Abstract
Purpose/Aim
Meniscus tears are a common injury to the knee associated with the development of osteoarthritis. Gene expression in the injured meniscus may be associated with early degeneration in the articular cartilage. The purpose of this study was to test the hypothesis that gene expression in meniscus tears is associated with early degenerative changes in the articular cartilage at the time of partial meniscectomy.
Materials and Methods
Torn meniscus was removed at the time of partial meniscectomy in 63 patients without radiographic osteoarthritis. Meniscal mRNA expression was measured by quantitative PCR for multiple molecular markers of osteoarthritis and cartilage homeostasis. The presence of early degenerative changes in the knee was recorded by X-ray (N=63), magnetic resonance imaging (MRI, N=48) and arthroscopy (N=63). Gene expression was tested for correlation with the presence/absence of degenerative changes after adjusting for age, sex and body mass index.
Results
Overall gene expression varied significantly with degenerative changes based on X-ray (P=0.047) and MRI (P=0.018). The linear combination of gene variation was also significant. However, only adiponectin (ADIPOQ) (P=0.015) was expressed at a significantly lower level in patients with chondrosis on MRI while the expression of ADIPOQ (P=0.035) and resistin (RETN) (P=0.017) was higher in patients with early degenerative changes on X-ray.
Conclusions
There is an overall association of gene expression in meniscal tears to early degenerative changes in the knee, but only a limited number of specific genes demonstrate this relationship. The roles of adiponectin and resistin in knee injury and osteoarthritis deserve further study.
Keywords: meniscus tear, partial meniscectomy, osteoarthritis, chondrosis, gene expression
INTRODUCTION
Meniscal tears are a common injury in the knee (1–3). They are likely to be an important early event in the initiation and propagation of osteoarthritis (OA) in the knee (4–7) and are known to predispose individuals to develop knee OA (8–12). Among athletes at the NFL combine, partial meniscectomy has been shown to be associated with degenerative changes in the articular cartilage (13) and predict a shorter length of career (14), potentially related to degeneration in the knee. While the relationship between meniscus injury and OA is well established, the exact biological underpinning of this connection is not well understood.
A recent study has demonstrated differences in gene expression of inflammation and OA-related markers in the torn meniscus at the time of arthroscopic partial meniscectomy (APM) based on patient age and injury pattern (15). A follow up study demonstrated that expression of OA- and obesity-related genes in the torn meniscus also has some relationship with patient body mass index (BMI) (16). Most recently, a study looking at the transcriptome-wide gene expression signatures in a limited number of patients failed to demonstrate a strong relationship between chondrosis as determined by arthroscopy and molecular markers at the time of APM (17). No other studies to date investigate how gene expression in the torn meniscus may relate to the presence of early chondrosis in the articular cartilage at the time of APM as measured by arthroscopy or magnetic resonance imaging (MRI). However, a recent study that has characterized meniscal pathology from OA and non-OA patients reported that certain gross, histologic, biochemical and gene expression changes are associated with radiographic scores in the knee (18).
The present study was designed to use X-ray, MRI and arthroscopic findings to assess the degree of degeneration in the knee and to investigate its relationship with the gene expression profile in the torn meniscus. We hypothesize that gene expression profile in the torn meniscus is associated with the presence of early degenerative changes in the knee (chondrosis), based on pre-operative MRI, radiographs, and arthroscopic findings. The relationship between meniscal gene expression and chondrosis could shed further light on the biological bridge between molecular changes in the meniscus and early degenerative changes in the articular cartilage.
MATERIALS AND METHODS
Meniscal surgery and grading of chondrosis
The study protocol was approved by the study site Institutional Review Board. Informed consent was obtained from all individual participants included in the study. Patients (N=63) diagnosed with symptomatic meniscus tears without advanced OA (i.e. moderate or advanced joint space narrowing or diffuse degenerative changes), concomitant ligament injury or autoimmune disorders were recruited for this study. The demographic data of the study population is summarized in Table 1. Meniscus tears were treated arthroscopically with resection of the dysfunctional fragment of the torn portion.
Table 1.
Characteristics of study patients
| Procedure (Total N) |
Chondrosis | N | Meniscus | N | Location | N | Mean age (range) years | BMI (Kg/m2) ±SD |
Sex F/M |
|---|---|---|---|---|---|---|---|---|---|
| Arthroscopy and X-ray (63 each) |
No | 40 | Medial | 55 | White-White | 43 | 43.2 (12.9–73.0) |
27.18±5.43 | 14/26 |
| Yes | 23 | Lateral | 8 | Red-White | 20 | 52.1 (34.2–67.2) |
29.33±4.33 | 6/17 | |
| MRI (48) |
No | 11 | Medial | 42 | White-White | 34 | 35.1 (12.9–53.8) |
28.02±4.87 | 5/6 |
| Yes | 37 | Lateral | 6 | Red-White | 14 | 48.7 (16.0–73.0) |
27.65±5.36 | 11/26 |
MRI = magnetic resonance imaging, BMI = body mass index, SD = standard deviation, F = female, m = male
The majority of patients in this cohort (48 out of 63) had undergone MRI evaluation at our study institution using a 1.5T machine. All of the patients had X-rays at our institution read by full-time academic musculoskeletal radiologists. Each knee was classified as normal (no radiographic evidence for degenerative changes) or positive for degeneration (radiographic degenerative changes such as mild joint space narrowing, osteophytes or sclerosis). All MRIs were read by full-time academic musculoskeletal radiologists to grade chondrosis. Each knee was classified as normal (no MRI evidence for chondrosis) or positive for chondrosis (any MRI finding of chondrosis in the knee). A bone bruise without overlying changes in the articular cartilage was not considered as evidence for chondrosis.
The arthroscopic findings were recorded with regard to changes in the articular cartilage based on a standard diagnostic arthroscopy performed as part of each surgery. Each knee was classified as normal (no arthroscopic evidence for chondrosis) or positive for chondrosis (any arthroscopic finding of at least Grade 2 articular cartilage change in the knee) using a modified Outerbridge scoring system (19).
Tissue processing and RNA isolation
The specimens were collected at the time of APM. They were handled using a previously published technique (15). Briefly, the anonymous specimens were transported to the laboratory from the operating room in sterile screw cap containers containing phosphate-buffered saline (PBS, Thermo Fisher Scientific, Rockford, IL, USA). The tissues were weighed and then washed twice with PBS to get rid of any blood cells and debris to avoid influence of contaminants on gene expression profile. The blot-dried tissues were put in 50 ml Falcon tubes and 1 ml of TRIzol reagent (Invitrogen, Carlsbad, CA, USA) was added for each 50–100 mg of the tissue wet weight and stored at −80°C until used for total RNA extraction according to previously described methods (16). RNA preparations were analyzed on a Nanodrop spectrophotometer (NanoDrop 2000, Thermo Fisher Scientific, Pittsburg, PA, USA) to measure concentration. Quality was assessed by 260/280 and 260/230 ratios as measures of protein and organic solvent contamination. RNA was stored at −80°C until used for analysis.
