Abstract
CD4+ cytotoxic T lymphocytes (CD4-CTLs) have been reported to play a protective role in several viral infections. However, little is known in humans about the biology of CD4-CTL generation, their functional properties, and heterogeneity, especially in relation to other well-described CD4+ memory T cell subsets. We performed single-cell RNA sequencing in more than 9000 cells to unravel CD4-CTL heterogeneity, transcriptional profile, and clonality in humans. Single-cell differential gene expression analysis revealed a spectrum of known transcripts, including several linked to cytotoxic and costimulatory function that are expressed at higher levels in the TEMRA (effector memory T cells expressing CD45RA) subset, which is highly enriched for CD4-CTLs, compared with CD4+ T cells in the central memory (TCM) and effector memory (TEM) subsets. Simultaneous T cell antigen receptor (TCR) analysis in single cells and bulk subsets revealed that CD4-TEMRA cells show marked clonal expansion compared with TCM and TEM cells and that most of CD4-TEMRA were dengue virus (DENV)–specific in donors with previous DENV infection. The profile of CD4-TEMRA was highly heterogeneous across donors, with four distinct clusters identified by the single-cell analysis. We identified distinct clusters of CD4-CTL effector and precursor cells in the TEMRA subset; the precursor cells shared TCR clonotypes with CD4-CTL effectors and were distinguished by high expression of the interleukin-7 receptor. Our identification of a CD4-CTL precursor population may allow further investigation of how CD4-CTLs arise in humans and, thus, could provide insights into the mechanisms that may be used to generate durable and effective CD4-CTL immunity.
INTRODUCTION
After exposure to pathogens, naive CD4+ T helper (TH) lymphocytes differentiate into memory and effector TH cell subsets: tissue-resident memory cells, which are mainly retained in the tissues, and central memory (TCM) and effector memory (TEM) cells, which recirculate between the blood and lymphoid organs or tissues, respectively (1, 2). In addition, TH cell subsets have been classified on the basis of their cytokine profile and functional properties into TH1, TH2, TH17, TH* (TH1/TH17), regulatory T cell, and follicular helper (TFH) T cell subsets (3, 4). Although T lymphocytes with cytotoxic function (CTLs) are predominantly restricted to conventional major histocompatibility complex (MHC) class I–restricted CD8+ T lymphocytes, the existence of MHC class II–restricted TH cells with cytotoxic potential (CD4-CTLs) in humans, nonhuman primates, and mice has been reported for many decades (5, 6). However, compared with the other TH subsets, the molecular and epigenetic mechanisms that drive the differentiation, maintenance, and function of human CD4-CTLs are poorly understood, mainly because of the lack of precise definition of the nature of this subset in humans.
CD4-CTLs were initially reported in humans with chronic viral infections such as human cytomegalovirus (hCMV), HIV, dengue virus (DENV), and hepatitis C virus (5, 7–15). CD4-CTLs have also been detected in mouse lungs as early as 1 week after acute influenza viral infection (16, 17). The magnitude of the CD4-CTL response has been associated with better clinical outcomes in both acute and chronic viral infections, implying that CD4-CTLs are an important component of the protective immune responses to viral infections (6). Furthermore, expansion of CD4-CTLs has been observed in donors carrying human leukocyte antigen (HLA) alleles associated with protection from severe dengue disease (7). Thus, eliciting a strong CD4-CTL response is considered an important goal of vaccination against certain viral infections (16, 18–20). The highly effective yellow fever vaccine has been shown to elicit a strong CD4-CTL response, which is required for protection against fatal infection in mouse models (18). CD4-CTLs have also been linked to protective antitumor immune responses, especially in virally induced tumors (18).
Given the importance of CD4-CTLs in acquired cellular immunity, we present here the single-cell transcriptomic and T cell antigen receptor (TCR) analysis of circulating human CD4-CTLs. CD4-CTLs were highly enriched in the effector memory T cells expressing CD45RA (CD4-TEMRA) subset and displayed notable intra- and interdonor heterogeneity. We show that the magnitude of CD4-TEMRA response is linked to the degree of clonal expansion and cytotoxicity profile of CD4-CTLs. Besides a comprehensive definition of the transcriptional program of conventional CD4-CTL effectors, we identified precursor cells sharing TCR clonotypes with CD4-CTL effectors that were distinguished by higher expression of the interleukin-7 receptor (IL-7R).
RESULTS
Cytotoxicity-related genes are enriched in the CD4-TEMRA subset
Human CD4-CTLs are enriched in the CD4-TEMRA subset (defined as CD3+CD4+CD45RA+CCR7− cells) (Fig. 1A), most notably in donors with previous DENV and CMV infection (5, 7–10). To capture the extent of cellular heterogeneity among human CD4-CTLs, we performed single-cell RNA sequencing (RNA-seq) in more than 9000 cells isolated ex vivo from the TEMRA subset and, as a control, in TH subsets that contain relatively fewer CD4-CTLs, such as TEM (CD3+CD4+CD45RA−CCR7− cells) and TCM (CD3+CD4+CD45RA−CCR7+ cells) subsets (Fig. 1A and table S1) (21). Using complementary methods of single-cell differential gene expression analysis [SCDE and MAST analysis, see Supplementary Materials and Methods and in (22, 23)], we compared the full-length transcriptome of CD4+ T lymphocytes present in TEMRA, TEM, and TCM subsets from three donors with previous DENV infection, carrying the HLA allele (DRB1*0401) previously reported to be protective against severe dengue disease (Fig. 1B and table S1) (7, 24).
Fig. 1. Cytotoxicity-related transcripts are enriched in the CD4-TEMRA sub-set.
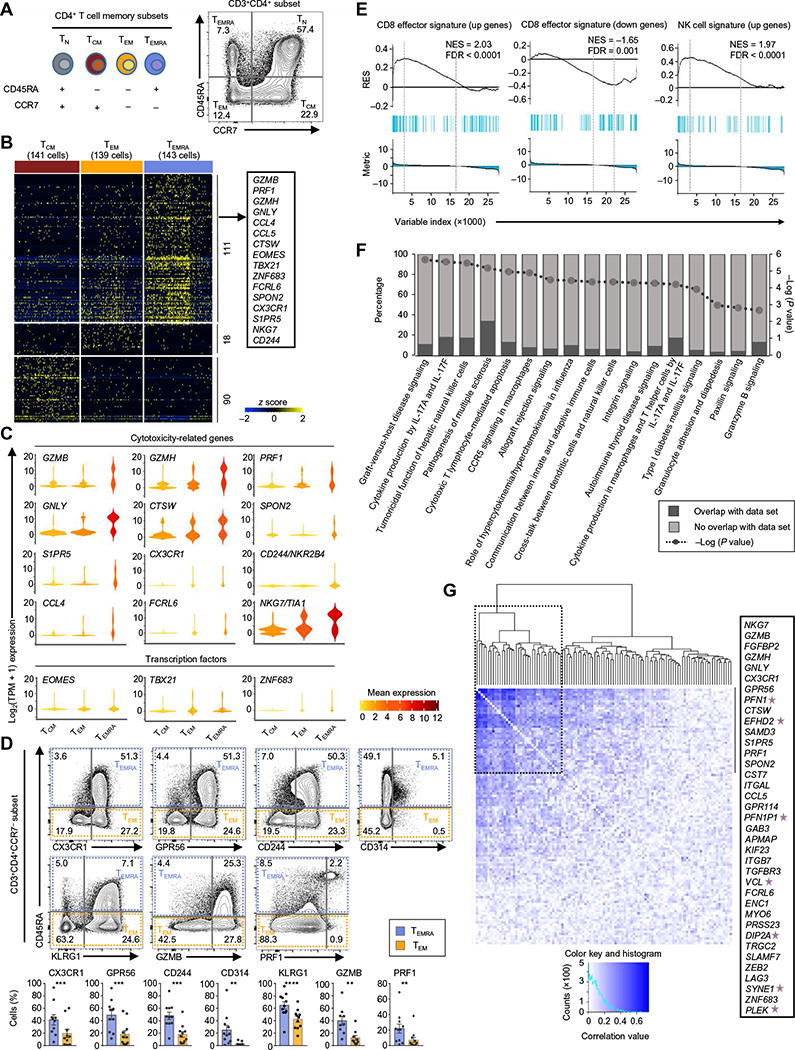
(A) Schematic representation of the CD4+ T cell subsets and their surface markers used for the study. (B) Single-cell RNA-seq analysis showing row-wise z scores of normalized TPM (transcripts per million) counts of cells in each subset (indicated at the top) for each differentially expressed transcript (rows) obtained by pairwise comparison of TEMRA versus TEM, TEMRA versus TCM, and TEM versus TCM [single-cell differential expression (SCDE) and model-based analysis of single-cell transcriptomics (MAST) analysis, Benjamini-Hochberg adjusted P < 0.05 and ≥2-fold change). (C) Violin plots show the single-cell expression pattern of the indicated TEMRA-enriched transcripts (cytotoxicity-related and TFs) in the indicated subsets. The shapes represent the distribution of cells based on their log2(TPM + 1) expression values (y axis). The color scale represents the mean expression. (D) The contour plots show the surface expression of CD45RA, CX3CR1, GPR56, CD244, CD314, and KLRG1 and intracellular expression of GZMB and PRF1 in singlet-gated CD3+CD4+CCR7− cells. The numbers denote the percentage of cells in each quadrant. Bar graphs beneath show the average percentages. Error bars are mean ± SEM from 9 (GZMB), 10 (CX3CR1), and 11 (other indicated proteins) donors. **P < 0.001, ***P < 0.0005, and ****P < 0.0001 from Student’s paired two-tailed t test. (E) GSEA enrichment plots for the indicated gene sets in the transcriptome of TEMRA versus TEM and TCM (see Supplementary Materials and Methods). The top portion of the plot shows the running enrichment score (RES) for the gene set as the analysis walks down the ranked list of genes and reflects the degree to which the gene set is overrepresented at the top or bottom of the ranked list of genes. The middle portion of the plot shows where the members of the gene set (indicated as blue lines) appear in the ranked list of genes. The bottom portion of the plot shows the value of the ranking metric. NES, normalized enrichment score; FDR, false discovery rate. (F) IPA of canonical pathways enriched in the TEMRA subset; P values calculated by Fisher exact test (see Supplementary Materials and Methods). (G) Spearman correlation plot showing the coexpression of the 111 TEMRA-enriched transcripts. Dashed black box shows a cluster of transcripts showing high correlation; the list of these transcripts is shown in the text box on the right, and stars highlight the genes not previously reported to have cytotoxic function.
We found 111 “TEMRA-enriched” transcripts with significantly higher mean expression in single cells from the TEMRA subset compared with the TEM and TCM subsets (see Supplementary Materials and Methods, Fig. 1B, and table S2). These included several transcripts linked to the cytotoxic function of CD8+ T lymphocytes and natural killer (NK) cells such as GZMB, PRF1, GZMH, GNLY, CCL4, CTSW, FCRL6, SPON2, CX3CR1, S1PR5, NKG7, and CD244 (Fig. 1, B and C) (6, 25); we confirmed the expression of some of these transcripts (CX3CR1, GPR56, CD244, CD314, KLRG1, GZMB, and PRF1) at the protein level (Fig. 1D). Gene set enrichment analysis (GSEA) (26) and ingenuity pathway analysis (IPA) of TEMRA-enriched transcripts also revealed significant overrepresentation of cytotoxicity signature genes in the TEMRA subset (Fig. 1, E and F). Transcripts encoding transcription factors (TFs) related to CTL function such as ZNF683 (Hobit), and Eomes and T-bet (encoded by TBX21) were also expressed at higher levels in single cells from the TEMRA subset (Fig. 1, B and C). ZNF683 has recently been shown to identify human CD4-CTLs (24), and Eomes and T-bet appear to be important in the development of CD4-CTLs (27, 28). These results confirm that human CD4-CTLs are highly enriched in the TEMRA subset. Coexpression analysis of TEMRA-enriched transcripts also revealed a number of genes (PFN1, PFN1P1, EFHD2, VCL, DIP2A, SYNE1, and PLEK) (29–32) whose expression was highly correlated with cytotoxicity signature genes, suggesting that the products of these genes may also play important roles in the development or function of CD4-CTLs (Fig. 1G).
