The CD163 long-range scavenger receptor cysteine-rich (SRCR) repeat was prepared in a Drosophila system and was crystallized using 20% PEG 4000, 0.15 M potassium sodium tartrate tetrahydrate pH 8.5. X-ray crystallographic characterization of this long-range SRCR repeat will provide structural and functional information for the scavenger receptor superfamily.
Keywords: scavenger receptor, long-range scavenger receptor cysteine-rich repeat, SRCR repeat, CD163, X-ray crystallography
Abstract
Scavenger receptors (SRs) play critical roles in various physiological and pathological pathways. One of them, CD163, is a multifunctional endocytic receptor and is characterized by a long-range scavenger receptor cysteine-rich (SRCR) repeat. However, the structural and functional details of this long-range SRCR repeat have not yet been elucidated. In this study, the CD163 long-range SRCR repeat was expressed in Drosophila Schneider 2 cells. The recombinant protein was homogeneous after purification by metal-affinity, cation-exchange and size-exclusion chromatography. Single crystals were obtained using 20% PEG 4000, 0.15 M potassium sodium tartrate tetrahydrate pH 8.5 and diffracted to 3.30 Å resolution. As the first view of a long-range SRCR repeat, this work lays the structural basis for a deep understanding of SRs and their multiple functions.
1. Introduction
Scavenger receptors (SRs), which are multi-domain membrane-associated proteins, were originally identified to recognize and internalize modified lipoproteins, for example oxidized low-density lipoprotein (Greaves & Gordon, 2009 ▸). They were subsequently demonstrated to play critical physiological and pathological roles, including in pathogen clearance, lipid transport, homeostasis and immunity (Canton et al., 2013 ▸). On the basis of their sequences and structural features, SRs are divided into ten classes (classes A–J; Zani et al., 2015 ▸). The class I SRs consist of CD163 (Law et al., 1993 ▸), CD163-like homologue (CD163-L1; Gronlund et al., 2000 ▸), WC1 (Mackay et al., 1986 ▸; Wijngaard et al., 1992 ▸) and SCART1 (Holm et al., 2009 ▸), which are characterized by a long-range repeat of five consecutive scavenger receptor cysteine-rich (SRCR) domains with a small linker domain between the second and third SRCR domains (Van Gorp, Delputte et al., 2010 ▸).
Among them, CD163 is specifically expressed in monocytes and macrophages (Sarrias et al., 2004 ▸; Areschoug & Gordon, 2009 ▸). CD163 primarily functions as a haemoglobin–haptoglobin SR in haem oxidative stress (Kristiansen et al., 2001 ▸; Graversen et al., 2002 ▸). In addition, it serves as a tumour necrosis factor-like weak inducer of apoptosis (TWEAK) SR (Bover et al., 2007 ▸; Moreno et al., 2009 ▸) and an erythroblast adhesion receptor (Fabriek et al., 2007 ▸). Moreover, it is utilized as a signalling sensor for bacteria (Fabriek et al., 2009 ▸) and as an invasion receptor for Porcine reproductive and respiratory syndrome virus (PRRSV; Calvert et al., 2007 ▸) and Simian hemorrhagic fever virus (SHFV; Caì et al., 2015 ▸).
