Abstract
The key question of precision medicine is whether it is possible to find clinically actionable granularity in diagnosing disease and classifying patient risk. The advent of next generation sequencing and the widespread adoption of electronic health records (EHRs) have provided clinicians and researchers a wealth of data and made possible the precise characterization of individual patient genotypes and phenotypes. Unstructured text — found in biomedical publications and clinical notes — is an important component of genotype and phenotype knowledge. Publications in the biomedical literature provide essential information for interpreting genetic data. Likewise, clinical notes contain the richest source of phenotype information in EHRs. Text mining can render these texts computationally accessible and support information extraction and hypothesis generation. This chapter reviews the mechanics of text mining in precision medicine and discusses several specific use cases, including database curation for personalized cancer medicine, patient outcome prediction from EHR-derived cohorts, and pharmacogenomic research. Taken as a whole, these use cases demonstrate how text mining enables effective utilization of existing knowledge sources and thus promotes increased value for patients and healthcare systems. Text mining is an indispensable tool for translating genotype-phenotype data into effective clinical care that will undoubtedly play an important role in the eventual realization of precision medicine.
Keywords: Precision medicine, Text mining, Genotype, Phenotype, EHR, NLP, Database curation, Cancer, Outcome prediction, Pharmacogenomics, Biomedical literature, Value
1.1 Introduction
The precision medicine ideal is that data about genes, environment, and lifestyle can enable optimal patient care by allowing physicians to customize each person’s treatment to reflect these unique health determinants. Governments and healthcare organizations around the globe have taken interest in this ideal, with the recent notable instance of the United States Precision Medicine Initiative (PMI) announced by President Barack Obama in January 2015 [1]. Prior to President Obama’s announcement, many countries, including China, the UK, Iceland, Japan, Canada and others, had established infrastructures for precision medicine research through the development of biobanks (repositories of patient DNA with accompanying databases that link the medical history and lifestyle information of donors to their biologic samples) [2]. Precision medicine is thus a global hope founded in the belief that it is possible to harness big data in healthcare and biology to promote health and relieve suffering.
The core challenge of precision medicine (PM) is that of classification: is it possible to discern differences between individuals in a heterogeneous population that can guide treatment decisions and support improved care? Which people, for example, are going to develop cancer? What medications will treat their cancers most effectively? What is it about patient A that makes her fundamentally distinct from patient B, and how should doctors tailor the care of these patients to reflect these distinctions? These questions have always been relevant to clinical practice, but a tradeoff has always existed between increasing information and decreasing clinical utility of that information. Precision medicine is a relevant concept now because technology has advanced in key areas of medicine to such a degree that it is possible that precise classification of individuals may indeed enable clinically effective, personalized treatment.
The last decade witnessed the birth of two key sources of data with great promise to enable the precise classification of individuals for medical care: the sequencing of the human genome with accompanying improvements in sequencing technology, and the widespread adoption of the electronic health record (EHR). For both these data sources, much of the information required to conduct precision medicine is contained within unstructured, written texts such as the biomedical literature and clinical notes. In its current state, this information is not computable; hence “unlocking” this information via natural language processing (NLP) is an essential and truly exciting area of study.
This chapter is about text mining for precision medicine (TM for PM). Text mining is a subfield of NLP dedicated to enabling computational analysis of text-locked data. The text mining workflow generally involves identification of specific entities in surface text such as diseases, genes, or relational terms and the deep normalization of these entities to standardized ontologies. Data thus processed become the input values for a variety of computations. There are two core functions of text mining: (1) information extraction and (2) hypothesis generation via relationship extraction.
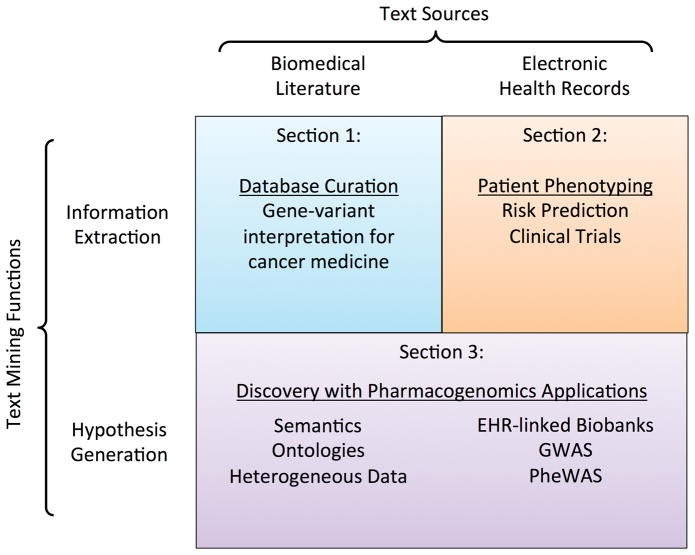
In this chapter (see Figure 1), we devote two sections to the information extraction functionality of text mining. The first section addresses mining biomedical literature for the purpose of assisting database curation in personalized cancer medicine. The second addresses mining EHRs for the purpose of cohort identification. The third section of this chapter explores the role of TM for PM as a vehicle for hypothesis generation and ties the previous two sections together with a discussion of methods of using biomedical literature and EHR texts to conduct pharmacogenomic research.
Figure 1.
The structure of this chapter reflects the two core functions of text mining and the two foremost text sources related to precision medicine. Section 1 discusses how text mining published literature can facilitate curation of genotype-phenotype databases for support of personalized cancer medicine. Section 2 discusses how text mining is useful in defining patient phenotypes from EHRs. Section 3 is about using text mining of both text sources for hypothesis generation in pharmacogenomics.
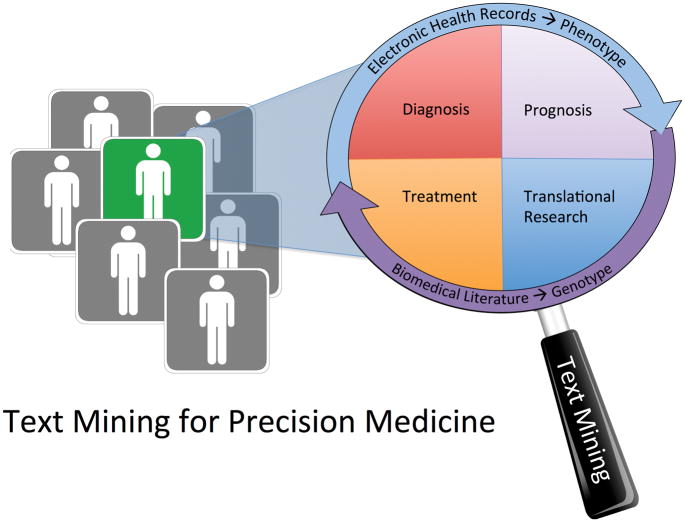
Two key terms to any discussion of precision medicine are the terms “genotype” and “phenotype”. These terms can be confusing to people who are new to genetics research because the meaning of both terms is contextual. Genotype, for example, can refer to the entirety of an individual’s unique assortment of genes, or it can refer to a specific variant of a single gene that distinguishes an individual from others. Likewise the term phenotype can be defined as broadly or as narrowly as context demands (a specific disease, such as age-related macular degeneration (AMD), could be considered a phenotype, but within a group of people with AMD, the presence or absence of specific findings such as aberrant blood vessel growth could also be considered a phenotype). In this chapter, we discuss the biomedical literature as an authoritative source of genotype information, and we discuss electronic health records as a dynamic resource of human phenotypes (see Figure 2).
Figure 2.
Text mining brings unstructured information into focus to characterize genotypes and phenotypes in precision medicine.
2.1 TM for PM: Personalized Cancer Medicine and Database Curation
One area where PM has already demonstrated value and great promise is the field of cancer medicine. This is why the near-term focus of the United States Precision Medicine Initiative is cancer diagnosis and treatment [1].
Cancer is a collection of diseases, all of which involve the development of a population of cells in the body that gain the potential to replicate infinitely. Cancers are typically named after the location of the cells that have undergone this transformation (e.g. “breast cancer” if the altered cells were initially breast cells). Cancer of any form is a disease of the genome [3, 4]. The changes that lead to cancer development occur within the DNA sequence and result in the removal of physiologic protections against cancer (loss of function mutations) and the production of new stimuli that promote cellular growth (gain of function mutations). Doctors are hopeful that genomics-driven precision medicine will be particularly effective in treating cancer because of the genetic nature of the disease process [1, 5] and because of the demonstrated effectiveness of therapies directed precisely at the genomic alterations that cause cancer [6].
Text mining has an intuitive place in the conceptual framework for the implementation of personalized cancer medicine, which involves (1) characterization of the “driver” mutations in a given patient’s tumor; and (2) identification of the drugs that will best counteract the effects of those driver mutations [5]. Both of these steps are information extraction tasks, which is a key function of text mining. Additionally, much of the information needed for performing these two tasks is contained within the biomedical literature. This section will discuss the current issues and science behind text mining for assisting database curation in personalized cancer medicine.