CD4-CTLs show marked clonal expansion
Given that memory CD4-CTLs are mainly generated after exposure to certain viruses such as DENV or CMV (5, 7–10), we expected to see a more restricted TCR repertoire, that is, greater clonal expansion in the CD4-TEMRA subset compared with CD4+ T lymphocytes in the TEM or TCM subsets, which harbor a more common memory pool. We performed parallel analysis of the TCR repertoire in single cells by decoding the full-length transcriptome profiles generated by the Smart-seq2 assay (33, 34). Using the TraCeR software (35), we reconstructed the TCRβ chains in 41 to 89% of single cells, the TCRα chain in 31 to 81%, and both chains in 22 to 70% of cells across all memory subsets (table S3). As expected, a greater clonal expansion was observed in the TEMRA subset compared with other subsets, as shown by highly interconnected clonotype network graphs for single cells from donor #6 (Fig. 2A). Furthermore, the analysis of single cells that shared TCRα or TCRβ chain clonotypes showed that more than 50% of cells in the TEMRA subset were clonally expanded (Fig. 2B, left and middle). To address the rare possibility of independent cells sharing one of the TCR chains, we also analyzed single cells that shared both TCRα and TCRβ chain clonotypes and found that ~46% of cells in the TEMRA subset (CD4-CTL–enriched) were clonally expanded compared with only ~5% and none of the cells in TEM and TCM subsets, respectively (Fig. 2B, right, and table S4). Together, these results suggested a highly restricted TCR repertoire in the TEMRA subset (Fig. 2, A and B). The clonally expanded cells had a higher mean expression of cytotoxic signature genes (TEMRA-enriched gene set) (Fig. 2, C and D), suggestive of greater effector potential (6, 25, 36).
Fig. 2. CD4-TEMRA cells show marked clonal expansion.
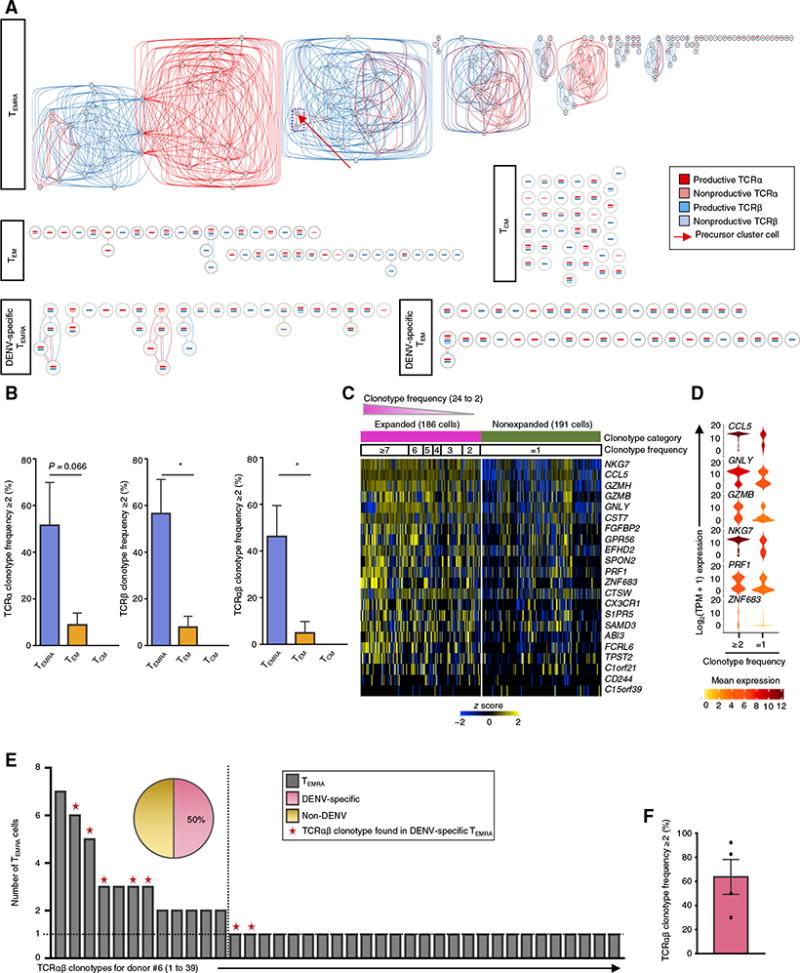
(A) Clonotype network graphs of cells from the TEMRA, TEM, and TCM subset and DENV-specific TEMRA and DENV-specific TEM cells from donor #6. Each circle represents a single cell; the reconstructed TCRα and TCRβ sequences for each cell are depicted as red- and blue-colored bars, respectively, inside each circle. Dark- and light-colored bars represent productive and nonproductive TCRs, respectively. Red connecting lines indicate shared TCRα sequences; blue lines indicate shared TCRβ sequences. Red arrow indicates a precursor cell. Purple dashed line indicates a precursor cell and an effector cell sharing the same unique TCRα and TCRβ chain clonotype. (B) Bar graph shows the percentage of cells with TCRα (left), TCRβ (middle), and TCRαβ (right) chain clonotype frequency ≥2 (clonally expanded cells) in TEMRA, TEM, and TCM subsets. Error bars are mean ± SEM from three donors with the same HLA allele. *P < 0.05 from Student’s paired two-tailed t test. (C) Single-cell RNA-seq analysis shows the row-wise z score of normalized TPM counts for the indicated TEMRA-enriched transcripts (SCDE and MAST analysis comparing TEMRA cells with clonotype frequency ≥2 or =1, Benjamini-Hochberg adjusted P < 0.05 and ≥2-fold change) in clonally expanded cells (unique TCRα and TCRβ chain clonotype frequency ≥2; magenta) and nonexpanded cells (unique TCRα and TCRβ chain clonotype frequency =1; green). The cells are arranged on the basis of the clonotype frequency, and the numbers on the top in black boxes represent clonotype frequency. (D) Violin plots show the single-cell expression pattern of the indicated cytotoxicity-related transcripts in clonally expanded versus nonexpanded TEMRA cells. The shapes represent the distribution of cells based on their log2(TPM + 1) expression values (y axis). The color scale represents the mean expression. (E) Bar graph shows the number of TEMRA cells with a unique TCRα and TCRβ chain clonotype for donor #6. Stars in red color indicate the clonotypes shared by DENV-specific TEMRA. Pie chart shows the percentage of expanded TEMRA cells that share the unique TCRα and TCRβ chain clonotypes of DENV-specific TEMRA cells (pink). (F) Bar graph shows the percentage of the expanded TEMRA cells that share TCRαβ clonotypes of DENV-specific TEMRA cells. Each dot represents a donor. Error bars are mean ± SEM from four donors.
To probe the pathogen specificity of clonally expanded CD4-TEMRA cells (CD4-CTL–enriched), we first determined the TCR clonotype of single cells in the CD4-TEMRA subset that responded ex vivo to a pool of DENV-specific peptides (7, 37) from four donors with previous DENV infection (Fig. 2, E and F, fig. S1, and table S4). Next, we asked what fraction of these DENV-specific TCR clonotypes was present in the general pool of cells present in the TEMRA subset from the same donor. On an average, 64% (n = 4) of the clonally expanded cells in the general TEMRA population carried the DENV-specific TCR clonotypes, and in all donors, one or both of the top two clonally expanded clonotypes in the TEMRA population were always DENV-specific (Fig. 2, E and F, fig. S1, and table S4), which suggested that most of the clonally expanded cells in the TEMRA population in these individuals were specific for DENV.
CD4-CTLs (CD4-TEMRA cells) are heterogeneous across donors
We next asked whether the clonality and transcriptome of CD4-CTLs differed between donors with and without previous DENV infection or among donors across different geographical locations [Sri Lanka (Asia) and the Americas (Nicaragua and San Diego, California)]. We observed a wide range in the proportion (0 to 88%; median, 44%) of clonally expanded cells across the 12 donors with no major differences in their proportion when classifying donors based on previous DENV infection status or geographical location [Sri Lanka (Asia) versus the Americas; table S1]. When we compared the percentage of clonally expanded cells between donors with higher versus lower proportion of CD4+ T lymphocytes in the TEMRA subset (classified as TEMRAhigh or TEMRAlow donors), we observed greater clonal expansion in TEMRAhigh donors (Fig. 3A). Consistent with this finding, there was a positive correlation between the proportion of CD4+ T lymphocytes in the TEMRA subset and the percentage of clonally expanded cells, suggesting that donors with a larger TEMRA pool have a greater degree of clonal expansion (Fig. 3B).
Fig. 3. CD4-TEMRA cells are heterogeneous across donors.
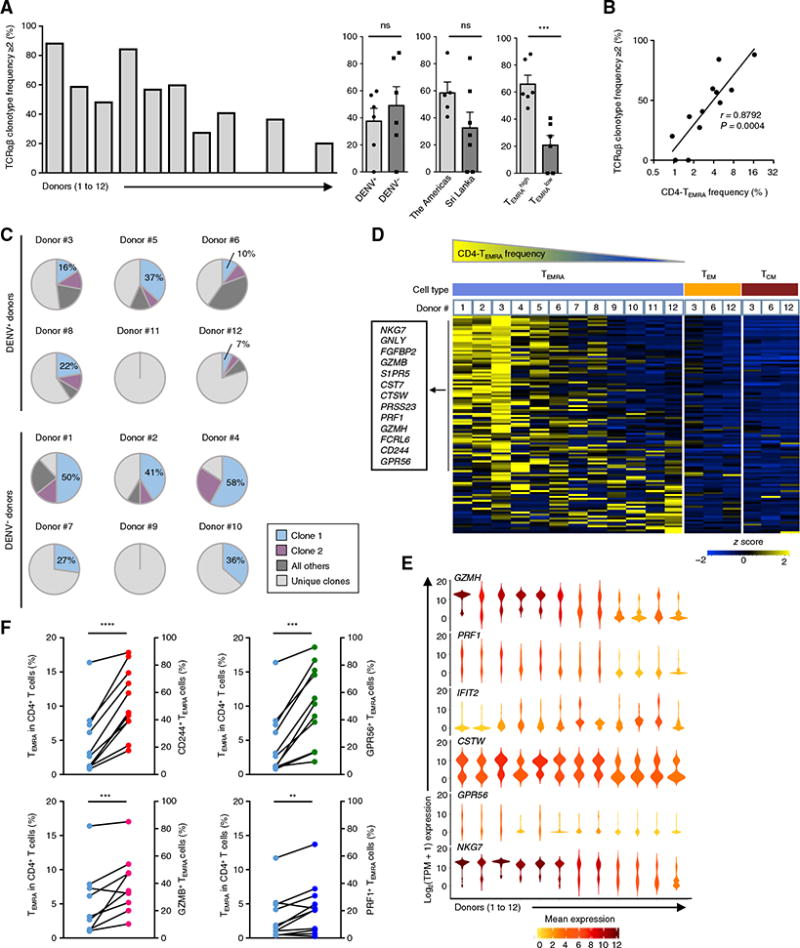
(A) Bar graph shows the percentage of clonally expanded TEMRA cells (cells having a unique TCRα and TCRβ chain clonotype with frequency ≥2) across 12 donors (left) and between donors classified on the basis of their status of previous DENV infection (DENV+ and DENV−), geographical location [Sri Lanka, Nicaragua, and San Diego (the Americas)], and CD4-TEMRA proportion (TEMRAhigh, donors #1 to #6; TEMRAlow, donors #7 to #12). Each dot represents data from a single donor. Error bars are mean ± SEM. ***P = 0.001 from Student’s unpaired two-tailed t test; ns, not significant. (B) Correlation between the proportions of the CD4-TEMRA subset in CD4+ T cells and clonally expanded TEMRA cells (cells having a unique TCRα and TCRβ chain clonotype with frequency ≥2). r, Spearman correlation coefficient; P, by Student’s two-tailed paired t test. (C) Pie charts show the percentage of TEMRA cells with the first (clone 1) and second (clone 2) most frequent unique TCRα and TCTβ chain clonotype, as well as other clonally expanded cells (all others) and the rest of the TCRα and TCRβ chain clonotypes present in only one cell (unique clones). (D) Single-cell RNA-seq analysis of TEMRA (blue), TEM (yellow), and TCM (brown) cells shows the row-wise normalized mean TPM counts of 111 TEMRA-enriched transcripts [from Fig. 1 (B and C)] (rows) for each donor (column). The panel above the heat map identifies the cell type and donor number (#1 to #12, ordered on the basis of the frequency of the CD4-TEMRA subset). (E) Violin plots show the single-cell expression pattern of the indicated transcripts in the TEMRA subset across the 12 donors. The shapes represent the distribution of cells based on their log2(TPM + 1) expression values (y axis). The color scale represents the mean expression. (F) Matched data from the same donors showing the proportion of TEMRA cells in the CD4+ T cell subset (blue) and the proportion of CD244+ (red), GPR56+ (green), GZMB+ (pink), and PRF1+ (dark blue) TEMRA cells from 9 to 11 donors. Each dot represents a donor. **P < 0.001, ***P < 0.0005, and ****P < 0.0001 from Student’s paired two-tailed t test.