Quantification of gene expression in meniscus
A total of 150–200 ng of isolated RNA was first treated with DNase I to remove traces of contaminating DNA (Invitrogen, Carlsbad, CA, USA). The DNase I treated RNA was then reverse-transcribed to synthesize complementary DNA (Invitrogen, Carlsbad, CA, USA) as per the manufacturer’s instructions. Custom-designed primers (15, 16) were obtained from Invitrogen (Carlsbad, CA, USA) for a panel of genes. Selection of these genes was based on their potential known role in cartilage homeostasis, OA, extracellular matrix degradation and obesity despite little or no information on their role in meniscus tears. The expression these genes was quantified by quantitative PCR on a 7500 Fast Real Time PCR System (Applied Biosystems, Foster City, CA, USA) using SYBR Green PCR master mix (Applied Biosystems, Foster City, CA, USA) according to manufacturer’s protocols. Samples were amplified with an initial activation step at 95°C for 10 min, followed by 40 cycles of denaturation at 95°C for 15 seconds and annealing at 60°C for one minute. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) acted as an endogenous reference gene for normalization of fluorescence thresholds (Ct) values for target genes. GAPDH is a conventionally used internal control for normalization because within-tissue variation of GAPDH mRNA expression levels is generally small. While we did not compare GAPDH to other housekeeping genes, the expression of GAPDH was stable across samples.
Statistical analysis
The relationship between the presence and absence of chondral damage and target gene expression levels was examined using a multivariate general linear model with log10-transformed gene expression values as the dependent variables and age, sex, BMI, and chondrosis (absent or present) as the independent variables:
This analysis was carried out separately for X-ray, MRI and arthroscopic findings. The probability of an effect of chondrosis on the collection of gene expression traits is given by Wilk’s Lambda and its associated F-ratio. Multivariate expression differences between patients with and without chondrosis are described by the canonical vector representing the linear combination of expression traits that best discriminates between the two groups. These coefficients are determined by the mean difference between groups divided by the residual variance/covariance matrix so that the significance of differences along highly variable within-group dimensions are downgraded relative to the same absolute difference along lowly variable dimensions. By this operation, canonical coefficient values account for observed within-group variation in all other traits when considering any one trait of interest. For example, if the residual correlation between two traits is strongly positive, under the null model of no difference, we expect that both traits would increase jointly for patients with chondrosis. When one trait expresses at a higher level than this null expectation while a second trait expresses at a lower than expected level, the traits will have positive and negative canonical coefficients, respectively. This linear combination of traits may be significantly different from the null model because of its unexpected combination of expression traits while neither trait is significantly different between groups by itself (20). Probabilities of chondrosis effects on individual gene expression traits were also obtained to interpret significant multivariate results. Associations among the different measures of chondrosis were performed with a Pearson’s Chi-square statistic using the Phi coefficient as the association parameter.
RESULTS
Sixty-three meniscus tissues were collected at the time of clinically indicated APM from patients with a known meniscus tear based on MRI (Table 1). The patient cohort included 36 females and 27 males with an average age of 45.8 ± 13.7 years. All but 2 tears occurred in the posterior horn, and most involved the white-white zone of the medial meniscus. The radiographic data was available for 63 individuals, 23 with some degree of early joint space narrowing and/or osteophyte formation (Fig. 1B) and 40 with no degenerative changes evident on the X-ray (Fig. 1A). The MRI findings were available from 48 subjects, 37 with chondrosis (Fig. 1C–D) and 11 without. All 63 subjects have arthroscopic data: 23 subjects with chondrosis (Fig. 1 F) and 40 subjects without (Fig. 1E).
Fig. 1.
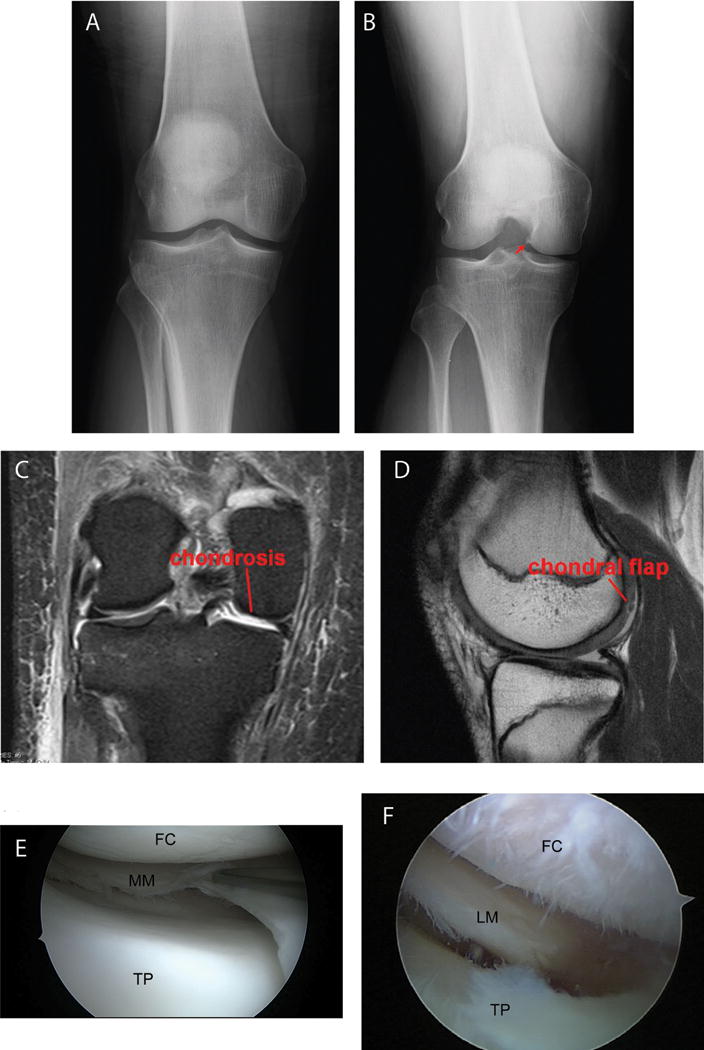
Examples of presence/absence of degenerative changes based on X-rays, MRI and arthroscopy. X-rays: (A) no OA; (B) joint space narrow and osteophyte formation (arrow) as evidence for early OA. MRI: (C) chondrosis; (D) chondral flap. Arthroscopy: (E) no chondrosis; (F) chondrosis. FC = femoral condyle, TP = tibia plateau, MM = medial meniscus, LM = lateral meniscus
Gene expression variation with degenerative changes measured by X-ray
We observed that the expression of all the genes, when taken together, was found to vary significantly (P=0.047) with the presence of degenerative changes as measured by X-ray (Table 2). Cytokines, transcription factors and adipokine genes were predominantly expressed at higher level with degenerative changes but only ADIPOQ (P=0.035) and RETN (P=0.017) were shown to express at significantly different levels in patients with X-ray based chondrosis than in patients without X-ray chondrosis regardless of the expression levels of other traits. The multivariate significance was also affected by the higher than expected expression of IL-1α, IL-1β, and MMP-13 and lower than expected expression of TNFα and ADAMTS-5 when accounting for expression of all the other traits based on the canonical vector describing expression differences related to chondrosis (Table 3). The residual correlations between expression traits, representing the correlations independent of age, sex, BMI, and chondrosis, are nearly always positive, varying between low to intermediate positive correlations to very high positive correlations. Hence, the highly varying multivariate dimension is all genes having higher or lower expression jointly. The observed canonical coefficients mix relatively high positive and low negative coefficients, a dimension with relatively low within-group variation.