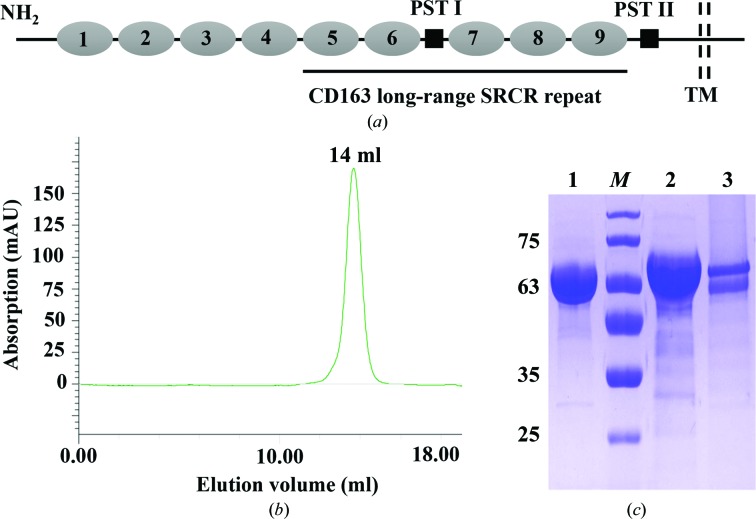
As a type I membrane glycoprotein, CD163 contains nine SRCR domains (SRCR1–9) in its large extracellular region, followed by a single transmembrane domain and a short cytoplasmic tail (Fig. 1 ▸ a; Van Gorp, Van Breedam et al., 2010 ▸). The long-range SRCR repeat of CD163 spans from the fifth SRCR domain (SRCR5) to the ninth SRCR domain (SRCR9). Additionally, a proline–serine–threonine (PST)-rich motif (PST I) is located between the sixth (SRCR6) and seventh (SRCR7) SRCR domains (Fig. 1 ▸ a; Van Gorp, Van Breedam et al., 2010 ▸). Despite numerous studies focusing on CD163, the structural and functional details of CD163, especially its long-range SRCR repeat, are currently not clear, hindering deep understanding of CD163 and SRs.
Figure 1.
Production of recombinant CD163 long-range SRCR repeat. (a) Schematic diagram of CD163 and its long-range SRCR repeat. CD163 consists of nine SRCR domains (1–9), two proline–serine–threonine (PST)-rich motifs (PST I and II), a transmembrane domain (TM) and a cytoplasmic tail. The CD163 long-range SRCR repeat spans from SRCR5 to SRCR9. (b) Size-exclusion chromatography of the CD163 long-range SRCR repeat on a calibrated Superdex 200 10/300 GL column. The target protein eluted at a retention volume of 14 ml, corresponding to approximately 67 kDa. (c) Characterization of recombinant CD163 long-range SRCR repeat by 12% SDS–PAGE after three-step purification. Lane 1, the CD163 long-range SRCR repeat under nonreducing conditions. Lane M, molecular-mass marker (labelled in kDa). Lane 2, the CD163 long-range SRCR repeat under reducing conditions. Lane 3, PNGase F-treated CD163 long-range SRCR repeat. PNGase F (about 36 kDa) is not obvious in the figure.
In this study, recombinant CD163 long-range SRCR repeat was expressed in the Drosophila Schneider 2 (S2) insect-cell system. The target protein was then purified and single crystals were successfully obtained and diffracted to 3.30 Å resolution. Further structural and functional studies are being carried out.
2. Materials and methods
2.1. Macromolecule production
The cDNA encoding the CD163 long-range SRCR repeat (residues 477–1028; the numbering is according to UniProt entry Q2VL90) was amplified from full-length porcine CD163 cDNA (GenBank accession No. JX292263) by a sticky-end polymerase chain reaction (PCR) using the primers listed in Table 1 ▸. The PCR product was isolated and inserted into the expression vector pMT/Bip/V5-His A between the BglII and MluI sites. The construct was transformed into the competent Escherichia coli strain DH5α and then screened on a Luria–Bertani (LB) plate (0.5% yeast extract, 1% tryptone, 1% NaCl, 2% agar) containing 100 µg ml−1 ampicillin (Invitrogen, Carlsbad, California, USA) to select positive colonies. After colony PCR, the recombinant expression vector was verified by Shanghai Sangon Biotech Co. Ltd (Shanghai, People’s Republic of China) and transfected into Drosophila S2 cells by the Cellfectin II reagent according to the manufacturer’s instructions (Invitrogen). The stably transfected