Abstract
We characterized the C9orf72 hexanucleotide repeat expansion (RE) mutation in amyotrophic lateral sclerosis (ALS) patients of 2 distinct origins, Ashkenazi and North Africa Jews (AJ, NAJ), its frequency, and genotype-phenotype correlations. In AJ, 80% of familial ALS (fALS) and 11% of sporadic ALS carried the RE, a total of 12.9% of all AJ-ALS compared to 0.3% in AJ controls (odds ratio [OR] = 44.3, p < 0.0001). In NAJ, 10% of fALS and 9% of sporadic ALS carried the RE, a total of 9.1% of all NAJ-ALS compared to 1% in controls (OR = 9.9, p = 0.0006). We identified a risk haplotype shared among all ALS patients, although an association with age at disease onset, fALS, and dementia were observed only in AJ. Variations were identified downstream the repeats. The risk haplotype and these polymorphisms were at high frequencies in alleles with 8 repeats or more, suggesting sequence instability. The different genotype-phenotype correlations and OR, together with the large range in age at onset, suggest that other modifiers and risk factors may affect penetrance and phenotype in ALS.
1. Introduction
The G4C2 hexanucleotide repeat expansion in C9orf72 gene is the most common genetic mutation that causes amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD). Since first identified (DeJesus-Hernandez et al., 2011; Renton et al., 2011), the mutation was reported in different cohorts around the world with various frequencies among familial ALS (fALS), sporadic ALS (sALS), FTD, and ethnic groups (Abramycheva et al., 2015; Beck et al., 2013; Borghero et al., 2014; Byrne et al., 2012; Chen et al., 2016; Gijselinck et al., 2012; Hubers et al., 2014; Itzcovich et al., 2016; Majounie et al., 2012; Scotter et al., 2017; Simon-Sanchez et al., 2012; Snowden et al., 2012; Stewart et al., 2012; Umoh et al., 2016; Vrabec et al., 2015). Common to all reports is the observation that the expansion of the repeat is more frequent in fALS. Moreover, the expansion is more frequent in ALS patients with dementia, as well as patients affected with FTD. In most studies, a risk haplotype was identified, although some rare cases of expansions on a nonrisk haplotype were reported.
The normal function of C9orf72 protein is still unclear but evidence supports roles in membrane trafficking and autophagy (Ciura et al., 2016; Farg et al., 2014; Sellier et al., 2016; Sullivan et al., 2016; Webster et al., 2016; Yang et al., 2016). Recently, it was shown that the normal C9orf72 protein localizes to processing bodies and is recruited to stress granules upon stress-related stimuli (Maharjan et al., 2017). Moreover, knockdown of C9orf72 completely abolishes stress granule formation and accelerates cell death.
We report here for the first time a high frequency of the G4C2 nucleotide repeat expansion in C9orf72 in a large cohort of ALS patients of Ashkenazi (AJ) and North Africa Jewish (NAJ) origins, which are genetically homogeneous populations and already proved valuable to study the genetics of neurodegenerative disorders, such as Parkinson's disease (Gan-Or et al., 2008; Orr-Urtreger et al., 2007). We further identified the mutation to be on a common, unstable risk haplotype and evaluated genotype-phenotype correlations.
2. Material and methods
2.1. Samples
DNA was extracted from the peripheral blood using standard protocols. Samples included 459 unrelated Jewish ALS patients followed at the ALS Clinic at Tel Aviv Sourasky Medical Center, Tel Aviv, Israel (Table 1). All patients had a diagnosis of clinically definite or probable ALS according to the revised El Escorial criteria (Brooks et al., 2000). The recruitment interval spanned from 2004 to 2016. Included in this cohort are 22 patients with known mutations in OPTN (Goldstein et al., 2016), PFN1 (Wu et al., 2012), and SOD1. Of the 459 patients, 349 were AJ (76%), 76 were Jews of Moroccan descent (16.6%, MJ), and 34 were of other North Africa origins (7.4%, NAJ-nm).
Table 1.
The ALS patients screened for the C9orf72 expansion mutation included Jews of Ashkenazi origin (AJ-ALS), Morocco (MJ-ALS) and other North Africa origins (NAJ-ALS).
| AJ-ALS | MJ-ALS | NAJ-ALS | Total | |
|---|---|---|---|---|
|
| ||||
| Number of unrelated patients (% of total) | 349 (76.0) | 76 (16.6) | 34 (7.4) | 459 |
|
| ||||
| Age at onset, yrs (±SD) | 63.0 (±11.6) | 54.5 (±13.1) | 56.4 (±13.1) | 61.1 (±12.4) |
| Males (%) | 208 (59.6) | 44 (57.9) | 16 (47) | 268 (58.4) |
| Familial ALS (%) | 10a (2.9) | 7b (9.2) | 3c (8.8) | 20 (4.3) |
|
| ||||
| Disease Duration, monthsd (±SD), number of patients | 36.8 (±27.9), n=293 | 38.2 (±28.9) n=57 | 34.4 (±22.0) n=26 | 36.8 (±27.6) n=376 |
|
| ||||
| Site of onset (%) | ||||
| Bulbar | 95 (27.2) | 21 (27.6) | 12 (35.3) | 128 (27.9) |
| Limb | 254 (72.8) | 55 (72.4) | 22 (64.7) | 331 (72.1) |
Seventeen patients had a questionable positive family history and were not included in the percentage calculation and analysis for association with familial ALS.
Six patients had a questionable positive family history and were not included in the percentage calculation and analysis for association with familial ALS.
One patient had a questionable positive family history and was not included in the percentage calculation and analysis for association with familial ALS.
Disease duration was calculated only for patients deceased or with tracheotomy
A total of 900 control samples were analyzed, that included 671 anonymous DNA samples from young healthy individuals, aged 20–45 years, mostly women (300 MJ and 371 AJ), who underwent routine genetic screening tests and were randomly selected and 229 healthy elderly AJ (age range: 44–87 years; average age 68.3 ± 8.9 years; 92 males and 137 females) (Goldstein et al., 2016).
All patients underwent an interview to disclose ancestry, family history of ALS, dementia or other neurodegenerative diseases, age at onset (AAO) defined as the age when the first symptoms appeared, and affected site at disease onset (limb or bulbar). Disease duration, defined as time from first symptoms to death or tracheotomy, was recorded for all patients, as well as the existence of cognitive or behavioral disturbances.
