Abstract
During an acute infection, antigenic stimulation leads to activation, expansion, and differentiation of naïve CD8 + T cells, first into cytotoxic effector cells and eventually into long-lived memory cells. T cell antigen receptors (TCRs) detect antigens on antigen-presenting cells (APCs) in the form of antigenic peptides bound to major histocompatibility complex I (MHC-I)-encoded molecules and initiate TCR signal transduction network. This process is mediated by phosphorylation of many intracellular signaling proteins. Protein O-GlcNAc modification is another post-translational modification involved in this process, which often has either reciprocal or synergistic roles with phosphorylation. In this study, using a chemoenzymatic glycan labeling technique and proteomics analysis, we compared protein O-GlcNAcylation of murine effector and memory-like CD8+ T cells differentiated in vitro. By quantitative proteomics analysis, we identified 445 proteins that are significantly regulated in either effector- or memory-like T cell subsets. Furthermore, qualitative and quantitative analysis identified highly regulated protein clusters that suggest involvement of this post-translational modification in specific cellular processes. In effector-like T cells, protein O-GlcNAcylation is heavily involved in transcriptional and translational processes that drive fast effector T cells proliferation. During the formation of memory-like T cells, protein O-GlcNAcylation is involved in a more specific, perhaps more targeted regulation of transcription, mRNA processing, and translation. Significantly, O-GlcNAc plays a critical role as part of the “histone code” in both CD8+ T cells subgroups.
Graphical Abstract
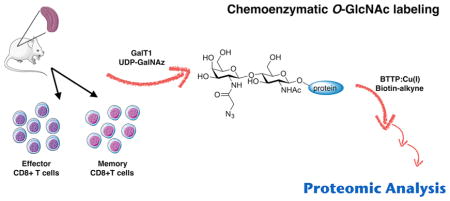
CD8+ T cells are central players in cell-mediated immunity. Following an acute infection, antigenic stimulation leads to activation, expansion, and differentiation of naïve CD8 + T cells into effector cells. Armed effector cells secrete cytotoxins, such as perforin and granzymes which are stored in specialized lytic granules, and cytokines (e.g., IFN-γ and TNF-α),1,2 to provide effective pathogen control. The T-cell response peaks around days 7-10 after a high proliferation phase largely driven by interleukin-2 (IL-2). A contraction phase follows this response peak, after which only a small fraction of CD8+ T cells survive (5–10% of the antigen-specific cells present at the peak of the effector response) leading to long-lived memory cells.3,4 Memory CD8+ T cells are maintained via homeostatic proliferation driven by cytokines IL-7 and IL-15.5–8 For these reasons, IL-2 or IL-7/IL-15 is often used in in vitro activation assays to generate effector- or memory-like CD8+ T cells, respectively.
T-cell activation via the engagement of the TCR results in a cascade of phosphorylation events on lymphocyte protein tyrosine kinase (Lck), immunoreceptor tyrosine-based activation motifs (ITAMs), or the zeta-chain associated protein kinase (ZAP-70), which promote recruitment and phosphor-ylation of the downstream adaptor or scaffold proteins that trigger T cell phenotypic changes.1 Having a reciprocal or synergistic relationship with protein phosphorylation, protein O-GlcNAc modification is another dynamic modulator of cellular signaling pathways.9,10 O-GlcNAcylation adds β-D-N-acetylglucosamine to serine or threonine residues of nuclear and cytoplasmic proteins.11 The addition and removal of this monosaccharide is a reversible process catalyzed by two enzymes, O-GlcNAc transferase (OGT) and O-GlcNAcase (OGA), respectively.12 Being dynamic in nature, coupled with its ability to quickly respond to metabolic changes, and its interplay with phosphorylation, positions this form of glycosylation to integrate environmental information and participate in multiple regulatory pathways. Functional roles identified for O-GlcNAc modification include nutrient sensing, protein stabilization, and transcriptional regulation.13 Aberrant O-GlcNAc modification has been implicated in pathologies of metabolic14 and neurodegenerative diseases,15,16 as well as in cancers17 and autoimmunity.18
However, the role of O-GlcNAc in the immune system has barely been explored. Early studies using in vitro activated splenocytes and T cell hybridomas with Concanavalin A and Phorbol 12-myristate 13-acetate revealed an increase in O-GlcNAcylation upon T-cell activation.19 Notch, TCR, and c-Myc were later found to be key regulators of T-cell protein O-GlcNAcylation via regulation of glucose and glutamine transport.20 It is also known that OGT is essential for T cell activation.21 A current model proposes that activation of T cells through the TCR leads to the association of transcription factors, the nuclear factor of activated T cells (NFAT) and the nuclear factor-κB (NF-κB), to OGT, resulting in their modification by O-GlcNAc. Glycosylation enhances their nuclear translocation, leading to increased transcriptional activation of many genes including IL-2.21–23
Despite the clear relevance of protein O-GlcNAcylation in T-cell biology, we have identified only one published study reporting the profiling of O-GlcNAcylated proteins in T cells. In 2016, using tetraacetylated N-Azidoacetyl galactosamine (Ac4GalNAz) as the metabolic substrate, the Davis laboratory characterized O-GlcNAcylated glycoproteins in in vitro activated human T cells.24 This study confirmed that T-cell activation resulted in a global elevation of O-GlcNAc levels, and in the absence of O-GlcNAc, IL-2 production and proliferation were compromised.24 Although this study identified more than 200 O-GlcNAc modified proteins, many of which have functional relevance to T-cell activation, identification was by no means exhaustive. Importantly, metabolic labeling using Ac4GalNAz as a substrate is complicated by nonspecific incorporation of this unnatural sugar into other sectors of glycans, as well as competition with the natural pool of UDP-GlcNAc, leading to an inaccurate representation of abundance based on the different activities of OGT and OGA for different substrates.25,26 In addition, to the best of our knowledge, no comprehensive profiling of protein O-GlcNAcylation in memory T cells has ever been reported in the literature.