Table 2.
The gene expression in torn meniscus varies by presence or absence of chondrosis in cartilage as determined by X-ray (P=0.047)
| (N=63) | No Chondrosis (N=40) | Chondrosis (N=23) | P | ||
|---|---|---|---|---|---|
| Gene | Least Square Mean | 95% Confidence Interval (upper limit–lower limit) | Least Square Mean | 95% Confidence Interval (upper limit–lower limit) | |
| IL1A | 0.0008 | 0.0019–0.0004 | 0.0019 | 0.0058–0.0007 | 0.220 |
| IL1B | 0.0009 | 0.0021–0.0004 | 0.0012 | 0.0035–0.0004 | 0.722 |
| IL6 | 0.0015 | 0.0034–0.0007 | 0.0021 | 0.0064–0.0007 | 0.607 |
| TNFA | 0.0004 | 0.0008–0.0002 | 0.0003 | 0.0008–0.0001 | 0.683 |
| ADAMTS4 | 0.0037 | 0.0064–0.0021 | 0.0021 | 0.0043–0.0010 | 0.217 |
| ADAMTS5 | 0.0065 | 0.0110–0.0038 | 0.0052 | 0.0105–0.0026 | 0.613 |
| BMP2 | 0.0022 | 0.0038–0.0013 | 0.0017 | 0.0036–0.0008 | 0.583 |
| MMP1 | 0.0017 | 0.0034–0.0009 | 0.0015 | 0.0036–0.0006 | 0.750 |
| MMP3 | 0.0102 | 0.0200–0.0052 | 0.0060 | 0.0148–0.0024 | 0.348 |
| MMP9 | 0.0009 | 0.0021–0.0004 | 0.0009 | 0.0025–0.0003 | 0.891 |
| MMP13 | 0.0035 | 0.0069–0.0018 | 0.0044 | 0.0107–0.0018 | 0.700 |
| IL8 | 0.0022 | 0.0044–0.0011 | 0.0020 | 0.0049–0.0008 | 0.823 |
| CCL3 | 0.0015 | 0.0031–0.0007 | 0.0009 | 0.0023–0.0003 | 0.389 |
| CCL3L1 | 0.0009 | 0.0018–0.0004 | 0.0005 | 0.0012–0.0002 | 0.291 |
| CXCL1 | 0.0006 | 0.0013–0.0003 | 0.0006 | 0.0015–0.0002 | 0.913 |
| CXCL3 | 0.0011 | 0.0022–0.0005 | 0.0006 | 0.0017–0.0002 | 0.392 |
| CXCL6 | 0.0010 | 0.0025–0.0004 | 0.0005 | 0.0018–0.0002 | 0.397 |
| CCL20 | 0.0011 | 0.0023–0.0006 | 0.0009 | 0.0022–0.0004 | 0.644 |
| COL1A1 | 0.0396 | 0.1258–0.0124 | 0.0323 | 0.1520–0.0069 | 0.833 |
| COL2A1 | 0.0068 | 0.0145–0.0032 | 0.0058 | 0.0161–0.0021 | 0.808 |
| ACAN | 0.0012 | 0.0038–0.0004 | 0.0026 | 0.0118–0.0006 | 0.426 |
| NFκB1A | 0.0050 | 0.0094–0.0026 | 0.0058 | 0.0135–0.0025 | 0.779 |
| NFκB2 | 0.0013 | 0.0023–0.0007 | 0.0018 | 0.0039–0.0008 | 0.475 |
| IκBA | 0.0038 | 0.0069–0.0021 | 0.0043 | 0.0095–0.0020 | 0.806 |
| ADIPOQ | 0.0002 | 0.0005–0.0001 | 0.0009 | 0.0027–0.0003 | 0.035 |
| APLN | 0.0028 | 0.0059–0.0014 | 0.0066 | 0.0177–0.0025 | 0.165 |
| LEP | 0.0004 | 0.0009–0.0002 | 0.0004 | 0.0010–0.0001 | 0.900 |
| RETN | 0.0001 | 0.0003–0.0000 | 0.0007 | 0.0021–0.0002 | 0.017 |
P values in bold face represent statistically significant differences
Table 3.
Standardized Canonical coefficients and residual correlations between gene expression traits in patients with and without chondrosis based on X-ray
| Trait | Standardized canonical coefficient |
IL1A | IL1B | IL6 | TNFA |
ADAMTS 4 |
ADAMTS 5 |
BMP2 | MMP1 | MMP3 | MMP9 | MMP13 | IL8 | CCL3 | CCL3L1 | CXCL1 | CXCL3 | CXCL6 | CCL20 | COL1A1 | COL2A1 | ACAN | NFKB1A | NFKB2 | IKBA | ADIPOQ | APLN | LEP | RETN |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IL1A | 1.91 | 1 | |||||||||||||||||||||||||||
| IL1B | 1.23 | 0.73 | 1 | ||||||||||||||||||||||||||
| IL6 | 0.33 | 0.77 | 0.77 | 1 | |||||||||||||||||||||||||
| TNFA | −1.01 | 0.84 | 0.77 | 0.72 | 1 | ||||||||||||||||||||||||
| ADAMTS4 | 0.37 | 0.51 | 0.47 | 0.54 | 0.49 | 1 | |||||||||||||||||||||||
| ADAMTS5 | −1.16 | 0.4 | 0.31 | 0.37 | 0.28 | 0.75 | 1 | ||||||||||||||||||||||
| BMP2 | −0.07 | 0.57 | 0.63 | 0.72 | 0.55 | 0.59 | 0.53 | 1 | |||||||||||||||||||||
| MMP1 | −0.07 | 0.35 | 0.46 | 0.29 | 0.43 | 0.39 | 0.18 | 0.16 | 1 | ||||||||||||||||||||
| MMP3 | −0.8 | 0.31 | 0.38 | 0.38 | 0.21 | 0.37 | 0.44 | 0.43 | 0.31 | 1 | |||||||||||||||||||
| MMP9 | −0.55 | 0.58 | 0.67 | 0.67 | 0.62 | 0.54 | 0.45 | 0.54 | 0.27 | 0.19 | 1 | ||||||||||||||||||
| MMP13 | 1.43 | 0.43 | 0.51 | 0.56 | 0.48 | 0.63 | 0.64 | 0.52 | 0.4 | 0.43 | 0.53 | 1 | |||||||||||||||||
| IL8 | −0.4 | 0.36 | 0.51 | 0.51 | 0.41 | 0.43 | 0.17 | 0.3 | 0.55 | 0.3 | 0.35 | 0.39 | 1 | ||||||||||||||||
| CCL3 | −0.82 | 0.77 | 0.79 | 0.72 | 0.75 | 0.47 | 0.32 | 0.57 | 0.34 | 0.21 | 0.56 | 0.53 | 0.37 | 1 | |||||||||||||||
| CCL3L1 | −0.32 | 0.69 | 0.81 | 0.72 | 0.72 | 0.46 | 0.25 | 0.65 | 0.39 | 0.29 | 0.65 | 0.51 | 0.47 | 0.78 | 1 | ||||||||||||||
| CXCL1 | −0.38 | 0.64 | 0.77 | 0.77 | 0.69 | 0.57 | 0.38 | 0.56 | 0.33 | 0.26 | 0.63 | 0.62 | 0.51 | 0.68 | 0.67 | 1 | |||||||||||||