S2 cells were selected in Schneider’s insect medium (Sigma–Aldrich, St Louis, Missouri, USA) supplemented with 10% heat-inactivated foetal bovine serum (Gibco, Grand Island, Nebraska, USA), antibiotics (100 U ml−1 penicillin and 100 µg ml−1 streptomycin) and 25 µg ml−1 blasticidin S (Invitrogen). The stably transfected cells were then grown in Sf-900 II serum-free medium (Invitrogen) and induced with 0.75 mM CuSO4 for 5 d at a constant temperature of 298 K and 120 rev min−1 in a shaker incubator according to a previous publication (Hou et al., 2016 ▸). The culture medium was harvested by centrifugation at 1000 rev min−1 for 10 min. After a second centrifugation at 10 000 rev min−1 for 40 min and clarification by filtration, the supernatant containing the target protein was applied onto a GE Ni Sepharose Excel column pre-equilibrated with 20 mM Tris–HCl pH 8.0, 150 mM NaCl. The target protein was eluted with 20 mM Tris–HCl pH 8.0, 150 mM NaCl, 200 mM imidazole. Subsequently, the elution solution was dialyzed and purified using a GE Mono S 5/50 GL cation-exchange prepacked column on a Bio-Rad BioLogic system. The target protein was eluted with 20 mM citric acid pH 4.6, 400 mM sodium citrate. It was further purified using a calibrated GE Superdex 200 10/300 GL prepacked column on a Bio-Rad BioLogic system with 20 mM citric acid pH 4.6, 20 mM sodium citrate as the elution buffer (Fig. 1 ▸ b). The fraction containing the target protein was collected and concentrated to 6.07 mg ml−1 in 20 mM citric acid pH 4.6, 20 mM sodium citrate using a Millipore ultracentrifugation tube (Merck, Carrigtwohill, Cork, Ireland). The purity of the target protein was determined by 12% SDS–PAGE under reducing and nonreducing conditions stained with Coomassie Brilliant Blue (Fig. 1 ▸ c). The protein was also treated with peptide:N-glycosidase F (PNGase F) according to the manufacturer’s instructions (New England BioLabs, Ipswich, Massachusetts, USA) and subjected to SDS–PAGE analysis (Fig. 1 ▸ c).
Table 1. Macromolecule-production information.
| Source organism | Sus scrofa |
| DNA source | cDNA of porcine CD163 |
| Forward primer† | GATCTCCCAGGCTGGTTGGAGG |
| TCCCAGGCTGGTTGGAGG | |
| Reverse primer‡ | CGACGCGTTGAGCACGTCACAGCAGCA |
| Cloning vector | pMT/Bip/V5-His A |
| Expression vector | pMT/Bip/V5-His A |
| Expression host | Drosophila Schneider 2 (S2) cells |
| Complete amino-acid sequence of the construct produced§ | RSPRLVGGDIPCSGRVEVQHGDTWGTVCDSDFSLEAASVLCRELQCGTVVSLLGGAHFGEGSGQIWAEEFQCEGHESHLSLCPVAPRPDGTCSHSRDVGVVCSRYTQIRLVNGKTPCEGRVELNILGSWGSLCNSHWDMEDAHVLCQQLKCGVALSIPGGAPFGKGSEQVWRHMFHCTGTEKHMGDCSVTALGASLCSSGQVASVICSGNQSQTLSPCNSSSSDPSSSIISEENGVACIGSGQLRLVDGGGRCAGRVEVYHEGSWGTICDDSWDLNDAHVVCKQLSCGWAINATGSAHFGEGTGPIWLDEINCNGKESHIWQCHSHGWGRHNCRHKEDAGVICSEFMSLRLISENSRETCAGRLEVFYNGAWGSVGRNSMSPATVGVVCRQLGCADRGDISPASSDKTVSRHMWVDNVQCPKGPDTLWQCPSSPWKKRLASPSEETWITCANKIRLQEGNTNCSGRVEIWYGGSWGTVCDDSWDLEDAQVVCRQLGCGSALEAGKEAAFGQGTGPIWLNEVKCKGNETSLWDCPARSWGHSDCGHKEDAAVTCSTRTGHHHHHH |
The sticky-end BglII restriction sites are underlined.
The MluI restriction site is underlined.
