Abstract
Noncoding RNAs (ncRNAs) produced in live cells may better reflect intracellular ncRNAs for research and therapy. Attempts were made to produce biologic ncRNAs, but at low yield or success rate. Here we first report a new ncRNA bioengineering technology using more stable ncRNA carrier (nCAR) containing a pre-miR-34a derivative identified by rational design and experimental validation. This approach offered a remarkable higher level expression (40%–80% of total RNAs) of recombinant ncRNAs in bacteria and gave an 80% success rate (33 of 42 ncRNAs). New FPLC and spin-column based methods were also developed for large- and small-scale purification of milligrams and micrograms of recombinant ncRNAs from half liter and milliliters of bacterial culture, respectively. We then used two bioengineered nCAR/miRNAs to demonstrate the selective release of target miRNAs into human cells, which were revealed to be Dicer dependent (miR-34a-5p) or independent (miR-124a-3p), and subsequent changes of miRNome and transcriptome profiles. miRNA enrichment analyses of altered transcriptome confirmed the specificity of nCAR/miRNAs in target gene regulation. Furthermore, nCAR assembled miR-34a-5p and miR-124-3p were active in suppressing human lung carcinoma cell proliferation through modulation of target gene expression (e.g., cMET and CDK6 for miR-34a-5p; STAT3 and ABCC4 for miR-124-3p). In addition, bioengineered miRNA molecules were effective in controlling metastatic lung xenograft progression, as demonstrated by live animal and ex vivo lung tissue bioluminescent imaging as well as histopathological examination. This novel ncRNA bioengineering platform can be easily adapted to produce various ncRNA molecules, and biologic ncRNAs hold the promise as new cancer therapeutics.
Introduction
Noncoding RNAs such as miRNAs play important roles in the regulation of target gene expression underlying various cellular processes, and dysregulation of ncRNAs is highly associated with human diseases including cancer (Cech and Steitz, 2014; Rupaimoole and Slack, 2017; Thyagarajan et al., 2018). Although the actions of ncRNAs have been extensively studied, our knowledge might not fully encompass these molecules on a true biologic level. ncRNAs currently used for biomedical research are produced primarily by chemical synthesis and decorated extensively with a wide array of artificial modifications (Corey, 2007; Bramsen and Kjems, 2012; Khvorova and Watts, 2017). Such chemically engineered/synthesized ncRNA agents from different manufacturers and laboratories vary broadly in the types, sites, and degrees of artificial modifications and thus likely have distinct higher-order structures, physicochemical properties, biologic/pharmacological activities, and safety profiles. Therefore, chemoengineered ncRNA molecules may not represent the properties of natural ncRNAs produced within live cells (Ho and Yu, 2016; Pereira et al., 2017). Recognized as foreign invaders by specific factors such as toll-like receptors (Hornung et al., 2005; Robbins et al., 2009), synthetic ncRNAs have been well documented to cause off-target effects and induce immunogenicity, which also vary broadly with different chemical modifications. The recent termination of Phase I clinical study on synthetic miR-34a mimic (MRX34) (Beg et al., 2017), owing to high incidence of adverse immune responses, again testifies the body’s capability to distinguish chemoengineered RNAi agents as foreign. Therefore, biologic approaches such as in vitro transcription (Beckert and Masquida, 2011) and, especially, bioengineering in live cells (Ponchon and Dardel, 2007; Ponchon et al., 2009; Huang et al., 2013; Li et al., 2014, 2015, 2018; Chen et al., 2015; Wang et al., 2015; Pereira et al., 2016; Fang et al., 2017), are highly warranted to produce natural RNA molecules that should better represent cellular ncRNA properties for basic research and experimental therapy (Ho and Yu, 2016; Pereira et al., 2017).
