complex converts 2-oxoglutarate to succinyl-CoA and CO2. This complex, also known as the α-ketoglutarate dehydrogenase complex, is composed of OGDH (E1), dihydrolipoamide succinyltransferase (DLST, E2), and lipoamide dehydrogenase (DLDH, E3) (50). Structure-function analysis showed that DHTKD1 and OGDHL are highly homologous to OGDH (E1). It appears that OGDHL represents a more recently discovered isoform of OGDH, whereas DHTKD1 mainly differs from its homologs at the N-terminus and substrate-binding pocket (Supplemental Figure 1) (47). Both OGDHL and DHTKD1 have a high probability of localizing to the mitochondria and comprise the 2-oxo acid dehydrogenase and transketolase domains. The N-terminal part of E1 is known to control the heterologous protein-protein interactions (47, 50). Hence, the structural differences between E1 and the E1-like proteins (DHTKD1 and OGDHL) in these regions may point to the different interactions. A functional study also suggests that DHTKD1 and OGDHL are similar to the OGDH E1 component and play an important role in energy production (51). In vitro depletion of DHTKD1 via RNA interference led to reduced production of ATP, suggesting an essential role for DHTKD1 in normal mitochondrial function and energy production (32).
Compared with DHTKD1, OGDHL preserves all the structural features of E1 associated with the substrate specificity and has substrate specificity for 2-oxoadipate (47). The signatures of Ca2+-binding sites were found in the Ca2+-activated OGDH and OGDHL but not in DHTKD1 (47). The N-terminus of DHTKD1 is about 60 amino acid residues shorter than that of OGDHL; even a small deletion within this region in OGDHL may affect its interaction with another subunit and the substrate-binding pocket. Therefore, DHTKD1 is most probably a structural analogue of OGDH. These differences suggest changes in heterologous protein interactions and accommodation of more polar and/or bulkier structural analogs of 2-oxoglutarate. This analog is most likely 2-oxoadipate, as it was shown that DHTKD1 is involved in the decarboxylation of 2-oxoadipate to glutaryl-CoA. Furthermore, DHTKD1 mutations increased levels of 2-oxoadipate in fibroblasts and can be normalized upon lentiviral expression of the WT DHTKD1 mRNA (33), demonstrating that DHTKD1 codes for the enzyme mediating the last step in the L-lysine degradation pathway (33). Additionally, all patients in this study were found to carry mutations either in DHTKD1 or in OGDHL, suggesting that — despite their structural and functional similarity — there is no redundancy or functional compensation between them. Finally, the link between the enzymes (DHTKD1 and OGDHL) in the lysine metabolic pathway and EoE provides potential targets for EoE treatment.
Patients with the mutations in OGDHL presented with EoE and dysfunction of mitochondria. Notably, we observed a similar effect of the OGDHL c.406C>T, p.Pro136Ser mutation on mitochondrial function in patient fibroblasts. The patient’s clinical presentation, biochemical evidence in muscle biopsy, and fibroblast and morphologic studies all support mitochondrial dysfunction. Although other groups have shown that the loss of OGDHL is associated with increased AKT signaling and cell survival, albeit under baseline (i.e., noninflammatory) conditions (52), the molecular mechanism of how OGDHL-driven mitochondrial dysfunction contributes to EoE has yet to be determined. These findings, along with the variable phenotypes seen in the reported patients, underscore the genetic and phenotypic heterogeneity associated with mitochondriopathies and further suggest that the particular disease manifestation may be dictated by either a second genetic event or an environmental stimulus.
Mitochondrial dysfunction and the generation of ROS have been demonstrated to modify asthma-like responses in allergen-challenged mice (53, 54). Our data, as well as those from other groups, have shown increased ROS production in DHTKD1-deficient esophageal epithelial and hepatic carcinoma (HepG2) cell lines, respectively. Notably, DHTKD1 gene silencing in esophageal epithelial cells had a significant increase in ROS, which was in line with what has previously been reported in HepG2 cells (32). With regard to EoE, the increase in ROS production could heighten esophageal epithelial cell sensitivity to the Th2 cytokine milieu. For instance, IL-13 signaling through JAK1 and ERK can stimulate ROS generation in intestinal epithelial cells, which, in turn, can potentiate IL-13 activation of ERK and STAT6, forming a positive-feedback mechanism (55). Thus, DHTKD1 deficiency in family 443 may exacerbate disease flares and contribute to the altered EoE transcriptome gene expression. Our findings that the 36 EoE transcriptome genes specifically dysregulated in family 443 were significantly enriched in immune/inflammatory responses and that one-third of these genes were also regulated by IL-13 in an in vitro system support this hypothesis.