2.2. Standard protocol approvals, registrations, and patient consents
All participants provided informed consent before entering the study. All DNA samples were coded and tested in an anonymous manner. The Institutional and National Supreme Helsinki Institutional Review Board Committees for Genetics Studies approved the study protocol and the informed consent forms.
2.3. C9orf72 expansion genotyping
Determining the number of the G4C2 repeats was done as previously described (DeJesus-Hernandez et al., 2011; Renton et al., 2011). Briefly, allele determination was based on 2 polymerase chain reactions (PCRs), 1 using primers flanking the repeat and 1 using repeat-primed PCR methodology. Some primers were modified (Table A.1, Fig. A.1). An expansion allele was recorded when the number of repeats was 30 or more (C9Positive [C9Pos]). The rest of the alleles of less than 30 repeats were considered negative for the expansion (C9Negative [C9Neg]). Fragment analysis was conducted on an ABI 3130xl Genetic Analyzer, and peaks were visualized using GeneMapper 4.0 (Applied Biosciences).
2.4. Deletion/insertion characterization
Primers flanking the repeats were designed to sequence alleles suspicious of having deletions or insertions (Table A.1-B, primer pairs 1–3). Sequence was performed using BigDye V1.1 chemistry and an ABI 3130xl Genetic Analyzer.
2.5. SNP genotyping and haplotype analysis
One hundred thirty-nine AJ-ALS and 127 AJ controls were genotyped on Affymetrix Genome-Wide Human single nucleotide polymorphism (SNP) 6.0 array. Of the 139 AJ-ALS, 14 were C9orf72 carriers. The analysis was done using SNP & Variation Suite v8.2 (Golden Helix, Inc.). Genotypes on chromosome 9 were aligned to identify the minimal linkage disequilibrium (LD) interval and to determine the risk haplotype. Six nonredundant SNPs were sequenced in 6 MJ-ALS and 4 NAJ-nm-ALS patients, carriers of the C9orf72 expansion (Table A.1–C).
2.6. Statistical analysis
Unless otherwise mentioned, all calculations were carried out using SPSS software V.15 (SPSS Inc., Chicago, IL) with 2-sided significant tests. Independence of categorical variables was tested using the χ2 distribution. For small cell counts, a Fisher's exact test (2-tailed) was used. AAO and disease duration were tested for association with expansion using independent t-test. Mann-Whitney U-test was used to determine if the distribution of wild-type alleles is the same across categories of ethnicity and risk or nonrisk haplotype. Survival curve was generated using Kaplan-Meier test.
3. Results
3.1. Wild-type alleles in the Jewish population
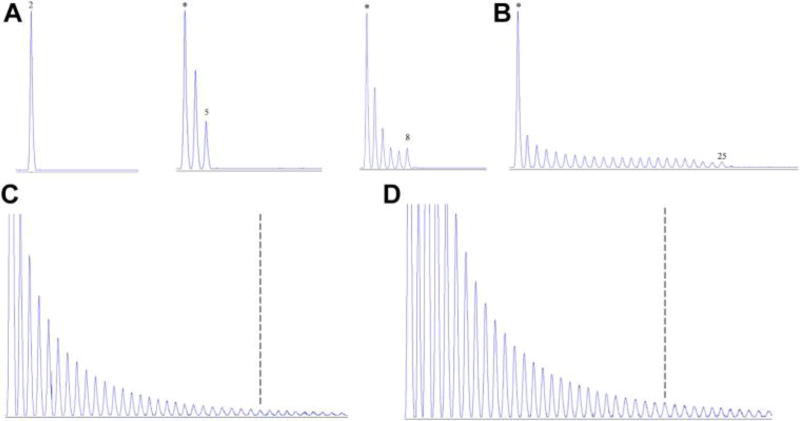
A total of 459 unrelated ALS patients and 900 ethnically matched controls were tested. Among the nonexpanded alleles, the most frequent number of repeats was 2, 5, and 8 in all groups (Fig. 1A, Table A.2, 60.0%, 11.1%, 12.7% of all alleles, respectively). The rest of the nonexpanded alleles were each observed at 0%–4.6% (Table A.2). Intermediate repeats (20–29 repeats) were rare and observed in 7 individuals (Fig. 1B, Table A.2). No difference in the nonexpanded allele distribution was observed between ALS and controls of Ashkenazi origin (p = 0.618) or of Moroccan origin (p = 0.759).
Fig. 1. C9orf72 G4C2 hexanucleotide repeat capillary-based sequence traces of the repeat-primed PCR (protocol 2).
(A) The 3 most common alleles in the Jewish population are 2, 5, and 8 repeats. * 1/4 The first peak represent 3 repeats when number of repeats is higher than 2. (B) Intermediate repeat size of 25 is observed. (C) Expanded allele is observed with a saw-tooth tail pattern that extends beyond 30 repeats (dotted line) with a 6-bp periodicity. (D) Expanded allele is observed in a sample that failed to give any PCR results using flanking primers, suggesting this individual carries 2 expanded alleles. Abbreviation: PCR, polymerase chain reaction.
3.2. C9orf72 expansion is strongly associated with ALS in the AJ population and is correlated with fALS, dementia, and earlier AAO
Of the 349 AJ-ALS, 45 carried the expansion (12.9%), compared to 2 AJ controls (Table 2, p = 2.04 × 10−18, odds ratio [OR] = 44.26, 95% confidence interval [CI]: 10.66–183.68, p < 0.0001; Fig. 1C). One patient was suggestive of having 2 expanded alleles because he showed an expansion pattern (Fig. 1D) with no PCR product when using the flanking primers, and primers incompatibility was excluded by using 9 different primer pairs (data not shown).
Table 2.
The association between C9orf72 expansion and ALS.
| Study group / Genotype |
No. Tested | Carriers of the C9orf72 expansion (30 repeats or more) |
Both alleles are less than 30 repeats |
p-value (Fisher's Exact test, df=1, two tailed) |
OR for heterozygous (95% CI, p-value of OR) |
|
|---|---|---|---|---|---|---|
| ALS patients: | AJ | 349 | 45a (12.9) | 304 (87.1) | p =2.04×10−18 | 44.26 (10.66–183.68, <0.0001) |
| MJ | 76 | 6 (7.9) | 70 (92.1) | p =0.0029 | 8.49 (2.07–34.76, 0.003) | |
| NAJ | 34 | 4b (11.8) | 30 (88.2) | p =0.0025 | 13.20 (2.82–61.78, 0.001) | |
| Total North Africa: MJ+NAJ | 110 | 10 (9.1) | 100 (90.9) | p =1.87×10−4 | 9.9 (2.67–36.69, 0.0006) | |
| Total (AJ, MJ, NAJ) | 459 | 55 (12.0) | 404 (88.0) | p =1.091×10−21 | 24.37 (9.68–61.33, <0.0001) | |
| Controlsc: | AJ | 600 | 2 (0.3) | 598 (99.7) | ||
| MJ | 300 | 3 (1.0) | 297 (99) | |||
| Total (AJ, MJ) | 900 | 5 (0.5) | 895 (99.5) | |||
One ALS patient had both alleles with more than 30 repeats.