2.2 Considerations for text mining in database curation
The biomedical literature helps clinicians and researchers interpret genetic information, and many databases in the cancer domain include literature references. Some prominent databases with literature curations include the Catalogue Of Somatic Mutations in Cancer (COSMIC), Online Mendelian Inheritance in Man (OMIM), ClinVar, ClinGen (the proposed manually curated component of ClinVar), SwissProt, the Human Gene Mutation Database (HGMD), the Pharmacogenomics Database (PharmGKB), and the Comparative Toxicogenomics Database (CTD). All the above databases are examples of genotype-phenotype databases[7]. The gold standard of quality in the curation of literature references for these databases is manual expert curation, but there is an indisputable need for text mining tools to assist in the curation process. Baumgartner et al elegantly illustrated this need by applying a found/fixed graph to examine the curation completeness of two databases – Entrez Gene and Swiss Prot. Ignoring the pace of new publications, they instead examined the numbers of missing entities within these two databases and compared the rate of generation of missing data annotations over time to the rate of resolution of these missing data points. They concluded that neither database would ever “catch up” to the pace of generation of information without changes to their curation processes [8]. The rate of biomedical discovery exceeds the curation capacity of these comprehensive resources.
Although it is true that the pace of article publication exceeds human curation capabilities, it is a fallacy to conclude that assimilation of all new information is necessary. In our experience, text mining applications are most likely to be adopted by domain experts such as clinicians, researchers and curation teams when the applications correctly limit the amount of information they return. Curators of databases may recognize a need to increase the pace and breadth of their curation efforts, but their intent is not to curate all new articles but rather to curate only the articles that further their institutional goals [9]. As an example, compare two high-quality genotype-phenotype databases, SwissProt and ClinVar. Both databases contain information about diseases associated with protein/gene sequence variations, but their institutional scope is different. The chief aim of SwissProt is to identify variants that alter protein function, while the chief aim of ClinVar is to provide evidence of the relationship between human genetic variants and phenotypes. Because of this difference, an article demonstrating a causative association between a variant and a given disease would likely rank higher in priority for curation in ClinVar (or its manually curated partner, ClinGen) than in SwissProt. To be useful to domain experts and database curators, text mining tools must balance (1) comprehensive analysis of the literature; with (2) filtering tools for ranking and identifying the most useful literature.
2.3 Text Mining in Database Curation
The curation workflow for genotype-phenotype databases involves three important steps where text mining can play a crucial role [12, 13]: (1) information retrieval and document triage; (2) named entity recognition and normalization; and (3) relation extraction. Figure 3 provides a schematic overview of this process. For an excellent treatment of the entire workflow, we direct readers to the review by Hirschman et al [10]. In the remainder of this section we will discuss the latter two aspects of the workflow.
Figure 3.
Genotype data permits incredibly deep classification of individuals. The biomedical literature contains a wealth of information regarding how to clinically interpret genetic knowledge. Text mining can facilitate expert curation of this information into genotype-phenotype databases.
An important step in curating genotype-phenotype databases is identifying relevant entities within text. A wide variety of entities are appropriate for text mining in precision medicine, including genes, gene variants, chemical/drug names, species, cohort types, diseases, or even other biological concepts such as analysis techniques or evidence levels. Tagging these entities is called named entity recognition (NER), and mapping tagged entities to standard vocabularies is called normalization. NER and normalization constitute the second step of the biocuration workflow. Mining named entities from text is challenging because of the variability of natural language expressions. We will discuss identification of three core entities for genotype-phenotype database curation: genes, variants, and diseases.
Genes
Several forms of natural language variation complicate gene NER: orthographical variations (e.g., “ESR1” vs “ESR-1”), morphological variations (e.g., “GHF-1 transcriptional factor” vs “GHF-1 transcription factor”), and abbreviations (e.g., “estrogen receptor alpha (ER)”). Ambiguity also arises in texts that discuss multiple species, since separate species frequently share genes of the same name with distinct sequences (e.g., ERBB2 can be either a human gene or mouse gene name). It is also possible for different genes to share the same name. For example, “AP-1” can refer to either the “jun proto-oncogene” (Entrez Gene: 3725) or the “FBJ murine osteosarcoma viral oncogene homolog” (Entrez Gene: 2353). GNormPlus is a state-of-the-art, open-source text mining system for gene NER and normalization [11]. GNormPlus utilizes multiple sophisticated text mining techniques and achieves a balanced performance in recall and precision. (Please see the text box, “Text Mining Performance Metrics” , in the last section of the chapter for more information about precision and recall.)
Text Box. Text Mining Performance Metrics.
The three most common evaluation metrics for text mining tools are precision, recall, and F1-score [109]. These metrics apply equally to tools for processing clinical text and published literature. Precision and recall are also common metrics for evaluating clinical diagnostic tests. We describe each below.
Precision is the text mining equivalent of the clinical metric of positive predictive value and is equal to the ratio of true positives to all positive values from a test. In lay terms, recall answers the question, “How likely is it that the results of this tool actually reflect what I was searching for?” Tools with high precision scores have low numbers of false positives and can thus be trusted to produce only correct results.
Recall is the text mining equivalent of the clinical metric of sensitivity and is equal to the proportion of all true positives that a test detects. In lay terms, recall answers the question,
“Can I rely on this tool to identify all relevant entities?” Tools with high recall scores have low numbers of false negatives and are thus most reliable.
A tradeoff exists between precision and recall such that it is possible to improve the precision of any tool at the expense of recall and vice versa. For this reason it is common in text mining evaluations to use a composite metric called F1-score, which is the harmonic mean of precision and recall.
Variants
Gene variants and mutations are not uniformly reported with standard nomenclature in biomedical texts so identification and normalization of variant mentions is challenging. Variant/mutation normalization is also complicated by the fact that nomenclature standards have evolved over time as researchers have gained additional insights into genetic complexities. The state-of-the-art tool for variant extraction is tmVar [12].
Diseases
Literature mentions of diseases involve frequent use of abbreviations, synonyms, morphological or orthographical variations, and inconsistent word ordering. DNorm is an open-source tool for disease NER and normalization that maps concepts to the MEDIC vocabulary using a pairwise learning to rank machine learning algorithm [13].
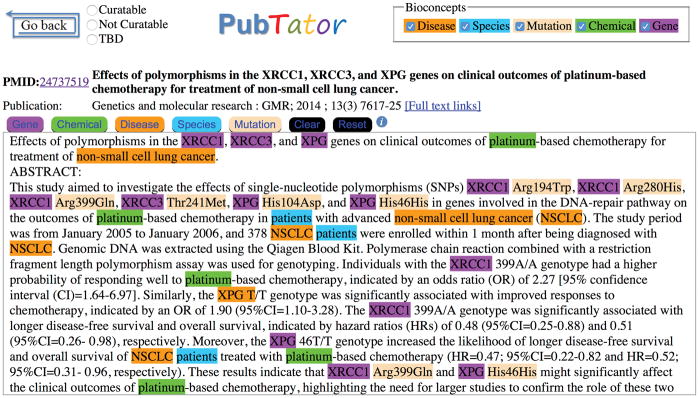
One of the most recent advancements in curation support is a tool produced by NCBI called PubTator [14] (see Figure 4). This web-based application incorporates multiple state-of-the-art tools, including all three NER tools discussed above, to support three curation tasks: document triage, NER and normalization, and relationship annotation. PubTator combines an an intuitive interface with comprehensive access to all references in PubMed, and is truly an excellent multipurpose tool for curation.
Figure 4.
This abstract includes examples of each of the five bio-entities that PubTator identifies. Note the correct identification of mentions to non-small cell lung cancer regardless of whether the text uses the full term or its abbreviation, NSCLC. Likewise, PubTator correctly interprets the term “patients” as a reference to a species, Homo sapiens. Although this abstract uses protein-level nomenclature to describe gene variants (e.g. “XRCC1 Arg399Gln”), the authors distinguish genotypes with nucleotides rather than amino acids (e.g. “the XRCC1 399A/A genotype”). This variability is an example of the challenges inherent to named entity recognition of gene mutations.
The identification of semantic relationships between entities is called relationship extraction and is considered the third step in the curation workflow. Conventionally, most relation extraction techniques have used co-occurrence metrics to relate two entities within the text (e.g. co-occurrence metrics make the assumption that if gene A and variant B are both in the same abstract, then variant B must be a variant of gene A). However, co-occurrence approaches ignore the textual content and result in many errors. For example, in variant-disease relationship extraction, co-occurrence methods will wrongly interpret negative results as support for an association. The high rate of false positives associated with co-occurrence methods significantly lowers the utility of these methods for genotype-phenotype database curation workflows. In response to these challenges, Singhal et al [15] developed a machine learning approach to extract disease-variant relations from biomedical text. Their machine learning approach learns patterns from the text to decide whether two entities co-occurring within the text have any stated relationship or association. The patterns are learned on six pre-defined features that capture both in-sentence and cross-sentence relation mentions. Negative findings within the text are taken into account using numeric sentiment descriptors. They demonstrate that machine learning delivers significantly higher performance than the previous co-occurrence-based state of the art [16].
2.4 Applications of text mining in personalized cancer medicine
Text mining plays an important role in the curation workflow of many cancer-related genotype-phenotype databases. For example, curators of SwissProt use a number of text-mining resources for document triage in their curation workflow, including TextPresso and iHOP [17]. In the Pharmacogenomics Database (PharmGKB) curators use an adaptation of the TextPresso tool, Pharmspresso, for information retrieval and document triage [18, 19]. The miRCancer database, which catalogs literature mentions of microRNA expression in cancer cells, uses a rule-based text mining approach to identify miRNA-cancer mentions for manual curation [20]. Two groups have used text mining to develop gene methylation databases for cancer research: MeInfoText [21] and PubMeth [22]. Still other groups have created cancer-specific databases using text mining. Examples of such databases include the Osteosarcoma Database [23] and the Dragon Database of Genes associated with Prostate Cancer (DDPC) [24].