| CXCL3 | −0.55 | 0.79 | 0.78 | 0.71 | 0.77 | 0.43 | 0.24 | 0.52 | 0.36 | 0.32 | 0.62 | 0.43 | 0.26 | 0.83 | 0.78 | 0.67 | 1 | ||||||||||||
| CXCL6 | 0.23 | 0.67 | 0.71 | 0.76 | 0.65 | 0.46 | 0.2 | 0.56 | 0.34 | 0.4 | 0.51 | 0.39 | 0.46 | 0.7 | 0.75 | 0.69 | 0.76 | 1 | |||||||||||
| CCL20 | −0.56 | 0.71 | 0.71 | 0.62 | 0.71 | 0.33 | 0.13 | 0.42 | 0.48 | 0.33 | 0.5 | 0.38 | 0.43 | 0.66 | 0.71 | 0.57 | 0.76 | 0.73 | 1 | ||||||||||
| COL1A1 | 0.06 | 0.16 | 0.33 | 0.25 | 0.23 | 0.32 | 0.3 | 0.1 | 0.41 | 0.29 | 0.22 | 0.53 | 0.56 | 0.1 | 0.18 | 0.32 | 0.08 | 0.08 | 0.3 | 1 | |||||||||
| COL2A1 | −0.58 | 0.29 | 0.39 | 0.24 | 0.34 | 0.07 | −0.2 | 0.02 | 0.41 | 0.03 | 0.12 | 0.16 | 0.42 | 0.11 | 0.31 | 0.38 | 0.3 | 0.23 | 0.41 | 0.5 | 1 | ||||||||
| ACAN | −0.25 | 0.41 | 0.44 | 0.36 | 0.42 | −0.03 | −0.2 | 0.22 | 0.18 | −0.05 | 0.27 | −0.24 | 0.02 | 0.31 | 0.38 | 0.28 | 0.47 | 0.48 | 0.44 | −0.26 | 0.24 | 1 | |||||||
| NFKB1A | 0.2 | 0.3 | 0.49 | 0.39 | 0.38 | 0.49 | 0.21 | 0.28 | 0.5 | 0.34 | 0.39 | 0.44 | 0.68 | 0.22 | 0.44 | 0.42 | 0.24 | 0.32 | 0.4 | 0.64 | 0.53 | 0.1 | 1 | ||||||
| NFKB2 | −0.4 | 0.53 | 0.62 | 0.55 | 0.48 | 0.51 | 0.35 | 0.38 | 0.53 | 0.51 | 0.49 | 0.4 | 0.62 | 0.41 | 0.56 | 0.56 | 0.49 | 0.57 | 0.57 | 0.37 | 0.39 | 0.37 | 0.69 | 1 | |||||
| IKBA | 0.47 | 0.35 | 0.57 | 0.29 | 0.45 | 0.27 | 0.13 | 0.23 | 0.57 | 0.32 | 0.32 | 0.26 | 0.62 | 0.21 | 0.46 | 0.37 | 0.31 | 0.31 | 0.45 | 0.62 | 0.63 | 0.21 | 0.77 | 0.69 | 1 | ||||
| ADIPOQ | 0.72 | 0.35 | 0.4 | 0.25 | 0.38 | 0.18 | 0.12 | 0.23 | 0.33 | 0.23 | 0.38 | 0.08 | 0.26 | 0.23 | 0.44 | 0.33 | 0.47 | 0.38 | 0.41 | −0.02 | 0.19 | 0.46 | 0.23 | 0.42 | 0.35 | 1 | |||
| APLN | 0.88 | 0.39 | 0.38 | 0.35 | 0.36 | 0.26 | 0.24 | 0.27 | 0.18 | 0.28 | 0.35 | 0.07 | 0.25 | 0.42 | 0.35 | 0.33 | 0.45 | 0.5 | 0.5 | −0.14 | −0.06 | 0.51 | 0.08 | 0.56 | 0.09 | 0.48 | 1 | ||
| LEP | −0.56 | 0.54 | 0.55 | 0.48 | 0.59 | 0.49 | 0.29 | 0.37 | 0.24 | 0.08 | 0.46 | 0.32 | 0.31 | 0.57 | 0.63 | 0.48 | 0.58 | 0.59 | 0.45 | 0.09 | 0.12 | 0.39 | 0.21 | 0.34 | 0.24 | 0.6 | 0.44 | 1 | |
| RETN | 0.2 | 0.41 | 0.49 | 0.39 | 0.47 | 0.15 | 0.05 | 0.22 | 0.1 | 0.02 | 0.46 | 0.13 | 0.08 | 0.34 | 0.44 | 0.45 | 0.55 | 0.38 | 0.44 | 0.09 | 0.19 | 0.46 | 0.07 | 0.36 | 0.27 | 0.52 | 0.35 | 0.46 | 1 |
Gene expression variation with chondrosis measured by MRI
In the multivariate analysis overall gene expression significantly varied with the presence of cartilage damage when measured by MRI (P=0.018) (Table 4). The majority of these genes were expressed slightly higher with chondrosis than without chondrosis but the expression pattern associated with chondrosis is complex. Several genes that have strong positive residual correlations have standardized canonical vector coefficients of opposite sign (Table 5). For example, expression of cytokines IL-1α, IL-1β, IL-6, and TNFα are all highly positively inter-correlated (r > 0.77) but IL-1α and IL-6 have strong positive loadings on the canonical vector while IL-1β and TNFα have strong negative loadings. This indicates that IL-1α and IL-6 expression increases in chondrosis (+ canonical coefficient) more than expected after taking into account all of the expression changes among other candidate genes while IL-1β and TNFα increase much less than expected in chondrosis (− canonical coefficient) given changes in other candidate genes. Extreme positive canonical coefficients are seen for IL-6, IL-1α, MMP-3, IL-8, CCL3L1, ACAN, IκBA, APLN, and RETN, indicating higher expression than expected given the residual correlations between traits while extreme negative coefficients are observed for IL-1β, TNFα, BMP-2, CXCL3, CCL20, NFκB1A, NFκB2, and ADIPOQ, indicating lower expression than expected given other traits. Multivariate significance here is due to the observation that expression traits with positive and negative canonical coefficients are all positively correlated (Table 5), indicating that this difference is in a relatively low variation dimension. Only ADIPOQ was shown to express at a significantly (P=0.015) lower level in patients with chondrosis than in patients without chondrosis regardless of the expression levels of other traits.