The functional studies presented herein have demonstrated a role for DHTKD1 and OGDHL in regulating mitochondrial function within the structural cells of the esophagus, specifically fibroblasts harboring the potentially novel DHTKD1 splicing mutation or OGDHL missense mutation esophageal epithelial cell lines deficient in DHTKD1. Mitochondrial defects, however, can affect additional cells types, such as mast cells and eosinophils, not analyzed in this study (27, 28, 30). In addition to the increase in ROS production, the loss of DHTKD1 resulted in increased expression of the mitochondrial gene RSAD2 that was increased by IL-13 through an as-yet undefined mechanism. RSAD2 is an EoE transcriptome gene that was upregulated by 6.9-fold in individuals from family 443 with active disease compared with NL individuals, representing a 2.5-fold increase compared with unrelated patients with active EoE. Interestingly, production of IL-4, IL-5, and IL-13 is attenuated in Rsad2-deficient CD4+ T cells after activation (56). Although our work on RSAD2 has been done in epithelial cells, it is interesting to speculate that RSAD2 could be involved early during disease onset (in epithelial cells or T cells). Thus, further studies investigating the biological impact of loss-of-function variants of DHTKD1 and OGDHL in esophageal epithelial cells, together with studies in key immune cells (e.g., mast cells, eosinophils, and Th2 cells), could provide additional insight into the pathologic effects of DHTKD1 regulation of mitochondrial function and the expression of Th2-promoting molecules such as RSAD2 in EoE.
We have also used WES to identify rare genetic variants in patients with EoE. Though disease causality has yet to be demonstrated for the de novo and compound heterozygous variants identified herein, we present plausible candidate genes functioning in many pathways germane to the pathophysiology of EoE (e.g., DOCK8, MYD88, ALOX15, IL5RA, LAMA5, TLR10, and WDR36). Furthermore, by linking the findings in genetically related EoE patients to a separate case of an EoE patient with a mitochondrial disorder, our data suggest mitochondrial dysfunction as a contributory pathway; this contributory pathway would undoubtedly interact with additional genetic and environmental factors that contribute to type 2 immunity, at least in the context of the esophagus.
In summary, our findings provide potentially novel insights into the diverse landscape of rare genetic variants present in EoE and underscore the level of genetic complexity of this rare disease and its related subphenotypes. Our findings call attention to an underlying genetic predisposition to EoE focused on rare variants in DHTKD1 and OGDHL and present evidence that mitochondrial dysfunction contributes to variable disease phenotype.
Methods
Cell lines.
Primary esophageal epithelial cells were isolated and cultured as previously described (9). The immortalized human esophageal epithelial cell line (EPC2) (provided by Anil Rustgi, University of Pennsylvania, Philadelphia, Pennsylvania, USA) was cultured as previously described (57, 58).
Patients.
All patients analyzed in this study were consented, and studies were approved by the CCHMC IRB. WES data from a total of 65 patients with physician-diagnosed EoE and 63 unaffected family members were assessed for rare, potentially damaging variants as described below. The patient characteristics of the index family (family 443) are described in Table 1.
WES.
WES was performed in the CCHMC Genetic Variation and Gene Discovery Core, the Oklahoma Medical Research Foundation (OMRF) Genomics facility, and Perkin Elmer. Libraries were prepared using the Illumina TruSeq capture kit and sequenced on an Illumina Hiseq2000 to generate 100-base, paired-end reads. Sequencing reads were aligned, and variant calls were made simultaneously following the Genome Analysis Toolkit (GATK) Best Practices using the GATK Unified Genotyper (59–61).
WES variant filtering.
Variant filtering was performed using SNP and Variation Suite (SVS) software (Golden Helix Inc.) (Figure 1). Filters included minimum read depths >15, genotype quality scores >20, and location within a region of segmental duplication defined as genomic regions of ≥1 kb exhibiting greater than 90% sequence similarity. Genotype calls were filtered using alternate allele ratios of <0.15 for homozygotes for the reference allele, 0.30–0.70 for heterozygotes, and >0.85 for homozygotes for the alternate allele. In addition to the public databases described below, variant frequencies were filtered against an internal CCHMC database of WES data from 357 patients with various diseases, including macrophage-activating syndrome, systemic lupus erythematosus, and juvenile idiopathic arthritis. Rare variants were filtered for using a MAF cutoff of <0.01 in the European ancestry populations from the 1,000 Genomes Project Phase 1 and ESP6500SI-V2 data sets (http://www.internationalgenome.org/category/phase-1/) (62, 63). A MAF threshold of <0.05 with rare variants was used to define compound heterozygous variants. Functional predictions were made using the database for nonsynonymous single-nucleotide variant functional predictions (dbNSFP) 2.3 (https://omictools.com/database-for-nonsynonymous-snps-functional-predictions-tool) (http://evs.gs.washington.edu/EVS/); rare variants were considered to be potentially damaging if they were predicted to not be tolerated or to not be benign in 4 of 5 algorithms (SIFT, PolyPhen2, MutationTaster, MutationAssessor, and Functional Analysis Through Hidden Markov Models [FATHMM]) (64–69).