One of the most prominent databases to use text mining to curate information related to precision cancer medicine is the Comparative Toxicogenomics Database (CTD), a publicly available database containing manually curated relationships between chemicals, genes, proteins, and diseases. CTD employs text mining to triage documents and identify entities in text for curation. They developed a metric called a document relevancy score to quantify how important a given literature reference might be to their curation goals, and they found that text mining reliably identifies articles that are most likely to provide the highest yield of new, relevant and biologically interpretable information [25]. CTD has also featured prominently in several BioCreative community challenges1. Track I of the BioCreative-2012 Workshop involved developing a document triage process with a web interface [27] for curation in CTD. Likewise, Track 3 of BioCreative IV involved developing web tools for performing named entity recognition on text passages using CTD’s controlled vocabulary [28]. More recently, the 2015 BioCreative V challenge included a chemical disease relation (CDR) task involving extraction of chemically induced diseases. The best systems in this task achieved an F1-score of 57% [29].
The need for using text mining in database curation is extremely strong. The interdisciplinary nature of precision cancer medicine and the volume of information relevant to customizing patient care necessitates the use of databases to integrate information and create broad access to it. The biomedical literature constitutes a relevant and authoritative source of information for such databases, and text mining can structure and summarize this information for rapid assimilation. As the science of text mining advances and tools become more robust and accurate, the immediate relevance of TM for PM will only increase.
3.1 TM for PM: EHRs and Phenotype Determination
The previous section discussed how text mining biomedical literature supports database curation and thus informs clinicians and researchers as they look deeply into individuals’ genotypes. In this next section, we consider a completely separate perspective of TM for PM — how text mining electronic health record (EHR) data can enable physicians to look broadly at an entire population by classifying patient phenotypes. Such patient phenotypes may be the most accurate means of representing the interplay of all the health determinants of precision medicine: genes, environment and lifestyle.
Consider the case of an actual clinical dilemma that occurred involving a teenage girl who was hospitalized for an autoimmune disorder called systemic lupus erythematosus (SLE or lupus)[30]. Several factors complicated this girl’s condition and predisposed her to forming blood clots. Although blood-thinning medications could protect against these clots, her providers were also concerned about paradoxical bleeding if they prescribed these medications. The key clinical question in this situation — whether to prescribe a blood thinner — was not readily answerable from published research studies or guidelines so her provider turned to her institution’s EHR and used its research informatics platform [31] to identify an ‘electronic cohort’ of pediatric patients who had in previous years experienced similar lupus exacerbations. From this cohort they identified a trend of complications from blood clots, which convinced them to administer anticoagulants.
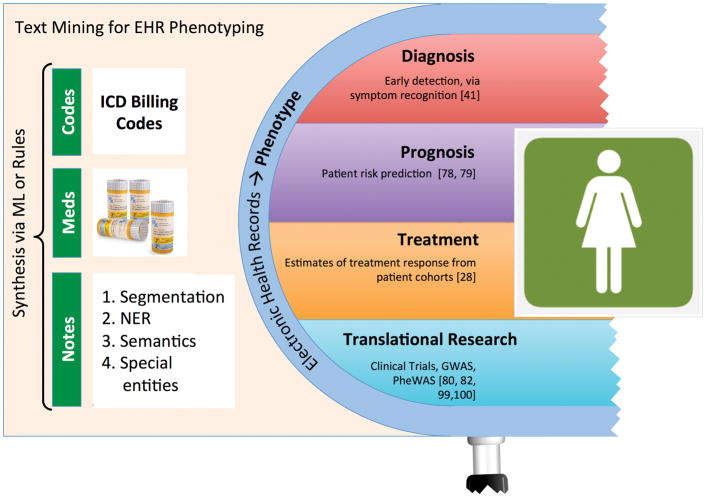
This story illustrates the potential of using EHR data to direct personalized care. The physicians in this case used EHR data to identify a group of similar patients with known outcomes. The outcome data then enabled them to estimate their patient’s risk and intervene to modify that risk. Although this analysis did not yield the statistical confidence of a formal clinical trial, the patient’s physicians felt that this EHR cohort analysis provided superior information than the alternatives — pooled opinion and physician recollection[30]. In a large healthcare system, EHR-derived cohorts can reflect the interplay of genes, environment, and lifestyle in the health outcomes of a specific group of patients. Many exciting applications of cohort identification from EHR data exist. This section will discuss two use cases: patient outcome prediction via patient similarity analytics, and cohort identification for clinical trials. Text mining is integral to the development of these applications because in many cases the richest and most diverse patient information in EHRs is contained in free, unstructured notes written by healthcare providers [32] (see Figure 5).
Figure 5.
EHRs are rich sources of phenotype information. Algorithms to extract phenotypes commonly incorporate text mining of clinical notes as well as billing codes and medications. In contrast to the deeply individual nature of genotype information, phenotype algorithms generate clinical insights by first looking broadly at aggregated populations of people with similar conditions and known health outcomes.
3.2 EHR phenotype determination
Identifying populations of people with shared health characteristics amounts to defining a phenotype. EHR data has been shown to be an effective source for comprehensive measurement of the phenotypic characteristics of populations [33]. Simply defined, EHRs are information systems that contain electronic records of all the key clinical and administrative data relevant to the medical history of each patient in a specific healthcare institution [34]. EHRs consist of both structured and unstructured data fields. Some of the key structured data fields include billing data, laboratory and vital signs, and medication records. Unstructured data is largely present in notes, of which there are two main types: clinical notes (e.g. history and physical notes or discharge summaries) and test results (e.g. radiology reports or pathology reports) [32]. EHR data (see Figure 6) is a promising source of phenotype information because (1) information in EHRs is relatively inexpensive to obtain since EHR data is generated as a byproduct of the healthcare process; (2) the scope of EHR data is vaster than the scope of any organized study — both in terms of the variety of pathology and in terms of longitudinal coverage; (3) the resolution of EHR data continually improves over time because additional encounters for any given individual lead to increased certainty regarding the presence or absence of a given diagnosis.
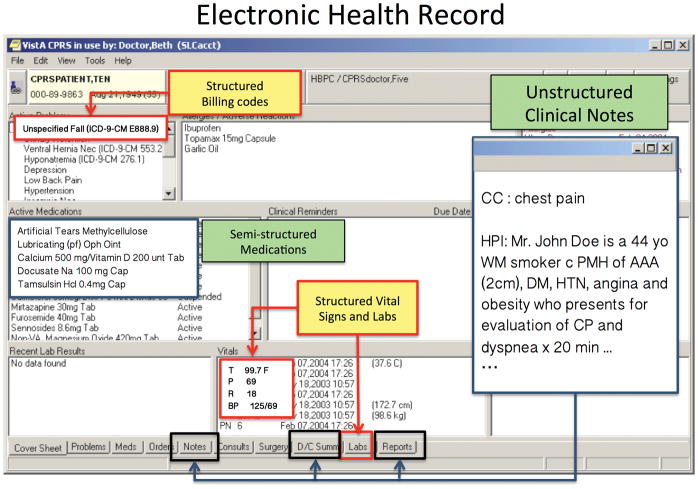
Figure 6.
The Veterans Information Systems and Technology Architecture (VISTA) is the most widely used EHR in the United States. Like most EHRs it contains structured data and unstructured text.
There are several noteworthy challenges inherent in using EHR data for precision medicine [35]. (1) EHR data consists of sensitive and highly confidential information with extensive legal protections [36] and real ethical pitfalls related to privacy [37]. (2) EHR data is incomplete for many reasons. One reason is that patients often utilize multiple healthcare systems (for example, a given patient may see specialists at different, unrelated hospitals), but separate systems do not share EHR data so information within EHRs can be fragmented [32]. (3) EHR data is complicated by multiple biases. The highest quality manner of collecting population phenotype data is through a prospective observational cohort, like the Framingham Heart Study (FHS) [38]. In comparison with such studies, EHR data mining, ranks much lower in an evidence hierarchy [39] and should be suspected of significant biases such as multiple confounders, selection bias (EHRs represent sick people more than healthy people), and measurement bias (e.g. documented physical exam findings may differ in reliability between physicians) [40].
In addition to the above challenges inherent to EHR data, the process of identifying patients with a specific diagnosis (i.e. defining a phenotype from an EHR) has its own attendant challenges. Sun et al illustrated these challenges in their work to identify people with the condition of high blood pressure (hypertension) from their institution’s EHR [41]. Physicians diagnose hypertension by observing blood pressures that consistently exceed a certain threshold over time. Even though many EHRs contain blood pressure measurements in structured formats, using blood pressure measurements alone to identify patients with the condition of hypertension is surprisingly inaccurate. This is because blood pressure measurements are inherently variable [42] (for example, blood pressures may rise in response to pain or anxiety) and are modifiable through treatment (i.e. the use of antihypertensive medication by people with the condition will result in normal blood pressure levels). Thus in the approach utilized by Sun et al to detect changes in HTN control status, detection models incorporating only aggregated blood pressure measurements identified many more false positives and false negatives than models that incorporated multiple features from the EHR.