In this study, we used a combination of chemoenzymatic glycan labeling and proteomic techniques to compare protein O-GlcNAcylation in in vitro differentiated murine effector- and memory-like CD8+ T cells. By qualitatively and quantitatively comparing protein O-GlcNAcylation from effector- and memory-like T cells, we found a few highly regulated protein clusters and pathways that are involved in the formation of T-cell effector or memory functions.
RESULTS AND DISCUSSION
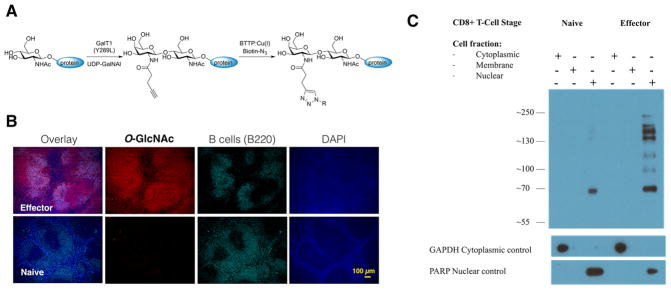
In Vivo Activated Effector CD8+ T Cells Elevate the O-GlcNAc Glycosylation Level
We used the well-established murine model of Listeria monocytogenes (LM) infection to study changes in O-GlcNAcylation upon activation of CD8+ T cells.27 Wild type C57BL/6J mice were infected with LM, and 7 days later, at the peak of T cell activation, we analyzed O-GlcNAcylation patterns in spleen sections using a two-step chemoenzymatic labeling method. This method is a slight modification of the protocol originally developed by Hsieh-Wilson and co-workers;28 it exploits a β1–4 galactosyltransferase mutant (GalT1 Y289L) to incorporate an N-(4-pentynoyl)-alkyne galactosamine (GalNAl) to the GlcNAc motif in O-GlcNAc modified proteins. The alkyne tag is then derivatized via the ligand accelerated copper(I)-catalyzed alkyneazide cycloaddition (CuAAC) with an azide-biotin probe for detection (Figure 1A).29–31 Applying this method to histological samples,32 we found that LM infection led to a significant elevation in O-GlcNAc expression in the spleen, and the increased O-GlcNAc was primarily found in the white pulp, a region which contains mainly lymphocytes such as T cells and B cells (Figure 1B, costained with B220 antibody for B-cells). Next, CD8+ T cells were enriched from spleens of naïve or infected mice, fractioned, and probed with an O-GlcNAc-specific antibody (RL2). Infection with LM led to an upregulation of global O-GlcNAc levels, and protein O-GlcNAcylation was primarily found and enriched in the nuclear fraction of CD8+ T cells (Figure 1C).
Figure 1.
Elevated O-GlcNAc levels for in vivo activated T cells. (A) Schematic representation of a two-step chemoenzymatic O-GlcNAc detection. Transferring of alkyne bearing galactose from UDP-GalNAl donor using GalT1(Y289L), followed by CuAAC using the BTTP-Cu(I) catalyst and an accelerated biotin-azide probe.29–31 (B) Chemoenzymatic staining of O-GlcNAc in 5 μm spleen FFPE tissue sections from naïve or 7-dpi with LM mice. Red = chemoenzymatic O-GlcNAc detection, cyan = B220 antibody B-cell detection, blue = DAPI nuclear stain. Scale bar = 100 μm. (C) Cell fractions from CD8+ T cells isolated from C57BL/6 naïve or 7-dpi with LM mice were probed for O-GlcNAc expression using antibody RL2. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and Poly (ADP-ribose) polymerase (PARP) were used as loading controls for the cytoplasmic and nuclear fractions respectively.
O-GlcNAc Levels Are Elevated for in Vitro Differ-entiated Effector and Memory-like CD8+ T Cells
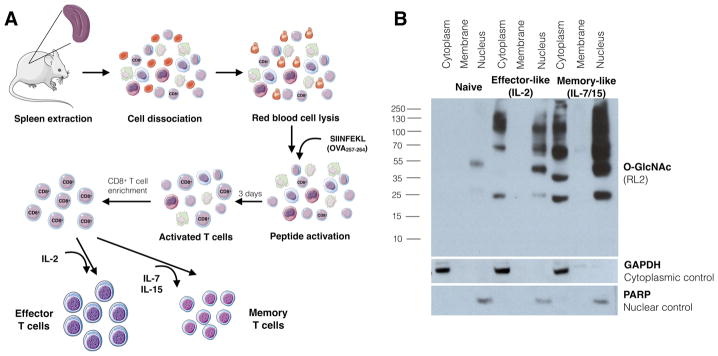
We subsequently assessed changes in protein O-GlcNAcylation status upon in vitro differentiation of naïve CD8 + T cells into effector- and memory-like cells using TCR transgenic OT-1 T cells that recognize the chicken ovalbumin (OVA) derived epitope OVA257-264 (SIINFEKL) in the context of the MHC class I H-2Kb.33 Splenocytes from OT-1 mice were incubated with OVA257–264 for 3 days. CD8+ T cells were then enriched and subjected to cytokine-mediated differentiation. IL-2 was used to generate effector-like T cells, whereas IL-7 and IL-15 exposure was used to yield memory-like T cells (Figure 2A).5 Activation and differentiation into effector- and memory-like T cells were confirmed by cell-surface activation marker staining and flow cytometry analysis (Figure S1). In naïve T cells, protein O-GlcNAcylation was only detectable in the nuclear fraction. Upon activation, an increase in O-GlcNAcylation was observed and became detectable in both cytoplasmic and nuclear fractions with higher levels of O-GlcNAcylation found in the nucleus. In effector-like CD8+ T cells, O-GlcNAc exhibited similar patterns of upregulation (Figure 2B) as reported for T-cell hybridomas and for in vitro anti-CD3/CD28 activated primary murine and human T cells.19,20,24 Interestingly, higher levels of cytoplasmic and nuclear O-GlcNAc were observed in memory-like CD8+ T cells compared to naïve untreated or effector-like CD8+ T cells, despite the fact that memory T cells are known to return to a more resting state similar to naïve cells.3 Memory-like CD8+ cells also expressed high levels of protein O-GlcNAcylation with high molecular weight (>130 kDa), which were less abundant in effector-like cells. These observations may suggest a permanent change that is unrelated to transient activation and may be related to the memory T cell priming that allows them to respond faster to new immune challenges.