DHTKD1 rare variant burden analysis.
Gene-based rare variant association testing was performed in SVS using the kernel-based adaptive cluster (KBAC) method with permutation testing (n = 1,000) as described by Liu and Leal (70): 43,482 rare, damaging variants identified as described above were compared between 37 unrelated patients with EoE and 379 control samples of European descent from the Phase 1 integrated variant release v3 Exome data from the 1000 Genome Project.
Whole-genome expression profiling.
RNA-seq was performed on patient biopsies from 6 healthy (normal) controls, 8 patients with active EoE, and 3 of the 4 affected individuals in family 443 (2 with active EoE, 1 with in EoE remission) as described previously (8). The EoE transcriptome was defined as those genes (n = 1,607) showing a fold change >2.0 between all patients with active EoE and normal controls at P < 0.05 (8). Analysis of the EoE transcriptome genes showing significantly altered levels in the 2 assessed individuals in family 443 with active EoE compared with the 8 patients with active EoE and the 6 normal controls was performed using a 1-way ANOVA with a Tukey post-hoc test and Benjamini-Hochberg FDR (P < 0.05, fold change >2.0). Sequencing reads were visualized. RNA-seq from esophageal epithelial cells grown at the ALI in the presence or absence of IL-13 (Peprotech, catalog 200-13) was performed as described (6, 9). Genes showing significant dysregulation (1-way ANOVA, P < 0.05, fold change > 2.0) after IL-13 treatment (n = 781 genes) were intersected with those altered in the affected family members (n = 33 genes), yielding 11 genes. All expression analyses and hierarchical clustering were performed using GeneSpring GX (Agilent Technologies). RNA-seq data are available in the GEO (GSE58640 and GSE65335).
Network analysis.
EoE transcriptome genes (n = 36) identified as significantly dysregulated in family 443 were analyzed for functional enrichment using ToppCluster (71) with a P value cutoff of 0.05. Gene-level network generation was performed using Cytoscape (72).
Sanger sequencing of DHTKD1 and OGDHL.
The 2 DHTKD1 variants (c.2500C>T, p.Arg834* and c.1897-1G>A) identified by WES were validated by PCR and Sanger sequencing at the CCHMC Genetic Variation and Gene Discovery Core. Chromatograms were analyzed using Sequencher software (Gene Codes Corporation). Similarly, the OGDHL variants were validated from the WES data or identified by PCR and Sanger sequencing at the CCHMC Genetic Variation and Gene Discovery Core. Chromatograms were analyzed using DNASTAR Lasergene (DNASTAR Inc.).
The structures of DHTKD1/OGDHL were built off of the homologous structure of sucA for E. coli complexed with AMP in a homodimer (73). The homology models were built using the homology modeling module of Rosetta; the sequence alignments were generated using HHblits and confirmed using the multiple sequence alignment server T-Coffee (74–76). The energy calculations of the destabilization of both internal structure and the protein-protein binding were calculated using the FoldX program (77). The potential phosphorylation and modifications sites of DHTKD1/OGDHL were checked in the eukaryotic linear motif (ELM) resource to identify potential motifs and were checked in the experimental database of known sites, Phosphosite (78, 79). The images were generated using PYMOL (The PyMOL Molecular Graphics System, Version 1.8, Schrödinger).
IHC staining.
IHC staining for DHTKD1 in esophageal biopsies was performed at the Pathology Research Core at CCHMC). Deparaffinized slides were pretreated with EDTA for 30 minutes prior to staining with rabbit anti-DHTKD1 (HPA037950) (MilliporeSigma) at a 1:50 dilution or rabbit IgG as a negative control (data not shown). Slides were developed using the Ventana ultraView Universal DAB Detection Kit (Ventana Medical Systems Inc.) and then counterstained with hematoxylin and bluing reagent.
Restriction fragment length polymorphism mapping (RFLP).
Using esophageal biopsy cDNA (generated as described below), a coding region of DHTKD1 spanning exons 14–16 was amplified by PCR and left untreated or treated with the restriction enzyme DdeI (New England Biolabs, catalog R0175S) for 1 hour at 37°C. Fragmentation patterns were analyzed by electrophoresis using a 1% agarose gel.
DHTKD1 gene silencing by shRNA.
Lentivirus production from the pGIPZ lentiviral plasmid (Thermo Fisher Scientific) containing an NSC shRNA or shRNA targeting DHTKD1 was performed at the CCHMC Viral Vector Core facility. Stably transduced EPC2 cells were generated by negative selection in puromycin, which was maintained in the culture media throughout all experiments. DHTKD1 knockdown efficiency was assessed by quantitative PCR (qPCR) as described (15) using the following primers: forward (5′ - GCTGGAGAATGTGCCTGAAA - 3′) and reverse (5′ - CCTCAAGTGAGGCCTCTTCC - 3′).