Because of the complexity of determining phenotypes from EHR data, a common convention is to use billing data such as International Classification of Disease (ICD) codes and Current Procedural Terminology (CPT) codes for representing phenotypes [32]. These billing codes are universal between healthcare systems and are available in structured formats. Yet billing data alone is also insufficient for accurate representation of disease phenotypes [43]. The best performing approaches to identifying EHR phenotypes incorporate multiple data fields, including text mining [44]. Wei et al demonstrated the benefit of text mining in phenotype development in a study where they examined the advantages and utilities of billing codes, clinical notes (using text mining), and medications from EHRs in detection of ten separate disease phenotypes. They found that information collected from clinical notes through text mining offered the best average sensitivity (0.77) out of all individual components, whereas billing code data had a sensitivity of 0.67. They also found that the relevance of using text mining of clinical notes in identifying phenotypes varied by disease (e.g. 84% of all information in the EHR necessary for identifying rheumatoid arthritis was contained in clinical notes whereas only 2% of information needed for identifying atrial fibrillation was contained in notes) [45]. Ultimately, the highest F1-scores resulted from combinations of two or more separate EHR components, such as billing data and text mining. In general, incorporating text mining in phenotype algorithms results in improvements in both the precision and recall [32].
3.3 Phenotype extraction from EHR text
In this section, we present a classification of the text mining approaches used in information extraction for EHR phenotype development, and we provide a brief overview of phenotyping. For a more comprehensive treatment of the entire approach for identifying patient phenotype cohorts from EHRs, please see the review by Shivade et al [46].
Text in clinical notes differs from text in biomedical literature in several ways. The foundational difference is that biomedical literature contains formal, peer-reviewed writing whereas clinical notes are comparatively informal. Physicians write clinical notes with the goal of maximizing time efficiency so clinical notes contain heavy use of abbreviations, often only decipherable from contextual cues. For legal reasons, modification of existing clinical notes is not permitted, yet due to time constraints, physicians often submit notes with only cursory proofreading. Consequently, spelling errors and unconventional sentence structures are common. Lastly, clinical notes frequently contain copied and pasted sections of values or text such as lab findings or vital signs, which complicate parsing of notes [47].
The extraction of relevant information from EHR texts involves four steps: (1) text segmentation; (2) entity identification and normalization; (3) evaluating for semantic modifiers such as negation and possibility phrases; (4) extraction of special entities from text.
Text segmentation. Free text from clinical notes needs to be segmented into fundamental units called “tokens” before any further processing takes place. Text segmentation is done using NLP parsers and tokenizers. cTAKES and the GATE framework are examples of open source NLP parsers designed for handling clinical text [48]. Several commercial options are also available.
NER and normalization. The most common approach for entity identification is to map tokens from segmented text into a target dictionaries such as SNOMED-CT, ICD, UMLS etc. Tools for mapping concepts in biomedical literature such as MetaMap can also process tokenized clinical texts [49, 50], but other clinically customized tools exist [51, 52] such as the HITex system [53] and the Knowledge Map Concept Identifier [53, 54].
Evaluation for semantic modifiers. Words that modify the semantic meaning of sentences are important for accurate phenotyping. For example, consider the significance of the word “no” in the following list that might appear in a provider’s note for a patient with chest pain: “no history of heart palpitations, dizziness/fainting, or tobacco use.” In a similar sense, identifying possibility phrases or status keywords such as “confirmed”, “possible”, “probable” etc. is very helpful.
Special entity extraction. Other important entities in free text include special keywords such as numerical measurements and units [55], dates and time-related words (e.g. “before”, “during” or “after”)[56] and phenotype-specific keywords [57].
3.3 Phenotype algorithm development
The entities extracted from clinical texts serve as inputs along with other structured EHR data such as billing codes and medications for phenotype algorithms. It is also worth acknowledging that several studies have looked at external sources such as imaging data [58, 59], drug characterization databases [60] and scientific articles from biomedical literature [61] to extract EHR phenotypes. Regardless of the data types used, phenotype algorithm development for EHRs involves identifying relevant features and then synthesizing those features — either through the application of expert-derived rules or through ML.
Rule application
For some conditions, clear clinical guidelines exist to guide phenotype algorithm development. The breathing disorder asthma is one such condition [62]. One way of defining a phenotype algorithm is to incorporate these clinical guidelines into a set of rules that can guide identification of patients with a given condition. Wu et al took this approach in modifying the asthma diagnostic criteria and developing a set of rules to identify patients with definite and probable asthma [43]. Other conditions also lend to rule-based algorithms. Nguyen et al [63] used a set of rules to build a classification system to identify lung cancer stages from free text data from pathology reports. Schmiedeskamp et al [64] used a set of empirical rules to combine ICD-9-CM codes, labs and medication data to classify patients with nosocomial Clostridium difficile infection. A few studies such as those by Kho et al [65], Klompas et al [66], Trick et al [67] and Mathias et al [68]] have used guidelines published by organizations such as the American Diabetes Association, the Centers for Disease Control and Prevention, the American Cancer Society and other trusted organizations to develop rule-based algorithms.
Machine Learning
As opposed to rule-based techniques where expert knowledge determines parameter significance, ML techniques identify patterns (not necessarily rules) from data. ML techniques are ideal for extracting phenotypes when rules are either not available or not comprehensive. There are two steps to ML phenotype algorithm development: feature selection and model building. “Feature selection” refers to the identification of parameters to use in ML algorithms. Establishing a sufficiently robust feature set prior to model building is important for achieving the best performance. We encourage the readers to read Bishop [69] for this topic.
Researchers have experimented with several ML models including probability [61, 70, 71], decision tree [58, 72, 73], discriminant [54, 70] and other types of ML models [52, 74, 75] to build phenotype categorization systems. In each case, the models essentially approach phenotyping as a classification or categorization problem with a positive class (the target phenotype) and a negative class (everything else). No consensus exists about which ML model is best for phenotype algorithm development. Wu et al, who developed the rule-based algorithm for identifying patients with asthma that we previously discussed, also developed an algorithm for asthma detection using ML. They chose a decision tree model and compared the results of their ML and rule-based algorithms. Both algorithms substantially outperformed a phenotyping approach using ICD codes alone, and the ML algorithm performed slightly better than the rule-based algorithm across all performance metrics. [43].
Phenotype extraction from EHRs is a challenging task that has become the rate-limiting step for applications of EHR phenotypes; nevertheless, broad, flexible phenotyping for point-of-care uses like patient outcome prediction is achievable. One factor that may aid in broad phenotype determination is the transferability of phenotype algorithms from one institute to another. The EMERGE network — a collaboration between healthcare systems with EHR-linked biobanks — demonstrated this transferability in a study where separate healthcare systems measured the performance of phenotype algorithms developed by a primary site at other sites within the network. They found that the majority of algorithms transferred well despite dramatic differences in EHR structures between sites. The majority of algorithms yielded PPV values above 90% [76]. Many phenotype algorithms are publicly available at the Phenotype Knowledgebase, an online repository of algorithms produced in partnership with the eMERGE Network and the NIH Collaboratory [77]. Other groups have investigated the possibility of automatic, high-throughput phenotype development. Yu et al employed a penalized logistic regression model to identify phenotypic features automatically and generated algorithms for identifying cases of rheumatoid arthritis and coronary artery disease cases using data obtained through text mining of EHRs. Their approach demonstrated comparable accuracy to algorithms trained with expert-curated features such as those in the previously mentioned study by the EMERGE network [78].
3.4 Patient outcome prediction through similarity analytics
Widespread interest exists in utilizing EHR phenotype data to produce point-of-care tools for clinicians [79]. One potential function is patient outcome prediction through similarity analytics. This is the use case of EHR phenotypes that we presented at the beginning of this section. Patient outcome prediction is a relatively new field of study. Consequently, even though text mining is an important component of the phenotype-determination algorithms that such prediction tools require, relatively few studies have examined the role of text mining in end-to-end risk prediction models. Most studies focus on either patient risk prediction or phenotype definition [46]. Regarding the former, Lee et al showed the potential benefit of phenotype-derived predictions in their work developing a patient similarity metric to quantify the degree of similarity between patients in an intensive care unit for the purpose of predicting 30-day mortality [80]. Their model demonstrated that using data from a relatively small (~100) subset of patients who possessed the greatest degree of similarity delivered the best predictive performance. One notable study has used text mining in an end-to-end fashion with patient outcome prediction: Cole et al used the NCBO Annotator to process clinical notes related to juvenile idiopathic arthritis (JIA) and employed this information in combination with billing code data to predict risk of developing uveitis (a vision-threatening complication of JIA) [81]. The field of patient similarity analytics using phenotype detection to drive outcome prediction is an exciting field of research where TM for PM promises great results.
3.5 Clinical Trial Recruitment
Another application of text mining for phenotype definition from EHRs is automated clinical trial eligibility screening. Precise phenotype cohort identification facilitates improvements in both the effectiveness (identifying the best patients) and efficiency (enrolling the most patients) of clinical trial recruitment. The key goals of this process are to (1) identify those populations who meet the inclusion criteria for a study, and (2) facilitate the most efficient workflow for enrollment of those patients into the correct trial [82]. Ni et al showed that a text-mining-based screening system could accomplish both goals [83]. They identified patient phenotypes using text mining of notes and billing code data to enable automated patient screening. At the same time, they obtained from ClinicalTrials.gov the narrative text of eligibility criteria of the trials being conducted at their institution and used NLP to extract pattern vectors for each clinical trial. They used these vectors to identify which trials would best fit a given patient. Ultimately, their process reduced workload of physicians in screening patients for clinical trials by 85%. Many other studies have shown similar benefits [82].
