Abstract
The nearly ubiquitous bacterial second messenger cyclic di-GMP is involved in a multitude of fundamental physiological processes such as sessility/motility transition and the switch between the acute and chronic infection status, combined with cell cycle control. The discovery of cyclic di-GMP, though, has been an example par excellence of scientific serendipity. We reiterate here its years-long discovery process as an activator of the cellulose synthase of the environmental bacterium Komagataeibacter xylinus and its consequences for follow-up research. Indeed, the discovery of cyclic di-GMP as a a ubiquitous second messenger contributed to the change in perception of bacteria as simple unicellular organisms just randomly building-up multicellular communities. The discovery of cyclic di-GMP also paved the way to the identification of other pro- and eukaryotic cyclic dinucleotide second messengers.
It has now been thirty years since Moshe Benziman and his group published their seminal Nature paper on the identification of the long-sought activator of the bacterial cellulose synthase as the cyclic dimeric (3′→5′) guanosine monophosphate (cyclic di-GMP) [1]. That paper signified the end of the long search for a low-molecular weight activator of in vitro cellulose biosynthesis, and, at the same time, marked the beginning of an entirely new area of research into the cellular role(s) of cyclic di-GMP and mechanisms of its action. The history of the discovery of cyclic di-GMP has been described in detail in several publications [2,3,4]. Here we present just a brief synopsis of this remarkable story that puts the current studies of cyclic di-GMP in a context.
Cellulose, poly-β-(1→4)-D-glucose, is probably the most abundant biopolymer on this planet. It is the key component of plant cell walls and has found numerous uses as firewood, lumber, paper, and sewing material [5]. In 1886, British scientist Adrian Brown showed that cellulose could be synthesized by certain bacteria [6]. In fact, plant cellulose synthase most likely has bacterial origin and was inherited by ancient plants from the cyanobacterial ancestors of their chloroplasts [7,8,9]. The process of cellulose biosynthesis by plants has long remained enigmatic and is still not fully understood. Accordingly, bacterial biosynthesis of cellulose, more amenable to experimental research, has been intensively studied throughout the 20th century. Some of such studies have been conducted in 1930s and 1940s at the Hebrew University of Jerusalem by Manfred Aschner (1901-1989) and Shlomo Hestrin (1914-1962) using the alpha-proteobacterium Acetobacter xylinum (current name, Komagataeibacter xylinus), which is an effective producer of pure microcrystalline cellulose fibers [10,11]. These studies were subsequently continued by Moshe Benziman (1928-2003) and his group [2].
The basic biochemistry of cellulose biosynthesis in algal, plant, and bacterial cells has been resolved by mid-1970s. It had been shown that the whole process begins with the glycolytic intermediate glucose-6-phosphate, which gets isomerized to glucose-1-phosphate. Glucose-1-phosphate then reacts with UTP, forming uridine-5′-diphosphate-α-D-glucose (UDP-glucose). UDP-glucose serves as a substrate for the membrane-bound cellulose synthase, which produces cellulose by transferring glucosyl residues from UDP-glucose to the growing β-D-1,4-glucan chain [12]. However, while whole cells of K. xylinus demonstrated robust production of cellulose, all attempts at purification of the active cellulose synthase were unsuccessful [3]. Even partly purified membrane fractions retained only about 0.2% of the cellulose synthase activity of the whole cells [12]. It was clear that the enzyme required membrane-bound and soluble component(s) to be active.
Benziman's group embarked on a long search for the conditions that would allow purification of an active enzyme. One milestone in this quest was the discovery of a specific activation of the enzyme fraction by micromolar amounts of GTP (Ka = 34 μM). To their surprise, a GTP analog guanosine 5′-[gamma-thio]triphosphate (GTPγS, which cannot be hydrolyzed to GDP and Pi) also proved to be an even more effective stimulator than GTP with an even lower (Ka = 17 μM). With the exception of these two, no other nucleotide or nucleotide derivative could serve as an effective activator of cellulose synthase [13]. Importantly, GDP, GMP, cGMP, guanosine 5′-[γ-thio]diphosphate and guanosine 5′-[(β,γ-imino]triphosphate were completely inactive. These observations suggested that the actual activator could be some derivative of GTP.
