Abstract
Antibodies play a crucial role in coupling the innate and adaptive immune responses against viral pathogens through their antigen binding domains and Fc-regions. Here, we describe how to measure the activation of Fc effector functions by monoclonal antibodies targeting the influenza virus hemagglutinin with the use of a genetically engineered Jurkat cell line expressing an activating type 1 Fc-FcγR. Using this method, the contribution of specific Fc-FcγR interactions conferred by immunoglobulins can be determined using an in vitro assay.
Keywords: Immunology, Issue 132,
Introduction
Immunity provided by the seasonal influenza vaccine has traditionally been assessed by the hemagglutination inhibition (HI) assay1, which measures the presence of antibodies that target the receptor binding site of the hemagglutinin. These HI-active antibodies can confer sterilizing immunity, but are generally narrow in their breadth of protection, providing immunity to only a select few strains of influenza virus. The isolation and characterization of broadly reactive monoclonal antibodies that recognize the hemagglutinin (HA) suggest the development of a universal influenza vaccine is within reach2,3,4,5,6,7,8. One of the major aims of a universal influenza vaccine is to induce a strong antibody response towards the conserved regions of the influenza virus9,10,11,12,13,14,15,16. Broadly reactive antibodies that recognize these conserved epitopes, composed of both neutralizing and non-neutralizing antibodies17,18, have been shown to require Fc-FcγR interactions for optimal protection in vivo and highlight the contribution of Fc-mediated immunity to the overall immune response to influenza virus19,20.
Correlates of protection are crucial in assessing immunity to infection. These metrics allow scientists and clinicians to estimate the efficacy of vaccines, the significance of the data, and the course of treatment. The only established correlate of protection against influenza virus infection is the hemagglutination inhibition assay. A titer of 1:40 is associated with a 50% reduction in the risk of illness1,21 – and reflects the presence of antibodies that inhibit agglutination by targeting the receptor binding site located on the globular head of the HA. However, it should also be noted that T-cell responses may be a better correlate of protection against influenza virus infection in the elderly. Interestingly, a recent report suggests that neuraminidase inhibition activity may be a better predictor of immunity to influenza22. Fortunately, typical in vitro assays such as neutralization or neuraminidase inhibition assays can measure HA stalk- or neuraminidase-specific antibodies. Most of these conventional in vitro assays, however, only take into account the function of the antigen binding region of the antibody and do not measure the role of the Fc region. Additionally, the contribution of non-neutralizing antibodies that protect via Fc receptor engagement in vivo are not detected17,18. In order to measure protection afforded by antibodies that induce Fc-mediated immunity such as antibody dependent cell mediated cytotoxicity (ADCC), a robust in vitro assay is needed.
The method described below assesses the ability of murine monoclonal antibodies to induce Fc-mediated functions through the use of a genetically modified Jurkat cell line expressing an activating murine type 1 FcR, FcγRIV. Antibody engagement of the FcγR transduces intracellular signaling that triggers nuclear factor of activated T-cells-mediated luciferase activity. The assay has several advantages over traditional techniques that require isolation and culture of primary effector cells and the use of flow cytometry to detect activation of Fc-mediated effector functions19,23,24,25,26. First, the protocol described here can easily be adapted to incorporate different viral targets, FcγRs, and antibodies from different species (human and murine). Second, the use of a modified Jurkat cell line expressing an FcγR with a luciferase reporter gene under a nuclear factor of activated T-cell (NFAT) allows for a large format assay that can be easily analyzed using a plate reader measuring luminescence. Here, the traditional activation of primary effector cells is replaced with the induction of NFAT-luciferase upon antibody engagement of an FcγR on the surface of the Jurkat cell. This 96-well plate format assay allows for the measurement of up to four different samples in triplicates with seven dilutions – a number of samples that could be cumbersome using traditional techniques. There are additional points to consider with this assay that may affect the design of experiments and the interpretation of data. The viral target must be expressed on the cell surface in order for antibody recognition. To mitigate this, the target antigen (e.g. an internal viral protein) may be directly coated on the surface of a plate. However, this has not yet been rigorously tested. Additionally, the modified Jurkat cell line expresses only one type of activating FcγR and does not express any inhibitory FcγRs, while primary cell lines express all receptors in a physiological context.