In most donors, we observed sharing of a single unique TCRα and TCRβ chain clonotype in a large fraction of CD4-TEMRA cells, as exampled in donor #4 [58% (11 of 19)], donor #1 [50% (21 of 42)], donor #2 [41% (24 of 58)], donor #5 [37% (11 of 30)], donor #8 [22% (6 of 27)], donor #3 [16% (4 of 25)], and donor #6 [10% (7 of 67)] of cells (Fig. 3C). Considering some of these donors were previously infected with DENV (DENV+ donors) raised the hypothesis that selection and expansion of such TCR clonotypes may be linked to the DENV infection. Even in donors without previous DENV infection (DENV− donors), we observed very high levels of clonal expansion (Fig. 3C), suggesting that other infections—perhaps CMV, which is common in the general population—may also contribute to the preferential expansion of some CD4-CTL clones.
Besides the heterogeneity in clonality, we also observed marked variability in the expression of TEMRA-enriched transcripts in CD4-TEMRA cells across the study donors (Fig. 3, D and E). For several cytotoxicity-related transcripts, the expression pattern was highly variable across the 12 donors (Fig. 3, D and E). CD4-TEMRA cells from donors with a larger preexisting TEMRA pool and greater clonal expansion displayed more cytotoxic features (Fig. 3D). In other words, cytotoxicity-related transcripts were expressed in a greater fraction of single cells or at higher mean levels; other TEMRA-enriched transcripts such as ZNF683, PRSS23, FCRL6, and IFIT2 also showed a similar pattern (Fig. 3, D and E). We confirmed at the protein level that greater proportion of CD4-TEMRA cells expressed cytotoxicity-related molecules CD244, GPR56, GZMB, and PRF1 in donors with larger preexisting TEMRA pool (Fig. 3F). Therefore, our combined transcriptomic, protein, and TCR analysis suggests that the CD4-TEMRA subset exhibits quantitative and qualitative differences across different donors irrespective of their DENV infection status or geographical location and, in many instances, related to the CD4-CTL function.
CD4-CTL effector cells revealed by single-cell analysis
We next asked whether the heterogeneity observed in the single-cell transcriptomes of CD4-TEMRA was due to the presence of multiple distinct subsets. Four clusters were revealed by unbiased clustering of CD4-TEMRA cells from the 12 study donors (917 cells) using the Seurat software (Fig. 4A) (38); other methods of clustering also revealed a similar pattern, and no major changes were introduced by technical and batch effects (see figs. S2 and S3, table S1, and Supplementary Materials and Methods) (39). The proportion of cells in each cluster varied greatly among donors, with marked differences observed between donors from Sri Lanka and the Americas (Fig. 4B, left), suggesting that the nature and type of infections may shape the molecular profiles of CD4-CTLs. Clonal expansion was observed more frequently in cells from clusters 1 and 2 (Fig. 4B, right, and table S4).
Fig. 4. Four distinct subsets of CD4-CTLs revealed by single-cell RNA-seq analysis.
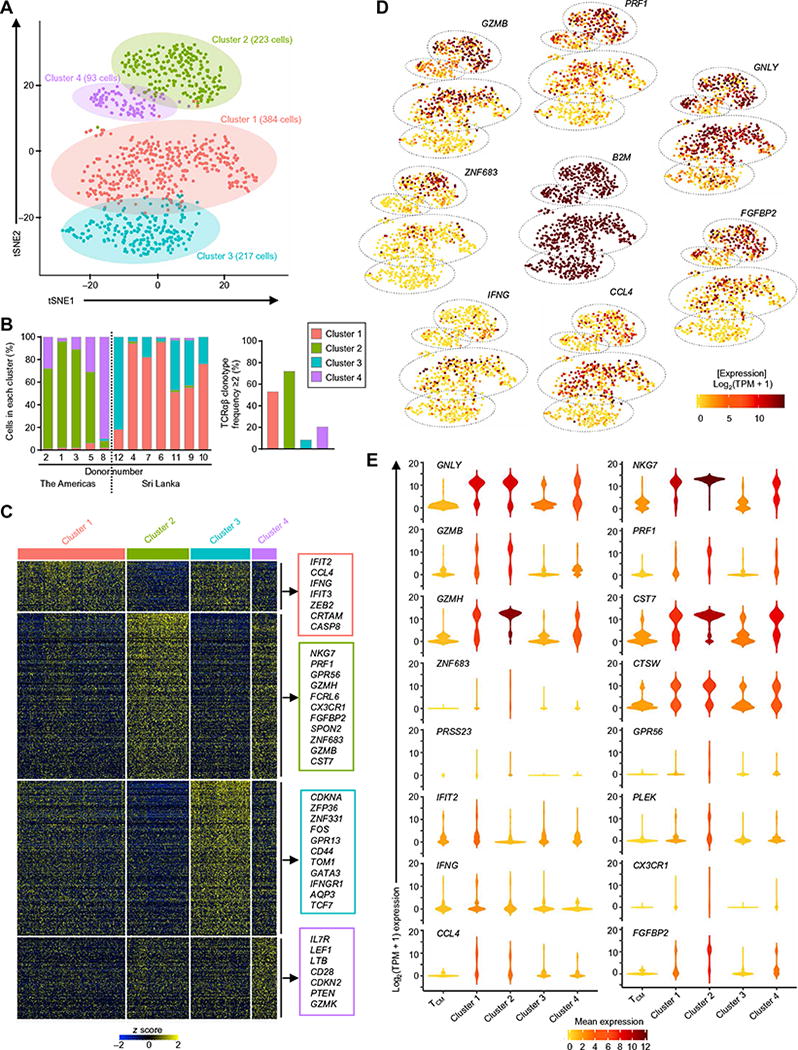
(A) t-Distributed stochastic neighbor embedding (tSNE) two-dimensional (2D) plot of single-cell RNA-seq data (n = 917 cells, based on 1000 most variable transcripts) shows four distinct clusters of cells in the TEMRA subset. (B) Bar graph shows the distribution of cells across four clusters in each indicated donor (donor numbers match those in table S1) classified on the basis of geographical location (the Americas and Sri Lanka, left) and the percentage of clonally expanded cells (cells having a unique TCRα and TCRβ chain clonotype with frequency ≥2) in each cluster (right). (C) Single-cell RNA-seq analysis shows row-wise z scores of mean-normalized TPM counts of cells in each cluster for each differentially expressed transcripts (rows) obtained by pairwise comparison of each cluster versus the rest of the clusters (Seurat analysis, Benjamini-Hochberg adjusted P < 0.05) for the four clusters. (D) tSNE 2D plots of single-cell RNA-seq data show the expression of the indicated differentially expressed transcripts (SCDE and MAST analysis, Benjamini-Hochberg adjusted P < 0.05 and ≥2-fold change). B2M is shown as a control housekeeping gene. (E) Violin plots show the single-cell expression pattern of the indicated differentially expressed transcripts across four TEMRA clusters along with TCM. The shapes represent the distribution of cells based on their log2(TPM + 1) expression values (y axis). The color scale represents the mean expression.
Single-cell differential gene expression analysis among the four clusters revealed notable differences in their molecular profiles (Fig. 4C, fig. S4, A to D, and table S5). IPA and differential expression analysis of the transcripts enriched in clusters 1 and 2 compared with clusters 3 and 4 showed significant overrepresentation of genes encoding products related to cytotoxicity, such as GZMB, GZMH, PRF1, GNLY, NKG7, ZNF683, and FGFBP2 (figs. S4, C and D, and S5 and table S5). Among these two clusters (1, 2), cluster 2 shows further enrichment for the cytotoxicity-related genes (figs. S4A and S5A), which suggested that cells in cluster 2 had a higher cytotoxic potential. Pairwise comparison of the clusters (fig. S4D and table S5) showed differences in the enrichment of cytotoxicity-related genes among clusters 1 and 2; although several of them were expressed by both the clusters (GZMB, GZMH, GNLY, NKG7, FASLG, and CASP10), many were expressed in a cluster-specific manner (Fig. 4, D and E, fig. S4D, and table S5). On the basis of the differential expression pattern of cytotoxicity-related transcripts in cluster 1 (FADD, IFNG, TNF, IFIT2, LMNA, CD69, FOS, JUN, DUSP1, and DUSP2) and cluster 2 (FGFBP2, SPON2, CX3CR1, GPR56, and PRF1), we hypothesize that their cytotoxic function is preferentially mediated by the FAS/death receptor and perforin pathways, respectively, which requires further functional verification (Fig. 4, D and E, fig. S4, A and D, and table S5).
Cells in clusters 1 and 2 expressed higher levels of KLRG1 (40–42) and CX3CR1 (7, 43) transcripts, which are linked to effector status of memory cells, and lower levels of CD27 and CD28 transcripts, which encode costimulation molecules (Figs. 4E and 5, A and B) (44, 45). These results indicate that cells in clusters 1 and 2 have a terminal effector state and hence are CD4-CTL effectors with high cytotoxic potential. Genes linked to cell survival and CD4-CTL function such as PRSS23, SPON2, TCF7 (encodes for TCF1), CRTAM, and ZNF683 (encodes for Hobit) (24, 46–51) were also highly expressed in single cells from clusters 1 and 2 (Fig. 4E). The role of ZNF683 (Hobit) (24, 46, 49) and other transcripts (PRSS23, SPON2, and TCF7) (47, 48) in the survival and long-term persistence of these high cytotoxic terminal effector cells deserves further investigation (45).
Fig. 5. CD4-CTL precursor cells in the TEMRA subset.