Figure 2.
Expression of elevated levels of O-GlcNAc by in vitro differentiated CD8+ T cells. (A) Workflow for in vitro CD8+ T cell differentiation. Total splenocytes from OT-1 mice are exposed to SIINFEKL peptide for 3 days, followed by CD8+ T cell enrichment and treatment with either IL-2 to generate effector-like T-cells or IL-15 and IL-7 to generate memory-like T cells. (B) Cell fractions from OT-1 CD8+ T cells, naïve or in vitro activated and differentiated into effector-like or memory-like T cells, analyzed for O-GlcNAc expression with RL2 antibody.
Identification of Protein O-GlcNAcylation Differences between Effector- and Memory-like CD8+ T Cells
We then proceeded to enrich O-GlcNAc-modified proteins from these in vitro differentiated effector and memory-like CD8+ T cells using chemoenzymatic glycan labeling with a similar procedure as shown in Figure 1A in which UDP-GalNAz was used as the nucleotide donor since it has been proved to be more effective for protein labeling in solution (Figure S2). The ligand-assisted CuAAC was used to introduce a biotin tag to O-GlcNAc modified proteins, followed by enrichment with immobilized streptavidin and on-bead digestion with trypsin. Tryptic digested peptides from both effector and memory-like T cell pull-downs were labeled by either a light or heavy dimethyl tag using previously published methods.34 Negative controls for both effector-like and memory-like cells were prepared in the same manner, except the GalT1 enzyme was omitted during the chemoenzymatic labeling step. To set the threshold of background signals, negative controls were labeled with a medium dimethyl tag. Effector (labeled with heavy dimethyl tag), memory (labeled with light dimethyl tag), and negative control peptides were analyzed simultaneously by mass spectrometry using previously published protocols.34 In total, six analyses, three biological replicates in technical duplicates, were performed. Acquired mass data were searched against the Uniprot mouse (Mus musculus) reference database (2016-01-01) using ProLuCID.35 The identified results were filtered by 1% protein level FDR and then quantified by comparing heavy, medium, and light signals. Each protein identification was assigned a calculated confidence score. We detected a total of 1904 proteins and then refined our results by removing 1459 proteins that were not significantly (p > 0.05) enriched in memory- and effector-like samples when compared to negative controls.
Our analysis identified 445 unique proteins; 116 proteins of these were identified with two or more quantitation values and thus assigned to the high-confidence group. Table 1 shows a selection of relevant proteins from this group. The full list and analysis of identified proteins can be found in the Supporting Information Tables S1 and S2, and raw data can be found in the SI spreadsheet). Within the high confidence group, two proteins were found with higher abundance in effector-like T cells. Nineteen were found primarily in memory-like T cells, and the rest were found in both groups without significant enrichment for one or the other. Table 1 also shows the cellular location and protein function assigned to each protein, as well as relevant pathways the proteins are part of as reported in the Ingenuity Knowledge Database. On the basis of the known cross-talk between O-GlcNAc and phosphorylation, we have included a report on whether each protein is known to be phosphorylated according to UniProt records. Remarkably, 81% (94/116) of the high confidence identified proteins and 72% of the total identified (219/445) are known to be phosphorylated.
Table 1.
Selected O-GlcNAc Enriched Proteins Identified in Glycoproteomics Analysis with High Confidencea
| protein | gene | Det | ref | Loc | function | Phos | pathways |
|---|---|---|---|---|---|---|---|
| plastin-2 | Lcp1 | E | - | Cyt | other | Y | - |
| 40S ribosomal protein S23 | Rps23 | E | 36 | Cyt | translation regulator | - | EIF2 signaling; mTOR signaling; eIF4 and p70S6K signaling |
| cell division cycle and apoptosis regulator protein 1 | Ccar1 | M | 24, 36 | Nuc | transcription regulator | Y | - |
| cathepsin B | Ctsb | M | 36 | Cyt | peptidase | - | autophagy; inflammasome pathway; phagosome maturation |
| hexokinase-1 | Hk1 | M | - | Cyt | kinase | - | GDP-glucose biosynthesis; glucose and glucose-1-phosphate degradation |
| E3 ubiquitin-protein ligase HUWE1 | Huwe1 | M | 36–38 | Nuc | transcription regulator | Y | - |
| DNA mismatch repair protein Msh6 | Msh6 | M | - | Nuc | enzyme | Y | colorectal cancer metastasis signaling; mismatch repair in eukaryotes |
| nuclear pore complex protein Nup214 | Nup214 | M | 24, 36 | Nuc | transporter | Y | - |
| nucleoporin p58/p45 | Nup58 | M | 24, 38, 39 | - | - | ||
| phosphatidylinositol-binding clathrin assembly protein | Picalm | M | 24, 36, 38 | Cyt | other | Y | clathrin-mediated endocytosis signaling |
| pleckstrin homology domain- containing, family A member 5 | Plekha5 | M | 38 | Cyt | other | - | - |
| protein phosphatase 1 regulatory subunit 12A | Ppp1r12a | M | 24, 36, 38 | Cyt | phosphatase | Y | CCR3 signaling in eosinophils; chemokine signaling; ERK/MAPK signaling; HIPPO signaling |
| RNA-binding protein 27 | Rbm27 | M | 24, 36, 38 | Nuc | other | Y | - |
| deoxynucleoside triphosphate triphosphohydrolase SAMHD1 | Samhd1 | M | 36 | Nuc | enzyme | Y | - |
| SAP30-binding protein | Sap30bp | M | 24, 36, 38 | Nuc | transcription regulator | Y | - |
| splicing factor 1 | Sf1 | M | 24, 36 | Nuc | other | Y | - |
| Transcription factor Sp1 | Sp1 | M | 36 | Nuc | transcription regulator | Y | ErbB2-ErbB3 signaling; estrogen-dependent breast cancer signaling; IL-10 signaling |
| spectrin beta chain, nonerythrocytic 1 | Sptbn1 | M | 24, 36, 40 | P.M. | other | - | sertoli cell-sertoli cell junction signaling |
| transcription initiation factor TFIID subunit 6 | Taf6 | M | 24, 36 | Nuc | transcription regulator | Y | assembly of RNA polymerase II complex; receptor signaling |