One from Algier, one from Egypt, one from Tunis and one from Egypt/Tunis
Control groups are in Hardy-Weinberg equilibrium
Abbreviations: ALS= amyotrophic lateral sclerosis; CI= confidence interval; OR= odds ratio; AJ= Ashkenazi Jews; MJ= Moroccan Jews; NAJ= North Africa Jews that are not of Morocco descent.
Of the 10 AJ-ALS with evident familial history (fALS), 8 were C9orf72 mutation carriers (Fisher's exact test, p = 1.57 × 10−6, Table 3). Three of them were previously reported (Majounie et al., 2012). Additional 3 patients with a family history of dementia were C9Pos.
Table 3.
Genotype-phenotype correlations of the C9orf72 expansion in Jewish ALS patients.
| AJ-ALS | MJ-ALS | All North-Africa Jewish ALS (MJ+NAJ) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| C9 expansion (≥30) (n=45) |
No expansion (<30) (n=304) |
p-valuea (χ2) | C9 expansion (≥30) |
No expansion (<30) |
p-valuea | C9 expansion (≥30) |
No expansion (<30) |
p-valuea | ||
| ALS classb | fALS (%) | 8 (80) | 2 (20) | p=1.57×10−6 | 1 (14.3) | 6 (85.7) | p=0.452 | 1 (10) | 9 (90) | p=1.00 |
| sALS (%) | 37 (10.9) | 302 (89.1) | 5 (7.2) | 64 (92.8) | 9 (9) | 91 (91) | ||||
| Dementiac | Dementia (%) | 7 (29.2) | 17 (70.8) | p=0.012 (6.29) | 2 (28.6) | 5 (71.4) | p=0.094 | 2 (18.2) | 9 (81.8) | p=0.265 |
| No dementia (%) | 37 (11.5) | 285 (88.5) | 4 (5.9) | 64 (94.1) | 8 (8.2) | 90 (91.8) | ||||
| Gender | Male | 24 (11.5) | 184 (88.5) | p=0.359 (0.84) | 2 (4.5) | 42(95.6) | p=0.233 | 3 (5) | 57 (95) | p=0.181 |
| Female | 21 (14.9) | 120 (85.1) | 4 (12.5) | 28 (87.5) | 7 (14) | 43 (71.6) | ||||
| Site of onset | Limb | 29 (11.4) | 225 (88.6) | p=0.178 (1.81) | 2 (3.6) | 53 (96.4) | p=0.046 | 5 (6.5) | 72 (93.5) | p=0.163 |
| Bulbar | 16 (16.8) | 79 (83.2) | 4 (19.0) | 17 (81.0) | 5 (15.2) | 28 (84.8) | ||||
| Age at onset (± SD) | 58.7 (±9.6) | 63.6 (±11.8) | p=0.008 (assuming equal variance) | 53.0 (±8.0) | 54.6 (±13.5) | p=0.77 (assuming equal variance) | 54.1 (±6.2) | 55.2 (±13.6) | p=0.647 (assuming unequal variance) | |
| Disease duration in months, (± SD), for deceased or with tracheotomy only | 36.9 (±22.7) | 36.8 (±28.6) | p=0.973 (assuming equal variance) | 32.8 (±7.5) | 38.8 (±30.4) | p=0.265 (assuming unequal variance) | 32.6 (±6.9) | 37.4 (±28.0) | p=0.253 (assuming unequal variance) | |
χ2 test (or Fisher's Exact test when numbers 5 or lower) was done on categorical variants. Independent t-test was done on AAO and Disease duration
See comments a–c in Table 1.
Excluding cases with unclear status of dementia.
In patients, a significant association with dementia was detected (Pearson χ2 = 6.29, df = 1, p = 0.012, Table 3), as well as with earlier AAO (58.7 ± 9.6 years in C9Pos compared to 63.6 ± 11.8 years in C9Neg [<30 repeats, t-test, p = 0.008]). C9orf72 expansion mutation did not correlate with gender, site of disease onset, or disease duration (Table 3). C9Pos did not show a significant shorter survival from the disease onset (median survival 37 months) than the C9Neg (median survival 33 months; p = 0.987, p = 0.458, p = 0.642 for log-rank, Breslow, and Tarone-Ware tests, respectively; with 13.3% and 14.8% censored of C9Pos and C9Neg, respectively).
3.3. C9orf72 expansion is strongly associated with ALS in the NAJ population
Of the MJ-ALS, 6 carried the expansion (7.9%), compared to 3 in MJ controls (OR = 8.49, 95% CI: 2.07–34.76, p = 0.003, Table 2). Four C9Pos ALS patients were observed in North Africa patients who are not of Moroccan origin (11.8%, OR = 13.2, 95% CI: 2.82–61.78, p = 0.001, Table 2). When looking at all these 10 North Africa ALS patients, the OR is 9.9 with p = 0.0006 (Table 2). In MJ-ALS, a tendency toward a correlation between the G4C2 expansion and dementia was suspected, as 28.6% of ALS with dementia were C9Pos (p = 0.094, Fisher's exact test), and only a weak significant correlation with site of onset was found (p = 0.046, Fisher's exact test, Table 3). These 2 observations do not exist when looking at the entire cohort of all North Africa patients (Table 3).