Further, activation by GTP could only be seen in the membrane fraction obtained in the presence of 20% polyethylene glycol (PEG-4000). It became clear that GTP interacted with some additional protein factors that were associated with the membrane-bound cellulose synthase only in the presence of PEG-4000. The presence of this protein factor and GTP activated cellulose synthesis almost 200-fold and achieved the rates that were as much as 40% of those obtained with whole cells [13]. Armed with this understanding, Aloni and colleagues succeeded in solubilizing the active cellulose synthase complex. The digitonin-solubilized enzyme still contained the GTP-interacting protein, still retained its capability to respond to GTP, and had essentially the same catalytic and regulatory properties as the membrane-bound form [14].
The next step was to characterize the GTP-binding protein and figure out whether it was an enzyme. This protein was found to bind to an agarose-hexane-GTP column and could be eluted by GTP. It was shown that this protein indeed acted on GTP, converting it to some guanine-containing activating factor. This factor was a low molecular mass, heat-stable compound that could be radioactively labeled when derived from [8-3H]GTP and [α-32P]GTP but not from [γ-32P]GTP. In the presence or absence of this compound, GTP, GDP, GMP, cGMP, ppGppp, GppppG, GpppppG, and guanosine 3′-diphosphate-S-diphosphate were all checked for their ability to stimulate cellulose synthase activity. Neither of them showed any effect, indicating the activating factor was a previously unknown guanylate derivative [15]. Using chemical analysis, the relative ratios of guanine, ribose and phosphate in this molecule were shown to be 1:1:1, whereas enzymatic analysis suggested the presence of 2′-5′ or 3′-5′ phosphodiester bonds. So, while its precise structure remained to be determined, the activating factor emerged as a cyclic nucleotide composed of GMP residues with 2′-5′ or 3′-5′ phosphodiester linkages [15].
In their final effort to characterize the activator molecule, Benziman and colleagues used DEAE-Sephadex chromatography to show that the analyzed compound consisted of no more than two GMP moieties, whereas its sensitivity to T1 endonuclease indicated that these two GMP moieties were linked by a 3′-5′phosphodiester bond. Mass-spectroscopic measurements estimated the molecular weight of this compound to be 690, which corresponded to the molecular weight of a cyclic diguanylic acid. Finally, the chemically synthesized cyclic bis(3′→5′) diguanylic acid was shown to stimulate cellulose synthase activity and have the same properties as the native activator in a variety of chemical and enzymatic tests [1].
Curiously, despite its importance and novelty, the Nature paper by Ross and colleagues reporting the identification of cyclic di-GMP as the activator of cellulose synthase [1] has not attracted much attention. In the 12 years, from 1987 till 2000, cyclic di-GMP has been mentioned in only 10 papers, all but one of which came from the Benziman's laboratory. Intriguingly though, these papers addressed several of the fundamental questions in cyclic dinucleotide second messenger signaling, equally important and still unresolved today. Benziman and his close collaborators touched upon the generality of cyclic di-GMP signaling in bacteria by demonstrating activation of a cellulose synthase in an α-proteobacterium different than K. xylinus [16], see also [17];identification of diguanylate cyclases and phosphodiesterases [18], see [19,20]; the presence of a second unrelated phosphodiesterase to break down pGpG [1], see [21,22], and took up the quest for the determination of the molecular basis of the enzymatic activity of cyclic di-GMP turnover proteins [23]. Equally important, determination of the cyclic di-GMP concentration in the bacterial cell [24,25] and the nonlinear correlation between cyclic di-GMP concentrations and physiological output [18,26] came up in the course of the analysis of regulation of cellulose biosynthesis by three different diguanylate cyclases in K. xylinus. Furthermore, the biological impact of chemically synthesized cyclic di-GMP analogues was tested [27,28,29] and screens for inhibitors of cyclic di-GMP diguanylate cyclases were initiated [30,31]. Last, but not least, interkingdom crosstalk of cyclic di-GMP was addressed [32,33,34] among other issues.