Efforts have been made to produce biologic ncRNAs via fermentation, whereas at a low yield or success rate. The use of p19 RNA-binding protein (Huang et al., 2013) offers a way to produce fully processed siRNAs in Escherichia coli; however, the low yield (e.g., 10–80 μg/l bacterial culture) makes this method impractical for the production of milligram quantities of RNAi agents. Utilization of tRNA scaffold (Ponchon and Dardel, 2007; Ponchon et al., 2009) may facilitate large-scale (e.g., up to 20 mg/l fermentation) production of ncRNAs; nevertheless, adoption of this method revealed that less than 20% of target ncRNAs could actually be expressed at isolatable levels (e.g., >3% of total RNAs) (Chen et al., 2015). Our recent efforts to produce recombinant pre-miRNA (mir) have demonstrated ample expression (10%–20% of total RNAs) of a hybrid ncRNA molecule in E. coli, namely pre-miR-34a and tRNAMet fused Sephadex aptamer (MSA) (MSA/mir-34a) (Chen et al., 2015). Although MSA/mir-34a can be used as a versatile scaffold, the success rate for producing isolatable target ncRNAs is still less than 30%, and the dependence on Sephadex aptamer is unknown.
Here, we first report the development of a more stable and reliable ncRNA carrier (nCAR) for rapid production of milligrams of target ncRNA agents at an enormously higher expression level (40%–80% of total RNAs) and success rate (∼80%; 33 of 42 target ncRNAs). We further established a readily adaptable pipeline for both small- and large-scale production of bioengineered ncRNAs, which we applied to generate a collection of ready-to-use biologic ncRNA molecules. Using two nCAR/miRNAs as examples, we further demonstrate that bioengineered miRNA agents, with intrinsic RNA properties for specific processing to target miRNAs, were able to selectively rewrite miRNome profiles and alter transcriptome of human cells, leading to antiproliferative properties against human lung carcinoma cells in vitro and antitumor activities in xenograft mouse models in vivo.
Materials and Methods
Bacterial Culture
DH5α (Thermo Fisher Scientific, Rockford, IL) and HST08 (Clontech Laboratories, Mountain View, CA) were grown at 37°C in LB supplemented with 100 μg/ml ampicillin for plasmid preparation and amplification. To produce RNA, 2×YT media supplemented with 100 μg/ml ampicillin was used for enriched growth of HST08 E. coli.
Human Cell Culture
Human lung carcinoma cell lines A549 H23, H1650, H1299, and H1975 were purchased from American Type Culture Collection (Manassas, VA). HEK293T and Dicer-KO (4–25) cell lines were kindly provided by Prof. Bryan R. Cullen (Duke University, Durham, NC) (Bogerd et al., 2014). Luciferase and green fluorescent protein (GFP)-expressing A549-Luc-GFP cells were generated by transduction with pCCLc-Luc-EGFP lentiviral constructs (Vector Core, UC Davis Medical Center, Sacramento, CA). Lung carcinoma cell lines were maintained in RPMI 1640 supplemented with 10% fetal bovine serum and 293T cells were maintained in DMEM supplemented with 10% fetal bovine serum and gentamicin (10 μg/ml), both grown at 37°C in a humidified atmosphere with 5% CO2 and 95% air.
Construction of ncRNA Expression Plasmids
Sequences of individual miRNAs and pre-miR-34a were obtained from miRBase (http://www.mirbase.org/), while siRNA and RNA aptamer sequences were gathered from previously reported studies (Supplemental Table S1). Inserts coding target ncRNA sequences (Supplemental Table S2) were generated by PCR amplification using primers (IDT, San Diego, CA) (Supplemental Table S2). Amplicons were annealed into pBSTNAV (Ponchon et al., 2009) linearized by SacII and EagI (New England Biolabs, Ipswich, MA) via Seamless Recombination (Clontech Laboratories). Plasmids were propagated in DH5α cells and confirmed by sequencing analyses (Genscript, Piscataway, NJ).
Expression of Recombinant RNAs in E. coli
HST08 competent cells were transformed with target plasmids and then grown either on small scale (5–50 ml) or large scale (500–600 ml). Total RNAs were isolated by phenol extraction, quantitated with a NanoDrop 2000 Spectrophotometer (Thermo Fisher Scientific), and analyzed by denaturing urea (8 M) polyacrylamide (8%) gel electrophoresis (PAGE). Images were acquired with ChemiDoc MP Imaging System (Bio-Rad, Hercules, CA). Band intensities were used to estimate roughly target RNA expression relative to total RNAs. Precise calculation of the fraction of target RNA in total RNAs was achieved by dividing the area of target RNA peak by total areas of all peaks in chromatograph during FPLC purification, as shown below.