3.4. C9orf72 expansion mutations in the Ashkenazi Jews and North Africa Jews share a risk haplotype
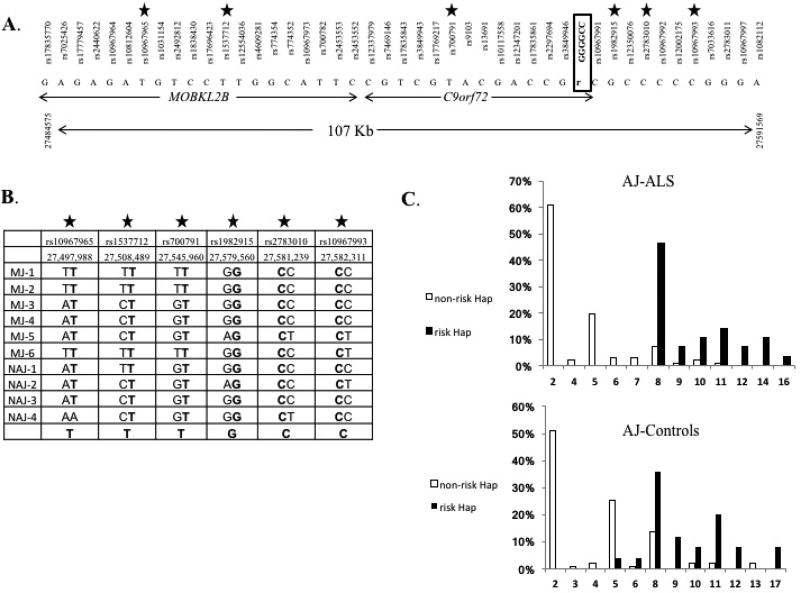
We analyzed whole-genome SNP genotypes of 139 AJ-ALS and 127 matched controls. Of the AJ-ALS, 14 carried the C9orf72 expansion mutation. We tested to see if they carry the original Finnish risk haplotype (Laaksovirta et al., 2010; Majounie et al., 2012). A 107-Kb minimal LD interval was identified (chr9:27484575–27591569) that included 44 informative SNPs, encompassed the complete C9orf72 gene (Fig. 2A), and was the same as the reported Finnish haplotype. A minimal number of 6 nonredundant SNPs determining the risk haplotype were genotyped in all 6 MJ-ALS and 4 NAJ-nm-ALS C9Pos patients (Fig. 2A). All these 10 patients carried the risk haplotype, with the exception of 1 patient from Egypt who was homozygous to the nonrisk allele at the proximal end of the LD interval (rs10967965, Fig. 2B) reducing the LD to 92.8 Kb. This suggests a common ancestor to all Jewish C9Pos ALS patients from Ashkenazi and North Africa origins.
Fig. 2. C9orf72 gene region haplotype analysis.
(A) 107-Kb risk haplotype is shared among AJ-ALS patients carrying the C9orf72 expansion, between rs17835770 and rs1082112 (107 Kb), that includes the complete C9orf72 gene. Marked with starts are the 6 nonredundant SNPs that define the risk haplotype. (B) Haplotype analysis of the 10 ALS patients of North Africa origin carrying the C9orf72 expansion. All patients carry the same risk haplotype as observed in the AJ-ALS, marked with bold letters and highlighted in the last row, with the exception of 1 patient (NAJ-nm-4) that is homozygous to the nonrisk allele at the proximal end of the LD interval (rs10967965, AA, Chr9:27,497,988) reducing the LD to 92.8 Kb. (C) Repeat allele frequencies for risk and nonrisk haplotypes in the AJ-ALS patients and AJ controls. The repeat sizes are higher for the risk haplotype in both groups and are highly significant (p < 0.0001). Abbreviation: AJ-ALS, ALS patients screened for the C9orf72 expansion mutation in Jews of Ashkenazi origin; ALS, amyotrophic lateral sclerosis; Hap, haplotype; LD, linkage disequilibrium; NAJ-nm, North Africa Jews that are not of Morocco descent.
3.5. The C9orf72 background risk haplotype of Ashkenazi
Of the AJ-ALS patients who were C9Neg (n = 125), 28 carried the risk haplotype (22.4%). Of the 127 controls which were C9Neg, 25 carried the risk haplotype (19.7%). For all 252 individuals, we scored the allele with the highest number of repeats and evaluated the correlation between these alleles and the risk haplotype. In the ALS group, all 28 that carried the risk haplotype had 8 repeats or more (100%) compared to only 11.3% of patients without the risk haplotype (p < 0.0001, Mann-Whitney U test, Fig. 2C). In AJ controls, 23 of the 25 that carried the risk haplotype had 8 repeats or more (92%) compared to 20 of 102 that did not carry the risk haplotype (19.6%) (p < 0.0001, Mann-Whitney U test, Fig. 2C). The same significant result was observed when looking at all AJ (n = 252, data not shown).
3.6. The number of repeats in the unexpanded alleles is higher in MJ compared to AJ
Interestingly, among the control individuals, the distribution of the wild-type alleles was significantly different in MJ compared to AJ, with higher number of repeats in the MJ group (p < 0.0001, Mann-Whitney U test). This might explain why expansion is more frequent in the MJ controls (1.0%) than in the AJ controls (0.3%). This was also observed in the ALS groups (p = 0.001, Mann-Whitney U test).
We further explored the possibility that higher number of repeats in wild-type alleles also affects the overall lower AAO observed in MJ-ALS patients compared to AJ-ALS (p = 2.99 × 10−8, t-test), but we did not find a significant correlation to AAO, regardless of the patient's ethnicity (data not shown), suggesting that the difference in AAO between MJ-ALS and AJ-ALS is not related to higher number of repeats in nonexpanded C9orf72 alleles.
3.7. Sequence instability is significantly more frequent in chromosomes bearing at least 8 repeats
We observed 5 types of deletions/insertions downstream the repeat: (1) 12-bp deletion in a wild-type allele (n = 4, Fig. A.2-A and H) or in an expanded allele (n = 19, Fig. A.2-B and H); (2) 12-bp duplication (n = 2, Fig. A.2-C and H); (3) 11-bp duplication (n = 5, Fig. A.2-D and H); (4) indel of 11-bp deletion and 1-bp insertion (n = 24, Fig. A.2-E and H); and (5) 24-bp insertion (n = 1, Fig. A.2-F and H). Two samples did not show a pattern of repeats when using protocol 2 and did show an expanded allele when using protocol 3; therefore, their variation at the 3′ end was not determined (Fig. A.2-G1 and G2). Altogether, these insertions and deletions, localized to highly GC-rich region, were relatively rare (n = 55, 2.0% of all alleles) and mostly present on chromosomes bearing 8 repeats or more (45/55, 81.8%), suggesting a tendency toward sequence instability when number of repeats is at least 8. The 12-bp deletion and the indel (11-bp deletion and 1-bp insertion) were mostly observed in AJ (20/23 and 23/24, respectively), while the 11-bp duplication was unique to MJ and the 12-bp duplication unique to AJ. Furthermore, 31.1% (19/61) of the C9orf72 expansion alleles were adjacent to the 12-bp deletion, compared to only 36 alleles bearing variation of 2655 C9Neg alleles (χ2 = 251.95, df = 1, p < 0.0001). Finally, we did not find a significant difference in AAO or disease duration between AJ-ALS patients carrying the C9orf72 expansion mutation with the deletion at the 3′ end compared to those C9Pos that did not have the deletion (t-test, p = 0.72 and p = 0.29, respectively).