| ubiquitin-associated protein 2 | Ubap2 | M | 36, 38 | Cyt | other | Y | - |
| ubiquilin-2 | Ubqln2 | M | 36 | Nuc | other | Y | - |
| fructose-bisphosphate aldolase A | Aldoa | D | 36, 38 | Cyt | enzyme | Y | gluconeogenesis I; glycolysis I; sucrose degradation V(mammalian) |
| bifunctional glutamate/proline- tRNA ligase | Eprs | D | 24, 36 | Cyt | enzyme | Y | tRNA Charging |
| fermitin family homologue 3 | Fermt3 | D | - | Cyt | enzyme | Y | - |
| guanine nucleotide-binding protein G(i) subunit alpha-2 | Gnai2 | D | 38 | P.M. | enzyme | - | CCR5 signaling; chemokine signaling; IL-8 signaling; leukocyte extravasation signaling; |
| host cell factor 1 | Hcfc1 | D | 24, 36–40 | Nuc | transcription regulator | Y | - |
| histone H1.3 | Hist1h1d | D | 36 | Nuc | other | Y | granzyme A signaling; protein kinase a signaling |
| heat shock protein HSP 90-alpha | Hsp90aa1 | D | 36, 37 | Cyt | enzyme | Y | androgen signaling; glucocorticoid receptor signaling; HIF1α signaling; PI3K/AKT signaling |
| Ras GTPase-activating-like protein IQGAP1 | Iqgap1 | D | 24, 36 | Cyt | other | Y | Cdc42 signaling; IL-8 signaling; ignaling by Rho family GTPases |
| protein disulfide-isomerase | P4hb | D | - | Cyt | enzyme | - | hypoxia signaling in the cardiovascular system; role of tissue factor in cancer; unfolded protein response |
| proliferation-associated protein 2G4 | Pa2g4 | D | 36 | Nuc | transcription regulator | Y | cell cycle: G1/S checkpoint regulation; cyclins and cell cycle regulation |
| polyadenylate-binding protein 1 | Pabpc1 | D | 36, 38 | Cyt | translation regulator | Y | antiproliferative role of TOB in T cell signaling; regulation of eIF4 and p70S6K signaling |
| programmed cell death 6- interacting protein | Pdcd6ip | D | 24, 36 | Cyt | other | Y | 14-3-3-mediated signaling; mechanisms of viral exit from host cells |
| splicing factor, proline- and glutamine-rich | Sfpq | D | 24, 36, 38 | Nuc | other | Y | - |
| TAR DNA-binding protein 43 | Tardbp | D | 36 | Nuc | transcription regulator | Y | - |
| valine-tRNA ligase | Vars | D | 36 | Cyt | enzyme | Y | tRNA charging |
| 14-3-3 protein theta | Ywhaq | D | 24, 36 | Cyt | other | Y | cell cycle: G2/M DNA damage checkpoint regulation; Myc mediated apoptosis signaling |
| serine-tRNA ligase, cytoplasmic | Sars | D | - | Cyt | enzyme | Y | selenocysteine biosynthesis; tRNA charging |
Det (detection): E = effector (p < 0.05), M = memory (p < 0.05), D = detected. ref (reported references). Loc (cellular location): Nuc = nucleus, Cyt = cytoplasm, P.M. = plasma membrane. Phos (known phosphorylation): Y = yes, N = no.
An efficient way to validate our results is to cross-reference our identified proteins with published reports. We compared our results with six publications of O-GlcNAc glycoproteomics including the data set from the aforementioned human CD8+ T cell report,24 as well as data obtained from brain extracts,37,38,40 osteoblasts,39 and HEK 293 cells.36 These reports compile O-GlcNAc enriched proteins and directly O-GlcNAc modified peptides prepared using various enrichment protocols including immunoprecipitation, lectin enrichment, and metabolic labeling followed by either direct click-on resin or click reaction to an affinity probe for avidin enrichment. Greater than 65% (76/116) of the identified proteins with high confidence and 55% of total proteins (243/445) have been previously reported in other studies, further validating our data.
Our experiment recognized proteins that have been positively identified and particularly studied for their O-GlcNAc modification such as Sp1,41 which is also a critical element for the activation of lymphocytes,42 or Hcfc1, which acts as a key regulator of gluconeogenesis43 and has also been implicated in the progression of viral infections via its interaction with OGT.44 We also identified previously unreported proteins with critical roles such as the protein disulfide-isomerase (P4hb), which participates in the unfolded protein response pathway and has been shown to be differentially expressed in activated T cells with different functional phenotypes.45,46 Another example is the cytoplasmic serine tRNA synthetase (Sars), which is involved in tRNA charging; while the role of Sars has not been specifically explored in T cells, other tRNA synthetases have been implicated in immune diseases such as idiopathic inflammatory myophaty, interstitial lung disease, or rheumatoid arthritis. Furthermore, alternative functions for some tRNA synthetases have emerged in critical pathways to T cell biology such as the ERK signaling pathway or glucose metabolism.47
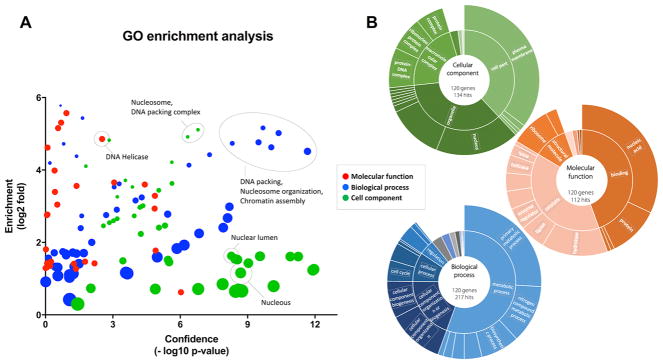
We further performed gene ontology enrichment analysis on the list of proteins identified with high confidence using both Panther48 and Enrichnet49 (Figure 3). Results from enrichment analysis are shown in Figure 3A with a plot displaying the protein clusters by the different categories (molecular function, biological processes, or cell compartment) according to the confidence of identification and the fold enrichment compared with the whole genome. The breakdown for the first two hierarchical levels of gene ontology analysis for each category can be seen in Figure 3B. Regarding cellular component localization of O-GlcNAc-enriched proteins, a large number of proteins belong to the organelles, particularly the nucleus, or to macromolecular complexes, primarily protein-DNA complexes or ribonucleoprotein complexes, as expected from previous publications. For molecular function, a large portion of proteins are classified as catalytic or binding function; while the former is evenly distributed among enzyme categories, most of the binding function is related to nucleic acid binding, in a similar fashion to the findings from human T cells proteomics results.24 The primary category for biological processes is related to metabolism; however, there are a significant number of proteins related to the organization of cellular components, cell cycle, and cell communication.