Another exciting application of precision EHR phenotyping related to trial recruitment is that of automated point-of-care clinical trial generation using EHRs [79]. Conducting randomized interventional studies using the existing infrastructure of the EHR involves building point-of-care tools into the EHR that will activate an enrollment process when a clinician is faced with a clinical decision where medical knowledge is insufficient to guide care [84] — for example, consider hypothetically the case study of the teen with lupus at the beginning of this section. In her situation, if an EHR trial was in place regarding the use of anticoagulation in a lupus exacerbation, and if the patient and her physician were truly ambivalent about what the right choice was, after she provided consent, the EHR would randomize the patient to one intervention or the other (i.e. give anticoagulants or withhold them). All subsequent follow-up would be purely observational through the data recorded in the course of the patient’s care. The randomized intervention in this type of study resolves some of the issues of confounding associated with observational data, and as such, this type of randomized observational study falls between observational studies and randomized trials in an evidence hierarchy. Vickers and Scardino proposed that this model might be applied in four areas: comparison of surgical techniques, ‘me too’ drugs, rare diseases, and lifestyle interventions [85]. Point-of-care clinical trials are an application of EHR phenotyping that might be the only cost-effective way to effectively study a large number of clinical questions related to precision medicine.
4.1 TM for PM: Hypothesis Generation
Please insert the Text Box - “Text Mining Performance Metrics” somewhere in this section
Text mining is useful for identifying genotypes and phenotypes from biomedical literature and EHRs. Yet information extraction is not the only function of TM for PM. Text mining tools can also generate hypotheses and by so doing support precision medicine research. In this section, we will address hypothesis generation using biomedical literature and EHR data related to one area of precision medicine — pharmacogenomics discovery.
Pharmacogenomics is the study of how genes affect drug response. In the context of this chapter, it may be helpful to view pharmacogenomics as a particular kind of genotype-phenotype relationship (i.e. consider response to a drug as a phenotype). Two areas of applied pharmacogenomics research where text mining tools for hypothesis generation have proved useful are drug repurposing (identifying new indications for existing, approved drugs) and drug adverse effect prediction.
Text mining tools for hypothesis generation universally function through identification of relationships between entities. These relationships can be semantically defined relationships, formal relationships in structured ontologies, or relationships across heterogeneous data sources [86]. Text mining tools that synthesize such relationships and successfully identify new information can increase research productivity and provide substantial savings in terms of opportunity cost. Hypothesis generating tools are particularly important in precision medicine because the large data sources associated with precision medicine encourage execution of multiple tests, which results in increased statistical penalties [7]. For example, although genetics researchers now have the capability of sequencing thousands of genes to identify genetic determinants of response to a particular drug, the analysis of each of these genes results in thousands of tests, each of which carries a specific probability of returning a false positive. Thus for every test that researchers perform, they must increase the value of their significance threshold, which in turn creates a bias preventing detection of rare variants and variants with small effect sizes. Well-formulated and supported hypotheses derived in silico from data such as biomedical literature grant researchers the ability to find support for a potential gene association before committing resources to test that association experimentally. As researchers are enabled to test fewer genes, the significance thresholds for discovery grow smaller, and the likelihood of discovering true associations increases.
4.2 Pharmacogenomic hypothesis generation from text mining biomedical literature
The world’s biomedical literature, when accessed via text mining, can become an incredibly rich database of multidimensional relationships, including core pharmacogenomic relationships such as those between genes, proteins, chemicals and diseases. Text mining makes such relationships computationally mappable and enables discovery of ‘hidden’ relationships that are not explicitly described in published literature and indeed, are not yet known. The validity of this conceptual approach to hypothesis generation was demonstrated in an early application of text mining that explored disease-chemical relationships to predict a benefit for using fish oil in Raynaud’s syndrome [87] and for using magnesium to treat migraines [88]. The discoveries hypothesized in these studies identified a straightforward form of relationship: if drug A is related to phenotype B in one body of literature, and disease C is related to phenotype B in a separate body of literature, drug A and disease C might be related to each other [89]. Reflecting an understanding of the complexity of biological disease processes, more recent approaches using text mining to generate hypotheses for precision medicine have explored drug-gene relationships, drug-protein interaction networks, and gene pathway relationships.
Regarding drug-gene relationships, Percha et al hypothesized that drug-drug interactions (DDIs) occur when different drugs interact with the same gene product. To prove this hypothesis, they extracted a network of gene-drug relationships from Medline (the indexed component of PubMed). Their work is notable for their extraction of the type of relationship between drugs and genes (e.g. “inhibit”, “metabolize”, etc.) as well as the gene and drug entities themselves. They verified their approach by predicting a number of known DDIs as well as several DDIs that were not reported in the literature [90]. In another work, Percha and Altman approached mapping the rich networks of drug-gene relationships in published literature by explicitly defining the ways in which gene-drug interactions are described in literature. They employed a novel algorithm, termed ensemble biclustering for classification, which they validated against manually curated sets from PharmGKB and DrugBank. Finally they applied it to Medline, creating a map of all drug-gene relationships in Medline with at least five mentions. This map contained 2898 pairs of drug-gene interactions that were novel to PharmGKB and DrugBank [91].
Drug-protein interactions (DPIs) are another form of relationship from which to generate pharmacogenomic hypotheses. Li et al used DPI data to create disease-specific drug-protein connectivity maps as a tool for drug repurposing. Their approach involved establishing connectivity maps between text-mined disease-drug relationships and an outside database of protein-drug interactions. They demonstrated the utility of this approach by applying their work to Alzheimer disease, where they used these maps to generate the hypotheses that prazosin, diltiazem and quinidine might have therapeutic effects in AD patients. By searching ClinicalTrials.gov, they discovered that one of these drugs, prazosin, was already under investigation as a therapy for agitation and aggression in AD patients [92].
Biological pathways are sequences of interactions between biological entities such as genes, chemicals, and proteins that combine to exert a change in a cell. Because these pathways are essentially complex networks of relationships, they have great potential for hypothesis generation, yet pathways are necessarily high in order. As we discussed in the biocuration section of this chapter, mining high-order entities from text remains challenging. Tari et al made significant progress in this domain with an approach to construction of pharmacogenomic pathways [93]. They produced pharmacokinetic pathways for 20 separate drugs (pharmacokinetic pathways describe how the body processes a drug). They extracted molecular drug interaction facts from databases such as Drug Bank and PharmGKB and then expanded this information with text-mined data from Medline. The key contribution of their approach is their use of automated reasoning to sort these facts and construct pathways according to logical time points by assigning pre- and post-condition states to entities. A comparison between their automatically constructed pathways and manually curated pathways for the same drugs in PharmGKB revealed that the automated approach achieved 84% precision for the extraction of enzymes and 82% for the extraction of metabolites. Their system enabled them to propose an additional 24 extra enzymes and 48 metabolites that were not included in the manually curated resources.
4.3 Hypothesis generation from EHRs
Many applications of TM for PM using EHR text for hypothesis generation require the integration of phenotypic data with genetic data. Although this is uncommon in EHRs, it is possible with EHR-linked biobanks. Examples of such biobanks include the NUgene project at Northwestern University [94], the Personalized Medicine Research Population project of the Marshfield Clinic [95] and BioVu at Vanderbilt [96]. A fitting starting point to a discussion of TM for PM in hypothesis generation using EHR text is the experimental design of a genome-wide association study (GWAS), which detects disease-causing genetic variants [97]. GWA studies are traditionally conducted by enrolling patients and then obtaining their phenotype and genotype through physical exam and gene sequencing; however, these studies can also be conducted using EHR-linked genetic data in conjunction with EHR phenotyping.
In a GWAS, researchers compare genes of people with a disease (the ‘cases’) to genes of people without the disease (the ‘controls’). Gene variants that are found more commonly among cases than controls are evidence of an association between a variant and a disease if the difference reaches statistical significance. Because of the statistical hazards of multiple testing mentioned in the introduction to this section, significance thresholds in GWAS are often quite stringent [98]. Ritchie et al demonstrated the feasibility of using EHR-linked genetic data in performing GWAS [99]. Text mining is important in EHR-based GWAS since accurately defining case and control phenotypes is a prerequisite to distinguishing genetic associations. For example, in an EHR-based GWAS study regarding cardiac rhythm, Denny et al employed text mining to detect negated concepts and family history from all physician-generated clinical documents. They linked this text-mined data with electrocardiogram data, billing codes, and labs to define phenotypes for cases and controls. The inclusion of text mining in these phenotype algorithms resulted in substantial improvements in recall while maintaining a high precision. Ultimately, this approach identified a novel gene for an ion channel involved in cardiac conduction [32, 100].
Text mining-enabled GWAS studies using EHR-linked genetic data are a cost-efficient and flexible avenue of discovery because EHRs can support exploration of an incredibly dynamic array of phenotypes. For example, Kullo et al used NLP of clinical notes and medication lists to identify case and control phenotypes for an EHR-based GWAS that employed genetic data from a previous study about one phenotype (peripheral artery disease) to perform an EHR-based GWAS about a completely separate phenotype (red blood cell traits) and identified a new variant in a gene previously unknown to be related to RBC function while also successfully replicating results of previous dedicated GWAS about RBC function [101]. Although the biases inherent in EHR data limit the reliability of these findings, this in silico method of discovery demonstrates the utility of text mining in EHR notes to generate hypotheses through GWAS.