4. Discussion
We hereby describe for the first time the frequency of C9orf72 expansion mutation in a large cohort of ALS patients from different Jewish origins, demonstrating high carrier rates. Our study indicates that the C9orf72 expansion is the most common genetic cause of ALS in AJ (13%) but only second to the founder OPTN691_692insAG mutation in MJ-ALS patients (8% compared to 14.5%) (Goldstein et al., 2016). Mutations in C9orf72 and OPTN together explain 16% and 20% of all AJ and MJ-ALS, respectively. Because both genes are involved in autophagy, our data further emphasize the important contribution of this pathway to ALS. Our results help understand the genetic background of ALS in patients of Jewish origin and will further improve decision making during the genetic workup and genetic consultation of patients and families, strongly supporting the need to screen all ALS patients for the C9orf72 mutation.
Phenotype-genotype correlation showed different results depending on the ethnicity of the patients. In ALS patients of North Africa origin, we did not detect any significant correlation between the expansion and different clinical characteristics of the disease. This might be due to the significantly earlier AAO in NAJ ALS patients compared to the AJ and the worldwide average, and the factors causing this difference might mask the correlation. In AJ, the correlation to fALS was very high compared to other populations reported [80% compared to 0% and 5% in Middle Eastern and Asian patients, respectively, the smallest extreme (Majounie et al., 2012), and 46.7% and 57.9% in Belgian and Sardinia, respectively, the highest extreme (Gijselinck et al., 2012; Majounie et al., 2012)]. Even when including the sALS cases with familial dementia history with fALS, this percentage stays high (61.1%). Moreover, C9orf72 mutation is also high in sALS compared to other populations (11% in AJ and 9% in NAJ), compared to a 6.3% found in a cross-sectional study that included 11 different European countries and populations in the United States (Majounie et al., 2012).
Our results also point to the need of studying a well characterized clinical cohort of FTD Jewish patients to determine the frequency of C9orf72 expansion mutation in this group. A genome-wide study on these 2 closely related neurodegenerative diseases, in a genetically homogeneous group, might decipher the genetic factors that lead to the development of one phenotype over the other.
The AJ are considered a relatively homogeneous population due to historic bottle necks and religious constraints. However, even within such a homogeneous group, we found a large variation in AAO among carriers, aged from 32 to 73 years, as well as a very large difference in the length of disease duration, from 5 to 138 months. Another example for the phenotypic diversity is the coexistence of the motor neuron phenotype with dementia in only about 16% of the C9Pos patients. These facts suggest the presence of other modifiers, and the need for whole-genome analyses, in addition to further accurate determination of the expansion length, using other methods, as well as the determination of the methylation status of the region.
The C9orf72 expansion mutation among Jews shares a risk haplotype. Comparing the risk haplotype to previously reported risk haplotypes in other populations suggests common ancestor with the European haplotype (Laaksovirta et al., 2010; Majounie et al., 2012; Smith et al., 2013). This risk haplotype was found in 22.4% of AJ controls and in high frequencies in the European and African populations (Garcia-Redondo et al., 2013), suggesting an ancient haplotype. It is still debatable if the expansion occurred only once and was inherited as an unstable allele or if chromosomes bearing 8 repeats or more are more prone to expansion. We identified 1 AJ-ALS C9Pos patient with 32 repeats. Another case of ALS patient was previously described with 28 repeats on the risk haplotype chromosome (Garcia-Redondo et al., 2013). Although possible, it is less likely that such a large shrinking from hundreds of repeats took place. Therefore, different mutation events within this risk chromosome could have occurred independently, with yet unexplained mechanism. This model can also be supported by the finding that in the control MJ population, the number of repeats is significantly higher than in AJ and the frequency of expansion carriers is higher as well (1.0% compared to 0.3%), although we did not show anticipation or a higher percentage of C9-ALS in MJ compared to AJ.
We found several variations downstream the repeat, all within the first 23 bp after the repeat. This region is GC rich (the first 100 bp after the repeat has 87% GC content), and the instability is observed mostly on chromosomes bearing 8 repeats or more. Interestingly, the indel variation (11-bp deletion and 1-bp insertion), which we observed in AJ and 1 MJ, was previously reported as unique to samples of Portuguese origin in a large-scale multinational study (Nordin et al., 2017). Also of interest is that some of the variations were observed on chromosomes that had less than 8 repeats. It might be that these rare cases represent rare recombinations that occurred between chromosomes with and without the expansion. The effect of this variation on gene function, if at all, is yet to be determined.
One of our ALS patients carried both the C9orf72 expansion mutation and OPTN 691-692AG insertion mutation in heterozygous state. This is yet another example of double mutations (Lattante et al., 2015). The surprisingly high number of ALS cases carrying the OPTN 691-692AG insertion mutation or the C9orf72 expansion mutation, and more specifically, the high frequency (1%) of each of these mutations in the controls, suggest this cohort as a good model to shed a light on the penetrance of these mutations and to find additional genetic modifiers of ALS, or protective alleles, and by that hinting to potential therapeutic interventions.
Supplementary Material
Acknowledgments
The authors thank Tova Naiman for her technical assistance. The authors are grateful to the patients and their families who participated in this study.
This work was supported by Adelis Foundation, by ALS Association (grant number 47717), and by Kahn Foundation.
Footnotes
Disclosure statement
Bryan J Traynor has a patent pending on the clinical testing and therapeutic intervention for the hexanucleotide repeat expansion of C9orf72; Avi Orr-Urtreger received research support from ALS Association and from Kahn and Adelis Foundations; and Vivian E Drory received research support from ALS Association and from Adelis Foundation. Other authors report no conflicts of interest.