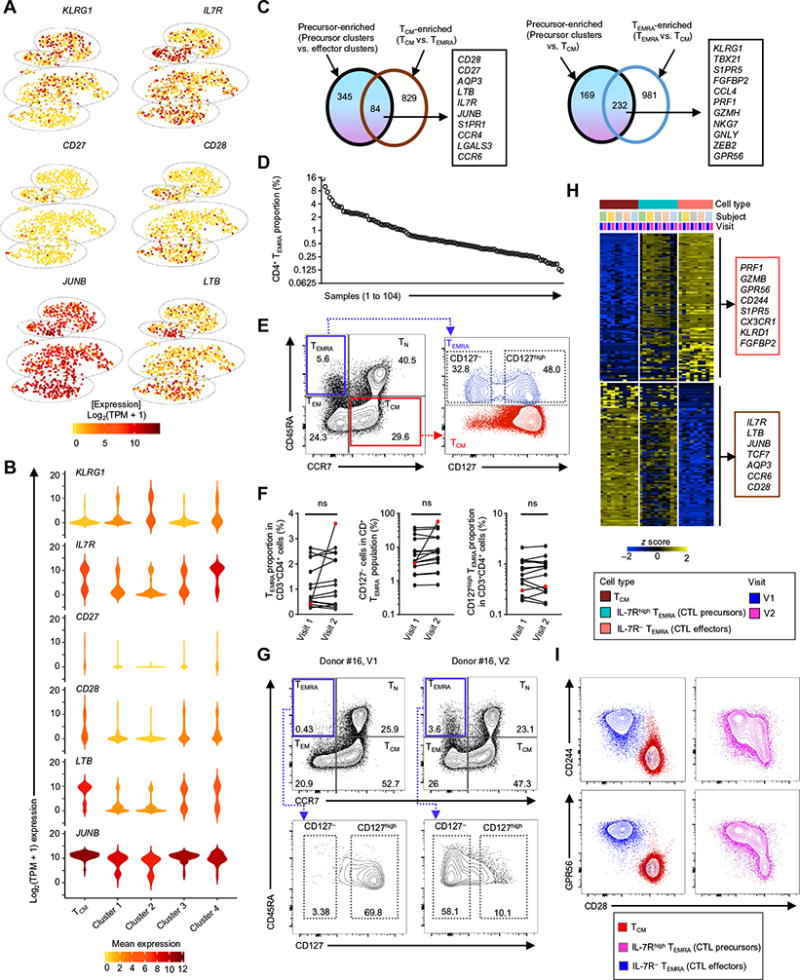
(A) tSNE 2D plots of single-cell RNA-seq data show the expression of the indicated differentially expressed transcripts between the four clusters (SCDE and MAST analysis, adjusted P < 0.05 and ≥2-fold change). (B) Violin plots show the single-cell expression pattern of the indicated differentially expressed transcripts across four TEMRA clusters along with TCM. Color scale shows the log2(TPM + 1) expression values (A) and mean expression values (B). The shapes represent the distribution of cells based on their log2(TPM + 1) expression values (y axis) (B). (C) Venn diagram comparing the transcripts enriched in precursor clusters (clusters 3 and 4) versus effector clusters (clusters 1 and 2) and transcripts enriched in TCM versus TEMRA (left); transcripts enriched in precursor clusters (clusters 3 and 4) versus TCM and transcripts enriched in TEMRA versus TCM (right) (gene list obtained from SCDE and MAST analysis, Benjamini-Hochberg adjusted P < 0.05 and ≥2-fold change). Examples of overlapping genes are shown to the right. (D) Percentage of the CD4-TEMRA subset in the CD4+T cell population across 104 samples from 89 individuals. (E) Contour plots show the surface expression of CD45RA and CCR7 (left) or CD127 (IL-7R) (right) in live and singlet-gated CD3+CD4+ T cells obtained from PBMCs (left). Contour plots show coexpression of CD45RA and CD127 in TEMRA and TCM subsets (right). The gating strategy shows CD127− and CD127high proportion. (F) Proportion of total CD4-TEMRA (left), CD127− CD4-TEMRA (middle), and CD127high CD4-TEMRA cells (right) in CD4+ T cells from 15 donors with two longitudinal samples (V1 and V2). Donor #16, marked in red color, shows marked increase in the proportion of CD4-TEMRA and CD127− CD4-TEMRA (effectors) cells from V1 to V2. (G) Contour plots show the surface expression of CD45RA and CCR7 (top) or CD127 (bottom) in live and singlet-gated CD3+CD4+ cells obtained from PBMCs of donor #16 at two time points (168 days apart) [V1 (left) and V2 (right)]. Bottom panels show the expression of CD127 in the CD4-TEMRA subset for the same donor. ns, not significant; from Student’s two-tailed paired t test. Numbers inside the contour plots show the proportion of the indicated cell type. (H) Bulk RNA-seq analysis of the IL-7Rhigh TEMRA subset (CD4-CTL precursors), IL-7R− TEMRA (CD4-CTL effectors), and TCM cells shows the row-wise normalized TPM counts of top 200 (100 up-regulated and down-regulated, based on fold change) differentially expressed transcripts obtained by pairwise comparison of IL-7R− TEMRA (CD4-CTL effectors) versus TCM from the five donors (DESeq2 analysis, Benjamini-Hochberg adjusted P < 0.05 and ≥2-fold change). The panel above the heat map identifies the cell type, donor, and visit. (I) Contour plots show the coexpression of CD244 or GPR56 with CD28 in TCM (red), IL-7Rhigh TEMRA (CD4-CTL precursors) (magenta), and IL-7R− TEMRA (CD4-CTL effectors) (blue) in singlet-gated CD3+CD4+ T lymphocytes.
The TEMRA subset includes CD4-CTL precursor cells
We next examined the transcriptional profiles of cells in clusters 3 and 4 to evaluate their clonal origin and functional relationship to the other cell populations. Compared with cells in clusters 1 and 2 (referred as CD4-CTL effectors), those in clusters 3 and 4 expressed lower levels of KLRG1 transcripts and higher levels of transcripts encoding IL-7R (IL7R or CD127), which is known to play an important role in mediating the long-term homeostatic survival of naive and memory T lymphocytes (Fig. 5A) (52–55). The mean expression of IL7R transcripts in cluster 4 cells was even higher than that in TCM cells (Fig. 5, A and B). The opposing expression pattern of KLRG1 and IL7R transcripts in clusters 3 and 4 is similar to that observed in memory precursor effector cells (IL-7Rhigh and KLRG1−) described in murine models of resolving acute infections (52, 56). Transcripts encoding CD27 and CD28 costimulation molecules, lymphotoxin β (LTB), and JUNB TF were also expressed at higher levels in clusters 3 and 4, similar to the pattern observed in TCM cells (Fig. 5, B and C). However, cells in both clusters 3 and 4 were still markedly different from TCM cells in that they expressed several transcripts that are specifically enriched in TEMRA cells (KLRG1, TBX21, S1PR5, FGFBP2, CCL4, PRF1, GZMH, GNLY, NKG7, ZEB2, and GPR56), albeit at lower levels compared with CD4-CTL effectors (cells in clusters 1 and 2) (Figs. 4E and 5C). On the basis of these results, we hypothesize that cells in clusters 3 and 4 likely represent memory precursor cells for the cells present in cluster 1 and 2 that have a terminal CD4-CTL effector phenotype.
Given that surface expression of IL-7R (CD127) can be readily determined by flow cytometry, we looked for the presence of potential CD4-CTL precursors within the CD4-TEMRA subset from an independent cohort of healthy donors. For these studies, we capitalized on the La Jolla Institute cohort of 89 healthy donors, of which 15 donors had longitudinal samples (two time points) (104 samples in total). As expected, there was a large variation (0.12 to 15.2%) in the proportion of the CD4-TEMRA subset across the study donors (Fig. 5D). On the basis of the surface expression pattern of IL-7R, cells in the CD4-TEMRA subset were classified into IL-7Rhigh and IL-7R− (Fig. 5E). The expression level of IL-7R on IL-7Rhigh TEMRA cells was similar to that observed in TCM and TN (naive T cell) CD4+ T lymphocytes (Fig. 5E and fig. S5B). These cells (IL-7Rhigh TEMRA cells) may represent the CD4-CTL precursors defined by our single-cell transcriptome analysis. In longitudinal samples obtained at 3- to 6-month intervals, the proportion of cells in the CD4-TEMRA subset was quite stable (Fig. 5F); however, in one donor, we noted a marked (>8-fold) expansion of the CD4-TEMRA subset; this expansion was mainly confined to the IL-7R− TEMRA cells (CD4-CTL effectors), and not the IL-7Rhigh TEMRA (CD4-CTL precursors), perhaps reflecting an expansion of effectors in response to an infection that occurred during the interval period (Fig. 5, F and G).
To further confirm that the IL-7Rhigh TEMRA subset (CD4-CTL precursors) shared the molecular profile of both TCM (long-term memory cells) and cytotoxic cells, we isolated IL-7Rhigh TEMRA and IL-7R− TEMRA (CD4-CTL effectors) along with TCM and TEM subsets and performed RNA-seq in two longitudinal samples from five donors (Fig. 5H). Consistent with our single-cell cluster analysis (Fig. 5C), the IL-7Rhigh TEMRA subset shared molecular features of both TCM (memory precursor cells) and cytotoxic cells (IL-7R− TEMRA, CD4-CTL effectors) that were stable over two longitudinal visits (Fig. 5H and table S6). The IL-7Rhigh TEMRA subset (CD4-CTL precursors) expressed both cytotoxicity-related molecules (GPR56 and CD244) and a co-stimulatory molecule (CD28, TCM-enriched) at the protein level (Fig. 5I). Together, these results show that cells in the IL-7Rhigh TEMRA subset have properties of memory precursor cells and cytotoxic cells.
CD4-CTL precursors share TCRs with CD4-CTL effectors
To verify our hypothesis, we analyzed the overlap in the TCR clonotypes of CD4-CTL effectors (clusters 1 and 2) with those from CD4-CTL precursors (clusters 3 and 4). In a total of 5 of 12 donors, TCR clonotypes were shared between CD4-CTL precursors and effectors (Figs. 2A and 6A, fig. S6A, and table S4). As examples, in donor #1, 23 of 80 TEMRA cells had the same TCRα chain clonotype (fig. S6A) as a single cell from precursor cluster (cluster 3); in donor #12, four TCRβ chain clonotypes were shared between CD4-CTL precursors and effectors (Fig. 6A, clones highlighted in dashed lines with the precursor cells indicated with red arrows). A similar pattern was observed in donors #3, #5, and #6 (Figs. 2A and 6A and table S4). To further support our inference, we constructed cell-state hierarchy maps using the Sincell software (39) for cells in the TCM, TEM, and TEMRA subsets and observed that CD4-CTL effectors clustered closest to the CD4-CTL precursors (Fig. 6B and fig. S6B). Together, these data lend support to the hypothesis that CD4-CTL effectors were mainly generated in vivo from distinct CD4-CTL precursor cells, although the possibility of arising from memory TEM or TCM cells cannot be excluded.
Fig. 6. CD4-CTL effectors share TCR clonotypes with CD4-CTL precursors.