Esophageal epithelial cell culture system.
The culture system has been described previously (15). Briefly, untreated EPC2 cells or EPC2 cells expressing shRNAs were grown to confluence on semipermeable membranes. Cells were then grown submerged in high-calcium (1.8 mM) media for 5 days, after which the media from the upper chamber was removed; cells were then grown for an additional 6 days at the ALI, untreated or in the presence of IL-13 (100 ng/ml).
Mitochondrial function testing.
Mitochondrial function (basal and maximal respiration rates, ATP production) in shRNA-expressing EPC2 cells (20,000 cells per well) and primary esophageal fibroblasts (10,000 per well) was assessed using the XF Cell Mito Stress Test Kit Assay according to the manufacturer’s protocol (Seahorse Bioscience) and as described (80). Briefly, ATP production measurements were done using Seahorse XF analyzer after treating cells with carbonyl cyanide-4-(trifluromethoxy) phenylhydrazone (FCCP), and ATP production rate was calculated based on the oxygen consumption after FCCP treatment, thus capturing ATP production rate. H2O2 production was measured in shRNA-expressing EPC2 cells using the Amplex Red Hydrogen Peroxide/Peroxidase Assay Kit (Invitrogen) as described (16).
qPCR.
RNA isolated from esophageal biopsies and the ALI samples were used for cDNA synthesis (iScript, Bio-Rad). qPCR analysis using SYBR Green was performed (Bio-Rad). Primers used for amplification of (and normalization to) the housekeeping gene GAPDH have been described previously (15). Normalized data were assessed for statistical significance using an unpaired, two-tailed t test with Welch’s correction and a significance threshold of P < 0.05.
Statistics.
Statistical analysis was performed using Prism 6.0 software. Comparison between 2 groups of data was done using a 2-tailed t test, and group comparisons were performed using ordinary 1-way ANOVA with Tukey’s or Dunn’s correction for multiple comparisons where appropriate. A statistical probability of P < 0.05 was considered significant. Dot plots were plotted showing both error bars, ± SEM or SD.
Study approval.
This study involving human subjects was conducted under an approved IRB (Cincinnati Children’s Hospital Medical Center) protocol number 2008-0090. An informed consent form was received from the subjects prior to inclusion in the study. Study participants were also made aware that their involvement in the study was voluntary and their declination to participate did not interfere with their standard of care.
Author contributions
MER conceived the study, interpreted data, and edited the manuscript. TH conceived OGDHL-related ideas and experiments, wrote the manuscript, analyzed and interpreted data, and edited the manuscript. JDS analyzed WES and RNA-seq data, performed experiments, interpreted results, and wrote the manuscript. KKC conducted experiments, analyzed and interpreted data, and wrote and edited the manuscript. TW analyzed and interpreted WES data, interpreted RNA-seq data, and edited the manuscript. XW, YP, and QW performed Sanger sequencing and interpreted data. AC performed 3-D modeling. EMS assisted with sample processing. MHC, JPA, and PEP assisted with patient selection and sample acquisition. PJD, BJA, LCK, KMK, and JBH analyzed and interpreted WES data.
Supplementary Material
Acknowledgments
We would like to thank the NHLBI GO ESP and its ongoing studies that produced and provided exome variant calls for comparison: the Lung GO Sequencing Project (HL-102923), the WHI Sequencing Project (HL-102924), the Broad GO Sequencing Project (HL-102925), the Seattle GO Sequencing Project (HL-102926), and the Heart GO Sequencing Project (HL-103010). This work was supported in part by NIH U19 AI070235, NIH R01 DK076893, NIH R01 DK107502, NIH P30 AR070549, PHS grant P30 DK0789392, NIH R01 AI124355, NIH R37 A1045898, NIH K24DK100303, NIH U01 HG0088666, NIH U01 AI130830, P30 AR070599, NIH R01 AI024717, the US Department of Veterans Affairs, the Thrasher Research Fund Early Career Award (NR-0171), the Campaign Urging Research for Eosinophilic Disease (CURED) Foundation, the Buckeye Foundation, the Sunshine Charitable Foundation and its supporters, Denise A. Bunning and David G. Bunning, and the Cincinnati Children’s Hospital Research Foundation. We also thank Shawna Hottinger for editorial assistance.
Version 1. 04/19/2018
Electronic publication
Funding Statement
This work was supported in part by NIH U19 AI070235, NIH R01 DK076893, NIH R01 DK107502, NIH P30 AR070549, PHS Grant P30 DK0789392, NIH R01 AI124355, NIH R37 A1045898, NIH K24DK100303, NIH U01 HG0088666, NIH U01 AI130830, P30 AR070599, NIH R01 AI024717, the US Department of Veterans Affairs, the Thrasher Research Fund Early Career Award (NR-0171), the Campaign Urging Research for Eosinophilic Disease (CURED) Foundation, the Buckeye Foundation, the Sunshine Charitable Foundation and its supporters, Denise A. Bunning and David G. Bunning, and the Cincinnati Children’s Hospital Research Foundation.