References
- Abramycheva NY, Lysogorskaia EV, Stepanova MS, Zakharova MN, Kovrazhkina EA, Razinskaya OD, Smirnov AP, Maltsev AV, Ustyugov AA, Kukharsky MS, Khritankova IV, Bachurin SO, Cooper-Knock J, Buchman VL, Illarioshkin SN, Skvortsova VI, Ninkina N. C9ORF72 hexanucleotide repeat expansion in ALS patients from the Central European Russia population. Neurobiol. Aging. 2015;36:2908.e5–2908.e9. doi: 10.1016/j.neurobiolaging.2015.07.004. [DOI] [PubMed] [Google Scholar]
- Beck J, Poulter M, Hensman D, Rohrer JD, Mahoney CJ, Adamson G, Campbell T, Uphill J, Borg A, Fratta P, Orrell RW, Malaspina A, Rowe J, Brown J, Hodges J, Sidle K, Polke JM, Houlden H, Schott JM, Fox NC, Rossor MN, Tabrizi SJ, Isaacs AM, Hardy J, Warren JD, Collinge J, Mead S. Large C9orf72 hexanucleotide repeat expansions are seen in multiple neurodegenerative syndromes and are more frequent than expected in the UK population. Am. J. Hum. Genet. 2013;92:345–353. doi: 10.1016/j.ajhg.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borghero G, Pugliatti M, Marrosu F, Marrosu MG, Murru MR, Floris G, Cannas A, Parish LD, Occhineri P, Cau TB, Loi D, Ticca A, Traccis S, Manera U, Canosa A, Moglia C, Calvo A, Barberis M, Brunetti M, Pliner HA, Renton AE, Nalls MA, Traynor BJ, Restagno G, Chio A ITALS-GEN and SARDINALS Consortia. Genetic architecture of ALS in Sardinia. Neurobiol. Aging. 2014;35:2882.e7–2882.e12. doi: 10.1016/j.neurobiolaging.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks BR, Miller RG, Swash M, Munsat TL World Federation of Neurology Research Group on Motor Neuron Diseases. El Escorial revisited: revised criteria for the diagnosis of amyotrophic lateral sclerosis. Amyotroph Lateral Scler. Other Motor Neuron Disord. 2000;1:293–299. doi: 10.1080/146608200300079536. [DOI] [PubMed] [Google Scholar]
- Byrne S, Elamin M, Bede P, Shatunov A, Walsh C, Corr B, Heverin M, Jordan N, Kenna K, Lynch C, McLaughlin RL, Iyer PM, O’Brien C, Phukan J, Wynne B, Bokde AL, Bradley DG, Pender N, Al-Chalabi A, Hardiman O. Cognitive and clinical characteristics of patients with amyotrophic lateral sclerosis carrying a C9orf72 repeat expansion: a population-based cohort study. Lancet Neurol. 2012;11:232–240. doi: 10.1016/S1474-4422(12)70014-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Lin Z, Chen X, Cao B, Wei Q, Ou R, Zhao B, Song W, Wu Y, Shang HF. Large C9orf72 repeat expansions are seen in Chinese patients with sporadic amyotrophic lateral sclerosis. Neurobiol. Aging. 2016;38:217.e15–217.e22. doi: 10.1016/j.neurobiolaging.2015.11.016. [DOI] [PubMed] [Google Scholar]
- Ciura S, Sellier C, Campanari ML, Charlet-Berguerand N, Kabashi E. The most prevalent genetic cause of ALS-FTD, C9orf72 synergizes the toxicity of ATXN2 intermediate polyglutamine repeats through the autophagy pathway. Autophagy. 2016;12:1406–1408. doi: 10.1080/15548627.2016.1189070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NA, Flynn H, Adamson J, Kouri N, Wojtas A, Sengdy P, Hsiung GY, Karydas A, Seeley WW, Josephs KA, Coppola G, Geschwind DH, Wszolek ZK, Feldman H, Knopman DS, Petersen RC, Miller BL, Dickson DW, Boylan KB, Graff-Radford NR, Rademakers R. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farg MA, Sundaramoorthy V, Sultana JM, Yang S, Atkinson RA, Levina V, Halloran MA, Gleeson PA, Blair IP, Soo KY, King AE, Atkin JD. C9ORF72, implicated in amytrophic lateral sclerosis and frontotemporal dementia, regulates endosomal trafficking. Hum. Mol. Genet. 2014;23:3579–3595. doi: 10.1093/hmg/ddu068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan-Or Z, Giladi N, Rozovski U, Shifrin C, Rosner S, Gurevich T, Bar-Shira A, Orr-Urtreger A. Genotype-phenotype correlations between GBA mutations and Parkinson disease risk and onset. Neurology. 2008;70:2277–2283. doi: 10.1212/01.wnl.0000304039.11891.29. [DOI] [PubMed] [Google Scholar]
- Garcia-Redondo A, Dols-Icardo O, Rojas-Garcia R, Esteban-Perez J, Cordero-Vazquez P, Munoz-Blanco JL, Catalina I, Gonzalez-Munoz M, Varona L, Sarasola E, Povedano M, Sevilla T, Guerrero A, Pardo J, Lopez de Munain A, Marquez-Infante C, de Rivera FJ, Pastor P, Jerico I, de Arcaya AA, Mora JS, Clarimon J, Group, C.O.S.S. Gonzalo-Martinez JF, Juarez-Rufian A, Atencia G, Jimenez-Bautista R, Moran Y, Mascias J, Hernandez-Barral M, Kapetanovic S, Garcia-Barcina M, Alcala C, Vela A, Ramirez-Ramos C, Galan L, Perez-Tur J, Quintans B, Sobrido MJ, Fernandez-Torron R, Poza JJ, Gorostidi A, Paradas C, Villoslada P, Larrode P, Capablo JL, Pascual-Calvet J, Goni M, Morgado Y, Guitart M, Moreno-Laguna S, Rueda A, Martin-Estefania C, Cemillan C, Blesa R, Lleo A. Analysis of the C9orf72 gene in patients with amyotrophic lateral sclerosis in Spain and different populations worldwide. Hum. Mutat. 2013;34:79–82. doi: 10.1002/humu.22211. [DOI] [PubMed] [Google Scholar]