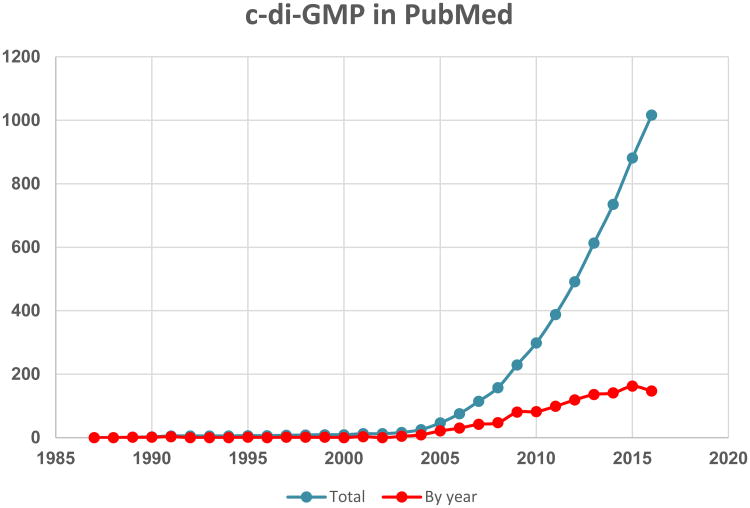
In this century, however, the situation has changed dramatically (Fig. 1). Based on independent observations around the beginning of the century [35,36,37,38], Benziman's legacy left the identification of GGDEF and EAL domains as diguanylate cyclases and phosphodiesterases [36,39,40,41], signals that regulate cyclic di-GMP turnover proteins [42], cyclic di-GMP receptors [43,44], and the widespread physiological impact of the ubiquitous second messenger cyclic di-GMP as a major sessility/motility life style, infection style and cell cycle regulator in Bacteria [45,46,47]. to be discovered by others. Unfortunately, and perhaps to his personal disappointment, Benziman's work could not achieve his other goal—to find a way to regulate and enhance cellulose biosynthesis in plants. As we know today, plant cellulose synthases do not require allosteric regulation by cyclic di-GMP [9].
Figure 1.
The history of the c-di-GMP field in publications. The number of papers containing the word ”di-GMP” or ”diGMP” or ”cyclic diguanylate” in PubMed are plotted over time, from 1987 to 2016.
The GGDEF and EAL domains that had been discovered by Benziman and coworkers in the diguanylate cyclases and phosphodiesterases involved in cyclic di-GMP turnover [18], have been found in multiple copies a variety of diverse bacteria [48,49,50]. Accordingly, cellulose biosynthesis has been detected in a wide variety of bacteria, including model organisms Escherichia coli and Salmonella typhimurium [51,52]. It soon became clear that, along with curli fimbriae, cyclic di-GMP-regulated cellulose production plays a key role in biofilm formation their pathogenic relatives from Escherichia, Salmonella, Citrobacter, Enterobacter, and Klebsiella genera [53]. These findings opened the flood gates, with hundreds of papers on cyclic di-GMP published every year (Fig. 1).