Figure 3.
Gene ontology analysis of reliable O-GlcNAc-enriched proteins. (A) Plot of identification confidence vs fold-enrichment by gene ontology for protein clusters according to molecular function (red), biological processes (blue), or cell component (green). (B) Gene ontology with first two levels of analysis for reliably O-GlcNAc-enriched proteins according to cellular component, molecular function, or biological process.
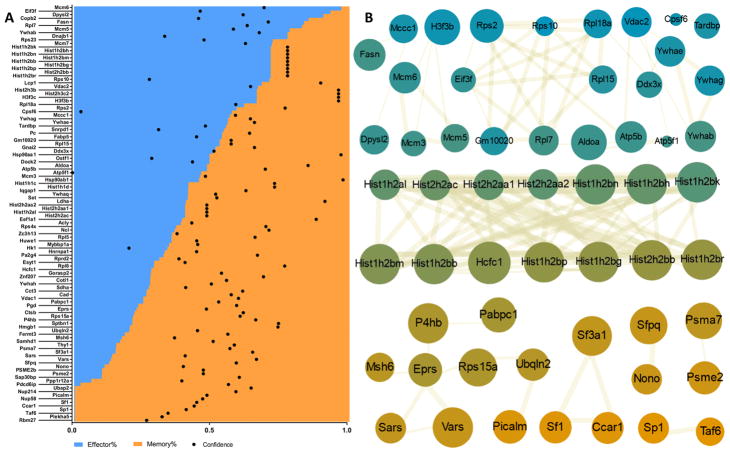
While confident assignment of higher abundance to effector or memory groups could only be done for 21 of the reliably identified proteins, the majority of proteins were detected in both groups. Figure 4A shows the percentage distribution for signal intensity between effector-like (blue) and memory-like (orange) groups when normalized to 100%. We performed a K-mean clustering based on the STRING-DB correlation scores of each protein pair and identified protein clusters in each group that begin to suggest relevant functions of protein O-GlcNAcylation for effector-like T cells, memory-like T cells, or for both cell types (Figure 4B). Several protein clusters were found to be specifically enriched in effector T cells, including the ribosomal protein cluster (Rps2, Rps10, Rpl15, Rpl18a, and Rpl7), minichromosome maintenance (MCM) protein complex cluster (Mcm3, Mcm5, and Mcm6), and the 14-3-3 protein family (Ywhae, Ywhag, and Ywhab). The MCM protein complexes are known to be involved in initiating genome replication,50,51 and both ribosomal and MCM clusters possibly reflect the fast proliferation rates of effector T cells. The 14-3-3 protein family is known for multiple cellular functions including cell cycle regulation, signaling, and possibly T cell activation.52 While the regulation of activity by O-GlcNAc modification of 14-3-3 ligands has been explored,53,54 there is still no evidence on how the direct modification of these proteins affects their activity.
Figure 4.
Contributions from memory/effector groups and network analysis of high-confidence identified proteins. (A) Plot of high-confidence identified proteins sorted by normalized signal contribution from highest effector-like sample percentage to highest memory-like contribution. Overlaid dots reflect normalized confidence of identification as defined by x axis. (B) STRING network analysis. Color of nodes defined by their percentage of memory (orange)/effector (blue) detection. Node size is proportional to the abundance, and connector thickness reflects strength of the known interaction. Main networks identified for primarily memory, effector, or both groups.
One of the predominant clusters found in memory-like T cells contains several tRNA synthetases (Sars, Eprs, and Vars). While the main function of these proteins is tRNA charging with their respective amino acids for use during protein synthesis, new studies have found that some members of the tRNA synthetase family have secondary, nontranslational functions that are critical for cellular processes such as protein synthesis and signaling, and that the switch between functions is mediated by phosphorylation.55,56 While unravelling the role of O-GlcNAc modification for these proteins in the context of memory T cell formation will require targeted research, the potential impact of this post-translational modification is quite clear. Another memory-related cluster involves splicing factor (sf1), cell division and apoptosis regulator protein 1 (ccar1), and splicing factor 3A subunit 1 (sf3a1), a network that suggests regulation of mRNA processing.
The main cluster identified for equal contributions between effector- and memory-like T cells consisted primarily of histones, perhaps unsurprisingly since O-GlcNAc is considered now part of the histone code that helps regulate higher-order chromatin structure and epigenetic memory.57 Overall, the analysis depicted in Figure 4 suggests that protein O-GlcNAcylation specifically enriched in effector-like CD8+ T cells are heavily involved in the transcription and translation processes that drive fast proliferation. It also suggests that the O-GlcNAc modification has a more specific, perhaps more targeted regulation of transcription, mRNA processing, and translation during memory T cell formation, while maintaining its critical role as part of the “histone code” in both T cell subgroups.