Small- and Large-Scale Purification of nCAR/sRNAs
Target BERA was purified on small scale using spin columns: RNA Clean & Concentrator and Select-a-Size DNA Clean & Concentrator (Zymo Research, Irvine, CA). RNA was isolated following the protocols suggested for >200 nt (RNA Clean & Concentrator) sequentially followed by a 50-bp cutoff protocol (Select-a-Size DNA Clean & Concentrator). Large-scale purification of target BERA was performed with a NGC QUEST 10PLUS fast protein liquid chromatography system (Bio-Rad) equipped with an anion exchange Enrich-Q 10 × 100 column (Bio-Rad). Separation of nCAR/sRNA from total RNAs was achieved on an Enrich-Q 10 × 100 column that was first equilibrated with Buffer A (10 mM sodium phosphate, pH 7.0) at a constant flow rate of 2.5 ml/min for 2 minutes and then followed by a gradient elution at the same flow rate: 55% Buffer B (Buffer A + 1 M sodium chloride, pH 7.0) for 4.8 minutes, 55%–75% Buffer B for 20.4 minutes, and then 100% Buffer B for 9.6 minutes. FPLC traces were monitored at 260/280 nm using a UV-visible detector, and individual fractions were collected for urea-PAGE analyses. After confirmation of purity and expected size, fractions containing target BERA were pooled, precipitated by ethanol, desalted, and concentrated/dissolved in nuclease-free water with an Amicon ultra-2 ml centrifugal filter (30 kDa; EMD Millipore, Billerica, MA).
Determination of RNA Purity
RNA purity was quantitated by HPLC analysis as previously described (Wang et al., 2015), and endotoxin activity was determined using the Pyrogent-5000 kinetic LAL assay (Lonza, Walkersville, MD) by following manufacturer’s instructions prior to in vitro and in vivo functional studies. The majority of nCAR/sRNAs purified by FPLC were >98% pure (Table 1) with minimal endotoxin levels (<1 EU/μg RNA).
TABLE 1.
Yields and purities of individual BERAs produced on a large scale using the nCAR platform and isolated by FPLC method
| nCAR | Yield | Purity |
|---|---|---|
| mg RNA/l fermentation | %; by HPLC | |
| miR-27a-3p | 10.1 | 99.4 |
| miR-27b-3p | 10.2 | 99.4 |
| miR-451a-5p | 18.8 | 98.9 |
| ApoB-siRNA | 17.0 | 99.0 |
| anti-miR-126-3p | 20.0 | 99.2 |
| ICAM1-siRNA | 12.0 | 98.6 |
| miR-298-5p | 15.4 | 97.2 |
| miR-519c-5p | 16.0 | 99.3 |
| miR-122-5p | 9.80 | 87.0 |
| Nrf2-siRNA | 18.7 | 98.3 |
| miR-126-3p | 11.6 | 99.8 |
| miR-144-5p | 10.6 | 98.7 |
| ARV7-siRNA | 17.0 | 99.3 |
| GFP-siRNA | 12.0 | 99.3 |
| let-7c-5p | 8.70 | 92.4 |
| miR-127-3p | 3.83 | 95.5 |
| miR-34a-5p | 16.6 | 98.3 |
| miR-124-3p | 8.34 | 98.3 |
| miR-328-3p | 14.8 | 99.7 |
| anti-miR-451b-5p | 11.3 | 99.3 |
| MSA | 5.72 | 99.6 |
| anti-miR-21-5p-3 | 7.48 | 98.2 |
| miR-33a-5p | 7.54 | 96.0 |
| anti-miR-21-5p-2 | 9.41 | 96.9 |
| anti-miR-122-5p | 7.67 | 99.3 |
| scrm-5 | 7.54 | 53.5 |
| scrm-4 | 7.43 | 98.2 |
| scrm-3 | 8.54 | 99.3 |
| anti-miR-21-5p | 11.7 | 99.4 |
| miR-34a/TPA3′+TPA5′ | 18.2 | 99.5 |
| miR-888/pegaptanib3′ | 16.0 | 98.3 |
| miR-34a/EpCAMA3′ | 12.7 | 99.0 |
RNA Sequencing and Data Analyses
293T and Dicer-KO cells were transfected with 20 nM of nCAR/miR-34a-5p, nCAR/miR-124-3p, or control tRNA. Cells were treated in triplicate and processed and sequenced separately. Total RNAs were isolated using TRIzol-chloroform protocol (Abcam, Cambridge, MA) at 48 hours posttransfection and RNA integrity was assessed by 1% standard agarose gel electrophoresis.