Although several cyclic di-GMP binding mechanisms had already been detected [54], it has taken much longer to uncover the exact mechanism of cellulose synthase activation by cyclic di-GMP, even after the identification of the cyclic di-GMP-binding PilZ domain at the C-terminus of the membrane-bound cellulose synthase subunit BcsA [43,55]. Only after the elucidation of the crystal structure of the bacterial cellulose synthase complex [56], it became clear that it contains a conserved gating loop that blocks access of UDP-glucose to the active site. Upon cyclic di-GMP binding to the PilZ domain, the gating loop moves away from the active site cleft and allows the proper functioning of the enzyme [17]. The discovery of cyclic di-GMP was subsequently followed by the serendipitous detection of prokaryotic cyclic di-AMP and cyclic GAMP, two more cyclic dinucleotides with distinct physiological roles and phylogenetic distribution [57,58]. Intriguingly, although synthesized by distinct enzyme families, these cyclic dinucleotides seem to be connected through enzyme promiscuity as variants of cyclic di-GMP synthesizing GGDEF domain proteins have recently been shown to produce cyclic GAMP [59]. Of note, the eukaryotic version of cyclic GAMP synthase seems to be a central component of the innate immune surveillance system [60,61]. Finally, to close the circle, cyclicyclic di-GMP has even reached the eukaryotic world to be involved in cell differentiation in the social amoeba Dictyostelium discoideum [62]. We will curiously await the next surprises that this signaling molecule will provide for us
Acknowledgments
We thank Dr. Dorit Amikam for helpful comments. UR is supported by the Swedish Research Council Natural Sciences and Engineering, the Karolinska Institutet and Petrus and Augusta Hedlund Foundation; MYG is supported by the NIH Intramural Research Program at the U.S. National Library of Medicine.
References
- 1.Ross P, Weinhouse H, Aloni Y, Michaeli D, Weinberger-Ohana P, Mayer R, Braun S, de Vroom E, van der Marel GA, van Boom JH, Benziman M. Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature. 1987;325:279–281. doi: 10.1038/325279a0. [DOI] [PubMed] [Google Scholar]
- 2.Ross P, Mayer R, Benziman M. Cellulose biosynthesis and function in bacteria. Microbiol Rev. 1991;55(1):35–58. doi: 10.1128/mr.55.1.35-58.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Delmer DP. Structure and biosynthesis of cellulose Part II: Biosynthesis. In: Kung SD, Yang SF, editors. Discoveries in Plant Biology. Vol. 3. World Scientific Publishing Co; Singapore - Hackensack, NJ - London: 2000. pp. 199–216. [Google Scholar]
- 4.Amikam D, Weinhouse H, Galperin MY. Moshe Benziman and the discovery of cyclic di-GMP. In: Wolfe AJ, Visick KL, editors. The Second Messenger Cyclic Di-GMP. ASM Press; Washington, DC: 2010. pp. 11–23. [Google Scholar]
- 5.Römling U, Galperin MY. Bacterial cellulose biosynthesis: diversity of operons, subunits, products and functions. Trends Microbiol. 2015;23(9):545–557. doi: 10.1016/j.tim.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown AJ. On acetic ferment which forms cellulose. J Chem Soc Transactions (London) 1886;49:432–439. [Google Scholar]
- 7.Nobles DR, Romanovicz DK, Brown RM., Jr Cellulose in cyanobacteria. Origin of vascular plant cellulose synthase? Plant Physiol. 2001;127(2):529–542. [PMC free article] [PubMed] [Google Scholar]
- 8.Nobles DR, Brown RM., Jr The pivotal role of cyanobacteria in the evolution of cellulose synthases and cellulose synthase-like proteins. Cellulose. 2004;11:437–448. [Google Scholar]