Conclusion
In this study, we have compared the O-GlcNAc modification of murine effector- and memory-like CD8+ T cells differentiated in vitro using chemoenzymatic glycan labeling and proteomics analysis. We have identified 445 unique proteins, of which >70% are known to be phosphorylated and 45% of which have not been previously reported in O-GlcNAc enrichment studies. By comparing identified proteins from both effector and memory-like T cells qualitatively and quantitatively, we have found some highly regulated protein clusters that suggest involvement of this post-translational modification in particular processes for different types of CD8+ T cells, such as mRNA processing for memory T cells and transcription and translation for effector T cells. To the best of our knowledge, this is the first comprehensive profiling of O-GlcNAc-enriched proteins in memory-like T cells. Overall, our findings confirm the importance of O-GlcNAc modification in T cell biology and identify new modified proteins in CD8+ T cell subsets that can begin unravelling the specific mechanisms by which O-GlcNAc affects T-cell biology.
Supplementary Material
Acknowledgments
This work was performed at The Scripps Research Institute with the financial support from the National Institutes of Health to P.W. (GM093282, GM113046) and to J.R.Y. (MH067880, GM103533). We thank the Histology Core Facility at TSRI for providing access to their equipment and support.
Footnotes
Notes
The authors declare no competing financial interest.
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acschem-bio.7b00869.
Supporting Tables S1 and S2, Figures S1 and S2, and detailed experimental procedures (PDF) Full data set for identified and excluded proteins with replicates (XLSX)
References
- 1.Smith-Garvin JE, Koretzky GA, Jordan MS. T Cell Activation. Annu Rev Immunol. 2009;27:591–619. doi: 10.1146/annurev.immunol.021908.132706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Janas ML, Groves P, Kienzle N, Kelso A. IL-2 Regulates Perforin and Granzyme Gene Expression in CD8+ T Cells Independently of Its Effects on Survival and Proliferation. J Immunol. 2005;175:8003–8010. doi: 10.4049/jimmunol.175.12.8003. [DOI] [PubMed] [Google Scholar]
- 3.Kaech SM, Ahmed R. Memory CD8+ T cell differentiation: initial antigen encounter triggers a developmental program in naïve cells. Nat Immunol. 2001;2:415–422. doi: 10.1038/87720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaech SM, Wherry EJ, Ahmed R. Effector and Memory T-Cell Differentiation: Implications for Vaccine Development. Nat Rev Immunol. 2002;2:251–262. doi: 10.1038/nri778. [DOI] [PubMed] [Google Scholar]
- 5.Boyman O, Purton JF, Surh CD, Sprent J. Cytokines and T-cell homeostasis. Curr Opin Immunol. 2007;19:320–326. doi: 10.1016/j.coi.2007.04.015. [DOI] [PubMed] [Google Scholar]
- 6.Goldrath AW, Luckey CJ, Park R, Benoist C, Mathis D. The molecular program induced in T cells undergoing homeostatic proliferation. Proc Natl Acad Sci U S A. 2004;101:16885–16890. doi: 10.1073/pnas.0407417101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sarkar S, Kalia V, Haining WN, Konieczny BT, Subramaniam S, Ahmed R. Functional and genomic profiling of effector CD8 T cell subsets with distinct memory fates. J Exp Med. 2008;205:625–640. doi: 10.1084/jem.20071641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carrio R, Bathe OF, Malek TR. Initial antigen encounter programs CD8+ T cells competent to develop into memory cells that are activated in an antigen-free, IL-7- and IL-15-rich environment. J Immunol. 2004;172:7315–7323. doi: 10.4049/jimmunol.172.12.7315. [DOI] [PubMed] [Google Scholar]
- 9.Bond MR, Hanover JA. A little sugar goes a long way: The cell biology of O-GlcNAc. J Cell Biol. 2015;208:869–880. doi: 10.1083/jcb.201501101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zeidan Q, Hart GW. The intersections between O-GlcNAcylation and phosphorylation: implications for multiple signaling pathways. J Cell Sci. 2010;123:13–22. doi: 10.1242/jcs.053678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Torres CR, Hart GW. Topography and Polypeptide Distribution of Terminal N-Acetylglucosamine Residues on the Surfaces of Intact Lymphocytes. J Biol Chem. 1984;259:2208–2217. [PubMed] [Google Scholar]
- 12.Vocadlo DJ. O-GlcNAc processing enzymes: Catalytic mechanisms, substrate specificity, and enzyme regulation. Curr Opin Chem Biol. 2012;16:488–497. doi: 10.1016/j.cbpa.2012.10.021. [DOI] [PubMed] [Google Scholar]
- 13.Vaidyanathan K, Durning S, Wells L. Functional O-GlcNAc modifications: Implications in molecular regulation and pathophysiology. Crit Rev Biochem Mol Biol. 2014;49:140–163. doi: 10.3109/10409238.2014.884535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Springhorn C, Matsha TE, Erasmus RT, Essop MF. Exploring leukocyte O-GlcNAcylation as a novel diagnostic tool for the earlier detection of type 2 diabetes mellitus. J Clin Endocrinol Metab. 2012;97:4640–4649. doi: 10.1210/jc.2012-2229. [DOI] [PubMed] [Google Scholar]
- 15.Zhu Y, Shan X, Yuzwa SA, Vocadlo DJ. The emerging link between O-GlcNAc and Alzheimer disease. J Biol Chem. 2014;289:34472–34481. doi: 10.1074/jbc.R114.601351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu F, Shi J, Tanimukai H, Gu J, Gu J, Grundke-Iqbal I, Iqbal K, Gong CX. Reduced O-GlcNAcylation links lower brain glucose metabolism and tau pathology in Alzheimer’s disease. Brain. 2009;132:1820–1832. doi: 10.1093/brain/awp099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dennis RJ, Taylor EJ, Macauley MS, Stubbs KA, Turkenburg JP, Hart SJ, Black GN, Vocadlo DJ, Davies GJ. Nat Struct Mol Biol. 2006;13:365–371. doi: 10.1038/nsmb1079. [DOI] [PubMed] [Google Scholar]