We have previously used this assay to demonstrate that epitope specificity in a polyclonal context plays a crucial role in regulating Fc-mediated effector functions and that optimal activation of antibody-dependent cell-mediated response requires two points of contact27,28. Here, we describe a method that assesses the ability of stalk-specific monoclonal antibodies to engage and activate the murine activating FcγRIV. Although there are exceptions29,30, a large number of broadly reactive antibodies target the antigenically conserved stalk region of the hemagglutinin and thus play an important role in the development of a universal influenza vaccine.
Protocol
The experiments preformed in this manuscript were done following Icahn School of Medicine’s institutional code of ethics and regulations.
1. Expression of Influenza Virus Hemagglutinin via Transfection or Infection
Transfection:
Plate Human Embryonic Kidney (HEK 293T) cells at a density of 2 x 104 cells/well in a white tissue culture treated 96-well plate and let it sit for 4 h in a 37 °C incubator (with 5% CO2).
Transfect cells with 100 ng of DNA coding for viral hemagglutinin and 0.2 μL of transfection reagent per well in a total volume of 50 μL. NOTE: Both plasmid and transfectant reagent are diluted in 1x Opti-Minimum Essential Media (MEM) Reduced Serum Medium.
After incubating plates in a 37 °C incubator with 5% CO2 for 18 h, remove transfection media.
2. Infection
Plate A549 or Madin Darby canine kidney cells at a density of 2 x 104 cells/well in a white tissue culture treated 96-well plate and grow for at least 18 h in a 37 °C incubator with 5% CO2.
Dilute virus in 1x MEM and infect cells at a multiplicity of infection of 3, 1, or 0.33 for 1 h in a 37 °C incubator with 5% CO2. Total volume should equal 100 μL. NOTE:10x MEM is diluted with water to make a 1x MEM working stock for virus dilution mentioned above.
After infection, remove the inoculum and add 100 μL of 1x MEM into the plate in the absence of trypsin.
Grow infected cells for 18 to 24 h in a 37 °C incubator with 5% CO2.
3. Assessing the FcRg/NFAT-mediated Activation of Luciferase Activity by Monoclonal Antibodies or Sera
First, replace growth media of transfected or infected cells with 25 µL of assay media (1x RPMI 1640 supplemented with 4% (vol/vol) low IgG fetal bovine sera).
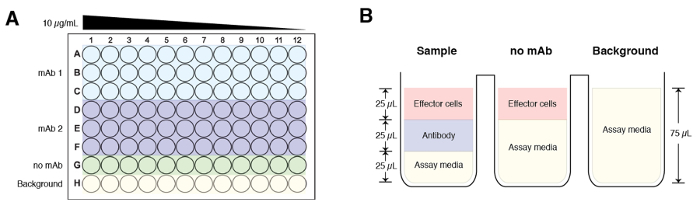
In a separate 96-well plate, perform four-fold dilutions of the antibodies of interest starting at 30 μg/mL using the assay media. Perform dilutions in triplicates and an example plate layout is illustrated in Figure 1A. Make sure to include a control that excludes antibody (no antibody control) as well as a background control (assay buffer alone). NOTE: For both the ‘no antibody’ and background controls, add 25 µL of assay buffer instead of antibody.
Transfer 25 μL of the serially diluted antibodies from Step 3.2 to corresponding wells on plate of transfected cells or infected cells.
Incubate plate in a 37 °C incubator (with 5% CO2) for 15 min.
Add 25 μL of the appropriate modified effector Jurkat cell expressing murine FcgRIV or human FcgRIIIa (75,000 of cells/well). Total volume for each well should be 75 µL and the starting final concentration of the antibody is at 10 µg/mL (Figure 1B). NOTE: For the background control, add 25 µL of assay buffer instead of effector cells. To examine antibody-dependent cell-mediated phagocytosis, a modified Jurkat cell expressing the human FcgRIIa can be used.
Incubate plate in a 37 °C incubator with 5% CO2 for 6 h.