Additional vector residues and the 6×His tag residues are underlined.
2.2. Crystallization
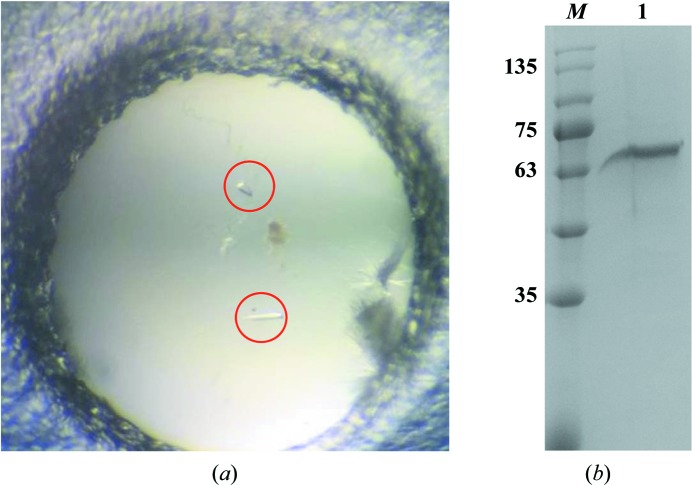
Crystallization of the purified CD163 long-range SRCR repeat was screened using more than 1000 conditions from commercial kits from Hampton Research. Preliminary crystallization screening was performed using the sitting-drop vapour-diffusion method in 48-well plates, with each well containing 100 µl reservoir solution and each drop consisting of 1 µl protein solution (6.07 mg ml−1) mixed with 1 µl reservoir solution. Initial crystals appeared using 20% PEG 3350, 0.2 M potassium sodium tartrate tetrahydrate. After optimization, well formed crystals of the CD163 long-range SRCR repeat were obtained using a reservoir solution consisting of 20% PEG 4000, 0.15 M potassium sodium tartrate tetrahydrate pH 8.5 (Fig. 2 ▸ a). The washed crystals were analysed by 12% SDS–PAGE to confirm whether the crystals consisted of the target protein (Fig. 2 ▸ b). In a search for cryoprotectants (glycerol, 2-methyl-2,4-pentanediol, ethylene glycol and sucrose), the crystals were found to be stable in reservoir solution containing 20%(v/v) glycerol. Detailed information on crystallization is given in Table 2 ▸.
Figure 2.
Crystallization of recombinant CD163 long-range SRCR repeat. (a) Crystals of the CD163 long-range SRCR repeat (circled in red). (b) SDS–PAGE analysis of the washed crystals. Lane M, molecular-mass marker (labelled in kDa). Lane 1, crystals of the CD163 long-range SRCR repeat dissolved in water.
Table 2. Crystallization of the CD163 long-range SRCR repeat.
| Method | Sitting-drop vapour diffusion |
| Plate type | 48-well plate |
| Temperature (K) | 298 |
| Protein concentration (mg ml−1) | 6.07 |
| Buffer composition of protein solution | 20 mM citric acid pH 4.6, 20 mM sodium citrate |
| Composition of reservoir solution | 0.15 M potassium sodium tartrate tetrahydrate pH 8.5, 20% PEG 4000 |
| Volume and ratio of drop | 2 µl (1:1 ratio of protein solution and reservoir solution) |
| Volume of reservoir (µl) | 100 |
2.3. Data collection and processing
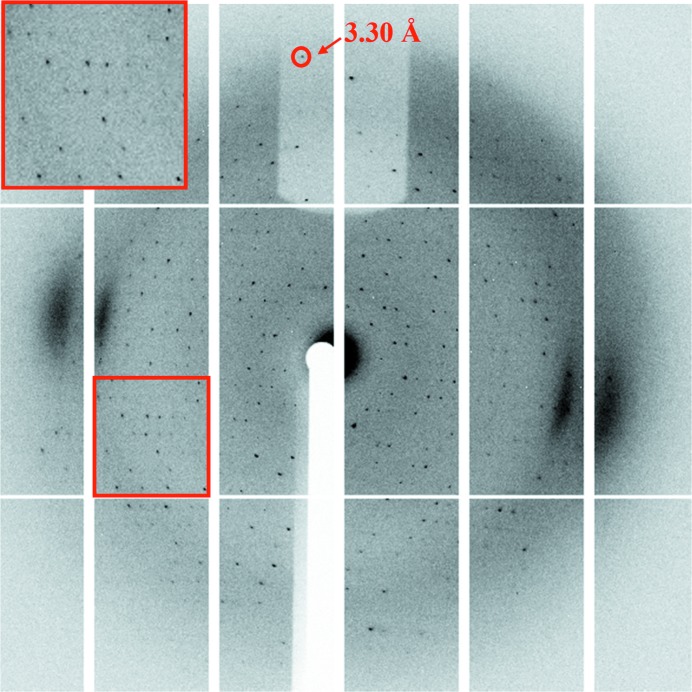
Prior to data collection, a crystal was picked up in a 0.05–0.1 mm cryoloop (Hampton Research), immediately soaked in cryoprotectant solution for 10–20 s and flash-cooled in liquid nitrogen. Diffraction experiments were conducted at 100 K using a PILATUS 6M detector on beamline BL18U (λ = 0.979 Å) at the National Center for Protein Sciences Shanghai (NCPSS) and Shanghai Synchrotron Radiation Facility (SSRF). A total of 360 images were collected at a crystal-to-detector distance of 300 mm with 0.1 s exposure for every 1° oscillation frame (Fig. 3 ▸). Diffraction data sets were processed using the HKL-3000 package (Minor et al., 2006 ▸). Data-collection information and processing statistics are summarized in Table 3 ▸.