Table 4.
The gene expression varies by presence or absence of chondrosis as determined by MRI (P=0.018)
| (N=48) | No Chondrosis (N=11) | Chondrosis (N=37) | P | ||
|---|---|---|---|---|---|
| Gene | Least Square Mean | 95% Confidence Interval (upper limit–lower limit) | Least Square Mean | 95% Confidence Interval (upper limit–lower limit) | |
| IL1A | 0.0009 | 0.0046–0.0002 | 0.0011 | 0.0028–0.0005 | 0.822 |
| IL1B | 0.0010 | 0.0048–0.0002 | 0.0013 | 0.0029–0.0005 | 0.827 |
| IL6 | 0.0007 | 0.0035–0.0001 | 0.0023 | 0.0055–0.0010 | 0.210 |
| TNFA | 0.0004 | 0.0016–0.0001 | 0.0004 | 0.0009–0.0002 | 0.946 |
| ADAMTS4 | 0.0029 | 0.0086–0.0010 | 0.0033 | 0.0060–0.0018 | 0.844 |
| ADAMTS5 | 0.0046 | 0.0140–0.0015 | 0.0058 | 0.0106–0.0031 | 0.729 |
| BMP2 | 0.0016 | 0.0043–0.0006 | 0.0019 | 0.0034–0.0011 | 0.715 |
| MMP1 | 0.0022 | 0.0071–0.0007 | 0.0021 | 0.0039–0.0011 | 0.902 |
| MMP3 | 0.0111 | 0.0386–0.0032 | 0.0073 | 0.0145–0.0037 | 0.569 |
| MMP9 | 0.0010 | 0.0047–0.0002 | 0.0012 | 0.0029–0.0005 | 0.793 |
| MMP13 | 0.0021 | 0.0080–0.0005 | 0.0047 | 0.0098–0.0022 | 0.307 |
| IL8 | 0.0017 | 0.0057–0.0005 | 0.0029 | 0.0056–0.0015 | 0.459 |
| CCL3 | 0.0007 | 0.0029–0.0002 | 0.0015 | 0.0033–0.0007 | 0.368 |
| CCL3L1 | 0.0003 | 0.0013–0.0001 | 0.0009 | 0.0019–0.0005 | 0.189 |
| CXCL1 | 0.0003 | 0.0013–0.0001 | 0.0008 | 0.0018–0.0004 | 0.239 |
| CXCL3 | 0.0010 | 0.0038–0.0003 | 0.0009 | 0.0018–0.0004 | 0.819 |
| CXCL6 | 0.0004 | 0.0020–0.0001 | 0.0010 | 0.0024–0.0004 | 0.347 |
| CCL20 | 0.0011 | 0.0036–0.0003 | 0.0009 | 0.0018–0.0005 | 0.878 |
| COL1A1 | 0.0269 | 0.2461–0.0029 | 0.0721 | 0.2424–0.0214 | 0.450 |
| COL2A1 | 0.0082 | 0.0343–0.0020 | 0.0068 | 0.0150–0.0031 | 0.828 |
| ACAN | 0.0015 | 0.0144–0.0002 | 0.0022 | 0.0075–0.0006 | 0.775 |
| NFKB1A | 0.0069 | 0.0204–0.0023 | 0.0066 | 0.0119–0.0036 | 0.940 |
| NFKB2 | 0.0010 | 0.0027–0.0004 | 0.0017 | 0.0029–0.0010 | 0.382 |
| IKBA | 0.0049 | 0.0137–0.0018 | 0.0048 | 0.0084–0.0027 | 0.954 |
| ADIPOQ | 0.0023 | 0.0112–0.0005 | 0.0002 | 0.0005–0.0001 | 0.015 |
| APLN | 0.0035 | 0.0146–0.0008 | 0.0033 | 0.0071–0.0015 | 0.931 |
| LEP | 0.0005 | 0.0018–0.0001 | 0.0005 | 0.0010–0.0002 | 0.969 |
| RETN | 0.0001 | 0.0008–0.0000 | 0.0003 | 0.0008–0.0001 | 0.445 |
P value in bold face represents a statistically significant difference
Table 5.
Standardized Canonical coefficients and residual correlations between gene expression traits in patients with and without chondrosis based on MRI
| Trait | Standardized canonical coefficient |
IL1A | IL1B | IL6 | TNFA |
ADAMTS 4 |
ADAMTS 5 |
BMP2 | MMP1 | MMP3 | MMP9 | MMP13 | IL8 | CCL3 | CCL3L1 | CXCL1 | CXCL3 | CXCL6 | CCL20 | COL1A1 | COL2A1 | ACAN | NFKB1A | NFKB2 | IKBA | ADIPOQ | APLN | LEP | RETN |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IL1A | 1.03 | 1 | |||||||||||||||||||||||||||
| IL1B | −2.89 | 0.79 | 1 | ||||||||||||||||||||||||||
| IL6 | 2.45 | 0.83 | 0.81 | 1 | |||||||||||||||||||||||||
| TNFA | −2.85 | 0.84 | 0.77 | 0.81 | 1 | ||||||||||||||||||||||||
| ADAMTS4 | 0.79 | 0.57 | 0.39 | 0.57 | 0.51 | 1 | |||||||||||||||||||||||
| ADAMTS5 | −0.03 | 0.44 | 0.36 | 0.41 | 0.33 | 0.83 | 1 | ||||||||||||||||||||||
| BMP2 | −2.07 | 0.62 | 0.64 | 0.71 | 0.62 | 0.56 | 0.51 | 1 | |||||||||||||||||||||
| MMP1 | 0.55 | 0.29 | 0.37 | 0.26 | 0.33 | 0.31 | 0.26 | 0.07 | 1 | ||||||||||||||||||||
| MMP3 | 1.05 | 0.32 | 0.35 | 0.42 | 0.24 | 0.36 | 0.46 | 0.42 | 0.29 | 1 | |||||||||||||||||||
| MMP9 | 0.09 | 0.6 | 0.66 | 0.7 | 0.6 | 0.56 | 0.53 | 0.56 | 0.26 | 0.21 | 1 | ||||||||||||||||||
| MMP13 | 0.94 | 0.44 | 0.5 | 0.59 | 0.44 | 0.67 | 0.67 | 0.51 | 0.4 | 0.46 | 0.56 | 1 | |||||||||||||||||
| IL8 | 0.99 | 0.42 | 0.39 | 0.49 | 0.38 | 0.33 | 0.23 | 0.22 | 0.47 | 0.32 | 0.32 | 0.44 | 1 | ||||||||||||||||
| CCL3 | 0.73 | 0.72 | 0.77 | 0.72 | 0.76 | 0.53 | 0.39 | 0.67 | 0.3 | 0.21 | 0.57 | 0.51 | 0.35 | 1 | |||||||||||||||