- Gijselinck I, Van Langenhove T, van der Zee J, Sleegers K, Philtjens S, Kleinberger G, Janssens J, Bettens K, Van Cauwenberghe C, Pereson S, Engelborghs S, Sieben A, De Jonghe P, Vandenberghe R, Santens P, De Bleecker J, Maes G, Baumer V, Dillen L, Joris G, Cuijt I, Corsmit E, Elinck E, Van Dongen J, Vermeulen S, Van den Broeck M, Vaerenberg C, Mattheijssens M, Peeters K, Robberecht W, Cras P, Martin JJ, De Deyn PP, Cruts M, Van Broeckhoven C. A C9orf72 promoter repeat expansion in a Flanders-Belgian cohort with disorders of the frontotemporal lobar degeneration-amyotrophic lateral sclerosis spectrum: a gene identification study. Lancet Neurol. 2012;11:54–65. doi: 10.1016/S1474-4422(11)70261-7. [DOI] [PubMed] [Google Scholar]
- Goldstein O, Nayshool O, Nefussy B, Traynor BJ, Renton AE, Gana-Weisz M, Drory VE, Orr-Urtreger A. OPTN 691_692insAG is a founder mutation causing recessive ALS and increased risk in heterozygotes. Neurology. 2016;86:446–453. doi: 10.1212/WNL.0000000000002334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubers A, Marroquin N, Schmoll B, Vielhaber S, Just M, Mayer B, Hogel J, Dorst J, Mertens T, Just W, Aulitzky A, Wais V, Ludolph AC, Kubisch C, Weishaupt JH, Volk AE. Polymerase chain reaction and Southern blot-based analysis of the C9orf72 hexanucleotide repeat in different motor neuron diseases. Neurobiol. Aging. 2014;35:1214.e1–1214.e6. doi: 10.1016/j.neurobiolaging.2013.11.034. [DOI] [PubMed] [Google Scholar]
- Itzcovich T, Xi Z, Martinetto H, Chrem-Mendez P, Russo MJ, de Ambrosi B, Uchitel OD, Nogues M, Silva E, Rojas G, Bagnatti P, Amengual A, Campos J, Rogaeva E, St George-Hyslop P, Allegri R, Sevlever G, Surace EI. Analysis of C9orf72 in patients with frontotemporal dementia and amyotrophic lateral sclerosis from Argentina. Neurobiol. Aging. 2016;40:192.e13–192.e15. doi: 10.1016/j.neurobiolaging.2016.02.001. [DOI] [PubMed] [Google Scholar]
- Laaksovirta H, Peuralinna T, Schymick JC, Scholz SW, Lai SL, Myllykangas L, Sulkava R, Jansson L, Hernandez DG, Gibbs JR, Nalls MA, Heckerman D, Tienari PJ, Traynor BJ. Chromosome 9p21 in amyotrophic lateral sclerosis in Finland: a genome-wide association study. Lancet Neurol. 2010;9:978–985. doi: 10.1016/S1474-4422(10)70184-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lattante S, Ciura S, Rouleau GA, Kabashi E. Defining the genetic connection linking amyotrophic lateral sclerosis (ALS) with frontotemporal dementia (FTD) Trends Genet. 2015;31:263–273. doi: 10.1016/j.tig.2015.03.005. [DOI] [PubMed] [Google Scholar]
- Maharjan N, Kunzli C, Buthey K, Saxena S. C9ORF72 regulates stress granule formation and its deficiency impairs stress granule assembly, hyper-sensitizing cells to stress. Mol. Neurobiol. 2017;54:3062–3077. doi: 10.1007/s12035-016-9850-1. [DOI] [PubMed] [Google Scholar]
- Majounie E, Renton AE, Mok K, Dopper EG, Waite A, Rollinson S, Chio A, Restagno G, Nicolaou N, Simon-Sanchez J, van Swieten JC, Abramzon Y, Johnson JO, Sendtner M, Pamphlett R, Orrell RW, Mead S, Sidle KC, Houlden H, Rohrer JD, Morrison KE, Pall H, Talbot K, Ansorge O, Chromosome 9-ALS/FTD Consortium, The French research network on FTLD/FTLD/ ALS, The ITALSGEN Consortium. Hernandez DG, Arepalli S, Sabatelli M, Mora G, Corbo M, Giannini F, Calvo A, Englund E, Borghero G, Floris GL, Remes AM, Laaksovirta H, McCluskey L, Trojanowski JQ, Van Deerlin VM, Schellenberg GD, Nalls MA, Drory VE, Lu CS, Yeh TH, Ishiura H, Takahashi Y, Tsuji S, Le Ber I, Brice A, Drepper C, Williams N, Kirby J, Shaw P, Hardy J, Tienari PJ, Heutink P, Morris HR, Pickering-Brown S, Traynor BJ. Frequency of the C9orf72 hexanucleotide repeat expansion in patients with amyotrophic lateral sclerosis and frontotemporal dementia: a cross-sectional study. Lancet Neurol. 2012;11:323–330. doi: 10.1016/S1474-4422(12)70043-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordin A, Akimoto C, Wuolikainen A, Alstermark H, Forsberg K, Baumann P, Pinto S, de Carvalho M, Hubers A, Nordin F, Ludolph AC, Weishaupt JH, Meyer T, Grehl T, Schweikert K, Weber M, Burkhardt C, Neuwirth C, Holmoy T, Morita M, Tysnes OB, Benatar M, Wuu J, Lange DJ, Bisgard C, Asgari N, Tarvainen I, Brannstrom T, Andersen PM. Sequence variations in C9orf72 downstream of the hexanucleotide repeat region and its effect on repeat-primed PCR interpretation: a large multinational screening study. Amyotroph. Lateral Scler. Frontotemporal Degener. 2017;18:256–264. doi: 10.1080/21678421.2016.1262423. [DOI] [PubMed] [Google Scholar]
- Orr-Urtreger A, Shifrin C, Rozovski U, Rosner S, Bercovich D, Gurevich T, Yagev-More H, Bar-Shira A, Giladi N. The LRRK2 G2019S mutation in Ashkenazi Jews with Parkinson disease: is there a gender effect? Neurology. 2007;69:1595–1602. doi: 10.1212/01.wnl.0000277637.33328.d8. [DOI] [PubMed] [Google Scholar]
- Renton AE, Majounie E, Waite A, Simon-Sanchez J, Rollinson S, Gibbs JR, Schymick JC, Laaksovirta H, van Swieten JC, Myllykangas L, Kalimo H, Paetau A, Abramzon Y, Remes AM, Kaganovich A, Scholz SW, Duckworth J, Ding J, Harmer DW, Hernandez DG, Johnson JO, Mok K, Ryten M, Trabzuni D, Guerreiro RJ, Orrell RW, Neal J, Murray A, Pearson J, Jansen IE, Sondervan D, Seelaar H, Blake D, Young K, Halliwell N, Callister JB, Toulson G, Richardson A, Gerhard A, Snowden J, Mann D, Neary D, Nalls MA, Peuralinna T, Jansson L, Isoviita VM, Kaivorinne AL, Holtta-Vuori M, Ikonen E, Sulkava R, Benatar M, Wuu J, Chio A, Restagno G, Borghero G, Sabatelli M, ITALSGEN Consortium. Heckerman D, Rogaeva E, Zinman L, Rothstein JD, Sendtner M, Drepper C, Eichler EE, Alkan C, Abdullaev Z, Pack SD, Dutra A, Pak E, Hardy J, Singleton A, Williams NM, Heutink P, Pickering-Brown S, Morris HR, Tienari PJ, Traynor BJ. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scotter EL, Smyth L, Bailey JA, Wong CH, de Majo M, Vance CA, Synek BJ, Turner C, Pereira J, Charleston A, Waldvogel HJ, Curtis MA, Dragunow M, Shaw CE, Smith BN, Faull RL. C9ORF72 and UBQLN2 mutations are causes of amyotrophic lateral sclerosis in New Zealand: a genetic and pathologic study using banked human brain tissue. Neurobiol. Aging. 2017;49:214.e1–214.e5. doi: 10.1016/j.neurobiolaging.2016.06.019. [DOI] [PubMed] [Google Scholar]