Footnotes
Conflict of interest: MER is a consultant for N.K.T. Therapeutics, PulmOne Therapeutics, Spoon Guru, Celgene, Shire, Astra Zeneca, and Novartis and has an equity interest in the first three listed and Immune Pharmaceuticals and royalties from reslizumab (Teva Pharmaceuticals). MER is an inventor of several patents, owned by Cincinnati Children’s Hospital.
Reference information: JCI Insight. 2018;3(8):e99922. https://doi.org/10.1172/jci.insight.99922.
Contributor Information
Kiran KC, Email: kiran.kc@cchmc.org.
Xinjian Wang, Email: Xinjian.Wang@cchmc.org.
Ting Wen, Email: ting.wen@cchmc.org.
Adam Chamberlin, Email: achamberlin@ambrygen.com.
J. Pablo Abonia, Email: pablo.abonia@cchmc.org.
Yanyan Peng, Email: Yanyan.Peng@cchmc.org.
Qiang Wu, Email: bbmcwq@163.com.
Phillip J. Dexheimer, Email: Phillip.Dexheimer@cchmc.org.
Taosheng Huang, Email: taosheng.huang@cchmc.org.
Marc E. Rothenberg, Email: rothenberg@cchmc.org.
References
- 1.Liacouras CA, et al. Eosinophilic esophagitis: updated consensus recommendations for children and adults. J Allergy Clin Immunol. 2011;128(1):3–20.e6; quiz 21. doi: 10.1016/j.jaci.2011.02.040. [DOI] [PubMed] [Google Scholar]
- 2.O’Shea KM, et al. Pathophysiology of Eosinophilic Esophagitis. Gastroenterology. 2018;154(2):333–345. doi: 10.1053/j.gastro.2017.06.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alexander ES, et al. Twin and family studies reveal strong environmental and weaker genetic cues explaining heritability of eosinophilic esophagitis. J Allergy Clin Immunol. 2014;134(5):1084–1092.e1. doi: 10.1016/j.jaci.2014.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sherrill JD, Rothenberg ME. Genetic dissection of eosinophilic esophagitis provides insight into disease pathogenesis and treatment strategies. J Allergy Clin Immunol. 2011;128(1):23–32. doi: 10.1016/j.jaci.2011.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kottyan LC, Rothenberg ME. Genetics of eosinophilic esophagitis. Mucosal Immunol. 2017;10(3):580–588. doi: 10.1038/mi.2017.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kottyan LC, et al. Genome-wide association analysis of eosinophilic esophagitis provides insight into the tissue specificity of this allergic disease. Nat Genet. 2014;46(8):895–900. doi: 10.1038/ng.3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rothenberg ME, et al. Common variants at 5q22 associate with pediatric eosinophilic esophagitis. Nat Genet. 2010;42(4):289–291. doi: 10.1038/ng.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sherrill JD, et al. Analysis and expansion of the eosinophilic esophagitis transcriptome by RNA sequencing. Genes Immun. 2014;15(6):361–369. doi: 10.1038/gene.2014.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blanchard C, et al. Coordinate interaction between IL-13 and epithelial differentiation cluster genes in eosinophilic esophagitis. J Immunol. 2010;184(7):4033–4041. doi: 10.4049/jimmunol.0903069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sleiman PM, et al. GWAS identifies four novel eosinophilic esophagitis loci. Nat Commun. 2014;5:5593. doi: 10.1038/ncomms6593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sherrill JD, et al. Variants of thymic stromal lymphopoietin and its receptor associate with eosinophilic esophagitis. J Allergy Clin Immunol. 2010;126(1):160–5.e3. doi: 10.1016/j.jaci.2010.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davis BP, et al. Eosinophilic esophagitis-linked calpain 14 is an IL-13-induced protease that mediates esophageal epithelial barrier impairment. JCI Insight. 2016;1(4):e86355. doi: 10.1172/jci.insight.86355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Litosh VA, Rochman M, Rymer JK, Porollo A, Kottyan LC, Rothenberg ME. Calpain-14 and its association with eosinophilic esophagitis. J Allergy Clin Immunol. 2017;139(6):1762–1771.e7. doi: 10.1016/j.jaci.2016.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blanchard C, et al. IL-13 involvement in eosinophilic esophagitis: transcriptome analysis and reversibility with glucocorticoids. J Allergy Clin Immunol. 2007;120(6):1292–1300. doi: 10.1016/j.jaci.2007.10.024. [DOI] [PubMed] [Google Scholar]
- 15.Sherrill JD, et al. Desmoglein-1 regulates esophageal epithelial barrier function and immune responses in eosinophilic esophagitis. Mucosal Immunol. 2014;7(3):718–729. doi: 10.1038/mi.2013.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zuo L, et al. IL-13 induces esophageal remodeling and gene expression by an eosinophil-independent, IL-13R alpha 2-inhibited pathway. J Immunol. 2010;185(1):660–669. doi: 10.4049/jimmunol.1000471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rothenberg ME, et al. Intravenous anti-IL-13 mAb QAX576 for the treatment of eosinophilic esophagitis. J Allergy Clin Immunol. 2015;135(2):500–507. doi: 10.1016/j.jaci.2014.07.049. [DOI] [PubMed] [Google Scholar]