Figure 3.
Representative diffraction image obtained from crystals of the CD163 long-range SRCR repeat. The inset shows a magnification of the diffraction pattern in the red box.
Table 3. Data collection and processing.
Values in parentheses are for the outer shell.
| Diffraction source | BL18U, SSRF |
| Wavelength (Å) | 0.979 |
| Temperature (K) | 100 |
| Detector | PILATUS 6M |
| Crystal-to-detector distance (mm) | 300 |
| Rotation range per image (°) | 1 |
| Total rotation range (°) | 360 |
| Exposure time per image (s) | 0.1 |
| Space group | C2 |
| a, b, c (Å) | 125.24, 59.37, 83.85 |
| α, β, γ (°) | 90.00, 117.81, 90.00 |
| Resolution range (Å) | 50.00–3.30 (3.42–3.30) |
| Total No. of reflections | 55439 |
| No. of unique reflections | 8479 |
| Completeness (%) | 99.8 (99.8) |
| Multiplicity | 6.5 (6.5) |
| 〈I/σ(I)〉 | 16.7 (3.4) |
| R r.i.m. | 0.076 |
3. Results and discussion
The lack of structural studies of long-range SRCR repeats is speculated to arise from the difficulty in preparing the large target protein with so many disulfide linkages. Recombinant CD163 long-range SRCR repeat (SRCR5–9, with 19 disulfide bonds; Fig. 1 ▸ a) was expressed in Drosophila S2 cells and purified by metal-affinity, cation-exchange and size-exclusion chromatography, yielding approximately 2 mg target protein per litre of insect-cell culture. We attempted to purify the target protein by anion-exchange chromatography (GE Mono Q 5/50 GL prepacked column) since its theoretical isoelectric point (pI) is about 5.71. However, we were unable to obtain protein with increased purity (data not shown). During size-exclusion chromatography, we also made efforts to purify the target protein using a high-pH buffer (20 mM Tris–HCl pH 8.0, 20 mM NaCl). Although the protein was stable in this buffer, it was inappropriate for subsequent crystallization (data not shown). As a result, we utilized a low-pH buffer (20 mM citric acid pH 4.6, 20 mM sodium citrate) during purification. The elution volume of the target protein was 14 ml on the calibrated gel-filtration column, with a molecular weight near 67 kDa (Fig. 1 ▸ b). As shown in Fig. 1 ▸(c), there was a difference between the bands corresponding to the purified protein under reducing (lane 2) and nonreducing (lane 1) conditions, which implies that disulfide bonds may affect the positions of the bands. In addition, the apparent molecular mass of the target protein was not consistent with the