- Sellier C, Campanari ML, Julie Corbier C, Gaucherot A, Kolb-Cheynel I, Oulad-Abdelghani M, Ruffenach F, Page A, Ciura S, Kabashi E, Charlet-Berguerand N. Loss of C9ORF72 impairs autophagy and synergizes with polyQ Ataxin-2 to induce motor neuron dysfunction and cell death. EMBO J. 2016;35:1276–1297. doi: 10.15252/embj.201593350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon-Sanchez J, Dopper EG, Cohn-Hokke PE, Hukema RK, Nicolaou N, Seelaar H, de Graaf JR, de Koning I, van Schoor NM, Deeg DJ, Smits M, Raaphorst J, van den Berg LH, Schelhaas HJ, De Die-Smulders CE, Majoor-Krakauer D, Rozemuller AJ, Willemsen R, Pijnenburg YA, Heutink P, van Swieten JC. The clinical and pathological phenotype of C9ORF72 hexanucleotide repeat expansions. Brain. 2012;135(Pt 3):723–735. doi: 10.1093/brain/awr353. [DOI] [PubMed] [Google Scholar]
- Smith BN, Newhouse S, Shatunov A, Vance C, Topp S, Johnson L, Miller J, Lee Y, Troakes C, Scott KM, Jones A, Gray I, Wright J, Hortobagyi T, Al-Sarraj S, Rogelj B, Powell J, Lupton M, Lovestone S, Sapp PC, Weber M, Nestor PJ, Schelhaas HJ, Asbroek AA, Silani V, Gellera C, Taroni F, Ticozzi N, Van den Berg L, Veldink J, Van Damme P, Robberecht W, Shaw PJ, Kirby J, Pall H, Morrison KE, Morris A, de Belleroche J, Vianney de Jong JM, Baas F, Andersen PM, Landers J, Brown RH, Jr, Weale ME, Al-Chalabi A, Shaw CE. The C9ORF72 expansion mutation is a common cause of ALSþ/-FTD in Europe and has a single founder. Eur. J. Hum. Genet. 2013;21:102–108. doi: 10.1038/ejhg.2012.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snowden JS, Rollinson S, Thompson JC, Harris JM, Stopford CL, Richardson AM, Jones M, Gerhard A, Davidson YS, Robinson A, Gibbons L, Hu Q, DuPlessis D, Neary D, Mann DM, Pickering-Brown SM. Distinct clinical and pathological characteristics of frontotemporal dementia associated with C9ORF72 mutations. Brain. 2012;135(Pt 3):693–708. doi: 10.1093/brain/awr355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart H, Rutherford NJ, Briemberg H, Krieger C, Cashman N, Fabros M, Baker M, Fok A, DeJesus-Hernandez M, Eisen A, Rademakers R, Mackenzie IR. Clinical and pathological features of amyotrophic lateral sclerosis caused by mutation in the C9ORF72 gene on chromosome 9p. Acta Neuropathol. 2012;123:409–417. doi: 10.1007/s00401-011-0937-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan PM, Zhou X, Robins AM, Paushter DH, Kim D, Smolka MB, Hu F. The ALS/FTLD associated protein C9orf72 associates with SMCR8 and WDR41 to regulate the autophagy-lysosome pathway. Acta Neuropathol. Commun. 2016;4:51. doi: 10.1186/s40478-016-0324-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umoh ME, Fournier C, Li Y, Polak M, Shaw L, Landers JE, Hu W, Gearing M, Glass JD. Comparative analysis of C9orf72 and sporadic disease in an ALS clinic population. Neurology. 2016;87:1024–1030. doi: 10.1212/WNL.0000000000003067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrabec K, Koritnik B, Leonardis L, Dolenc-Groselj L, Zidar J, Smith B, Vance C, Shaw C, Rogelj B, Glavac D, Ravnik-Glavac M. Genetic analysis of amyotrophic lateral sclerosis in the Slovenian population. Neurobiol. Aging. 2015;36:1601.e17–1601.e20. doi: 10.1016/j.neurobiolaging.2014.11.011. [DOI] [PubMed] [Google Scholar]
- Webster CP, Smith EF, Bauer CS, Moller A, Hautbergue GM, Ferraiuolo L, Myszczynska MA, Higginbottom A, Walsh MJ, Whitworth AJ, Kaspar BK, Meyer K, Shaw PJ, Grierson AJ, De Vos KJ. The C9orf72 protein interacts with Rab1a and the ULK1 complex to regulate initiation of autophagy. EMBO J. 2016;35:1656–1676. doi: 10.15252/embj.201694401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CH, Fallini C, Ticozzi N, Keagle PJ, Sapp PC, Piotrowska K, Lowe P, Koppers M, McKenna-Yasek D, Baron DM, Kost JE, Gonzalez-Perez P, Fox AD, Adams J, Taroni F, Tiloca C, Leclerc AL, Chafe SC, Mangroo D, Moore MJ, Zitzewitz JA, Xu ZS, van den Berg LH, Glass JD, Siciliano G, Cirulli ET, Goldstein DB, Salachas F, Meininger V, Rossoll W, Ratti A, Gellera C, Bosco DA, Bassell GJ, Silani V, Drory VE, Brown RH, Jr, Landers JE. Mutations in the profilin 1 gene cause familial amyotrophic lateral sclerosis. Nature. 2012;488:499–503. doi: 10.1038/nature11280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Liang C, Swaminathan K, Herrlinger S, Lai F, Shiekhattar R, Chen JF. A C9ORF72/SMCR8-containing complex regulates ULK1 and plays a dual role in autophagy. Sci. Adv. 2016;2:e1601167. doi: 10.1126/sciadv.1601167. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.