- 18.Shi Y, Tomic J, Wen F, Shaha S, Bahlo A, Harrison R, Dennis JW, Williams R, Gross BJ, Walker S, Zuccolo J, Deans JP, Hart GW, Spaner DE. Aberrant O-GlcNAcylation characterizes chronic lymphocytic leukemia. Leukemia. 2010;24:1588–1598. doi: 10.1038/leu.2010.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kearse KP, Hart GW. Lymphocyte activation induces rapid changes in nuclear and cytoplasmic glycoproteins. Proc Natl Acad Sci U S A. 1991;88:1701–5. doi: 10.1073/pnas.88.5.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Swamy M, Pathak S, Grzes KM, Damerow S, Sinclair LV, van Aalten DMF, Cantrell DA. Glucose and glutamine fuel protein O-GlcNAcylation to control T cell self-renewal and malignancy. Nat Immunol. 2016;17:712–720. doi: 10.1038/ni.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Golks A, Tran TTT, Goetschy JF, Guerini D. Requirement for O-linked N-acetylglucosaminyltransferase in lymphocytes activation. EMBO J. 2007;26:4368–4379. doi: 10.1038/sj.emboj.7601845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Golks A, Guerini D. The O-linked N-acetylglucosamine modification in cellular signalling and the immune system. “Protein modifications: beyond the usual suspects” review series. EMBO Rep. 2008;9:748–753. doi: 10.1038/embor.2008.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramakrishnan P, Clark PM, Mason DE, Peters EC, Hsieh-Wilson LC, Baltimore D. Activation of the transcriptional function of the NF-kappaB protein c-Rel by O-GlcNAc glycosylation. Sci Signaling. 2013;6:ra75. doi: 10.1126/scisignal.2004097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lund PJ, Elias JE, Davis MM. Global Analysis of O-GlcNAc Glycoproteins in Activated Human T Cells. J Immunol. 2016;197:3086–3098. doi: 10.4049/jimmunol.1502031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vocadlo DJ, Hang HC, Kim EJ, Hanover JA, Bertozzi CR. A chemical approach for identifying O-GlcNAc-modified proteins in cells. Proc Natl Acad Sci U S A. 2003;100:9116–21. doi: 10.1073/pnas.1632821100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boyce M, Carrico IS, Ganguli AS, Yu SH, Hangauer MJ, Hubbard SC, Kohler JJ, Bertozzi CR. Metabolic cross-talk allows labeling of O-linked beta-N-acetylglucosamine-modified proteins via the N-acetylgalactosamine salvage pathway. Proc Natl Acad Sci U S A. 2011;108:3141–3146. doi: 10.1073/pnas.1010045108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Condotta SA, Richer MJ, Badovinac VP, Harty JT. Advanced Immunology. 1. Elsevier Inc; 2012. Probing CD8 T cell responses with listeria monocytogenes infection. [DOI] [PubMed] [Google Scholar]
- 28.Khidekel N, Arndt S, Lamarre-Vincent N, Lippert A, Poulin-Kerstien KG, Ramakrishnan B, Qasba PK, Hsieh-Wilson LC. A Chemoenzymatic Approach toward the Rapid and Sensitive Detection of O-GlcNAc Posttranslational Modifications. J Am Chem Soc. 2003;125:16162–16163. doi: 10.1021/ja038545r. [DOI] [PubMed] [Google Scholar]
- 29.Jiang H, Zheng T, Lopez-Aguilar A, Feng L, Kopp F, Marlow FL, Wu P. Monitoring Dynamic Glycosylation in Vivo Using Supersensitive Click Chemistry. Bioconjugate Chem. 2014;25:698–706. doi: 10.1021/bc400502d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang W, Hong S, Tran A, Jiang H, Triano R, Liu Y, Chen X, Wu P. Sulfated Ligands for the Copper(I)-Catalyzed Azide-Alkyne Cycloaddition. Chem - Asian J. 2011;6:2796–2802. doi: 10.1002/asia.201100385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Soriano del Amo D, Wang W, Jiang H, Besanceney C, Yan AC, Levy M, Liu Y, Marlow FL, Wu P. Biocompatible Copper (I) Catalysts for in Vivo Imaging of Glycans. J Am Chem Soc. 2010;132:16893–16899. doi: 10.1021/ja106553e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rouhanifard SH, Lopez-Aguilar A, Wu P. CHoMP: A chemoenzymatic histology method using clickable probes. ChemBioChem. 2014;15:2667–2673. doi: 10.1002/cbic.201402433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clarke SR, Barnden M, Kurts C, Carbone FR, Miller JF, Heath WR. Characterization of the ovalbumin-specific TCR transgenic line OT-I: MHC elements for positive and negative selection. Immunol Cell Biol. 2000;78:110–117. doi: 10.1046/j.1440-1711.2000.00889.x. [DOI] [PubMed] [Google Scholar]
- 34.Boersema PJ, Raijmakers R, Lemeer S, Mohammed S, Heck AJR. Multiplex peptide stable isotope dimethyl labeling for quantitative proteomics. Nat Protoc. 2009;4:484–494. doi: 10.1038/nprot.2009.21. [DOI] [PubMed] [Google Scholar]
- 35.Xu T, Park SK, Venable JD, Wohlschlegel JA, Diedrich JK, Cociorva D, Lu B, Liao L, Hewel J, Han X, Wong CCL, Fonslow B, Delahunty C, Gao Y, Shah H, Yates JR. ProLuCID: An improved SEQUEST-like algorithm with enhanced sensitivity and specificity. J Proteomics. 2015;129:16–24. doi: 10.1016/j.jprot.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hahne H, Sobotzki N, Nyberg T, Helm D, Borodkin VS, van Aalten DMF, Agnew B, Kuster B. Proteome Wide Purification and Identification of O-GlcNAc-Modified Proteins Using Click Chemistry and Mass Spectrometry. JProt Res. 2013;12:927–936. doi: 10.1021/pr300967y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wells L, Vosseller K, Cole RN, Cronshaw JM, Matunis MJ, Hart GW. Mapping Sites of O-GlcNAc Modification Using Affinity Tags for Serine and Threonine Post-translational Modifications. Mol Cell Proteomics. 2002;1:791–804. doi: 10.1074/mcp.m200048-mcp200. [DOI] [PubMed] [Google Scholar]