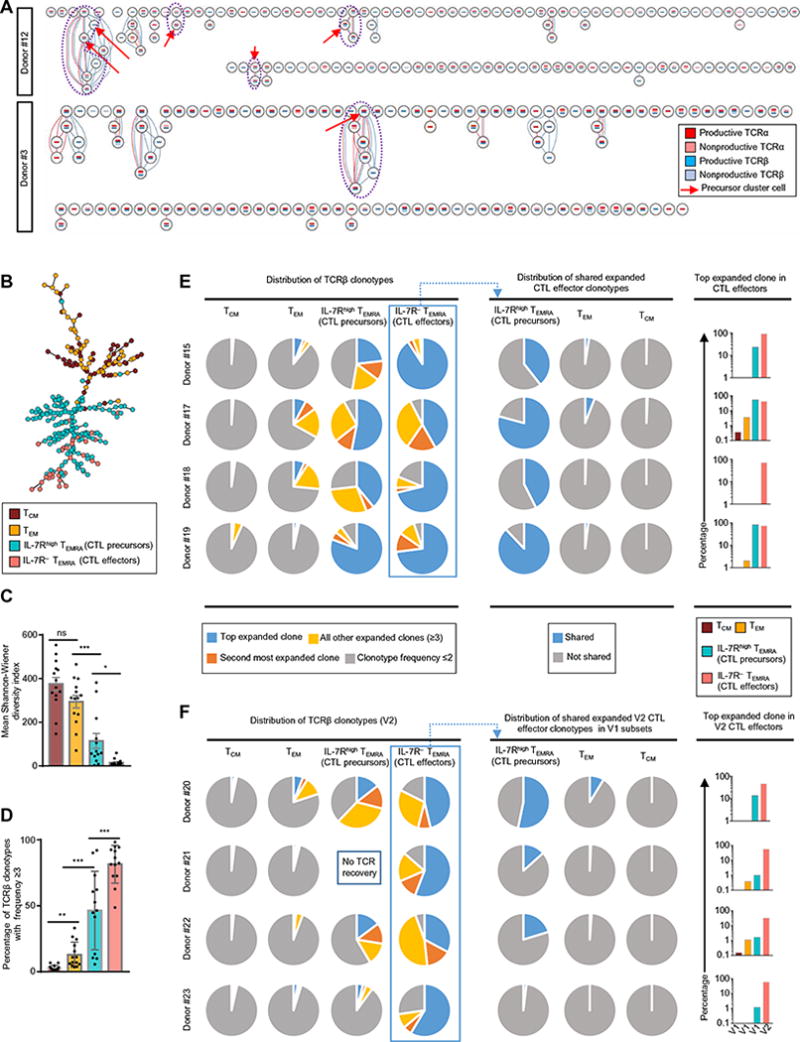
(A) Clonotype network graphs of single cells from TEMRA subsets from donors #12 (top) and #3 (bottom). Each circle represents a single cell; the reconstructed TCRα and TCRβ sequences for each cell are depicted as red- and blue-colored bars, respectively, inside each circle. Dark- and light-colored bars represent productive and nonproductive TCRs, respectively. Red connecting lines indicate shared TCRα sequences; blue lines indicate shared TCRβ sequences. Red arrow indicates a precursor cell. Purple dashed line indicates a cluster of cells with intermixed precursor and effector cells sharing the same clonotype. (B) Cell-state hierarchy map constructed for precursor cluster TEMRA (light green), effector cluster TEMRA (orange), TCM (brown), and TEM (yellow) cells from donor #12. The network shows connection (black line) between cells (circle). (C and D) TCR-seq assays in TCM, TEM, IL-7Rhigh TEMRA (CD4-CTL precursors), and the IL-7R− TEMRA (CD4-CTL effectors) subset from 14 donors; bar graphs show the Shannon-Wiener diversity index obtained using V(D)J tools (C) and percentage of expanded TCRβ clonotypes (clonotype frequency ≥3) (D); error bars are mean ± SEM from 13 to 14 donors. *P < 0.05, **P < 0.005, and ***P < 0.001 from Student’s paired two-tailed t test; ns, not significant. (E) Pie charts (left) show the distribution of TCRβ clonotypes based on clonal frequency: the most clonally expanded (top expanded clone; blue) and the next most clonally expanded (second most expanded clone; orange), as well as the rest of the expanded (≥3) (all other expanded clones; yellow) and nonexpanded clonotypes (frequency ≤2; gray). The pie charts (middle) show the distribution of shared (overlapping) expanded effector TCRβ clonotypes (frequency ≥3) within the other indicated subsets. The graphs (right) show the percentage overlap of the most expanded TCRβ clonotype from effector subset with the other indicated subsets. (F) Pie charts (left) show the distribution of TCRβ clonotypes based on clonal frequency: the most clonally expanded (top expanded clone; blue) and the next most clonally expanded (second most expanded clone; orange), as well as the rest of the expanded (≥3) (all other expanded clones; yellow) and nonexpanded clonotypes (frequency ≤2; gray) in V2 samples from the longitudinal samples. The pie charts (middle) show the distribution of shared (overlapping) expanded V2 effector clonotypes (frequency ≥3) within other subsets from V1 (V2 samples). The graphs (right) show the percentage overlap of the most expanded TCRβ clonotype from the effector subset at V2 with other subsets at V1.
To support our hypothesis that CD4-CTL effectors are mainly derived from the precursor population, we performed a unique molecular identifier (UMI)–based TCR sequencing (TCR-seq) assays (57) in TCM, TEM, IL-7Rhigh TEMRA (CD4-CTL precursors), and IL-7R− TEMRA (CD4-CTL effectors) subsets from 14 donors (table S7). Consistent with single-cell TCR analysis (Fig. 2, A and B), the CD4-CTL precursor and effector cells in the TEMRA subset had highly restricted TCR repertoire, compared with cells in the TCM and TEM subsets, as shown by lower Shannon-Wiener diversity index (Fig. 6C) (58), and contained higher percentage of expanded clonotypes (Fig. 6, D, E and F, left, and fig. S7, left). A larger fraction of the clonotypes in the CD4-CTL precursor cells relative to TEM and TCM was composed of the expanded clonotypes (frequency, ≥3) present in the CD4-CTL effector cells (Fig. 6E, middle, and fig. S7, middle). Further, the most expanded clonotype in the CD4-CTL effector cells was observed more often and at higher frequency in CD4-CTL precursor cells relative to TEM and TCM (Fig. 6E, right graphs, and fig. S7, right graphs).
We also performed TCR-seq in CD4+ T cell memory subsets from five donors who provided longitudinal samples obtained at 3- to 6-month intervals to assess the kinetics of precursor-effector relation. To better assess the relationship between effectors and putative precursors for each donor, we determined the proportion of expanded CD4-CTL effectors clonotypes from second visit (V2) that were present in the different CD4+ T cell memory subsets at the first visit (V1) (table S8). In four of the five donors analyzed, a greater fraction of the TCR clonotypes detected in CD4-CTL effectors at V2 was shared with cells in the precursor population (IL-7Rhigh TEMRA subset) relative to the TEM or TCM subset at V1 (Fig. 6F, middle, fig. S7, and table S8). A total of 12 of the CD4-CTL effector clonotypes from V2 were only found in CD4-CTL precursors at V1 but not in the TEM, TCM, or CD4-CTL effectors (table S8). Together, the kinetic data from the longitudinal samples provided stronger evidence to support the precursor-effector relationship and suggested that the IL-7Rhigh TEMRA subset is the predominant precursor/progenitor for the CD4-CTL effector cells.
Variable number of CD4-CTL precursors across donors
The proportion of CD4-CTL precursors varied greatly among donors, ranging from 3 to 92% of cells in the TEMRA subset (fig. S8A). To determine whether additional CD4-CTL subsets exist and to definitively assess the presence of CD4-CTL precursors, we analyzed single-cell transcriptomes of 10 times as many cells (>6000 cells) from two donors using the high-throughput 10× genomics platform. The sensitivity of the 10× genomics platform, which uses beads to capture transcripts and also sequences only their 3′ ends (59), is twofold lower than the Smart-seq2 protocol used to study full-length transcriptomes of the same donors (fig. S8, B and C). In donor #2, where 24 of 87 cells (28%) (fig. S8A) were previously assessed as CD4-CTL precursors, unbiased clustering of more than 3000 single-cell transcriptomes revealed two dominant clusters of ~2000 and ~1000 cells each, implying that sequencing more cells does not necessarily reveal additional clusters (Fig. 7A, left). These two clusters differentially expressed transcripts characteristic of CD4-CTL effectors (GZMB and NKG7) and precursors (IL7R, JUNB, and LTB), respectively (Fig. 7B, left, and table S9). Even in donor #1, where only 3 of 87 cells (3.4%) (fig. S8A) were previously found to be CD4-CTL precursors by full-length transcriptome analysis, we observed a similar proportion of CD4-CTL precursors [116 of 3664 cells (3.2%)] when analyzing more cells by the 10× genomics platform (Fig. 7, A and B, right, and table S9). At the single-cell level, CD4-CTL precursors in the CD4-TEMRA subset were distinguished by the high expression levels of IL7R transcripts. The expression pattern of molecules in CD4-CTL precursor and effector cells was further confirmed by single-cell RNA-seq analysis of purified population of IL-7Rhigh and IL-7R− TEMRA cells (Fig. 7C and table S1).
Fig. 7. Variable number of CD4-CTL precursors across donors.
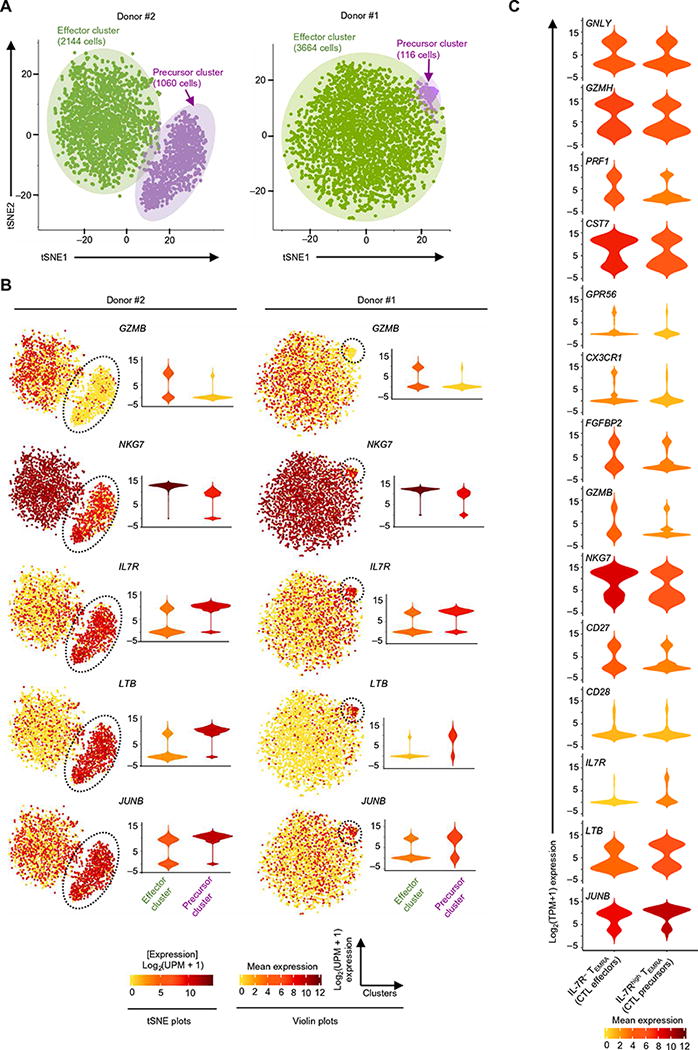
(A) tSNE 2D plot of single-cell RNA-seq data obtained using the Seurat software package for donors #2 (left) and #1 (right) from the 3′ transcriptome of single cells (RNA-seq carried out using the 10× genomics platform) for the most variable transcripts (1799, donor #2; 1569, donor #1). (B) tSNE 2D plots (left) and violin plots (right) of single-cell RNA-seq data show the expression [log2(UPM + 1) expression; UPM, UMI per million] of the indicated differentially expressed transcripts (SCDE and MAST analysis, Benjamini-Hochberg adjusted P < 0.05 and ≥2-fold change) for the indicated donors. Precursor cluster is marked with a black dashed line. The shapes in violin plots represent the distribution of cells based on their log2(UPM + 1) expression values (y axis). The color scale represents the expression of log2(UPM + 1) (tSNE 2D plots) and mean expression (violin plots). (C) Violin plots show the single-cell expression pattern of the indicated differentially expressed transcripts between IL-7Rhigh TEMRA (CD4-CTL precursors) and IL-7R− TEMRA (CD4-CTL effectors) subsets. The shapes represent the distribution of cells based on their log2(TPM + 1) expression values (y axis). Color scale shows the mean expression values.