One limitation of GWA studies is that the selection of cases and controls permits investigation of only one phenotype at a time and prevents the detection of gene variants that might predispose to multiple diseases. For example, it took two separate studies performed at different times to demonstrate that variants in the FTO gene predispose to both diabetes and to obesity. Text mining EHRs can enable discovery of such gene-disease relationships through an experimental modality called a phenome-wide association study (PheWAS), which is essentially the reverse design of a GWAS. In a PheWAS, the cases and controls are people with or without a specific gene variant that is suspected of causing disease. Comparison of a broad array of hundreds of disease phenotypes experienced by people with and without the variant allows the detection of multiple gene-disease associations and suggests etiologic relationships between disease types. The first PheWAS studies used only billing code data to define phenotypes, but subsequent studies have shown that using text mining of clinical notes in addition to billing data improves the significance of results [32].
PheWAS studies are a powerful hypothesis-generating application of TM for PM. Moore et al, noting that PheWAS enable discovery of multiple phenotypes associated with single genes, used clinical trial data from the AIDS Clinical Trials Group (ACTG) to explore phenotypes related to drug adverse effects in AIDS therapies. Their first published work established baseline associations between clinical measurements and patient genotypes, replicating 20 known associations and identifying several that were novel in HIV-positive cohorts [102]. In other areas of medicine, Rzhetsky et al used a PheWAS approach to hypothesize a relationship between autism, bipolar, and schizophrenia [103]. Likewise, Shameer et al performed a PheWAS and demonstrated that gene variants that affect characteristics of platelets also have an association with heart attack and autoimmune diseases [104]. Each of these findings identifies a potential avenue for pharmacogenomic therapy and demonstrates the potential of text mining EHRs for hypothesis generation.
5.1 TM for PM Conclusion: Value in Healthcare
One final concept that merits discussion is that of value. Value in healthcare is defined as health outcomes achieved per dollar spent [105]. Every medical intervention, including precision medicine and TM for PM, should be weighed in terms of this framework. How much will precision medicine benefit patients and at what cost?
In many circumstances value actually opposes the implementation of precision medicine. For many conditions, increasing the granularity with which we understand our patients may result in benefits, but those benefits may be so slight that the cost of obtaining them renders the technology valueless [106]. In some settings the current therapies or diagnostics may be so effective and inexpensive that the costs of PM will not merit the marginal gains. Alternatively, even if PM does greatly enhance diagnosis and prediction of disease, if no effective therapies exist for that disease, the overall value of PM will be reduced [107]. It is difficult to predict in the early stages of adoption of PM which diseases and therapies will benefit from PM and which will not.
Value is also the tantalizing target of precision medicine. In the 2016 Precision Medicine Initiative Summit, which took place one year after the announcement of the PMI, United States President Barack Obama reviewed the status of the Precision Medicine Initiative and asserted the potential of precision medicine to produce efficient and cost-effective healthcare [108]. Many factors support this assertion. Regarding the numerator of the value equation (healthcare outcomes) it is likely that precision medicine will indeed increase prevention of many diseases and improve therapeutic options for diseases that are detected. Regarding the denominator (cost), two factors may lower the relative costs and favor its adoption: (1) human DNA is largely unvarying (with the exception of cancer) within a single individual throughout the lifespan. Therefore, although genetic sequence analysis may be initially expensive compared to other diagnostic tests, as our understanding increases of the role of genes in health and disease the repeated utility of sequence data will lower the comparative cost. (2) Data in electronic health records are already widely collected and stored so use of this data should require only minimal expense.
Text mining is a vehicle to obtain increased utility from existing information resources, and it offers several advantages in the precision medicine value equation. Mining biomedical literature, for example, can help streamline curation and can improve research efficiency through hypothesis generation. Likewise, mining EHR text facilitates the use of this underutilized source of important patient phenotype information and enables a host of useful applications. As far as TM for PM can demonstrate increased value, its merit and ultimate adoption into mainstream medicine is assured.
Footnotes
BioCreative is one of a number of community-wide competitions and collaborative efforts designed to assess and advance the state of the art in text mining for biomedicine. Past challenges have addressed many issues related to TM for PM. For more information regarding this unique aspect of the text mining community, please see the review by Huang and Lu [26]
References
- 1.Collins FS, Varmus H. A new initiative on precision medicine. N Engl J Med. 2015;372:793–795. doi: 10.1056/NEJMp1500523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Swede H, Stone CL, Norwood AR. National population-based biobanks for genetic research. Genet Med. 2007;9:141–149. doi: 10.1097/gim.0b013e3180330039. [DOI] [PubMed] [Google Scholar]
- 3.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 4.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 5.Garraway LA, Verweij J, Ballman KV. Precision oncology: an overview. J Clin Oncol. 2013;31:1803–1805. doi: 10.1200/JCO.2013.49.4799. [DOI] [PubMed] [Google Scholar]
- 6.Schwaederle M, Zhao M, Lee JJ, et al. Impact of Precision Medicine in Diverse Cancers: A Meta-Analysis of Phase II Clinical Trials. J Clin Oncol. 2015;33:3817–3825. doi: 10.1200/JCO.2015.61.5997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brookes AJ, Robinson PN. Human genotype-phenotype databases: aims, challenges and opportunities. Nat Rev Genet. 2015;16:702–715. doi: 10.1038/nrg3932. [DOI] [PubMed] [Google Scholar]
- 8.Baumgartner WA, Jr, Cohen KB, Fox LM, et al. Manual curation is not sufficient for annotation of genomic databases. Bioinformatics. 2007;23:i41–8. doi: 10.1093/bioinformatics/btm229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu Z, Hirschman L. Biocuration workflows and text mining: overview of the BioCreative 2012 Workshop Track II. Database. 2012;2012:bas043. doi: 10.1093/database/bas043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hirschman L, Burns GAPC, Krallinger M, et al. Text mining for the biocuration workflow. Database. 2012;2012:bas020. doi: 10.1093/database/bas020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wei C-H, Kao H-Y, Lu Z. GNormPlus: An Integrative Approach for Tagging Genes, Gene Families, and Protein Domains. Biomed Res Int. 2015;2015:918710. doi: 10.1155/2015/918710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wei C-H, Harris BR, Kao H-Y, Lu Z. tmVar: a text mining approach for extracting sequence variants in biomedical literature. Bioinformatics. 2013;29:1433–1439. doi: 10.1093/bioinformatics/btt156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leaman R, Islamaj Dogan R, Lu Z. DNorm: disease name normalization with pairwise learning to rank. Bioinformatics. 2013;29:2909–2917. doi: 10.1093/bioinformatics/btt474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wei C-H, Kao H-Y, Lu Z. PubTator: a web-based text mining tool for assisting biocuration. Nucleic Acids Res. 2013;41:W518–22. doi: 10.1093/nar/gkt441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singhal A, Simmons M, Lu Z. Text Mining for Precision Medicine: Automating disease mutation relationship extraction from biomedical literature. J Am Med Inform Assoc. doi: 10.1093/jamia/ocw041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doughty E, Kertesz-Farkas A, Bodenreider O, et al. Toward an automatic method for extracting cancer- and other disease-related point mutations from the biomedical literature. Bioinformatics. 2011;27:408–415. doi: 10.1093/bioinformatics/btq667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.UniProt. UniProt: Annotation Guidelines. [Google Scholar]
- 18.Garten Y, Altman RB. Pharmspresso: a text mining tool for extraction of pharmacogenomic concepts and relationships from full text. BMC Bioinformatics. 2009;10(Suppl 2):S6. doi: 10.1186/1471-2105-10-S2-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thorn CF, Klein TE, Altman RB. Pharmacogenomics and bioinformatics: PharmGKB. Pharmacogenomics. 2010;11:501–505. doi: 10.2217/pgs.10.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xie B, Ding Q, Han H, Wu D. miRCancer: a microRNA–cancer association database constructed by text mining on literature. Bioinformatics. 2013 doi: 10.1093/bioinformatics/btt014. [DOI] [PubMed] [Google Scholar]
- 21.Fang Y-C, Lai P-T, Dai H-J, Hsu W-L. MeInfoText 2.0: gene methylation and cancer relation extraction from biomedical literature. BMC Bioinformatics. 2011;12:471. doi: 10.1186/1471-2105-12-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ongenaert M, Van Neste L, De Meyer T, et al. PubMeth: a cancer methylation database combining text-mining and expert annotation. Nucleic Acids Res. 2008;36:D842–6. doi: 10.1093/nar/gkm788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poos K, Smida J, Nathrath M, et al. Structuring osteosarcoma knowledge: an osteosarcoma-gene association database based on literature mining and manual annotation. Database. 2014 doi: 10.1093/database/bau042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maqungo M, Kaur M, Kwofie SK, et al. DDPC: Dragon Database of Genes associated with Prostate Cancer. Nucleic Acids Res. 2011;39:D980–5. doi: 10.1093/nar/gkq849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davis AP, Wiegers TC, Johnson RJ, et al. Text mining effectively scores and ranks the literature for improving chemical-gene-disease curation at the comparative toxicogenomics database. PLoS One. 2013;8:e58201. doi: 10.1371/journal.pone.0058201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang C-C, Lu Z. Community challenges in biomedical text mining over 10 years: success, failure and the future. Brief Bioinform. 2016;17:132–144. doi: 10.1093/bib/bbv024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wiegers TC, Davis AP, Mattingly CJ. Collaborative biocuration--text-mining development task for document prioritization for curation. Database. 2012;2012:bas037. doi: 10.1093/database/bas037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arighi CN, Wu CH, Cohen KB, et al. BioCreative-IV virtual issue. Database. 2014 doi: 10.1093/database/bau039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wei C-H, Peng Y, Leaman R, et al. Overview of the BioCreative V chemical disease relation (CDR) task. Proceedings of the fifth BioCreative challenge evaluation workshop; Sevilla, Spain. 2015. [Google Scholar]
- 30.Frankovich J, Longhurst CA, Sutherland SM. Evidence-based medicine in the EMR era. N Engl J Med. 2011;365:1758–1759. doi: 10.1056/NEJMp1108726. [DOI] [PubMed] [Google Scholar]
- 31.Lowe HJ, Ferris TA, Hernandez PM, Weber SC. STRIDE--An integrated standards-based translational research informatics platform. AMIA Annu Symp Proc. 2009;2009:391–395. [PMC free article] [PubMed] [Google Scholar]
- 32.Denny JC. Chapter 13: Mining electronic health records in the genomics era. PLoS Comput Biol. 2012;8:e1002823. doi: 10.1371/journal.pcbi.1002823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kohane IS. Using electronic health records to drive discovery in disease genomics. Nat Rev Genet. 2011;12:417–428. doi: 10.1038/nrg2999. [DOI] [PubMed] [Google Scholar]
- 34.(2012) CMS.gov - EHR Overview.