Thaw and warm luciferase substrate buffer in a 37 °C water for about 1 h before use. NOTE: To make the luciferase substrate buffer, reconstitute lyophilized luciferase assay substrate in assay buffer provided. The reconstituted substrate buffer can be stored at -30 °C to -10 °C for up to six weeks.
Add 75 μL/well of luciferase substrate buffer to all wells except for the background control.
Read on a luminometer.
Calculate fold induction with the following equation: Fold induction = (Induced value-Background value) / (No mAb value-Background value).
Representative Results
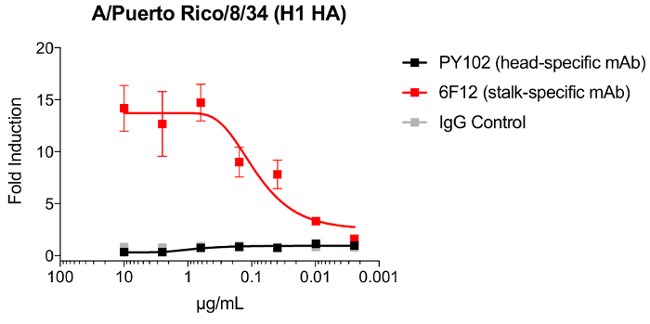
We demonstrated that a stalk-specific mAb, 6F12, but not head-specific a head-specific mAb, PY102, was able to activate primary natural killer cells by upregulating CD107a and interferon γ19. To model the ability of a stalk-specific antibody to activate primary human NK cells, we show that a stalk-specific mAb, 6F12, can robustly activate the modified Jurkat cell expressing the murine FcγRIV by ~14-fold. On the other hand, the head-specific mAb, PY102 and the control IgG do not (Figure 2)28.
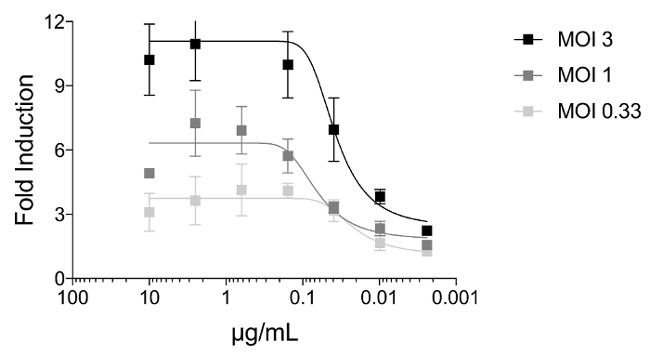
To investigate if the multiplicity of infection (MOI) can affect the induction of Jurkat cells expressing the murine FcγRIV, we infected MDCK cells with X31 (a reassortant influenza A virus expressing the hemagglutinin and neuraminidase of A/Hong Kong/1/68 (H3N2) virus with the internal segments of A/Puerto Rico/8/34 (H1N1)) with an MOI of 3, 1, or 0.33. Twenty four hours later, Jurkat cells expressing murine FcγRIV and mAb 9H10 were added unto the infected MDCK cells. In Figure 3, we show that cells infected with an MOI of 3 can induce the luminescence around 3-fold higher than MDCK cells infected with an MOI of 0.33.
Figure 1: A schematic of a sample plate layout and composition of the experimental groups. (A) The starting concentration for each antibody sample is at 10 µg/mL and serially diluted across the plate (column 1 to column 12) 4-fold. Each sample is done in triplicates. Row G is reserved for the no mAb control and Row H is background. (B) Final volume for each experimental group before addition of luciferase substrate buffer is 75 µL. Of note, the sample and ‘no mAb groups’ have 25 µL of effector cells added, while the background control groups have 75 µL of assay media. Please click here to view a larger version of this figure.
Figure 2: Induction of modified Jurkat cells expressing FcgR by a stalk monoclonal antibody. A549 cells were infected with A/Puerto Rico/8/34 (H1N1) with an MOI of 3. At 24 hours post infection, murine FcγRIV-expressing Jurkat cells in combination with PY102, 6F12 or a IgG control were added. Induction of Jurkat cells were assessed by luminescence signal generated by firefly luciferase under the NFAT response element. Error bars = standard deviations. Please click here to view a larger version of this figure.