- 9.Pear JR, Kawagoe Y, Schreckengost WE, Delmer DP, Stalker DM. Higher plants contain homologs of the bacterial celA genes encoding the catalytic subunit of cellulose synthase. Proc Natl Acad Sci USA. 1996;93(22):12637–12642. doi: 10.1073/pnas.93.22.12637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aschner M, Hestrin S. Fibrillar structure of cellulose of bacterial and animal origin. Nature. 1946;157:659. doi: 10.1038/157659b0. [DOI] [PubMed] [Google Scholar]
- 11.Hestrin S, Aschner M, Mager J. Synthesis of cellulose by resting cells of Acetobacter xylinum. Nature. 1947;159:64–65. doi: 10.1038/159064a0. [DOI] [PubMed] [Google Scholar]
- 12.Swissa M, Aloni Y, Weinhouse H, Benizman M. Intermediary steps in Acetobacter xylinum cellulose synthesis: studies with whole cells and cell-free preparations of the wild type and a celluloseless mutant. J Bacteriol. 1980;143(3):1142–1150. doi: 10.1128/jb.143.3.1142-1150.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aloni Y, Delmer DP, Benziman M. Achievement of high rates of in vitro synthesis of 1,4-b-D-glucan: activation by cooperative interaction of the Acetobacter xylinum enzyme system with GTP, polyethylene glycol, and a protein factor. Proc Natl Acad Sci USA. 1982;79(21):6448–6452. doi: 10.1073/pnas.79.21.6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aloni Y, Cohen R, Benziman M, Delmer D. Solubilization of the UDP-glucose:1,4-b-D-glucan 4-b-D-glucosyltransferase (cellulose synthase) from Acetobacter xylinum. A comparison of regulatory properties with those of the membrane-bound form of the enzyme. J Biol Chem. 1983;258(7):4419–4423. [PubMed] [Google Scholar]
- 15.Ross P, Aloni Y, Weinhouse C, Michaeli D, Weinberger-Ohana P, Mayer R, Benziman M. An unusual guanyl oligonucleotide regulates cellulose synthesis. in Acetobacter xylinum FEBS Lett. 1985;186(2):191–196. doi: 10.1016/0014-5793(85)80706-7. [DOI] [PubMed] [Google Scholar]
- 16.Amikam D, Benziman M. Cyclic diguanylic acid and cellulose synthesis in Agrobacterium tumefaciens. J Bacteriol. 1989;171(12):6649–6655. doi: 10.1128/jb.171.12.6649-6655.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morgan JL, McNamara JT, Zimmer J. Mechanism of activation of bacterial cellulose synthase by cyclic di-GMP. Nat Struct Mol Biol. 2014;21(5):489–496. doi: 10.1038/nsmb.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tal R, Wong HC, Calhoon R, Gelfand DH, Fear AL, Volman G, Mayer R, Ross P, Amikam D, Weinhouse H, Cohen A, Sapir S, Ohana P, Benziman M. Three cdg operons control cellular turnover of cyclic di-GMP in Acetobacter xylinum: genetic organization and occurrence of conserved domains in isoenzymes. J Bacteriol. 1998;180(17):4416–4425. doi: 10.1128/jb.180.17.4416-4425.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schmidt AJ, Ryjenkov DA, Gomelsky M. The ubiquitous protein domain EAL is a cyclic diguanylate-specific phosphodiesterase: enzymatically active and inactive EAL domains. J Bacteriol. 2005;187(14):4774–4781. doi: 10.1128/JB.187.14.4774-4781.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schirmer T. C-di-GMP synthesis: structural aspects of evolution, catalysis and regulation. J Mol Biol. 2016;428(19):3683–3701. doi: 10.1016/j.jmb.2016.07.023. [DOI] [PubMed] [Google Scholar]