| CCL3L1 | 3.21 | 0.67 | 0.75 | 0.65 | 0.74 | 0.39 | 0.29 | 0.66 | 0.27 | 0.26 | 0.64 | 0.49 | 0.33 | 0.77 | 1 | ||||||||||||||
| CXCL1 | 0.36 | 0.71 | 0.76 | 0.8 | 0.72 | 0.55 | 0.44 | 0.57 | 0.24 | 0.32 | 0.62 | 0.64 | 0.44 | 0.68 | 0.62 | 1 | |||||||||||||
| CXCL3 | −1.33 | 0.77 | 0.8 | 0.76 | 0.81 | 0.48 | 0.32 | 0.62 | 0.22 | 0.28 | 0.65 | 0.4 | 0.19 | 0.82 | 0.81 | 0.69 | 1 | ||||||||||||
| CXCL6 | 0.05 | 0.69 | 0.69 | 0.74 | 0.71 | 0.36 | 0.24 | 0.57 | 0.19 | 0.44 | 0.5 | 0.39 | 0.32 | 0.73 | 0.68 | 0.65 | 0.79 | 1 | |||||||||||
| CCL20 | −1.35 | 0.66 | 0.69 | 0.65 | 0.67 | 0.32 | 0.18 | 0.41 | 0.35 | 0.3 | 0.5 | 0.36 | 0.37 | 0.66 | 0.71 | 0.57 | 0.74 | 0.75 | 1 | ||||||||||
| COL1A1 | −0.25 | 0.13 | 0.2 | 0.22 | 0.13 | 0.34 | 0.37 | 0.03 | 0.37 | 0.28 | 0.2 | 0.52 | 0.56 | −0.01 | 0.08 | 0.26 | 0 | 0.01 | 0.22 | 1 | |||||||||
| COL2A1 | 0.3 | 0.24 | 0.29 | 0.18 | 0.24 | −0.02 | −0.2 | −0.05 | 0.33 | −0.01 | 0.05 | 0.09 | 0.33 | −0.04 | 0.21 | 0.26 | 0.2 | 0.05 | 0.28 | 0.43 | 1 | ||||||||
| ACAN | 2.56 | 0.4 | 0.4 | 0.28 | 0.42 | −0.16 | −0.2 | 0.22 | 0.05 | −0.08 | 0.19 | −0.3 | −0.21 | 0.21 | 0.26 | 0.13 | 0.4 | 0.35 | 0.4 | −0.39 | 0.14 | 1 | |||||||
| NFKB1A | −0.97 | 0.38 | 0.3 | 0.4 | 0.33 | 0.37 | 0.21 | 0.14 | 0.46 | 0.25 | 0.33 | 0.44 | 0.63 | 0.12 | 0.29 | 0.3 | 0.16 | 0.2 | 0.37 | 0.6 | 0.52 | 0.02 | 1 | ||||||
| NFKB2 | −2.96 | 0.63 | 0.57 | 0.61 | 0.5 | 0.39 | 0.38 | 0.32 | 0.5 | 0.46 | 0.54 | 0.42 | 0.54 | 0.33 | 0.43 | 0.47 | 0.41 | 0.43 | 0.55 | 0.3 | 0.3 | 0.33 | 0.63 | 1 | |||||
| IKBA | 2.37 | 0.33 | 0.42 | 0.25 | 0.33 | 0.09 | 0.15 | 0.13 | 0.44 | 0.22 | 0.27 | 0.19 | 0.51 | 0.03 | 0.31 | 0.21 | 0.17 | 0.14 | 0.28 | 0.53 | 0.56 | 0.13 | 0.69 | 0.62 | 1 | ||||
| ADIPOQ | −2.64 | 0.39 | 0.4 | 0.33 | 0.33 | 0.08 | 0.15 | 0.23 | 0.13 | 0.17 | 0.41 | 0.12 | 0.13 | 0.18 | 0.43 | 0.33 | 0.38 | 0.34 | 0.29 | −0.1 | 0.06 | 0.47 | 0.16 | 0.41 | 0.26 | 1 | |||
| APLN | 1.48 | 0.39 | 0.43 | 0.39 | 0.36 | 0.19 | 0.26 | 0.31 | 0.07 | 0.24 | 0.39 | 0.12 | 0.12 | 0.4 | 0.26 | 0.35 | 0.39 | 0.45 | 0.45 | −0.18 | −0.18 | 0.53 | 0.03 | 0.54 | −0.02 | 0.48 | 1 | ||
| LEP | 0.59 | 0.52 | 0.47 | 0.44 | 0.55 | 0.46 | 0.41 | 0.39 | 0.07 | 0.15 | 0.45 | 0.32 | 0.13 | 0.52 | 0.56 | 0.43 | 0.58 | 0.53 | 0.39 | −0.01 | −0.09 | 0.3 | 0.07 | 0.28 | 0.05 | 0.66 | 0.43 | 1 | |
| RETN | 1.53 | 0.37 | 0.42 | 0.39 | 0.35 | 0.07 | 0.08 | 0.25 | −0.13 | −0.04 | 0.44 | 0.14 | −0.12 | 0.14 | 0.3 | 0.38 | 0.44 | 0.26 | 0.3 | −0.04 | 0.04 | 0.42 | −0.06 | 0.3 | 0.11 | 0.6 | 0.34 | 0.39 | 1 |
Gene expression variation with chondrosis measured by arthroscopy
Overall, the expression of all the genes studied here did not vary significantly with the presence/absence of chondrosis as measured by arthroscopy (P=0.588). Since, overall gene expression was not statistically significant, the expression of individual genes whether or not statistically significant is not inconsistent with the null model of no difference.
Correlation of degenerative changes between X-ray, MRI and arthroscopy
There is no correlation between the presence of degenerative changes based on X-ray and MRI (X2 = 0.079, 1 degree of freedom, P = 0.779, Phi = 0.04). While 78% of patients with degenerative changes based on X-ray evaluation had a diagnosis of chondrosis from their MRI, 74% of those with no degenerative change as judged from the X-ray also were diagnosed with chondrosis based on their MRI. In contrast, the chondrosis scores measured by arthroscopy were significantly correlated with both MRI (X2 = 11.395, 1 degree of freedom, P = 0.007, Phi = 0.482) and X-ray (X2 = 6.194, 1 degree of freedom, P = 0.013, Phi = 0.031).
DISCUSSION
Our study shows an overall association between gene expression in meniscal tears and the presence of early chondrosis in the knee articular cartilage as measured by MRI and X-ray but limited association of specific genes. No relationship between gene expression and presence of chondrosis as measured by arthroscopy was found.