Figure 3: The multiplicity of infection affects induction of a stalk-specific antibody. MDCK cells were infected with X31 (H3N2) with an MOI of 3, 1, or 0.33. At 24 h post infection, FcgR-expressing Jurkat cells in combination with 9H10 (starting dilution of 10 µg/mL) added. Induction of Jurkat cells were assessed by luminescence signal generated by firefly luciferase under the NFAT response element. Error bars = standard deviations. Please click here to view a larger version of this figure.
Discussion
The method described here allows the user to measure the ability of an HA-specific monoclonal antibody to engage the murine FcγRIV. The target antigen, influenza virus hemagglutinin, is expressed on the surface of cells after infection by virus or transfection of plasmid DNA. The assay is further amenable to using different surface-expressed viral proteins in combination with other FcγRs (humans or murine). Additionally, we have used sera samples to assess the ability of antibodies in a polyclonal context to induce Fc effector functions (data not shown), which may provide more relevant information when studying the adaptive immune response induced by infection or vaccination. We additionally speculate that the assay can be modified to measure the induction of antibodies that recognize internal viral proteins by directly coating the plates with soluble protein. However, this methodology has not been rigorously tested.
Induction of antibody-dependent effector functions can be affected by several factors including but not limited to the effector to target ratio (number of effectors cells added per well), the concentration of antibody used, the length of incubation between effector and target cells, and the concentration of low IgG serum used in the assay buffer.
Historically, the standard assay to measure the antibody-dependent cellular cytotoxicity activity of antibodies or polyclonal sera involves primary natural killer (NK) cells harvested from donors. Engagement of FcγR and activation of the intracellular pathway is typically determined by the expression of activation markers CD107a and/or IFN-γ19,23,24,31. Although the classical assay uses more relevant effector cells as they have not been genetically engineered, donor variation due to allelic differences in the FcγR may affect NK cell activation and result in batch to batch variation. Alternative assays used as surrogates for ADCC activity include an FcγR dimer-based ELISA or a chromium release killing assay25. Though the use of an FcγR dimer-based ELISA is high throughput, the assay may not be physiologically relevant as this assay does not use effector cells32. While the chromium release assay measures direct killing, it is not high throughput and requires primary NK cells which again are donor dependent33. One recent method describe the use of murine T-cell hybridomas stably expressing chimeric FcγR-CD3ζ chain molecules and measures activation through the secretion of IL-2. The assay described is similar to the described method here, which makes use of a modified T-cell as a surrogate for an effector cell expressing a singular activating FcγR34. Alternatively, a cell line (NK92.05-CD16) expressing the FcγRIIIa can be used as the effector cell and upregulation of the degranulation antigen CD107a is used as an activation marker. However, the use of virions and viral peptides on ELISA plates on the latter protocol should be noted as proteins coated onto hydrophobic surfaces may not always recapitulate the physiological structure of antigens35.
The method described here is a 96-well format assay, makes use of physiologically relevant and natively expressed viral targets and can be modified for murine or human antibodies. The genetically modified Jurkat cells can be maintained in culture and cryopreserved36. Though currently the effector cells express only one activating FcγR, the many advantages of using this protocol described in this study should be taken into consideration when measuring antibody-dependent cell-mediated effector function.
Disclosures
Authors declare no conflicts of interest.
Acknowledgments
This project has been funded in part with federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under CEIRS contract P01AI097092-04S1 (P.E.L.).