Small RNAs.
Library construction was prepared by Beijing Genomics Institute (Shenzhen, China) where small RNAs less than 30 nt were collected through gel separation (15% urea-PAGE) starting with 1 μg of total RNA. Small RNA fragments were ligated to adenylated 3′ adapters annealed to unique barcodes, followed by the ligation of 5′ adapters and reverse transcription (RT). After synthesis of the first strand cDNA, the product was expanded by 15 cycles of PCR amplification. A second size selection operation was carried out to purify the PCR amplicons from nonspecific products, selecting 103- to 115-bp fragments by gel separation. After gel purification, PCR yield was quantified by Qubit 3.0 Fluorometer (Invitrogen, Carlsbad, CA) and samples were pooled to make a single strand DNA circle (ssDNA circle) to yield the final small RNA library. DNA nanoballs (DNBs) were generated with the ssDNA circle by rolling circle replication to enlarge fluorescent signals at the sequencing process. The DNBs were loaded into the patterned nanoarrays, and single-end reads of 50-bp were read on the BGISEQ-500 platform (Shenzhen, China). The FASTQ-formatted sequence data were analyzed using the miRDeep module (An et al., 2013) to obtain the read counts of known miRNAs. To compute the read counts derived from transfected nCAR/miR-34a-5p or nCAR/miR-124-3p, a PERL script was developed to map sequence reads from FASTQ-formatted sequence data to the corresponding constructor, followed by counting isoform reads in individual samples. Read counts of known miRNAs and sRNAs derived from nCAR/miR-34a-5p or nCAR/miR-124-3p among individual samples were thus used for the analysis of significantly, differentially expressed miRNAs (P < 0.05 and log2CPM > 6) between phenotype using EdgeR (Robinson et al., 2010).
Messenger RNAs.
mRNAs were purified using poly-T oligo-attached magnetic beads. Following purification, the mRNA was fragmented into small pieces using divalent cations under elevated temperature. The cleaved RNA fragments were copied into first-strand cDNA using random primers by reverse transcription, followed by second strand cDNA synthesis using DNA Polymerase I and RNase H. These cDNA fragments contain an additional “A” at the 3′ end to allow subsequent ligation of the adapter. The products were then purified and enriched with PCR amplification, quantified with Qubit and used to create DNB-based nanoarrays by rolling circle replication. Stepwise sequencing was performed using the combinatorial Probe-Anchor (cPAL) ligation approach and read on the BGISEQ-500 system. The FASTQ-formatted sequence data were analyzed using a BWA-RSEM-EdgeR workflow (Li and Durbin, 2009; Robinson et al., 2010; Li and Dewey, 2011), with sequence reads mapped to the reference human-genome assembly (Feb. 2009, GRCh37/hg19) with BWA software. Sequence read counts for individual genes were computed using RSEM, and the resulting read counts from individual samples were subjected to the detection of differentially expressed mRNAs (log2FC > 1.2 or log2FC < 0.8, P < 0.05, and log2CPM > 5) between phenotypes using EdgeR package. Networks, functions, and pathways analyses were generated using Ingenuity Pathway Analysis (Ingenuity Systems; Qiagen, Redwood City, CA), primarily based on experimentally demonstrated interactions in human and rodent studies.
Enrichment Analysis.
miRNA target enrichment analyses of significantly downregulated genes (log2FC < 0.8, P < 0.05, and log2CPM >5) were undertaken using miRNA targets predicted by TargetScan (Lewis et al., 2005), and enrichment P values were computed by Fisher’s exact test.