- 35.Hripcsak G, Albers DJ. Next-generation phenotyping of electronic health records. J Am Med Inform Assoc. 2013;20:117–121. doi: 10.1136/amiajnl-2012-001145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rosano G, Pelliccia F, Gaudio C, Coats AJ. The challenge of performing effective medical research in the era of healthcare data protection. Int J Cardiol. 2014;177:510–511. doi: 10.1016/j.ijcard.2014.08.077. [DOI] [PubMed] [Google Scholar]
- 37.Shoenbill K, Fost N, Tachinardi U, Mendonca EA. Genetic data and electronic health records: a discussion of ethical, logistical and technological considerations. J Am Med Inform Assoc. 2014;21:171–180. doi: 10.1136/amiajnl-2013-001694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Long MT, Fox CS. The Framingham Heart Study - 67 years of discovery in metabolic disease. Nat Rev Endocrinol. 2016 doi: 10.1038/nrendo.2015.226. [DOI] [PubMed] [Google Scholar]
- 39.Harris RP, Helfand M, Woolf SH, et al. Current methods of the US Preventive Services Task Force: a review of the process. Am J Prev Med. 2001;20:21–35. doi: 10.1016/s0749-3797(01)00261-6. [DOI] [PubMed] [Google Scholar]
- 40.Sessler DI, Imrey PB. Clinical Research Methodology 2: Observational Clinical Research. Anesth Analg. 2015;121:1043–1051. doi: 10.1213/ANE.0000000000000861. [DOI] [PubMed] [Google Scholar]
- 41.Sun J, McNaughton CD, Zhang P, et al. Predicting changes in hypertension control using electronic health records from a chronic disease management program. J Am Med Inform Assoc. 2014;21:337–344. doi: 10.1136/amiajnl-2013-002033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kawano Y. Diurnal blood pressure variation and related behavioral factors. Hypertens Res. 2011;34:281–285. doi: 10.1038/hr.2010.241. [DOI] [PubMed] [Google Scholar]
- 43.Wu ST, Sohn S, Ravikumar KE, et al. Automated chart review for asthma cohort identification using natural language processing: an exploratory study. Ann Allergy Asthma Immunol. 2013;111:364–369. doi: 10.1016/j.anai.2013.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Denny JC, Peterson JF, Choma NN, et al. Extracting timing and status descriptors for colonoscopy testing from electronic medical records. J Am Med Inform Assoc. 2010;17:383–388. doi: 10.1136/jamia.2010.004804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wei W-Q, Teixeira PL, Mo H, et al. Combining billing codes, clinical notes, and medications from electronic health records provides superior phenotyping performance. J Am Med Inform Assoc. 2015 doi: 10.1093/jamia/ocv130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shivade C, Raghavan P, Fosler-Lussier E, et al. A review of approaches to identifying patient phenotype cohorts using electronic health records. J Am Med Inform Assoc. 2014;21:221–230. doi: 10.1136/amiajnl-2013-001935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meystre SM, Savova GK, Kipper-Schuler KC, Hurdle JF. Extracting information from textual documents in the electronic health record: a review of recent research. Yearb Med Inform. 2008:128–144. [PubMed] [Google Scholar]
- 48.Savova GK, Masanz JJ, Ogren PV, et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc. 2010;17:507–513. doi: 10.1136/jamia.2009.001560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program. Proc AMIA Symp. 2001:17–21. [PMC free article] [PubMed] [Google Scholar]
- 50.Bejan CA, Xia F, Vanderwende L, et al. Pneumonia identification using statistical feature selection. J Am Med Inform Assoc. 2012;19:817–823. doi: 10.1136/amiajnl-2011-000752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McCowan IA, Moore DC, Nguyen AN, et al. Collection of cancer stage data by classifying free-text medical reports. J Am Med Inform Assoc. 2007;14:736–745. doi: 10.1197/jamia.M2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lehman L-W, Saeed M, Long W, et al. Risk stratification of ICU patients using topic models inferred from unstructured progress notes. AMIA Annu Symp Proc. 2012;2012:505–511. [PMC free article] [PubMed] [Google Scholar]
- 53.Zeng QT, Goryachev S, Weiss S, et al. Extracting principal diagnosis, co-morbidity and smoking status for asthma research: evaluation of a natural language processing system. BMC Med Inform Decis Mak. 2006;6:30. doi: 10.1186/1472-6947-6-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carroll RJ, Eyler AE, Denny JC. Naïve Electronic Health Record phenotype identification for Rheumatoid arthritis. AMIA Annu Symp Proc. 2011;2011:189–196. [PMC free article] [PubMed] [Google Scholar]
- 55.Garvin JH, DuVall SL, South BR, et al. Automated extraction of ejection fraction for quality measurement using regular expressions in Unstructured Information Management Architecture (UIMA) for heart failure. J Am Med Inform Assoc. 2012;19:859–866. doi: 10.1136/amiajnl-2011-000535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sohn S, Savova GK. Mayo clinic smoking status classification system: extensions and improvements. AMIA Annu Symp Proc. 2009;2009:619–623. [PMC free article] [PubMed] [Google Scholar]
- 57.Sohn S, Kocher J-PA, Chute CG, Savova GK. Drug side effect extraction from clinical narratives of psychiatry and psychology patients. J Am Med Inform Assoc. 2011;18(Suppl 1):i144–9. doi: 10.1136/amiajnl-2011-000351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mani S, Chen Y, Arlinghaus LR, et al. Early prediction of the response of breast tumors to neoadjuvant chemotherapy using quantitative MRI and machine learning. AMIA Annu Symp Proc. 2011;2011:868–877. [PMC free article] [PubMed] [Google Scholar]
- 59.Berty HL, Simon M, Chapman BE. A semi-automated quantification of pulmonary artery dimensions in computed tomography angiography images. AMIA Annu Symp Proc. 2012;2012:36–42. [PMC free article] [PubMed] [Google Scholar]
- 60.Liu M, Wu Y, Chen Y, et al. Large-scale prediction of adverse drug reactions using chemical, biological, and phenotypic properties of drugs. J Am Med Inform Assoc. 2012;19:e28–35. doi: 10.1136/amiajnl-2011-000699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhao D, Weng C. Combining PubMed knowledge and EHR data to develop a weighted bayesian network for pancreatic cancer prediction. J Biomed Inform. 2011;44:859–868. doi: 10.1016/j.jbi.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lung NH, Institute B, Others. Expert Panel Report 3 (EPR 3): guidelines for the diagnosis and management of asthma. Vol. 40 Bethesda: National Institutes of Health; 2007. [Google Scholar]
- 63.Nguyen AN, Lawley MJ, Hansen DP, et al. Symbolic rule-based classification of lung cancer stages from free-text pathology reports. J Am Med Inform Assoc. 2010;17:440–445. doi: 10.1136/jamia.2010.003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schmiedeskamp M, Harpe S, Polk R, et al. Use of International Classification of Diseases, Ninth Revision, Clinical Modification codes and medication use data to identify nosocomial Clostridium difficile infection. Infect Control Hosp Epidemiol. 2009;30:1070–1076. doi: 10.1086/606164. [DOI] [PubMed] [Google Scholar]
- 65.Kho AN, Hayes MG, Rasmussen-Torvik L, et al. Use of diverse electronic medical record systems to identify genetic risk for type 2 diabetes within a genome-wide association study. J Am Med Inform Assoc. 2012;19:212–218. doi: 10.1136/amiajnl-2011-000439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Klompas M, Haney G, Church D, et al. Automated identification of acute hepatitis B using electronic medical record data to facilitate public health surveillance. PLoS One. 2008;3:e2626. doi: 10.1371/journal.pone.0002626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Trick WE, Zagorski BM, Tokars JI, et al. Computer algorithms to detect bloodstream infections. Emerg Infect Dis. 2004;10:1612–1620. doi: 10.3201/eid1009.030978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mathias JS, Gossett D, Baker DW. Use of electronic health record data to evaluate overuse of cervical cancer screening. J Am Med Inform Assoc. 2012;19:e96–e101. doi: 10.1136/amiajnl-2011-000536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bishop CM. Pattern Recognition and Machine Learning. Springer Verlag; 2006. [Google Scholar]
- 70.Sesen MB, Kadir T, Alcantara R-B, et al. Survival prediction and treatment recommendation with Bayesian techniques in lung cancer. AMIA Annu Symp Proc. 2012;2012:838–847. [PMC free article] [PubMed] [Google Scholar]
- 71.Kawaler E, Cobian A, Peissig P, et al. Learning to predict post-hospitalization VTE risk from EHR data. AMIA Annu Symp Proc. 2012;2012:436–445. [PMC free article] [PubMed] [Google Scholar]