References
- Hobson D, Curry RL, Beare AS, Ward-Gardner A. The role of serum haemagglutination-inhibiting antibody in protection against challenge infection with influenza A2 and B viruses. J Hygiene. 1972;70(04):767–777. doi: 10.1017/s0022172400022610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Throsby M, et al. Heterosubtypic neutralizing monoclonal antibodies cross-protective against H5N1 and H1N1 recovered from human IgM+ memory B cells. PloS One. 2008;3(12):e3942. doi: 10.1371/journal.pone.0003942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekiert DC, et al. Antibody recognition of a highly conserved influenza virus epitope. Science. 2009;324(5924):246–251. doi: 10.1126/science.1171491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekiert DC, et al. A Highly Conserved Neutralizing Epitope on Group 2 Influenza A Viruses. Science. 2011;333(6044):843–850. doi: 10.1126/science.1204839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan GS, et al. A pan-H1 anti-hemagglutinin monoclonal antibody with potent broad-spectrum efficacy in vivo. J Virol. 2012;86(11):6179–6188. doi: 10.1128/JVI.00469-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan GS, et al. Characterization of a Broadly Neutralizing Monoclonal Antibody That Targets the Fusion Domain of Group 2 Influenza A Virus Hemagglutinin. J Virol. 2014;88(23):13580–13592. doi: 10.1128/JVI.02289-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesen RHE, et al. A common solution to group 2 influenza virus neutralization. PNAS. 2014;111(1):445–450. doi: 10.1073/pnas.1319058110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfus C, et al. Highly Conserved Protective Epitopes on Influenza B Viruses. Science. 2012;337(6100):1343–1348. doi: 10.1126/science.1222908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krammer F, Palese P, Steel J. Advances in Universal Influenza Virus Vaccine Design and Antibody Mediated Therapies Based on Conserved Regions of the Hemagglutinin. Curr Top Microb and Immunol. 2014. Chapter 408. [DOI] [PubMed]
- Impagliazzo A, et al. A stable trimeric influenza hemagglutinin stem as a broadly protective immunogen. Science. 2015. [DOI] [PubMed]
- Nachbagauer R, Krammer F. Clinical Microbiology and Infection. Clin Microb Infect. 2017. pp. 1–7. [DOI] [PMC free article] [PubMed]
- Krammer F. Novel universal influenza virus vaccine approaches. Current opinion in virology. 2016;17:95. doi: 10.1016/j.coviro.2016.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steel J, et al. Influenza virus vaccine based on the conserved hemagglutinin stalk domain. mBio. 2010;1(1) doi: 10.1128/mBio.00018-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krammer F, Pica N, Hai R, Margine I, Palese P. Chimeric hemagglutinin influenza virus vaccine constructs elicit broadly protective stalk-specific antibodies. J Virol. 2013;87(12):6542–6550. doi: 10.1128/JVI.00641-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krammer F, Pica N, Hai R, Tan GS, Palese P. Hemagglutinin Stalk-Reactive Antibodies Are Boosted following Sequential Infection with Seasonal and Pandemic H1N1 Influenza Virus in Mice. J Virol. 2012;86(19):10302–10307. doi: 10.1128/JVI.01336-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hai R, et al. Influenza viruses expressing chimeric hemagglutinins: globular head and stalk domains derived from different subtypes. J Virol. 2012;86(10):5774–5781. doi: 10.1128/JVI.00137-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunand CJH, et al. Both Neutralizing and Non-Neutralizing Human H7N9 Influenza Vaccine-Induced Monoclonal Antibodies Confer Protection. Cell Host and Microbe. 2016;19(6):800–813. doi: 10.1016/j.chom.2016.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan GS, et al. Broadly-Reactive Neutralizing and Non-neutralizing Antibodies Directed against the H7 Influenza Virus Hemagglutinin Reveal Divergent Mechanisms of Protection. PLoS Path. 2016;12(4):e1005578. doi: 10.1371/journal.ppat.1005578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiLillo DJ, Tan GS, Palese P, Ravetch JV. Broadly neutralizing hemagglutinin stalk-specific antibodies require FcγR interactions for protection against influenza virus in vivo. Nat Med. 2014;20(2):143–151. doi: 10.1038/nm.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiLillo DJ, Palese P, Wilson PC, Ravetch JV. Broadly neutralizing anti-influenza antibodies require Fc receptor engagement for in vivo protection. J Clin Invest. 2016;126(2):605–610. doi: 10.1172/JCI84428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter CW, Oxford JS. Determinants of immunity to influenza infection in man. Brit Med Bull. 1979;35(1):69–75. doi: 10.1093/oxfordjournals.bmb.a071545. [DOI] [PubMed] [Google Scholar]