The overall meniscal expression of important genes implicated in articular cartilage homeostasis and OA development was found to vary with the degree of articular surface chondrosis at a statistical significant level for X-ray (P=0.047) and MRI (P=0.018). Only a few genes showed a variation in expression at statistically significant levels when taken alone. For instance, two important adipokine genes adiponectin (ADIPOQ) and resistin (RETN), that have unaccounted roles in OA (5, 15, 19–21), were found to be differentially upregulated in the menisci from knees with degenerative changes on X-ray consistent with early stage OA (16, 22). While we did not find any correlation of extracellular matrix genes, and matrix metalloproteinases, with evidence for early degenerative changes, a study by Roller et al., (18) found that some of these gene transcripts in the meniscus were associated with advanced OA. MMP3, MMP13, COL1, COL3, and COL6 were moderately correlated, and MMP2 and COL2 were weakly correlated while MMP1 and VEGF were not at all correlated with OA score. One plausible explanation of the discrepancy between these findings and findings from our study is that these authors used menisci from OA and non-OA joints, while our samples were taken only from non-OA patients. Perhaps surprisingly, we observed that in univariate analysis of knees with changes on MRI only ADIPOQ was significantly (P=0.015) differentially expressed at a lower level in patients with chondrosis, opposite of our findings in knees with radiographic changes. One possible reason for this finding could be that an initial downregulation of ADIPOQ in the meniscus occurs with early chondrosis. This emphasizes the potential importance of timing with regards to injury and the development of chondrosis and osteoarthritis.
These findings corroborate with an earlier study in which only limited evidence was found for a relationship between early degenerative changes in the articular cartilage based on arthroscopy and gene expression in the injured meniscus (23). In the previous study, a global survey of gene expression in a small sample of patients showed that 49 genes were differentially regulated in knees with chondrosis compared to knees without chondrosis. When chondrosis was present in the knee, genes representing cell catabolism (cAMP catabolic process), and tissue and endothelial cell development were repressed while those involved in T cell differentiation and apoptosis were elevated. Another study has reported up-regulation of genes involved in inflammation and cytokine production and down-regulation of genes related to DNA repair processes in meniscal cells from knees with OA compared to meniscal cells from knees without OA (24).
Today, OA is viewed as a condition of the entire joint, with greater emphasis on the potential contribution of different types of tissue from the knee to the pathogenesis of the disease. Meniscus tears are associated with the development of OA (8–12) as approximately 50% of people with meniscal tears have radiographic evidence for OA 10–20 years post-injury (21). While meniscal injury is likely to be an important early event in the initiation and propagation of OA in the knee, it is unclear how degenerative changes in the menisci affect cartilage homeostasis (4–7). Therefore, the gene expression signatures in the injured meniscus could hold important information about the overall health of the knee joint in general. For example, the association of adipokine genes with chondrosis may have important implications for the infrapatellar fat pad and synovium, as well as articular cartilage, at the time of surgery and potentially stratify risk for future progression of OA in the knee.
Our study did not find a significant correlation between gene expression in the torn meniscus and chondrosis based on arthroscopic findings. There are several possible explanations for the lack of this correlation. First, categorizing knees based on grade 2 or higher chondrosis anywhere in the knee may not be optimal. It may be better to include only knees with higher degrees of chondrosis, or knees with chondrosis in the same compartment as the meniscus tear, or assess across the entire spectrum of chondrosis using standard grading (19). Second, arthroscopy may not be optimal for picking up early degenerative changes such as diffuse thinning which is not apparent at arthroscopy. Chu and colleagues (25) have shown that arthroscopic findings do not correlate with MRI findings and have advocated for the use of enhanced methods of evaluating articular cartilage such as optical coherence tomography. Finally, our cohort may not have had knees with degenerative changes advanced enough to find associated differences.
Another possibility could be that this study was not adequately powered to pick up changes on the level of individual genes. The multivariate significance was caused by both positive and negative expression level canonical coefficients for a series of genes that have highly positively correlated residual gene expression values. These gene expression values were not significantly different under chondrosis, when considered singly, but were expressed at higher or lower levels than expected given the expression levels for all other genes. Hence, a complex linear combination of gene expression levels including both positive and negative coefficients for positively correlated traits distinguishes gene expression between joints with and without chondrosis.
The sample size was not adequate to include a number of potentially important variables in the analysis such as the type of meniscus tear, mechanism of meniscus injury, and involvement of the medial or lateral menisci. Furthermore, 1.5 T MRIs are not the gold standard for assessing articular cartilage damage, although they are the clinical standard currently. As the cohort only includes patients with early chondrosis, adding patients with a broader spectrum of disease could shed further light on how these relationships change with OA. In this study, we used a candidate gene approach in which the election of genes was based on their known role in OA, inflammation, obesity and cartilage homeostasis. We could also perform entire transcriptome analysis in a large population, rather than focusing on targeted candidate genes as we have done for other studies in the past (17, 26–28) to circumvent the bias associated with the selection of candidate genes. Finally, a prospective study with longitudinal follow up could investigate the relationship of meniscal gene expression to future degeneration in the joint as the current study is a single time point cross sectional analysis which cannot assess progression over time.
In conclusion, our study demonstrates that early changes in the articular cartilage seen on MRI and X-ray have limited association with differences in gene expression of the injured meniscus. The roles of adiponectin and resistin in knee injury, particularly meniscal tears, and OA deserve further study. Additional research is needed to assess whether greater differences are seen in the meniscus from knees with more advanced OA.
Acknowledgments
Funding
This study was supported by an Orthopaedic Research and Education Foundation (OREF) grant to Dr. Brophy, by an R01AR063757 (Sandell) and by P30-AR057235 (Musculoskeletal Research Center, Sandell, P.I.) from the National Institutes of Arthritis and Musculoskeletal and Skin Diseases (NIH). Dr. Rai is supported through NIH Pathway to Independence Award (1K99AR064837) from National Institute of Arthritis and Musculoskeletal and Skin Diseases. The content of this publication is solely the responsibility of the authors and does not necessarily represent the official views of the NIH or the National Institute of Arthritis and Musculoskeletal and Skin Diseases.
Footnotes
Declaration of interest
Dr. Rai is one of the Guest Editors of the Special Issue of Connective Tissue Research on Meniscus. All other authors are not aware of any professional or financial affiliations that could be perceived as potential conflicts of interest that may be perceived to have biased the presentation.