DISCUSSION
CD4-CTLs have long been considered to be terminal effector cells derived from TEM cells after persistent or repeated (long-term) antigen stimulation in the context of certain viral infections, particularly CMV and DENV (7, 8, 14). Consistent with the notion, our single-cell transcriptome studies of the TEMRA subset did identify cells that have features of terminal CD4-CTL effector cells (KLRG1high and IL-7Rlow; clusters 1 and 2). However, we observed another population of cells in the TEMRA subset that displayed a molecular program (KLRG1low and IL-7Rhigh; clusters 3 and 4) indicative of memory precursor cells, intermixed with several features of CD4-CTLs (KLRG1, TBX21, S1PR5, FGFBP2, CCL4, PRF1, GZMH, GNLY, NKG7, ZEB2, and GPR56), albeit less prominently expressed than in the CD4-CTL effectors. Bulk and single-cell transcriptome analysis of the purified IL-7Rhigh and IL-7R− TEMRA subset further confirmed our observation from single-cell transcriptome analysis of total TEMRA subset. In several donors, we found cells in the CD4-CTL effector subset that shared their TCR clonotypes with cells in the precursor subset. TCR-seq analysis in longitudinal samples further confirmed this finding and provided more evidence for the relation between CD4-CTL precursors and effectors. Although the kinetic studies support the development of effectors from precursor cells, the nature of our studies in humans limits the definitive assessment of directionality; hence, we cannot completely exclude the possibility that the progenitor cell generated immediately after infection is the cytotoxic effector cell that eventually reverts. By defining the single-cell transcriptional program of CD4-CTL precursor cells, we have identified a number of previously unknown molecules potentially important for their differentiation and function and represent attractive targets for further validation studies.
We showed that surface expression of the IL-7R (CD127) in TEMRA cells defines a subset of cells with the expression of IL-7R as high as that observed in the TCM subset. Given the established role of the IL-7R signaling pathway in homeostatic T cell proliferation and survival, such IL-7Rhigh TEMRA cells are likely to represent the precursors of CD4-TEMRA. The isolation of a CD4-CTL precursor subset based on the surface expression of IL-7R would enable detailed epigenetic studies to define the nature and extent of CD4-CTL reprogramming in such precursor cells. These future studies are likely to provide insights into the molecular mechanisms that govern the early development, differentiation, and function of CD4-CTLs in humans as they transit from naive to memory precursor and effector cells.
The CD4-CTL effector cells displayed a number of known cytotoxicity molecules and previously unknown players (PFN1, PFN1P1, EFHD2, VCL, DIP2A, PLEK, and SYNE1) whose single-cell coexpression pattern suggests an important biological role for these molecules in CD4-CTL function. Further, their transcriptional program suggests that CD4-CTL effectors are terminally differentiated and likely short-lived because they express low levels of the costimulatory molecules CD27 and CD28 and high levels of KLRG1 transcripts (41, 52, 60–62). However, we observed marked clonal expansion of CD4-CTL effectors in several donors, suggesting that, compared with other conventional TH effector cells, different molecular mechanisms may operate in CD4-CTL effectors to promote their long-term survival. IL-7R− TEMRA CD4-CTL effectors highly expressed several molecules that are linked to cell survival such as CRTAM, ZNF683 (Hobit), PRSS23, SPON2, and TCF7 (encodes TCF1) (24, 46–51). Therefore, we hypothesize that these candidate molecules are likely to confer long-term survival properties to CD4-CTL effectors, which warrants further functional investigation in model organisms.
Overall, our single-cell transcriptomic studies in the human CD4-TEMRA population have uncovered an unprecedented level of heterogeneity, presumably created by the diverse nature of infections and the timing of exposures coupled with genetic diversity among our study donors. We identified a TEMRA subset of precursor CD4-CTLs, whose isolation and further characterization may open avenues for investigating the mechanisms that govern the generation of CD4-CTLs in humans. The stem-like virus-specific long-lived human memory T cell subset originating from TN cells has been described for CD8 compartment, where these cells share the molecular profiles of TN and TCM subsets (63). Considering the expansion of TEMRA in response to viral infections such as DENV and CMV, it is possible that IL-7Rhigh TEMRA subset (CD4-CTL precursors) may develop from such compartment. Understanding the origins and biology of potentially long-lived CD4-CTL precursors may pave the way for developing strategies to boost durable CD4-CTL immune responses after vaccination against viral infections and cancer. A comprehensive assessment of heterogeneity in pathogen- or vaccine epitope–specific CD4-CTLs by single-cell approaches is likely to yield insights into the nature of protective CD4-CTL response generated against specific pathogens or vaccines.
MATERIALS AND METHODS
Study design
The goal of this study was to use single-cell RNA-seq assay to capture the transcriptome of individual cells in CD4+ T cell memory subsets in human peripheral blood mononuclear cells (PBMCs). Details on the sample collection and processing are described in Supplementary Materials and Methods.
Flow cytometry
Human PBMCs were isolated and stained as described in Supplementary Materials and Methods.
Single-cell RNA-seq
Single-cell RNA-seq was performed as described previously (33, 34), with some modifications that are described in Supplementary Materials and Methods.
Single-cell RNA-seq and statistical analysis
Data processing and analysis were performed using R, Qlucore Omics, and GraphPad Prism, as described in Supplementary Materials and Methods.
TCR-seq and analysis
TCR-seq was performed and analyzed as described previously (57, 58). Details are described in Supplementary Materials and Methods.
Supplementary Material
Fig. S1. Most of the expanded TCR clonotypes are DENV-specific.
Fig. S2. TEMRA cells cluster into four distinct clusters.
Fig. S3. Batch analysis of TEMRA cells.
Fig. S4. TEMRA clusters express distinct set of transcripts.
Fig. S5. Clusters 1 and 2 are enriched for transcripts involved in cytotoxicity-related pathways.
Fig. S6. Precursor cells in TEMRA subset.
Fig. S7. CD4-CTL precursors share clonotypes with CD4-CTL effectors.
Fig. S8. Smart-seq2 is more sensitive compared with droplet-based approach (10× genomics).
Table S12. List of gene sets used for GSEA (for data in Fig. 1E).
Table S2. List of differentially expressed genes between memory subsets [for data in Fig. 1 (B and C)].
Table S4. TCRα and TCRβ chain reconstruction from full-length single-cell transcriptome of every single cell sequenced from the 15 donors.
Table S5. List of differentially expressed genes between clusters (for data in Fig. 4C and fig. S4).
Table S6. List of differentially expressed genes between TCM and CD4-CTL effectors (for data in Fig. 5H).
Table S7. TCRα and TCRβ chain sequences derived from TCR-seq analysis of 14 donors [for data in Fig. 6 (E and F) and fig. S7].
Table S8. TCRα and TCRβ chain clonotype sharing derived from TCR-seq analysis of longitudinally collected samples from five donors (for data in Fig. 6F and fig. S7).
Table S9. List of differentially expressed genes between the two clusters in donors #1 and #2 [for data in Fig. 7 (A and B)].
Table S1. Summary of study donors.
Table S3. Summary of TCRα and TCRβ chains recovered from full-length single-cell transcriptomes of all single cells in the 15 donors.
Table S10. List of antibodies used in the study.
Table S11. Primers used for TCR-seq.
Acknowledgments
We thank the members of the Vijayanand laboratory for the critical reading of the manuscript and discussions and the Human Immune Profiling Consortium team at the La Jolla Institute (LJI) for the valuable discussions. We thank S. Rosales, S. Liang, and D. Singh from Vijayanand’s group for the help with sequencing runs. We thank C. Kim, L. Boggeman, D. Hinz, and R. Simmons at the LJI Flow Cytometry Core for assisting with cell sorting. We thank C. Cerpas and A. Balmaseda for the preparation of PBMCs in Nicaragua. We thank D. M. Chudakov for sharing the detailed protocol for TCR-seq.
Funding: This work was supported by NIH grants [U19AI118626 and U19AI118610 (to P.V. and A.S.); R01HL114093 and R24AI108564 (to P.V.)], the William K. Bowes Jr. Foundation (to P.V.), and NIH contract nos. HHSN272200900042C and HHSN27220140045C (to A.S.).
Footnotes
Author contributions: V.S.P., G.S., A.S., B.P., and P.V. conceived the work. V.S.P. and P.V. designed the study and wrote the manuscript. V.S.P. designed and performed the experiments and analyzed the data under the supervision of P.V. A.M. assisted V.S.P. in the RNA-seq analysis and performed all R-based programming. J.C. assisted V.S.P. in the single-cell RNA-seq experiments. P.O. assisted V.S.P. and D.W. in the DENV peptide stimulation experiments. B.J.S. performed the IL-7R fluorescence-activated cell sorting experiments shown in Fig. 5 (D to G). E.H. and A.D.d.S. collected the blood samples in Nicaragua and Sri Lanka and determined the DENV serostatus, respectively.
Competing interests: All authors declare that they have no competing interests.
Data and materials availability: The sequence data sets reported in this paper have been deposited in the Gene Expression Omnibus with accession no. GSE106544.