- 18.Rochman M, et al. Neurotrophic tyrosine kinase receptor 1 is a direct transcriptional and epigenetic target of IL-13 involved in allergic inflammation. Mucosal Immunol. 2015;8(4):785–798. doi: 10.1038/mi.2014.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vernon N, Shah S, Lehman E, Ghaffari G. Comparison of atopic features between children and adults with eosinophilic esophagitis. Allergy Asthma Proc. 2014;35(5):409–414. doi: 10.2500/aap.2014.35.3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brown-Whitehorn TF, Spergel JM. The link between allergies and eosinophilic esophagitis: implications for management strategies. Expert Rev Clin Immunol. 2010;6(1):101–109. doi: 10.1586/eci.09.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abonia JP, et al. High prevalence of eosinophilic esophagitis in patients with inherited connective tissue disorders. J Allergy Clin Immunol. 2013;132(2):378–386. doi: 10.1016/j.jaci.2013.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neptune ER, et al. Dysregulation of TGF-beta activation contributes to pathogenesis in Marfan syndrome. Nat Genet. 2003;33(3):407–411. doi: 10.1038/ng1116. [DOI] [PubMed] [Google Scholar]
- 23.Gallo EM, et al. Angiotensin II-dependent TGF-β signaling contributes to Loeys-Dietz syndrome vascular pathogenesis. J Clin Invest. 2014;124(1):448–460. doi: 10.1172/JCI69666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aceves SS, Newbury RO, Dohil R, Bastian JF, Broide DH. Esophageal remodeling in pediatric eosinophilic esophagitis. J Allergy Clin Immunol. 2007;119(1):206–212. doi: 10.1016/j.jaci.2006.10.016. [DOI] [PubMed] [Google Scholar]
- 25.Aceves SS, Chen D, Newbury RO, Dohil R, Bastian JF, Broide DH. Mast cells infiltrate the esophageal smooth muscle in patients with eosinophilic esophagitis, express TGF-β1, and increase esophageal smooth muscle contraction. J Allergy Clin Immunol. 2010;126(6):1198–204.e4. doi: 10.1016/j.jaci.2010.08.050. [DOI] [PubMed] [Google Scholar]
- 26.Miles MV, et al. Acquired coenzyme Q10 deficiency in children with recurrent food intolerance and allergies. Mitochondrion. 2011;11(1):127–135. doi: 10.1016/j.mito.2010.08.010. [DOI] [PubMed] [Google Scholar]
- 27.Zhang B, Asadi S, Weng Z, Sismanopoulos N, Theoharides TC. Stimulated human mast cells secrete mitochondrial components that have autocrine and paracrine inflammatory actions. PLoS One. 2012;7(12):e49767. doi: 10.1371/journal.pone.0049767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang B, et al. Human mast cell degranulation and preformed TNF secretion require mitochondrial translocation to exocytosis sites: relevance to atopic dermatitis. J Allergy Clin Immunol. 2011;127(6):1522–31.e8. doi: 10.1016/j.jaci.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang L, et al. The Phosphatidylcholine Transfer Protein Stard7 is Required for Mitochondrial and Epithelial Cell Homeostasis. Sci Rep. 2017;7:46416. doi: 10.1038/srep46416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yousefi S, et al. Catapult-like release of mitochondrial DNA by eosinophils contributes to antibacterial defense. Nat Med. 2008;14(9):949–953. doi: 10.1038/nm.1855. [DOI] [PubMed] [Google Scholar]
- 31.Yang L, Lewkowich I, Apsley K, Fritz JM, Wills-Karp M, Weaver TE. Haploinsufficiency for Stard7 is associated with enhanced allergic responses in lung and skin. J Immunol. 2015;194(12):5635–5643. doi: 10.4049/jimmunol.1500231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu W, et al. DHTKD1 is essential for mitochondrial biogenesis and function maintenance. FEBS Lett. 2013;587(21):3587–3592. doi: 10.1016/j.febslet.2013.08.047. [DOI] [PubMed] [Google Scholar]
- 33.Danhauser K, et al. DHTKD1 mutations cause 2-aminoadipic and 2-oxoadipic aciduria. Am J Hum Genet. 2012;91(6):1082–1087. doi: 10.1016/j.ajhg.2012.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Q, et al. DOCK8 regulates lymphocyte shape integrity for skin antiviral immunity. J Exp Med. 2014;211(13):2549–2566. doi: 10.1084/jem.20141307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lambe T, et al. DOCK8 is essential for T-cell survival and the maintenance of CD8+ T-cell memory. Eur J Immunol. 2011;41(12):3423–3435. doi: 10.1002/eji.201141759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Q, et al. Combined immunodeficiency associated with DOCK8 mutations. N Engl J Med. 2009;361(21):2046–2055. doi: 10.1056/NEJMoa0905506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ngo VN, et al. Oncogenically active MYD88 mutations in human lymphoma. Nature. 2011;470(7332):115–119. doi: 10.1038/nature09671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Treon SP, et al. MYD88 L265P somatic mutation in Waldenström’s macroglobulinemia. N Engl J Med. 2012;367(9):826–833. doi: 10.1056/NEJMoa1200710. [DOI] [PubMed] [Google Scholar]