References
- 1.Brophy RH, Barnes R, Rodeo SA, Warren RF. Prevalence of musculoskeletal disorders at the NFL Combine–trends from 1987 to 2000. Medicine and science in sports and exercise. 2007;39(1):22–7. doi: 10.1249/01.mss.0000241637.52231.18. [DOI] [PubMed] [Google Scholar]
- 2.McCarthy MM, Voos JE, Nguyen JT, Callahan L, Hannafin JA. Injury profile in elite female basketball athletes at the Women’s National Basketball Association combine. The American journal of sports medicine. 2013;41(3):645–51. doi: 10.1177/0363546512474223. [DOI] [PubMed] [Google Scholar]
- 3.Swenson DM, Collins CL, Best TM, Flanigan DC, Fields SK, Comstock RD. Epidemiology of knee injuries among U.S. high school athletes, 2005/2006-2010/2011. Medicine and science in sports and exercise. 2013;45(3):462–9. doi: 10.1249/MSS.0b013e318277acca. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bennett LD, Buckland-Wright JC. Meniscal and articular cartilage changes in knee osteoarthritis: a cross-sectional double-contrast macroradiographic study. Rheumatology (Oxford) 2002;41(8):917–23. doi: 10.1093/rheumatology/41.8.917. [DOI] [PubMed] [Google Scholar]
- 5.Casscells SW. The torn or degenerated meniscus and its relationship to degeneration of the weight-bearing areas of the femur and tibia. Clinical orthopaedics and related research. 1978;(132):196–200. [PubMed] [Google Scholar]
- 6.Lewandrowski KU, Muller J, Schollmeier G. Concomitant meniscal and articular cartilage lesions in the femorotibial joint. Am J Sports Med. 1997;25(4):486–94. doi: 10.1177/036354659702500411. [DOI] [PubMed] [Google Scholar]
- 7.Noble J, Hamblen DL. The pathology of the degenerate meniscus lesion. The Journal of bone and joint surgery British volume. 1975;57(2):180–6. [PubMed] [Google Scholar]
- 8.Ding C, Martel-Pelletier J, Pelletier JP, Abram F, Raynauld JP, Cicuttini F, et al. Meniscal tear as an osteoarthritis risk factor in a largely non-osteoarthritic cohort: a cross-sectional study. The Journal of rheumatology. 2007;34(4):776–84. [PubMed] [Google Scholar]
- 9.Englund M. Meniscal tear–a feature of osteoarthritis. Acta orthopaedica Scandinavica Supplementum. 2004;75(312):1–45. backcover. [PubMed] [Google Scholar]
- 10.Englund M. The role of the meniscus in osteoarthritis genesis. Rheum Dis Clin North Am. 2008;34(3):573–9. doi: 10.1016/j.rdc.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 11.Englund M, Guermazi A, Lohmander LS. The meniscus in knee osteoarthritis. Rheumatic diseases clinics of North America. 2009;35(3):579–90. doi: 10.1016/j.rdc.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 12.Englund M, Guermazi A, Lohmander SL. The role of the meniscus in knee osteoarthritis: a cause or consequence? Radiologic clinics of North America. 2009;47(4):703–12. doi: 10.1016/j.rcl.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 13.Nepple JJ, Brophy RH, Matava MJ, Wright RW, Clohisy JC. Radiographic findings of femoroacetabular impingement in National Football League Combine athletes undergoing radiographs for previous hip or groin pain. Arthroscopy : the journal of arthroscopic & related surgery : official publication of the Arthroscopy Association of North America and the International Arthroscopy Association. 2012;28(10):1396–403. doi: 10.1016/j.arthro.2012.03.005. [DOI] [PubMed] [Google Scholar]
- 14.Brophy RH, Gill CS, Lyman S, Barnes RP, Rodeo SA, Warren RF. Effect of anterior cruciate ligament reconstruction and meniscectomy on length of career in National Football League athletes: a case control study. The American journal of sports medicine. 2009;37(11):2102–7. doi: 10.1177/0363546509349035. [DOI] [PubMed] [Google Scholar]
- 15.Brophy RH, Rai MF, Zhang Z, Torgomyan A, Sandell LJ. Molecular analysis of age and sex-related gene expression in meniscal tears with and without a concomitant anterior cruciate ligament tear. The Journal of bone and joint surgery American volume. 2012;94(5):385–93. doi: 10.2106/JBJS.K.00919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rai MF, Sandell LJ, Cheverud JM, Brophy RH. Relationship of age and body mass index to the expression of obesity and osteoarthritis-related genes in human meniscus. Int J Obes (Lond) 2013;37(9):1238–46. doi: 10.1038/ijo.2012.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rai MF, Patra D, Sandell LJ, Brophy RH. Transcriptome analysis of injured human meniscus reveals a distinct phenotype of meniscus degeneration with aging. Arthritis and rheumatism. 2013;65(8):2090–101. doi: 10.1002/art.37984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roller BL, Monibi FA, Stoker AM, Kuroki K, Bal BS, Cook JL. Characterization of knee meniscal pathology: correlation of gross, histologic, biochemical, molecular, and radiographic measures of disease. The journal of knee surgery. 2015;28(2):175–82. doi: 10.1055/s-0034-1376333. [DOI] [PubMed] [Google Scholar]
- 19.Outerbridge RE. The etiology of chondromalacia patellae. The Journal of bone and joint surgery British volume. 1961;43-B:752–7. doi: 10.1302/0301-620X.43B4.752. [DOI] [PubMed] [Google Scholar]
- 20.Manley BFJ. Multivariate Statistical Methods: A Primer. New York: Chapman & Hall; 1986. [Google Scholar]
- 21.McDermott ID, Amis AA. The consequences of meniscectomy. The Journal of bone and joint surgery British volume. 2006;88(12):1549–56. doi: 10.1302/0301-620X.88B12.18140. [DOI] [PubMed] [Google Scholar]
- 22.Quatman CE, Quatman-Yates CC, Schmitt LC, Paterno MV. The clinical utility and diagnostic performance of MRI for identification and classification of knee osteochondritis dissecans. The Journal of bone and joint surgery American volume. 2012;94(11):1036–44. doi: 10.2106/JBJS.K.00275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Benneker LM, Heini PF, Anderson SE, Alini M, Ito K. Correlation of radiographic and MRI parameters to morphological and biochemical assessment of intervertebral disc degeneration. Eur Spine J. 2005;14(1):27–35. doi: 10.1007/s00586-004-0759-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun Y, Mauerhan DR, Honeycutt PR, Kneisl JS, Norton JH, Hanley EN, Jr, et al. Analysis of meniscal degeneration and meniscal gene expression. BMC musculoskeletal disorders. 2010;11:19. doi: 10.1186/1471-2474-11-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chu CR, Williams A, Tolliver D, Kwoh CK, Bruno S, 3rd, Irrgang JJ. Clinical optical coherence tomography of early articular cartilage degeneration in patients with degenerative meniscal tears. Arthritis Rheum. 2010;62(5):1412–20. doi: 10.1002/art.27378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brophy RH, Tycksen ED, Sandell LJ, Rai MF. Changes in transcriptome-wide gene expression of anterior cruciate ligament tears based on time from injury. Am J Sports Med. :2016. doi: 10.1177/0363546516643810. In Press. [DOI] [PubMed] [Google Scholar]
- 27.Rai MF, Sandell LJ, Zhang B, Wright RW, Brophy RH. RNA Microarray Analysis of Macroscopically Normal Articular Cartilage from Knees Undergoing Partial Medial Meniscectomy: Potential Prediction of the Risk for Developing Osteoarthritis. PLoS One. 2016;11(5):e0155373. doi: 10.1371/journal.pone.0155373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rai MF, Patra D, Sandell LJ, Brophy RH. Relationship of gene expression in the injured human meniscus to body mass index: a biologic connection between obesity and osteoarthritis. Arthritis & rheumatology. 2014;66(8):2152–64. doi: 10.1002/art.38643. [DOI] [PMC free article] [PubMed] [Google Scholar]