- 21.Orr MW, Donaldson GP, Severin GB, Wang J, Sintim HO, Waters CM, Lee VT. Oligoribonuclease is the primary degradative enzyme for pGpG in Pseudomonas aeruginosa that is required for cyclic-di-GMP turnover. Proc Natl Acad Sci USA. 2015;112(36):E5048–5057. doi: 10.1073/pnas.1507245112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cohen D, Mechold U, Nevenzal H, Yarmiyhu Y, Randall TE, Bay DC, Rich JD, Parsek MR, Kaever V, Harrison JJ, Banin E. Oligoribonuclease is a central feature of cyclic diguanylate signaling in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2015;112(36):11359–11364. doi: 10.1073/pnas.1421450112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rao F, Qi Y, Chong HS, Kotaka M, Li B, Li J, Lescar J, Tang K, Liang ZX. The functional role of a conserved loop in EAL domain-based cyclic di-GMP-specific phosphodiesterase. J Bacteriol. 2009;191(15):4722–4731. doi: 10.1128/JB.00327-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weinhouse H, Sapir S, Amikam D, Shilo Y, Volman G, Ohana P, Benziman M. c-di-GMP-binding protein, a new factor regulating cellulose synthesis in Acetobacter xylinum. FEBS Lett. 1997;416(2):207–211. doi: 10.1016/s0014-5793(97)01202-7. [DOI] [PubMed] [Google Scholar]
- 25.Simm R, Morr M, Remminghorst U, Andersson M, Romling U. Quantitative determination of cyclic diguanosine monophosphate concentrations in nucleotide extracts of bacteria by matrix-assisted laser desorption/ionization-time-of-flight mass spectrometry. Anal Biochem. 2009;386(1):53–58. doi: 10.1016/j.ab.2008.12.013. [DOI] [PubMed] [Google Scholar]
- 26.Massie JP, Reynolds EL, Koestler BJ, Cong JP, Agostoni M, Waters CM. Quantification of high-specificity cyclic diguanylate signaling. Proc Natl Acad Sci USA. 2012;109(31):12746–12751. doi: 10.1073/pnas.1115663109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blommers MJ, Haasnoot CA, Walters JA, van der Marel GA, van Boom JH, Hilbers CW. Solution structure of the 3′-5′ cyclic dinucleotide d(pApA) A combined NMR, UV melting, and molecular mechanics study. Biochemistry. 1988;27(22):8361–8369. doi: 10.1021/bi00422a011. [DOI] [PubMed] [Google Scholar]
- 28.Hsu CYDD, Jones RA. Synthesis and physical characterization of bis 3′-5′ cyclic dinucleotides (Np-Np): RNA polymerase inhibitors. Nucleosides Nucleotides. 1985;4:377–389. [Google Scholar]
- 29.Witte G, Hartung S, Buttner K, Hopfner KP. Structural biochemistry of a bacterial checkpoint protein reveals diadenylate cyclase activity regulated by DNA recombination intermediates. Mol Cell. 2008;30(2):167–178. doi: 10.1016/j.molcel.2008.02.020. [DOI] [PubMed] [Google Scholar]
- 30.Ohana P, Delmer DP, Carlson RW, Glushka J, Azadi P, Bacic T, Benziman M. Identification of a novel triterpenoid saponin from Pisum sativum as a specific inhibitor of the diguanylate cyclase of Acetobacter xylinum. Plant Cell Physiol. 1998;39(2):144–152. doi: 10.1093/oxfordjournals.pcp.a029351. [DOI] [PubMed] [Google Scholar]
- 31.Lieberman OJ, Orr MW, Wang Y, Lee VT. High-throughput screening using the differential radial capillary action of ligand assay identifies ebselen as an inhibitor of diguanylate cyclases. ACS Chem Biol. 2014;9(1):183–192. doi: 10.1021/cb400485k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Amikam D, Steinberger O, Shkolnik T, Ben-Ishai Z. The novel cyclic dinucleotide 3′-5′ cyclic diguanylic acid binds to p21ras and enhances DNA synthesis but not cell replication in the Molt 4 cell line. Biochem J. 1995;311(3):921–927. doi: 10.1042/bj3110921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karaolis DK, Means TK, Yang D, Takahashi M, Yoshimura T, Muraille E, Philpott D, Schroeder JT, Hyodo M, Hayakawa Y, Talbot BG, Brouillette E, Malouin F. Bacterial c-di-GMP is an immunostimulatory molecule. J Immunol. 2007;178(4):2171–2181. doi: 10.4049/jimmunol.178.4.2171. [DOI] [PubMed] [Google Scholar]