- 72.Van den Bulcke T, Vanden Broucke P, Van Hoof V, et al. Data mining methods for classification of Medium-Chain Acyl-CoA dehydrogenase deficiency (MCADD) using non-derivatized tandem MS neonatal screening data. J Biomed Inform. 2011;44:319–325. doi: 10.1016/j.jbi.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 73.Mani S, Chen Y, Elasy T, et al. Type 2 diabetes risk forecasting from EMR data using machine learning. AMIA Annu Symp Proc. 2012;2012:606–615. [PMC free article] [PubMed] [Google Scholar]
- 74.Tatari F, Akbarzadeh-T M-R, Sabahi A. Fuzzy-probabilistic multi agent system for breast cancer risk assessment and insurance premium assignment. J Biomed Inform. 2012;45:1021–1034. doi: 10.1016/j.jbi.2012.05.004. [DOI] [PubMed] [Google Scholar]
- 75.Kim D, Shin H, Song YS, Kim JH. Synergistic effect of different levels of genomic data for cancer clinical outcome prediction. J Biomed Inform. 2012;45:1191–1198. doi: 10.1016/j.jbi.2012.07.008. [DOI] [PubMed] [Google Scholar]
- 76.Newton KM, Peissig PL, Kho AN, et al. Validation of electronic medical record-based phenotyping algorithms: results and lessons learned from the eMERGE network. J Am Med Inform Assoc. 2013;20:e147–e154. doi: 10.1136/amiajnl-2012-000896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.The Phenotype KnowledgeBase. PheKB; [Accessed 1 Mar 2016]. https://phekb.org/ [Google Scholar]
- 78.Yu S, Liao KP, Shaw SY, et al. Toward high-throughput phenotyping: unbiased automated feature extraction and selection from knowledge sources. J Am Med Inform Assoc. 2015;22:993–1000. doi: 10.1093/jamia/ocv034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schneeweiss S. Learning from big health care data. N Engl J Med. 2014;370:2161–2163. doi: 10.1056/NEJMp1401111. [DOI] [PubMed] [Google Scholar]
- 80.Lee J, Maslove DM, Dubin JA. Personalized mortality prediction driven by electronic medical data and a patient similarity metric. PLoS One. 2015;10:e0127428. doi: 10.1371/journal.pone.0127428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cole TS, Frankovich J, Iyer S, et al. Profiling risk factors for chronic uveitis in juvenile idiopathic arthritis: a new model for EHR-based research. Pediatr Rheumatol Online J. 2013;11:45. doi: 10.1186/1546-0096-11-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Köpcke F, Prokosch H-U. Employing computers for the recruitment into clinical trials: a comprehensive systematic review. J Med Internet Res. 2014;16:e161. doi: 10.2196/jmir.3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ni Y, Wright J, Perentesis J, et al. Increasing the efficiency of trial-patient matching: automated clinical trial eligibility pre-screening for pediatric oncology patients. BMC Med Inform Decis Mak. 2015;15:28. doi: 10.1186/s12911-015-0149-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.D’Avolio L, Ferguson R, Goryachev S, et al. Implementation of the Department of Veterans Affairs’ first point-of-care clinical trial. J Am Med Inform Assoc. 2012;19:e170–6. doi: 10.1136/amiajnl-2011-000623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Vickers AJ, Scardino PT. The clinically-integrated randomized trial: proposed novel method for conducting large trials at low cost. Trials. 2009;10:14. doi: 10.1186/1745-6215-10-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hahn U, Cohen KB, Garten Y, Shah NH. Mining the pharmacogenomics literature--a survey of the state of the art. Brief Bioinform. 2012;13:460–494. doi: 10.1093/bib/bbs018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Swanson DR. Fish oil, Raynaud’s syndrome, and undiscovered public knowledge. Perspect Biol Med. 1986;30:7–18. doi: 10.1353/pbm.1986.0087. [DOI] [PubMed] [Google Scholar]
- 88.Swanson DR. Migraine and magnesium: eleven neglected connections. Perspect Biol Med. 1988;31:526–557. doi: 10.1353/pbm.1988.0009. [DOI] [PubMed] [Google Scholar]
- 89.Swanson DR. Medical literature as a potential source of new knowledge. Bull Med Libr Assoc. 1990;78:29–37. [PMC free article] [PubMed] [Google Scholar]
- 90.Percha B, Garten Y, Altman RB. Biocomputing 2012. WORLD SCIENTIFIC; 2011. DISCOVERY AND EXPLANATION OF DRUG-DRUG INTERACTIONS VIA TEXT MINING; pp. 410–421. [PMC free article] [PubMed] [Google Scholar]
- 91.Percha B, Altman RB. Learning the Structure of Biomedical Relationships from Unstructured Text. PLoS Comput Biol. 2015;11:e1004216. doi: 10.1371/journal.pcbi.1004216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Li J, Zhu X, Chen JY. Building disease-specific drug-protein connectivity maps from molecular interaction networks and PubMed abstracts. PLoS Comput Biol. 2009;5:e1000450. doi: 10.1371/journal.pcbi.1000450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tari L, Anwar S, Liang S, et al. SYNTHESIS OF PHARMACOKINETIC PATHWAYS THROUGH KNOWLEDGE ACQUISITION AND AUTOMATED REASONING. Biocomputing. 2010:465–476. doi: 10.1142/9789814295291_0048. WORLD SCIENTIFIC address = year = 2012 edition =, year = 2012 edition = [DOI] [PubMed] [Google Scholar]
- 94.Ormond KE, Cirino AL, Helenowski IB, et al. Assessing the understanding of biobank participants. Am J Med Genet A. 2009;149A:188–198. doi: 10.1002/ajmg.a.32635. [DOI] [PubMed] [Google Scholar]
- 95.McCarty CA, Nair A, Austin DM, Giampietro PF. Informed consent and subject motivation to participate in a large, population-based genomics study: the Marshfield Clinic Personalized Medicine Research Project. Public Health Genomics. 2006;10:2–9. doi: 10.1159/000096274. [DOI] [PubMed] [Google Scholar]
- 96.Bowton EA, Collier SP, Wang X, et al. Phenotype-Driven Plasma Biobanking Strategies and Methods. J Pers Med. 2015;5:140–152. doi: 10.3390/jpm5020140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Visscher PM, Brown MA, McCarthy MI, Yang J. Five years of GWAS discovery. Am J Hum Genet. 2012;90:7–24. doi: 10.1016/j.ajhg.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Klein RJ, Zeiss C, Chew EY, et al. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308:385–389. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ritchie MD, Denny JC, Crawford DC, et al. Robust replication of genotype-phenotype associations across multiple diseases in an electronic medical record. Am J Hum Genet. 2010;86:560–572. doi: 10.1016/j.ajhg.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Denny JC, Ritchie MD, Crawford DC, et al. Identification of genomic predictors of atrioventricular conduction: using electronic medical records as a tool for genome science. Circulation. 2010;122:2016–2021. doi: 10.1161/CIRCULATIONAHA.110.948828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kullo IJ, Ding K, Jouni H, et al. A genome-wide association study of red blood cell traits using the electronic medical record. PLoS One. 2010 doi: 10.1371/journal.pone.0013011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Moore CB, Verma A, Pendergrass S, et al. Phenome-wide Association Study Relating Pretreatment Laboratory Parameters With Human Genetic Variants in AIDS Clinical Trials Group Protocols. Open Forum Infect Dis. 2015;2:ofu113. doi: 10.1093/ofid/ofu113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rzhetsky A, Wajngurt D, Park N, Zheng T. Probing genetic overlap among complex human phenotypes. Proc Natl Acad Sci U S A. 2007;104:11694–11699. doi: 10.1073/pnas.0704820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Shameer K, Denny JC, Ding K, et al. A genome- and phenome-wide association study to identify genetic variants influencing platelet count and volume and their pleiotropic effects. Hum Genet. 2014;133:95–109. doi: 10.1007/s00439-013-1355-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Porter ME. What Is Value in Health Care? N Engl J Med. 2010;363:2477–2481. doi: 10.1056/NEJMp1011024. [DOI] [PubMed] [Google Scholar]
- 106.Rubin R. Precision medicine: the future or simply politics? JAMA. 2015;313:1089–1091. doi: 10.1001/jama.2015.0957. [DOI] [PubMed] [Google Scholar]
- 107.Prasad V, Fojo T, Brada M. Precision oncology: origins, optimism, and potential. Lancet Oncol. 2016;17:e81–e86. doi: 10.1016/S1470-2045(15)00620-8. [DOI] [PubMed] [Google Scholar]
- 108.Remarks by the President in Precision Medicine Panel Discussion. [Accessed 2 Mar 2016];whitehouse.gov. 2016 https://www.whitehouse.gov/the-press-office/2016/02/25/remarks-president-precision-medicine-panel-discussion.
- 109.Huang J. Performance Measures of Machine Learning. University of Western Ontario; 2006. [Google Scholar]