- 38.Trinidad JC, Barkan DT, Gulledge BF, Thalhammer A, Sali A, Schoepfer R, Burlingame AL. Global Identification and Characterization of Both O-GlcNAcylation and Phosphorylation at the Murine Synapse. Mol Cell Proteomics. 2012;11:215–229. doi: 10.1074/mcp.O112.018366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nagel AK, Schilling M, Comte-Walters S, Berkaw MN, Ball LE. Identification of O-Linked N-Acetylglucosamine (O-GlcNAc)-modified Osteoblast Proteins by Electron Transfer Dissociation Tandem Mass Spectrometry Reveals Proteins Critical for Bone Formation. Mol Cell Proteomics. 2013;12:945–955. doi: 10.1074/mcp.M112.026633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Khidekel N, Ficarro SB, Peters EC, Hsieh-Wilson LC. Exploring the O-GlcNAc proteome: direct identification of O-GlcNAc-modified proteins from the brain. Proc Natl Acad Sci U S A. 2004;101:13132–13137. doi: 10.1073/pnas.0403471101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Donovan K, Alekseev O, Qi X, Cho W, Azizkhan-Clifford J. O-GlcNAc modification of transcription factor sp1 mediates hyperglycemia-induced VEGF-A upregulation in retinal cells. Investig Ophthalmol Vis Sci. 2014;55:7862–7873. doi: 10.1167/iovs.14-14048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lacroix I, Lipcey C, Imbert J, Kahn-Perles B. Sp1 Transcriptional Activity Is Up-regulated by Phosphatase 2A in Dividing T Lymphocytes. J Biol Chem. 2002;277:9598–9605. doi: 10.1074/jbc.M111444200. [DOI] [PubMed] [Google Scholar]
- 43.Ruan HB, Han X, Li MD, Singh JP, Qian K, Azarhoush S, Zhao L, Bennett AM, Samuel VT, Wu J, III, Yates JR, Yang X. O-GlcNAc Transferase/Host Cell Factor C1 Complex Regulates Gluconeogenesis by Modulating PGC-1α Stability. Cell Metab. 2012;16:226–237. doi: 10.1016/j.cmet.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Daou S, Mashtalir N, Hammond-martel I, Pak H, Yu H, Sui G, Vogel JL, Kristie TM, Affar EB. Crosstalk between O-GlcNAcylation and proteolytic cleavage regulates the host cell factor-1 maturation pathway. Proc Natl Acad Sci U S A. 2011;108:2747–2752. doi: 10.1073/pnas.1013822108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Waugh KA, Leach SM, Moore BL, Bruno TC, Buhrman JD, Slansky JE. Molecular Profile of Tumor-Specific CD8 + T Cell Hypofunction in a Transplantable Murine Cancer Model. J Immunol. 2016;197:1477–1488. doi: 10.4049/jimmunol.1600589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Verdeil G, Puthier D, Nguyen C, Schmitt-Verhulst A-M, Auphan-Anezin N. STAT5-Mediated Signals Sustain a TCR-Initiated Gene Expression Program toward Differentiation of CD8 T. J Immunol. 2006;176:4834–4842. doi: 10.4049/jimmunol.176.8.4834. [DOI] [PubMed] [Google Scholar]
- 47.Park SG, Schimmel P, Kim S. Aminoacyl tRNA syntehtases and their connections to disease. Proc Natl Acad Sci U S A. 2008;105:11043–11049. doi: 10.1073/pnas.0802862105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mi H, Muruganujan A, Casagrande JT, Thomas PD. Large-scale gene function analysis with the PANTHER classification system. Nat Protoc. 2013;8:1551–1566. doi: 10.1038/nprot.2013.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Glaab E, Baudot A, Krasnogor N, Schneider R, Valencia A. EnrichNet: Network-based gene set enrichment analysis. Bioinformatics. 2012;28:451–457. doi: 10.1093/bioinformatics/bts389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fardini Y, Dehennaut V, Lefebvre T, Issad T. O-GlcNAcylation: A new cancer hallmark? Front Endocrinol (Lausanne, Switz) 2013;4:1–14. doi: 10.3389/fendo.2013.00099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tye BK. MCM Proteins in DNA Replication. Annu Rev Biochem. 1999;68:649–686. doi: 10.1146/annurev.biochem.68.1.649. [DOI] [PubMed] [Google Scholar]
- 52.Morrison DK. The 14–3-3 proteins: integrators of diverse signaling cues that impact cell fate and cancer development. Trends Cell Biol. 2009;19:16–23. doi: 10.1016/j.tcb.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brimble S, Wollaston-Hayden EE, Teo CF, Morris AC, Wells L. The Role of the O-GlcNAc Modification in Regulating Eukaryotic Gene Expression. Curr Signal Transduction Ther. 2010;5:12–24. doi: 10.2174/157436210790226465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Özcan S, Andrali SS, Cantrell JEL. Modulation of transcription factor function by O-GlcNAc modification. Biochim Biophys Acta, Gene Regul Mech. 2010;1799:353–364. doi: 10.1016/j.bbagrm.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Paul M, Schimmel P. Essential Non-Translational Functions of tRNA Synthetases. Nat Chem Biol. 2013;9:145–153. doi: 10.1038/nchembio.1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fu G, Xu T, Shi Y, Wei N, Yang XL. tRNA-controlled nuclear import of a human tRNA synthetase. J Biol Chem. 2012;287:9330–9334. doi: 10.1074/jbc.C111.325902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hanover JA. Epigenetics Gets Sweeter: O-GlcNAc joins the “Histone Code. Chem Biol. 2010;17:1272–1274. doi: 10.1016/j.chembiol.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.