- 34.Danilchanka O, Mekalanos JJ. Cyclic dinucleotides and the innate immune response. Cell. 2013;154(5):962–970. doi: 10.1016/j.cell.2013.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jones HA, Lillard JW, Jr, Perry RD. HmsT, a protein essential for expression of the haemin storage (Hms+) phenotype of Yersinia pestis. Microbiology. 1999;145(8):2117–2128. doi: 10.1099/13500872-145-8-2117. [DOI] [PubMed] [Google Scholar]
- 36.Römling U, Rohde M, Olsen A, Normark S, Reinkoster J. AgfD, the checkpoint of multicellular and aggregative behaviour in Salmonella typhimurium regulates at least two independent pathways. Mol Microbiol. 2000;36(1):10–23. doi: 10.1046/j.1365-2958.2000.01822.x. [DOI] [PubMed] [Google Scholar]
- 37.Hecht GB, Newton A. Identification of a novel response regulator required for the swarmer-to-stalked-cell transition in Caulobacter crescentus. J Bacteriol. 1995;177(21):6223–6229. doi: 10.1128/jb.177.21.6223-6229.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tischler AD, Camilli A. Cyclic diguanylate (c-di-GMP) regulates Vibrio cholerae biofilm formation. Mol Microbiol. 2004;53(3):857–869. doi: 10.1111/j.1365-2958.2004.04155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barends TR, Hartmann E, Griese JJ, Beitlich T, Kirienko NV, Ryjenkov DA, Reinstein J, Shoeman RL, Gomelsky M, Schlichting I. Structure and mechanism of a bacterial light-regulated cyclic nucleotide phosphodiesterase. Nature. 2009;459(7249):1015–1018. doi: 10.1038/nature07966. [DOI] [PubMed] [Google Scholar]
- 40.Chan C, Paul R, Samoray D, Amiot NC, Giese B, Jenal U, Schirmer T. Structural basis of activity and allosteric control of diguanylate cyclase. Proc Natl Acad Sci USA. 2004;101(49):17084–17089. doi: 10.1073/pnas.0406134101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ausmees N, Mayer R, Weinhouse H, Volman G, Amikam D, Benziman M, Lindberg M. Genetic data indicate that proteins containing the GGDEF domain possess diguanylate cyclase activity. FEMS Microbiol Lett. 2001;204(1):163–167. doi: 10.1111/j.1574-6968.2001.tb10880.x. [DOI] [PubMed] [Google Scholar]
- 42.Chang AL, Tuckerman JR, Gonzalez G, Mayer R, Weinhouse H, Volman G, Amikam D, Benziman M, Gilles-Gonzalez MA. Phosphodiesterase A1, a regulator of cellulose synthesis in Acetobacter xylinum, is a heme-based sensor. Biochemistry. 2001;40(12):3420–3426. doi: 10.1021/bi0100236. [DOI] [PubMed] [Google Scholar]
- 43.Amikam D, Galperin MY. PilZ domain is part of the bacterial c-di-GMP binding protein. Bioinformatics. 2006;22(1):3–6. doi: 10.1093/bioinformatics/bti739. [DOI] [PubMed] [Google Scholar]
- 44.Sudarsan N, Lee ER, Weinberg Z, Moy RH, Kim JN, Link KH, Breaker RR. Riboswitches in eubacteria sense the second messenger cyclic di-GMP. Science. 2008;321(5887):411–413. doi: 10.1126/science.1159519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Simm R, Morr M, Kader A, Nimtz M, Romling U. GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility. Mol Microbiol. 2004;53(4):1123–1134. doi: 10.1111/j.1365-2958.2004.04206.x. [DOI] [PubMed] [Google Scholar]
- 46.Paul R, Weiser S, Amiot NC, Chan C, Schirmer T, Giese B, Jenal U. Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain. Genes Dev. 2004;18(6):715–727. doi: 10.1101/gad.289504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tischler AD, Camilli A. Cyclic diguanylate regulates Vibrio cholerae virulence gene expression. Infect Immun. 2005;73(9):5873–5882. doi: 10.1128/IAI.73.9.5873-5882.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Galperin MY, Natale DA, Aravind L, Koonin EV. A specialized version of the HD hydrolase domain implicated in signal transduction. J Mol Microbiol Biotechnol. 1999;1(2):303–305. [PMC free article] [PubMed] [Google Scholar]