calculated value of 60.61 kDa under nonreducing conditions (Fig. 1 ▸ c, lane 1). This is probably owing to potential N-linked glycosylation at Asn936 in the construct, since the band showed a significant change after deglycosylation using PNGase F (Fig. 1 ▸ c, lane 3). Based on the results above, the target protein was confirmed to exist as a monomer. Using purified CD163 long-range SRCR repeat, we generated crystals which had a good morphology despite their small volume (Fig. 2 ▸ a). The crystals were washed and analysed by SDS–PAGE, which gave a single band, confirming that the crystallized material was the target protein (Fig. 2 ▸ b). The crystals were flash-cooled in liquid nitrogen after the addition of 20% glycerol as a cryoprotectant and diffracted to 3.30 Å resolution (Fig. 3 ▸). Diffraction data sets were processed using the HKL-3000 package. We cut off the highest resolution shell at an I/σ(I) of 3.4, where the linear R factor and CC1/2 were 0.471 and 0.981, respectively. The crystal belonged to space group C2, with unit-cell parameters a = 125.24, b = 59.37, c = 83.85 Å (Table 3 ▸). The most probable Matthews coefficient was 2.32 Å3 Da−1 and corresponded to one protein molecule per asymmetric unit, with a solvent content of 47%. The detailed statistics of data collection are listed in Table 3 ▸.
The overall CD163 long-range SRCR repeat shares relatively low sequence identity (<40%) with other SRs of known structure. Furthermore, the different SRCR domains in this long-range SRCR repeat are heterogeneous in their sequences, disulfide-bond compositions and glycosylation. Therefore, although we have previously determined the crystal structure of CD163 SRCR5 (Ma, Jiang et al., 2017 ▸; Ma, Qiao et al., 2017 ▸), the current work is the first view of a long-range SRCR repeat and will be the largest SR structure to be determined to our knowledge. The results will lay a basis for a deep understanding of SRs and their multiple functions. Further structural and functional studies are being carried out and will be reported elsewhere.
Acknowledgments
We thank the staff of the BL18U beamline at the National Center for Protein Sciences Shanghai (NCPSS) and Shanghai Synchrotron Radiation Facility (SSRF), Shanghai, People’s Republic of China for assistance during data collection.
Funding Statement
This work was funded by National Natural Science Foundation of China grants 31490601 and 31602036. Basic and Advanced Technology Research Program of Henan Province grant 11230041322 to Songlin Qiao. Henan Academy of Agricultural Sciences grant [2017]76-22 to Rui Li. Natural Science Foundation of Henan Province grant 182300410001.