- Memoli MJ, et al. Evaluation of Antihemagglutinin and Antineuraminidase Antibodies as Correlates of Protection in an Influenza A/H1N1 Virus Healthy Human Challenge Model. mBio. 2016;7(2):e00417. doi: 10.1128/mBio.00417-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jegaskanda S, et al. Age-associated cross-reactive antibody-dependent cellular cytotoxicity toward 2009 pandemic influenza A virus subtype H1N1. J Infect Dis. 2013;208(7):1051–1061. doi: 10.1093/infdis/jit294. [DOI] [PubMed] [Google Scholar]
- Jegaskanda S, et al. Cross-Reactive Influenza-Specific Antibody-Dependent Cellular Cytotoxicity Antibodies in the Absence of Neutralizing Antibodies. J Immunol. 2013;190(4):1837–1848. doi: 10.4049/jimmunol.1201574. [DOI] [PubMed] [Google Scholar]
- Jegaskanda S, Wheatley AK, Kent SJ. ScienceDirectAntibody-dependent cellular cytotoxicity and influenza virus. Curr Opin Virol. 2017;22(C):89–96. doi: 10.1016/j.coviro.2016.12.002. [DOI] [PubMed] [Google Scholar]
- Srivastava V, et al. Identification of Dominant Antibody-Dependent Cell-Mediated Cytotoxicity Epitopes on the Hemagglutinin Antigen of Pandemic H1N1 Influenza Virus. J Virol. 2013;87(10):5831–5840. doi: 10.1128/JVI.00273-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W, et al. Epitope specificity plays a critical role in regulating antibody-dependent cell-mediated cytotoxicity against influenza A virus. PNAS. 2016;113(42):11931–11936. doi: 10.1073/pnas.1609316113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leon PE, et al. Optimal activation of Fc-mediated effector functions by influenza virus hemagglutinin antibodies requires two points of contact. PNAS. 2016. p. 201613225. [DOI] [PMC free article] [PubMed]
- Benjamin E, et al. A Broadly Neutralizing Human Monoclonal Antibody Directed against a Novel Conserved Epitope on the Influenza Virus H3 Hemagglutinin Globular Head. J Virol. 2014;88(12):6743–6750. doi: 10.1128/JVI.03562-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekiert DC, Kashyap AK, et al. Cross-neutralization of influenza A viruses mediated by a single antibody loop. Nature. 2013;488(7417):526–532. doi: 10.1038/nature11414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W, et al. Broadly Neutralizing Anti-Influenza Virus Antibodies: Enhancement of Neutralizing Potency in Polyclonal Mixtures and IgA Backbones. J Virol. 2015;89(7):3610. doi: 10.1128/JVI.03099-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristensen AB, et al. Antibody Responses with Fc-Mediated Functions after Vaccination of HIV-Infected Subjects with Trivalent Influenza Vaccine. J Virol. 2016;90(12):5724–5734. doi: 10.1128/JVI.00285-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg SB, Criswell BS, Six HR, Couch RB. Lymphocyte cytotoxicity to influenza virus-infected cells. II. Requirement for antibody and non-T lymphocytes. J Immunol (Baltimore, Md. : 1950) 1977;119(6):2100–2106. [PubMed] [Google Scholar]
- Corrales-Aguilar E, Trilling M, et al. A novel assay for detecting virus-specific antibodies triggering activation of Fcγ receptors. J Immunol Meth. 2013;387(1-2):21–35. doi: 10.1016/j.jim.2012.09.006. [DOI] [PubMed] [Google Scholar]
- de Vries RD, et al. Influenza virus-specific antibody dependent cellular cytoxicity induced by vaccination or natural infection. Vaccine. 2017;35(2):238–247. doi: 10.1016/j.vaccine.2016.11.082. [DOI] [PubMed] [Google Scholar]
- Cheng Z, et al. Development of a robust reporter-based ADCC assay with frozen, thaw-and-use cells to measure Fc effector function of therapeutic antibodies. J Immunol Meth. 2014;414(1):69–81. doi: 10.1016/j.jim.2014.07.010. [DOI] [PubMed] [Google Scholar]