- 49.Galperin MY. Conserved ‘hypothetical’ proteins: new hints and new puzzles. Comp Funct Genomics. 2001;2(1):14–18. doi: 10.1002/cfg.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Galperin MY, Nikolskaya AN, Koonin EV. Novel domains of the prokaryotic two-component signal transduction systems. FEMS Microbiol Lett. 2001;203(1):11–21. doi: 10.1111/j.1574-6968.2001.tb10814.x. [DOI] [PubMed] [Google Scholar]
- 51.Zogaj X, Nimtz M, Rohde M, Bokranz W, Römling U. The multicellular morphotypes of Salmonella typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol Microbiol. 2001;39(6):1452–1463. doi: 10.1046/j.1365-2958.2001.02337.x. [DOI] [PubMed] [Google Scholar]
- 52.Römling U. Molecular biology of cellulose production in bacteria. Res Microbiol. 2002;153(4):205–212. doi: 10.1016/s0923-2508(02)01316-5. [DOI] [PubMed] [Google Scholar]
- 53.Zogaj X, Bokranz W, Nimtz M, Römling U. Production of cellulose and curli fimbriae by members of the family Enterobacteriaceae isolated from the human gastrointestinal tract. Infect Immun. 2003;71(7):4151–4158. doi: 10.1128/IAI.71.7.4151-4158.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chou SH, Galperin MY. Diversity of cyclic ci-GMP-binding proteins and mechanisms. J Bacteriol. 2016;198(1):32–46. doi: 10.1128/JB.00333-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ryjenkov DA, Simm R, Romling U, Gomelsky M. The PilZ domain is a receptor for the second messenger c-di-GMP: the PilZ domain protein YcgR controls motility in enterobacteria. J Biol Chem. 2006;281(41):30310–30314. doi: 10.1074/jbc.C600179200. [DOI] [PubMed] [Google Scholar]
- 56.Morgan JL, Strumillo J, Zimmer J. Crystallographic snapshot of cellulose synthesis and membrane translocation. Nature. 2013;493(7431):181–186. doi: 10.1038/nature11744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Davies BW, Bogard RW, Young TS, Mekalanos JJ. Coordinated regulation of accessory genetic elements produces cyclic di-nucleotides for V. cholerae virulence. Cell. 2012;149(2):358–370. doi: 10.1016/j.cell.2012.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339(6121):786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hallberg ZF, Wang XC, Wright TA, Nan B, Ad O, Yeo J, Hammond MC. Hybrid promiscuous (Hypr) GGDEF enzymes produce cyclic AMP-GMP (3′, 3′-cGAMP) Proc Natl Acad Sci USA. 2016;113(7):1790–1795. doi: 10.1073/pnas.1515287113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ablasser A, Goldeck M, Cavlar T, Deimling T, Witte G, Rohl I, Hopfner KP, Ludwig J, Hornung V. cGAS produces a 2′-5′-linked cyclic dinucleotide second messenger that activates STING. Nature. 2013;498(7454):380–384. doi: 10.1038/nature12306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gao P, Ascano M, Wu Y, Barchet W, Gaffney BL, Zillinger T, Serganov AA, Liu Y, Jones RA, Hartmann G, Tuschl T, Patel DJ. Cyclic [G(2′,5′)pA(3′,5′)p] is the metazoan second messenger produced by DNA-activated cyclic GMP-AMP synthase. Cell. 2013;153(5):1094–1107. doi: 10.1016/j.cell.2013.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schaap P. Cyclic di-nucleotide signaling enters the eukaryote domain. IUBMB Life. 2013;65(11):897–903. doi: 10.1002/iub.1212. [DOI] [PMC free article] [PubMed] [Google Scholar]