SUPPLEMENTARY MATERIALS
immunology.sciencemag.org/cgi/content/full/3/19/eaan8664/DC1
Materials and Methods
References (64–73)
REFERENCES AND NOTES
- 1.Rosenblum MD, Way SS, Abbas AK. Regulatory T cell memory. Nat Rev Immunol. 2016;16:90–101. doi: 10.1038/nri.2015.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Swain SL, McKinstry KK, Strutt TM. Expanding roles for CD4+ T cells in immunity to viruses. Nat Rev Immunol. 2012;12:136–148. doi: 10.1038/nri3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O’Shea JJ, Paul WE. Mechanisms underlying lineage commitment and plasticity of helper CD4+ T cells. Science. 2010;327:1098–1102. doi: 10.1126/science.1178334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arlehamn CL, Seumois G, Gerasimova A, Huang C, Fu Z, Yue X, Sette A, Vijayanand P, Peters B. Transcriptional profile of tuberculosis antigen–specific T cells reveals novel multifunctional features. J Immunol. 2014;193:2931–2940. doi: 10.4049/jimmunol.1401151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheroutre H, Husain MM. CD4 CTL: Living up to the challenge. Semin Immunol. 2013;25:273–281. doi: 10.1016/j.smim.2013.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Juno JA, van Bockel D, Kent SJ, Kelleher AD, Zaunders JJ, Munier CML. Cytotoxic CD4 T cells—Friend or foe during viral infection? Front Immunol. 2017;8:19. doi: 10.3389/fimmu.2017.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weiskopf D, Bangs DJ, Sidney J, Kolla RV, De Silva AD, de Silva AM, Crotty S, Peters B, Sette A. Dengue virus infection elicits highly polarized CX3CR1+ cytotoxic CD4+ T cells associated with protective immunity. Proc Natl Acad Sci USA. 2015;112:E4256–E4263. doi: 10.1073/pnas.1505956112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Derhovanessian E, Maier AB, Hähnel K, Beck R, de Craen AJM, Slagboom EP, Westendorp RGJ, Pawelec G. Infection with cytomegalovirus but not herpes simplex virus induces the accumulation of late-differentiated CD4+ and CD8+ T-cells in humans. J Gen Virol. 2011;92:2746–2756. doi: 10.1099/vir.0.036004-0. [DOI] [PubMed] [Google Scholar]
- 9.Intlekofer AM, Takemoto N, Wherry EJ, Longworth SA, Northrup JT, Palanivel VR, Mullen AC, Gasink CR, Kaech SM, Miller JD, Gapin L, Ryan K, Russ AP, Lindsten T, Orange JS, Goldrath AW, Ahmed R, Reiner SL. Effector and memory CD8+ T cell fate coupled by T-bet and eomesodermin. Nat Immunol. 2005;6:1236–1244. doi: 10.1038/ni1268. [DOI] [PubMed] [Google Scholar]
- 10.Takemoto N, Intlekofer AM, Northrup JT, Wherry EJ, Reiner SL. Cutting edge: IL-12 inversely regulates T-bet and eomesodermin expression during pathogen-induced CD8+ T cell differentiation. J Immunol. 2006;177:7515–7519. doi: 10.4049/jimmunol.177.11.7515. [DOI] [PubMed] [Google Scholar]
- 11.Appay V, Zaunders JJ, Papagno L, Sutton J, Jaramillo A, Waters A, Easterbrook P, Grey P, Smith D, McMichael AJ, Cooper DA, Rowland-Jones SL, Kelleher AD. Characterization of CD4+ CTLs ex vivo. J Immunol. 2002;168:5954–5958. doi: 10.4049/jimmunol.168.11.5954. [DOI] [PubMed] [Google Scholar]
- 12.Zaunders JJ, Dyer WB, Wang B, Munier ML, Miranda-Saksena M, Newton R, Moore J, Mackay CR, Cooper DA, Saksena NK, Kelleher AD. Identification of circulating antigen-specific CD4+ T lymphocytes with a CCR5+, cytotoxic phenotype in an HIV-1 long-term nonprogressor and in CMV infection. Blood. 2004;103:2238–2247. doi: 10.1182/blood-2003-08-2765. [DOI] [PubMed] [Google Scholar]
- 13.Norris PJ, Moffett HF, Yang OO, Kaufmann DE, Clark MJ, Addo MM, Rosenberg ES. Beyond help: Direct effector functions of human immunodeficiency virus type 1-specific CD4+ T cells. J Virol. 2004;78:8844–8851. doi: 10.1128/JVI.78.16.8844-8851.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Leeuwen EMM, Remmerswaal EBM, Vossen MTM, Rowshani AT, Wertheim-van Dillen PME, van Lier RAW, ten Berge IJM. Emergence of a CD4+CD28-granzyme B+, cytomegalovirus-specific T cell subset after recovery of primary cytomegalovirus infection. J Immunol. 2004;173:1834–1841. doi: 10.4049/jimmunol.173.3.1834. [DOI] [PubMed] [Google Scholar]
- 15.Aslan N, Yurdaydin C, Wiegand J, Greten T, Ciner A, Meyer MF, Heiken H, Kuhlmann B, Kaiser T, Bozkaya H, Tillmann HL, Bozdayi AM, Manns MP, Wedemeyer H. Cytotoxic CD4+ T cells in viral hepatitis. J Viral Hepat. 2006;13:505–514. doi: 10.1111/j.1365-2893.2006.00723.x. [DOI] [PubMed] [Google Scholar]
- 16.Brown DM, Lee S, de la Luz Garcia-Hernandez M, Swain SL. Multifunctional CD4 cells expressing gamma interferon and perforin mediate protection against lethal influenza virus infection. J Virol. 2012;86:6792–6803. doi: 10.1128/JVI.07172-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McKinstry KK, Strutt TM, Kuang Y, Brown DM, Sell S, Dutton RW, Swain SL. Memory CD4+ T cells protect against influenza through multiple synergizing mechanisms. J Clin Invest. 2012;122:2847–2856. doi: 10.1172/JCI63689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Watson AM, Lam LK, Klimstra WB, Ryman KD. The 17D-204 vaccine strain-induced protection against virulent yellow fever virus is mediated by humoral immunity and CD4+ but not CD8+ T cells. PLOS Pathog. 2016;12:e1005786. doi: 10.1371/journal.ppat.1005786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vogel AJ, Brown DM. Single-dose CpG immunization protects against a heterosubtypic challenge and generates antigen-specific memory T cells. Front Immunol. 2015;6:327. doi: 10.3389/fimmu.2015.00327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Terahara K, Ishii H, Nomura T, Takahashi N, Takeda A, Shiino T, Tsunetsugu-Yokota Y, Matano T. Vaccine-induced CD107a+ CD4+ T cells are resistant to depletion following AIDS virus infection. J Virol. 2014;88:14232–14240. doi: 10.1128/JVI.02032-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mahnke YD, Brodie TM, Sallusto F, Roederer M, Lugli E. The who’s who of T-cell differentiation: Human memory T-cell subsets. Eur J Immunol. 2013;43:2797–2809. doi: 10.1002/eji.201343751. [DOI] [PubMed] [Google Scholar]
- 22.Finak G, McDavid A, Yajima M, Deng J, Gersuk V, Shalek AK, Slichter CK, Miller HW, McElrath MJ, Prlic M, Linsley PS, Gottardo R. MAST: A flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 2015;16:278. doi: 10.1186/s13059-015-0844-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kharchenko PV, Silberstein L, Scadden DT. Bayesian approach to single-cell differential expression analysis. Nat Methods. 2014;11:740–742. doi: 10.1038/nmeth.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oja AE, Vieira Braga FA, Remmerswaal EBM, Kragten NAM, Hertoghs KML, Zuo J, Moss PA, van Lier RAW, van Gisbergen KPJM, Hombrink P. The transcription factor hobit identifies human cytotoxic CD4+ T cells. Front Immunol. 2017;8:325. doi: 10.3389/fimmu.2017.00325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hidalgo LG, Einecke G, Allanach K, Halloran PF. The transcriptome of human cytotoxic T cells: Similarities and disparities among allostimulated CD4+ CTL, CD8+ CTL and NK cells. Am J Transplant. 2008;8:627–636. doi: 10.1111/j.1600-6143.2007.02128.x. [DOI] [PubMed] [Google Scholar]
- 26.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pearce EL, Mullen AC, Martins GA, Krawczyk CM, Hutchins AS, Zediak VP, Banica M, DiCioccio CB, Gross DA, Mao C-a, Shen H, Cereb N, Yang SY, Lindsten T, Rossant J, Hunter CA, Reiner SL. Control of effector CD8+ T cell function by the transcription factor eomesodermin. Science. 2003;302:1041–1043. doi: 10.1126/science.1090148. [DOI] [PubMed] [Google Scholar]
- 28.Eshima K, Chiba S, Suzuki H, Kokubo K, Kobayashi H, Iizuka M, Iwabuchi K, Shinohara N. Ectopic expression of a T-box transcription factor, eomesodermin, renders CD4+ Th cells cytotoxic by activating both perforin- and FasL-pathways. Immunol Lett. 2012;144:7–15. doi: 10.1016/j.imlet.2012.02.013. [DOI] [PubMed] [Google Scholar]
- 29.Witke W. The role of profilin complexes in cell motility and other cellular processes. Trends Cell Biol. 2004;14:461–469. doi: 10.1016/j.tcb.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 30.Chen H, Choudhury DM, Craig SW. Coincidence of actin filaments and talin is required to activate vinculin. J Biol Chem. 2006;281:40389–40398. doi: 10.1074/jbc.M607324200. [DOI] [PubMed] [Google Scholar]
- 31.Cohen DM, Kutscher B, Chen H, Murphy DB, Craig SW. A conformational switch in vinculin drives formation and dynamics of a talin-vinculin complex at focal adhesions. J Biol Chem. 2006;281:16006–16015. doi: 10.1074/jbc.M600738200. [DOI] [PubMed] [Google Scholar]
- 32.Hoppmann N, Graetz C, Paterka M, Poisa-Beiro L, Larochelle C, Hasan M, Lill CM, Zipp F, Siffrin V. New candidates for CD4 T cell pathogenicity in experimental neuroinflammation and multiple sclerosis. Brain. 2015;138:902–917. doi: 10.1093/brain/awu408. [DOI] [PubMed] [Google Scholar]
- 33.Picelli S, Faridani OR, Björklund ÅK, Winberg G, Sagasser S, Sandberg R. Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc. 2014;9:171–181. doi: 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- 34.Engel I, Seumois G, Chavez L, Samaniego-Castruita D, White B, Chawla A, Mock D, Vijayanand P, Kronenberg M. Innate-like functions of natural killer T cell subsets result from highly divergent gene programs. Nat Immunol. 2016;17:728–739. doi: 10.1038/ni.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stubbington MJT, Lönnberg T, Proserpio V, Clare S, Speak AO, Dougan G, Teichmann SA. T cell fate and clonality inference from single-cell transcriptomes. Nat Methods. 2016;13:329–332. doi: 10.1038/nmeth.3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takeuchi A, Badr MESG, Miyauchi K, Ishihara C, Onishi R, Guo Z, Sasaki Y, Ike H, Takumi A, Tsuji NM, Murakami Y, Katakai T, Kubo M, Saito T. CRTAM determines the CD4+ cytotoxic T lymphocyte lineage. J Exp Med. 2016;213:123–138. doi: 10.1084/jem.20150519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weiskopf D, Angelo MA, Grifoni A, O’Rourke PH, Sidney J, Paul S, Silva AD De, Phillips E, Mallal S, Premawansa S, Premawansa G, Wijewickrama A, Peters B, Sette A. HLA-DRB1 alleles are associated with different magnitudes of dengue virus–specific CD4+ T-cell responses. J Infect Dis. 2016;214:1117–1124. doi: 10.1093/infdis/jiw309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Satija R, Farrell JA, Gennert D, Schier AF, Regev A. Spatial reconstruction of single-cell gene expression data. Nat Biotechnol. 2015;33:495–502. doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Juliá M, Telenti A, Rausell A. Sincell: An R/Bioconductor package for statistical assessment of cell-state hierarchies from single-cell RNA-seq. Bioinformatics. 2015;31:3380–3382. doi: 10.1093/bioinformatics/btv368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hanke T, Corral L, Vance RE, Raulet DH. 2F1 antigen, the mouse homolog of the rat “mast cell function-associated antigen”, is a lectin-like type II transmembrane receptor expressed by natural killer cells. Eur J Immunol. 1998;28:4409–4417. doi: 10.1002/(SICI)1521-4141(199812)28:12<4409::AID-IMMU4409>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 41.Voehringer D, Koschella M, Pircher H. Lack of proliferative capacity of human effector and memory T cells expressing killer cell lectinlike receptor G1 (KLRG1) Blood. 2002;100:3698–3702. doi: 10.1182/blood-2002-02-0657. [DOI] [PubMed] [Google Scholar]
- 42.Blaser C, Kaufmann M, Pircher H. Virus-activated CD8 T cells and lymphokine-activated NK cells express the mast cell function-associated antigen, an inhibitory C-type lectin. J Immunol. 1998;161:6451–6454. [PubMed] [Google Scholar]