- 39.Fulkerson PC, Schollaert KL, Bouffi C, Rothenberg ME. IL-5 triggers a cooperative cytokine network that promotes eosinophil precursor maturation. J Immunol. 2014;193(8):4043–4052. doi: 10.4049/jimmunol.1400732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abdulnour-Nakhoul SM, et al. Alterations in junctional proteins, inflammatory mediators and extracellular matrix molecules in eosinophilic esophagitis. Clin Immunol. 2013;148(2):265–278. doi: 10.1016/j.clim.2013.05.004. [DOI] [PubMed] [Google Scholar]
- 41.Matoso A, et al. Expression microarray analysis identifies novel epithelial-derived protein markers in eosinophilic esophagitis. Mod Pathol. 2013;26(5):665–676. doi: 10.1038/modpathol.2013.41. [DOI] [PubMed] [Google Scholar]
- 42.Blanchard C, et al. Eotaxin-3 and a uniquely conserved gene-expression profile in eosinophilic esophagitis. J Clin Invest. 2006;116(2):536–547. doi: 10.1172/JCI26679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ritié L, et al. Abnormal Wnt and PI3Kinase signaling in the malformed intestine of lama5 deficient mice. PLoS ONE. 2012;7(5):e37710. doi: 10.1371/journal.pone.0037710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oosting M, et al. Human TLR10 is an anti-inflammatory pattern-recognition receptor. Proc Natl Acad Sci USA. 2014;111(42):E4478–E4484. doi: 10.1073/pnas.1410293111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lyons JJ, et al. Mendelian inheritance of elevated serum tryptase associated with atopy and connective tissue abnormalities. J Allergy Clin Immunol. 2014;133(5):1471–1474. doi: 10.1016/j.jaci.2013.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Desmet FO, Hamroun D, Lalande M, Collod-Béroud G, Claustres M, Béroud C. Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009;37(9):e67. doi: 10.1093/nar/gkp215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bunik VI, Degtyarev D. Structure-function relationships in the 2-oxo acid dehydrogenase family: substrate-specific signatures and functional predictions for the 2-oxoglutarate dehydrogenase-like proteins. Proteins. 2008;71(2):874–890. doi: 10.1002/prot.21766. [DOI] [PubMed] [Google Scholar]
- 48.Schymkowitz J, Borg J, Stricher F, Nys R, Rousseau F, Serrano L. The FoldX web server: an online force field. Nucleic Acids Res. 2005;33(Web Server issue):W382–W388. doi: 10.1093/nar/gki387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chin KC, Cresswell P. Viperin (cig5), an IFN-inducible antiviral protein directly induced by human cytomegalovirus. Proc Natl Acad Sci USA. 2001;98(26):15125–15130. doi: 10.1073/pnas.011593298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bunik V, Kaehne T, Degtyarev D, Shcherbakova T, Reiser G. Novel isoenzyme of 2-oxoglutarate dehydrogenase is identified in brain, but not in heart. FEBS J. 2008;275(20):4990–5006. doi: 10.1111/j.1742-4658.2008.06632.x. [DOI] [PubMed] [Google Scholar]
- 51.Goncalves RL, Bunik VI, Brand MD. Production of superoxide/hydrogen peroxide by the mitochondrial 2-oxoadipate dehydrogenase complex. Free Radic Biol Med. 2016;91:247–255. doi: 10.1016/j.freeradbiomed.2015.12.020. [DOI] [PubMed] [Google Scholar]
- 52.Sen T, et al. OGDHL is a modifier of AKT-dependent signaling and NF-κB function. PLoS One. 2012;7(11):e48770. doi: 10.1371/journal.pone.0048770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aguilera-Aguirre L, Bacsi A, Saavedra-Molina A, Kurosky A, Sur S, Boldogh I. Mitochondrial dysfunction increases allergic airway inflammation. J Immunol. 2009;183(8):5379–5387. doi: 10.4049/jimmunol.0900228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Khare A, et al. Mitochondrial H2O2 in Lung Antigen-Presenting Cells Blocks NF-κB Activation to Prevent Unwarranted Immune Activation. Cell Rep. 2016;15(8):1700–1714. doi: 10.1016/j.celrep.2016.04.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mandal D, Fu P, Levine AD. REDOX regulation of IL-13 signaling in intestinal epithelial cells: usage of alternate pathways mediates distinct gene expression patterns. Cell Signal. 2010;22(10):1485–1494. doi: 10.1016/j.cellsig.2010.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 56.Qiu LQ, Cresswell P, Chin KC. Viperin is required for optimal Th2 responses and T-cell receptor-mediated activation of NF-kappaB and AP-1. Blood. 2009;113(15):3520–3529. doi: 10.1182/blood-2008-07-171942. [DOI] [PubMed] [Google Scholar]