References
- Areschoug, T. & Gordon, S. (2009). Cell. Microbiol. 11, 1160–1169. [DOI] [PubMed]
- Bover, L. C., Cardó-Vila, M., Kuniyasu, A., Sun, J., Rangel, R., Takeya, M., Aggarwal, B. B., Arap, W. & Pasqualini, R. (2007). J. Immunol. 178, 8183–8194. [DOI] [PubMed]
- Caì, Y. et al. (2015). J. Virol. 89, 844–856. [DOI] [PMC free article] [PubMed]
- Calvert, J. G., Slade, D. E., Shields, S. L., Jolie, R., Mannan, R. M., Ankenbauer, R. G. & Welch, S. K. (2007). J. Virol. 81, 7371–7379. [DOI] [PMC free article] [PubMed]
- Canton, J., Neculai, D. & Grinstein, S. (2013). Nature Rev. Immunol. 13, 621–634. [DOI] [PubMed]
- Fabriek, B. O., Polfliet, M. M., Vloet, R. P., van der Schors, R. C., Ligtenberg, A. J., Weaver, L. K., Geest, C., Matsuno, K., Moestrup, S. K., Dijkstra, C. D. & van den Berg, T. K. (2007). Blood, 109, 5223–5229. [DOI] [PubMed]
- Fabriek, B. O., van Bruggen, R., Deng, D. M., Ligtenberg, A. J. M., Nazmi, K., Schornagel, K., Vloet, R. P. M., Dijkstra, C. D. & van den Berg, T. K. (2009). Blood, 113, 887–892. [DOI] [PubMed]
- Graversen, J. H., Madsen, M. & Moestrup, S. K. (2002). Int. J. Biochem. Cell Biol. 34, 309–314. [DOI] [PubMed]
- Greaves, D. R. & Gordon, S. (2009). J. Lipid Res. 50, S282–S286. [DOI] [PMC free article] [PubMed]
- Gronlund, J., Vitved, L., Lausen, M., Skjodt, K. & Holmskov, U. (2000). J. Immunol. 165, 6406–6415. [DOI] [PubMed]
- Holm, D., Fink, D. R., Grønlund, J., Hansen, S. & Holmskov, U. (2009). Mol. Immunol. 46, 1663–1672. [DOI] [PubMed]
- Hou, J., Li, R., Ma, H., Qiao, S. & Zhang, G. (2016). Front. Agr. Sci. Eng. 3, 65–71.
- Kristiansen, M., Graversen, J. H., Jacobsen, C., Sonne, O., Hoffman, H. J., Law, S. K. & Moestrup, S. K. (2001). Nature (London), 409, 198–201. [DOI] [PubMed]
- Law, S. K. A., Micklem, K. J., Shaw, J. M., Zhang, X.-P., Dong, Y., Willis, A. C. & Mason, D. Y. (1993). Eur. J. Immunol. 23, 2320–2325. [DOI] [PubMed]
- Ma, H., Jiang, L., Qiao, S., Zhi, Y., Chen, X.-X., Yang, Y., Huang, X., Huang, M., Li, R. & Zhang, G.-P. (2017). J. Virol. 91, e01897-16. [DOI] [PMC free article] [PubMed]
- Ma, H., Qiao, S., Huang, M., Jiang, L., Li, R. & Zhang, G. P. (2017). Chin. J. Struct. Chem. 36, 1409–1417.
- Mackay, C. R., Maddox, J. F. & Brandon, M. R. (1986). Eur. J. Immunol. 16, 19–25. [DOI] [PubMed]
- Minor, W., Cymborowski, M., Otwinowski, Z. & Chruszcz, M. (2006). Acta Cryst. D62, 859–866. [DOI] [PubMed]
- Moreno, J. A., Muñoz-García, B., Martín-Ventura, J. L., Madrigal-Matute, J., Orbe, J., Páramo, J. A., Ortega, L., Egido, J. & Blanco-Colio, L. M. (2009). Atherosclerosis, 207, 103–110. [DOI] [PubMed]
- Sarrias, M. R., Grønlund, J., Padilla, O., Madsen, J., Holmskov, U. & Lozano, F. (2004). Crit. Rev. Immunol. 24, 1–37. [DOI] [PubMed]
- Van Gorp, H., Delputte, P. L. & Nauwynck, H. J. (2010). Mol. Immunol. 47, 1650–1660. [DOI] [PubMed]
- Van Gorp, H., Van Breedam, W., Van Doorsselaere, J., Delputte, P. L. & Nauwynck, H. J. (2010). J. Virol. 84, 3101–3105. [DOI] [PMC free article] [PubMed]
- Wijngaard, P. L., Metzelaar, M. J., MacHugh, N. D., Morrison, W. I. & Clevers, H. C. (1992). J. Immunol. 149, 3273–3277. [PubMed]
- Zani, I. A., Stephen, S. L., Mughal, N. A., Russell, D., Homer-Vanniasinkam, S., Wheatcroft, S. B. & Ponnambalam, S. (2015). Cells, 4, 178–201. [DOI] [PMC free article] [PubMed]