- 43.Böttcher JP, Beyer M, Meissner F, Abdullah Z, Sander J, Höchst B, Eickhoff S, Rieckmann JC, Russo C, Bauer T, Flecken T, Giesen D, Engel D, Jung S, Busch DH, Protzer U, Thimme R, Mann M, Kurts C, Schultze JL, Kastenmüller W, Knolle PA. Functional classification of memory CD8+ T cells by CX3CR1 expression. Nat Commun. 2015;6:8306. doi: 10.1038/ncomms9306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Appay V, Dunbar PR, Callan M, Klenerman P, Gillespie GMA, Papagno L, Ogg GS, King A, Lechner F, Spina CA, Little S, Havlir DV, Richman DD, Gruener N, Pape G, Waters A, Easterbrook P, Salio M, Cerundolo V, McMichael AJ, Rowland-Jones SL. Memory CD8+ T cells vary in differentiation phenotype in different persistent virus infections. Nat Med. 2002;8:379–385. doi: 10.1038/nm0402-379. [DOI] [PubMed] [Google Scholar]
- 45.Appay V, Rowland-Jones SL. Lessons from the study of T-cell differentiation in persistent human virus infection. Semin Immunol. 2004;16:205–212. doi: 10.1016/j.smim.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 46.Braun J, Frentsch M, Thiel A. Hobit and human effector T-cell differentiation: The beginning of a long journey. Eur J Immunol. 2015;45:2762–2765. doi: 10.1002/eji.201545959. [DOI] [PubMed] [Google Scholar]
- 47.Chan HS, Chang SJ, Wang TY, Ko HJ, Lin YC, Lin KT, Chang KM, Chuang YJ. Serine protease PRSS23 is upregulated by estrogen receptor alpha and associated with proliferation of breast cancer cells. PLOS ONE. 2012;7:e30397. doi: 10.1371/journal.pone.0030397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schmid F, Wang Q, Huska MR, Andrade-Navarro MA, Lemm M, Fichtner I, Dahlmann M, Kobelt D, Walther W, Smith J, Schlag PM, Stein U. SPON2, a newly identified target gene of MACC1, drives colorectal cancer metastasis in mice and is prognostic for colorectal cancer patient survival. Oncogene. 2016;35:5942–5952. doi: 10.1038/onc.2015.451. [DOI] [PubMed] [Google Scholar]
- 49.Vieira Braga FA, Hertoghs KML, Kragten NAM, Doody GM, Barnes NA, Remmerswaal EBM, Hsiao C-C, Moerland PD, Wouters D, Derks IAM, vanS tijn A, Demkes M, Hamann J, Eldering E, Nolte MA, Tooze RM, ten Berge IJM, van Gisbergen KPJM, van Lier RAW. Blimp-1 homolog Hobit identifies effector-type lymphocytes in humans. Eur J Immunol. 2015;45:2945–2958. doi: 10.1002/eji.201545650. [DOI] [PubMed] [Google Scholar]
- 50.Jeannet G, Boudousquié C, Gardiol N, Kang J, Huelsken J, Held W. Essential role of the Wnt pathway effector Tcf-1 for the establishment of functional CD8 T cell memory. Proc Natl Acad Sci USA. 2010;107:9777–9782. doi: 10.1073/pnas.0914127107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou X, Yu S, Zhao DM, Harty JT, Badovinac VP, Xue HH. Differentiation and persistence of memory CD8+ T cells depend on T cell factor 1. Immunity. 2010;33:229–240. doi: 10.1016/j.immuni.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kaech SM, Tan JT, Wherry EJ, Konieczny BT, Surh CD, Ahmed R. Selective expression of the interleukin 7 receptor identifies effector CD8 T cells that give rise to long-lived memory cells. Nat Immunol. 2003;4:1191–1198. doi: 10.1038/ni1009. [DOI] [PubMed] [Google Scholar]
- 53.Ibegbu CC, Xu YX, Harris W, Maggio D, Miller JD, Kourtis AP. Expression of killer cell lectin-like receptor G1 on antigen-specific human CD8+ T lymphocytes during active, latent, and resolved infection and its relation with CD57. J Immunol. 2005;174:6088–6094. doi: 10.4049/jimmunol.174.10.6088. [DOI] [PubMed] [Google Scholar]
- 54.Bengsch B, Spangenberg HC, Kersting N, Neumann-Haefelin C, Panther E, von Weizsäcker F, Blum HE, Pircher H, Thimme R. Analysis of CD127 and KLRG1 expression on hepatitis C virus-specific CD8+ T cells reveals the existence of different memory T-cell subsets in the peripheral blood and liver. J Virol. 2007;81:945–953. doi: 10.1128/JVI.01354-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Robbins SH, Terrizzi SC, Sydora BC, Mikayama T, Brossay L. Differential regulation of killer cell lectin-like receptor G1 expression on T cells. J Immunol. 2003;170:5876–5885. doi: 10.4049/jimmunol.170.12.5876. [DOI] [PubMed] [Google Scholar]
- 56.Kaech SM, Cui W. Transcriptional control of effector and memory CD8+ T cell differentiation. Nat Rev Immunol. 2012;12:749–761. doi: 10.1038/nri3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shugay M, Britanova OV, Merzlyak EM, Turchaninova MA, Mamedov IZ, Tuganbaev TR, Bolotin DA, Staroverov DB, Putintseva EV, Plevova K, Linnemann C, Shagin D, Pospisilova S, Lukyanov S, Schumacher TN, Chudakov DM. Towards error-free profiling of immune repertoires. Nat Methods. 2014;11:653–655. doi: 10.1038/nmeth.2960. [DOI] [PubMed] [Google Scholar]
- 58.Shugay M, Bagaev DV, Turchaninova MA, Bolotin DA, Britanova OV, Putintseva EV, Pogorelyy MV, Nazarov VI, Zvyagin IV, Kirgizova VI, Kirgizov KI, Skorobogatova EV, Chudakov DM. VDJtools: Unifying post-analysis of T Cell receptor repertoires. PLOS Comput Biol. 2015;11:e1004503. doi: 10.1371/journal.pcbi.1004503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zheng GXY, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, Ziraldo SB, Wheeler TD, McDermott GP, Zhu J, Gregory MT, Shuga J, Montesclaros L, Underwood JG, Masquelier DA, Nishimura SY, Schnall-Levin M, Wyatt PW, Hindson CM, Bharadwaj R, Wong A, Ness KD, Beppu LW, Deeg HJ, McFarland C, Loeb KR, Valente WJ, Ericson NG, Stevens EA, Radich JP, Mikkelsen TS, Hindson BJ, Bielas JH. Massively parallel digital transcriptional profiling of single cells. Nat Commun. 2017;8:14049. doi: 10.1038/ncomms14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Boettler T, Panther E, Bengsch B, Nazarova N, Spangenberg HC, Blum HE, Thimme R. Expression of the interleukin-7 receptor alpha chain (CD127) on virus-specific CD8+ T cells identifies functionally and phenotypically defined memory T cells during acute resolving hepatitis B virus infection. J Virol. 2006;80:3532–3540. doi: 10.1128/JVI.80.7.3532-3540.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fuller MJ, Hildeman DA, Sabbaj S, Gaddis DE, Tebo AE, Shang L, Goepfert PA, Zajac AJ. Cutting edge: Emergence of CD127high functionally competent memory T cells is compromised by high viral loads and inadequate T cell help. J Immunol. 2005;174:5926–5930. doi: 10.4049/jimmunol.174.10.5926. [DOI] [PubMed] [Google Scholar]
- 62.Lang KS, Recher M, Navarini AA, Harris NL, Löhning M, Junt T, Probst HC, Hengartner H, Zinkernagel RM. Inverse correlation between IL-7 receptor expression and CD8 T cell exhaustion during persistent antigen stimulation. Eur J Immunol. 2005;35:738–745. doi: 10.1002/eji.200425828. [DOI] [PubMed] [Google Scholar]
- 63.Gattinoni L, Lugli E, Ji Y, Pos Z, Paulos CM, Quigley MF, Almeida JR, Gostick E, Yu Z, Carpenito C, Wang E, Douek DC, Price DA, June CH, Marincola FM, Roederer M, Restifo NP. A human memory T cell subset with stem cell-like properties. Nat Med. 2011;17:1290–1297. doi: 10.1038/nm.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Weiskopf D, Angelo MA, de Azeredo EL, Sidney J, Greenbaum JA, Fernando AN, Broadwater A, Kolla RV, De Silva AD, de Silva AM, Mattia KA, Doranz BJ, Grey HM, Shresta S, Peters B, Sette A. Comprehensive analysis of dengue virus-specific responses supports an HLA-linked protective role for CD8+ T cells. Proc Natl Acad Sci USA. 2013;110:E2046–E2053. doi: 10.1073/pnas.1305227110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kanakaratne N, Wahala WMPB, Messer WB, Tissera HA, Shahani A, Abeysinghe N, de Silva AM, Gunasekera M. Severe dengue epidemics in Sri Lanka, 2003–2006. Emerg Infect Dis. 2009;15:192–199. doi: 10.3201/eid1502.080926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fernandez RJ, Vazquez S. Serological diagnosis of dengue by an ELISA inhibition method (EIM) Mem Inst Oswaldo Cruz. 1990;85:347–351. doi: 10.1590/s0074-02761990000300012. [DOI] [PubMed] [Google Scholar]
- 67.Kraus AA, Messer W, Haymore LB, de Silva AM. Comparison of plaque- and flow cytometry-based methods for measuring dengue virus neutralization. J Clin Microbiol. 2007;45:3777–3780. doi: 10.1128/JCM.00827-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Swanstrom JA, Plante JA, Plante KS, Young EF, McGowan E, Gallichotte EN, Widman DG, Heise MT, de Silva AM, Baric RS. Dengue virus envelope dimer epitope monoclonal antibodies isolated from dengue patients are protective against Zika virus. mBio. 2016;7:e01123–e16. doi: 10.1128/mBio.01123-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gaublomme JT, Yosef N, Lee Y, Gertner RS, Yang LV, Wu C, Pandolfi PP, Mak T, Satija R, Shalek AK, Kuchroo VK, Park H, Regev A. Single-cell genomics unveils critical regulators of Th17 cell pathogenicity. Cell. 2015;163:1400–1412. doi: 10.1016/j.cell.2015.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zheng C, Zheng L, Yoo JK, Guo H, Zhang Y, Guo X, Kang B, Hu R, Huang JY, Zhang Q, Liu Z, Dong M, Hu X, Ouyang W, Peng J, Zhang Z. Landscape of infiltrating T cells in liver cancer revealed by single-cell sequencing. Cell. 2017;169:1342–1356. doi: 10.1016/j.cell.2017.05.035. [DOI] [PubMed] [Google Scholar]
- 71.Seumois G, Zapardiel-Gonzalo J, White B, Singh D, Schulten V, Dillon M, Hinz D, Broide DH, Sette A, Peters B, Vijayanand P. Transcriptional profiling of Th2 cells identifies pathogenic features associated with asthma. J Immunol. 2016;197:655–664. doi: 10.4049/jimmunol.1600397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ganesan AP, Clarke J, Wood O, Garrido-Martin EM, Chee SJ, Mellows T, Samaniego-Castruita D, Singh D, Seumois G, Alzetani A, Woo E, Friedmann PS, King EV, Thomas GJ, Sanchez-Elsner T, Vijayanand P, Ottensmeier CH. Tissue-resident memory features are linked to the magnitude of cytotoxic T cell responses in human lung cancer. Nat Immunol. 2017;18:940–950. doi: 10.1038/ni.3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Most of the expanded TCR clonotypes are DENV-specific.
Fig. S2. TEMRA cells cluster into four distinct clusters.
Fig. S3. Batch analysis of TEMRA cells.
Fig. S4. TEMRA clusters express distinct set of transcripts.
Fig. S5. Clusters 1 and 2 are enriched for transcripts involved in cytotoxicity-related pathways.
Fig. S6. Precursor cells in TEMRA subset.
Fig. S7. CD4-CTL precursors share clonotypes with CD4-CTL effectors.
Fig. S8. Smart-seq2 is more sensitive compared with droplet-based approach (10× genomics).
Table S12. List of gene sets used for GSEA (for data in Fig. 1E).
Table S2. List of differentially expressed genes between memory subsets [for data in Fig. 1 (B and C)].
Table S4. TCRα and TCRβ chain reconstruction from full-length single-cell transcriptome of every single cell sequenced from the 15 donors.
Table S5. List of differentially expressed genes between clusters (for data in Fig. 4C and fig. S4).
Table S6. List of differentially expressed genes between TCM and CD4-CTL effectors (for data in Fig. 5H).
Table S7. TCRα and TCRβ chain sequences derived from TCR-seq analysis of 14 donors [for data in Fig. 6 (E and F) and fig. S7].
Table S8. TCRα and TCRβ chain clonotype sharing derived from TCR-seq analysis of longitudinally collected samples from five donors (for data in Fig. 6F and fig. S7).
Table S9. List of differentially expressed genes between the two clusters in donors #1 and #2 [for data in Fig. 7 (A and B)].
Table S1. Summary of study donors.
Table S3. Summary of TCRα and TCRβ chains recovered from full-length single-cell transcriptomes of all single cells in the 15 donors.
Table S10. List of antibodies used in the study.
Table S11. Primers used for TCR-seq.