- 57.Andl CD, et al. Epidermal growth factor receptor mediates increased cell proliferation, migration, and aggregation in esophageal keratinocytes in vitro and in vivo. J Biol Chem. 2003;278(3):1824–1830. doi: 10.1074/jbc.M209148200. [DOI] [PubMed] [Google Scholar]
- 58.Harada H, et al. Telomerase induces immortalization of human esophageal keratinocytes without p16INK4a inactivation. Mol Cancer Res. 2003;1(10):729–738. [PubMed] [Google Scholar]
- 59.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43(5):491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Van der Auwera GA, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013;43:11.10.1–11.1033. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.1000 Genomes Project Consortium. et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491(7422):56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. NHLBI Exome Sequencing Project (ESP): Exome Variant Server. University of Washington. http://evs.gs.washington.edu/EVS. Accessed April 17, 2018.
- 64.Reva B, Antipin Y, Sander C. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res. 2011;39(17):e118. doi: 10.1093/nar/gkr407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schwarz JM, Rödelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods. 2010;7(8):575–576. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- 66.Shihab HA, et al. Predicting the functional, molecular, and phenotypic consequences of amino acid substitutions using hidden Markov models. Hum Mutat. 2013;34(1):57–65. doi: 10.1002/humu.22225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Adzhubei IA, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4(7):1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 69.Liu X, Jian X, Boerwinkle E. dbNSFP v2.0: a database of human non-synonymous SNVs and their functional predictions and annotations. Hum Mutat. 2013;34(9):E2393–E2402. doi: 10.1002/humu.22376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liu DJ, Leal SM. A novel adaptive method for the analysis of next-generation sequencing data to detect complex trait associations with rare variants due to gene main effects and interactions. PLoS Genet. 2010;6(10):e1001156. doi: 10.1371/journal.pgen.1001156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kaimal V, Bardes EE, Tabar SC, Jegga AG, Aronow BJ. ToppCluster: a multiple gene list feature analyzer for comparative enrichment clustering and network-based dissection of biological systems. Nucleic Acids Res. 2010;38(Web Server issue):W96–102. doi: 10.1093/nar/gkq418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Frank RA, Price AJ, Northrop FD, Perham RN, Luisi BF. Crystal structure of the E1 component of the Escherichia coli 2-oxoglutarate dehydrogenase multienzyme complex. J Mol Biol. 2007;368(3):639–651. doi: 10.1016/j.jmb.2007.01.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kim DE, Chivian D, Baker D. Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res. 2004;32(Web Server issue):W526–W531. doi: 10.1093/nar/gkh468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Remmert M, Biegert A, Hauser A, Söding J. HHblits: lightning-fast iterative protein sequence searching by HMM-HMM alignment. Nat Methods. 2011;9(2):173–175. doi: 10.1038/nmeth.1818. [DOI] [PubMed] [Google Scholar]
- 76.Erb I, Notredame C. How should we measure proportionality on relative gene expression data? Theory Biosci. 2016;135(1-2):21–36. doi: 10.1007/s12064-015-0220-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schymkowitz JW, Rousseau F, Martins IC, Ferkinghoff-Borg J, Stricher F, Serrano L. Prediction of water and metal binding sites and their affinities by using the Fold-X force field. Proc Natl Acad Sci USA. 2005;102(29):10147–10152. doi: 10.1073/pnas.0501980102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li J, et al. The Molecule Pages database. Nature. 2002;420(6916):716–717. doi: 10.1038/nature01307. [DOI] [PubMed] [Google Scholar]
- 79.Dinkel H, et al. ELM 2016--data update and new functionality of the eukaryotic linear motif resource. Nucleic Acids Res. 2016;44(D1):D294–D300. doi: 10.1093/nar/gkv1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Simon M, et al. Mutations of human NARS2, encoding the mitochondrial asparaginyl-tRNA synthetase, cause nonsyndromic deafness and Leigh syndrome. PLoS Genet. 2015;11(3):e1005097. doi: 10.1371/journal.pgen.1005097. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.