Reverse Transcription Quantitative Real-Time PCR
Cells were transfected with 20 nM of nCAR/miR-34a-5p, nCAR/miR-124-3p, or control using Lipofectamine 3000 (Life Technologies) and harvested at 48 hours posttransfection. Total RNAs were extracted using Direct-zol RNA isolation kit (Zymo Research), and 500 ng of total RNAs was used for cDNA synthesis with random hexamers or respective stem-loop primers (Supplemental Table S3). RT was conducted with NxGen M-MuLV reverse transcriptase (Lucigen, Middleton, WI), and qPCR analysis was carried out on a CFX96 Touch real-time PCR system (Bio-Rad) using quantitative RT-PCR master mix and respective primers (Supplemental Table S3). Levels of miRNA were normalized to U6 snRNA and levels of mRNA were normalized to 18S rRNA (N = 9) in corresponding samples, determined using the eq. 2–ΔΔCT. Reactions were run in triplicate in three independent experiments and similar results were obtained.
Cell Viability Assay
Cells were transfected with various doses of nCAR/miR-34a-5p, nCAR/miR-124-3p, or control tRNA. Cell viability was determined with CellTiter-Glo 2.0 assay kit (Promega, Madison, WI) at 72 hours posttransfection, according to manufacturer’s protocol. Luminescence was recorded using a SpectraMax Microplate Reader (Molecular Devices, Sunnyvale, CA) at an integration time of 250 ms. Inhibition was calculated by adjusting vehicle control to 0%, and dose-response curves were established by plotting inhibition versus RNA concentration. Data were fit into a normalized inhibitory dose-response model with variable slope (Y = 100/[1 + 10^((LogEC50 − X) × HillSlope)]); GraphPad Prism, San Diego, CA) for the estimation of EC50 and Hill slope values (Supplemental Table S4). All experiments were performed in triplicates.
Western Blot Assay
A549, 293T, and Dicer-KO 293T cells were seeded at 0.25 × 106 cells/well and treated with 30 nM nCAR/miR-34a-5p, nCAR/miR-124-3p, or tRNA control. Cultured cells were harvested at 48 hours and lysed with RIPA lysis buffer (Sigma Aldrich, St. Louis, MO) containing protease inhibitor cocktail (Roche Diagnostics, Mannheim, Germany). Total cell lysate (30 μg) was resolved by 10% SDS-PAGE gel, transferred to PVDF membranes (250 mA for 2 hours), and probed with target antibodies. Antibodies against CDK6 (C-21), cMET (C-28), Dicer (H-212), GAPDH (FL-335), and SIRT1 (H-300) were obtained from Santa Cruz Biotechnology (Santa Cruz, CA); p-STAT3 (Tyr705) and STAT3 (124H6) were acquired from Cell Signaling Technology (Beverly, MA); β-actin (AC-15) from Sigma-Aldrich; and MRP4 (M4I-10) from Abcam. Subsequently, secondary antibodies linked with peroxidase were anti-rabbit (Jackson ImmunoResearch Inc., West Grove, PA), anti-mouse IgG, or anti-rat IgG (Cell Signaling). Protein bands were captured by a luminescent image analyzer (ChemiDoc MP Imaging System).
Metastatic Lung Xenograft Mouse Models and Therapy Studies
All animal procedures were approved by the Institutional Animal Care and Use Committee of University of California, Davis, and all animal studies were conducted in accordance with the relevant national and international guidelines. Five-week-old female nonobese diabetic/severe combined immunodeficient (NOD/SCID) mice (NOD.CB17-Prkdcscid/J) were purchased from Jackson Laboratory (Bar Harbor, ME) and adaptively fed at least 1 week before experiments.
A549 metastatic xenograft mouse models were established by injecting 3.5 × 106 A549-luc/GFP cells via tail vein. Tumor growth was monitored using bioluminescence imaging by injecting D-luciferin potassium salt solution (150 mg/kg) intraperitoneally 10 minute postanesthesia. Images were acquired with the ChemiDoc MP Imaging System. Fourteen days after inoculation, 27 mice were randomly divided into three groups (N = 9 per group). Mice were treated intravenously with 30 μg of control tRNA, nCAR/miR-34a-5p, or nCAR/miR-124-3p formulated with in vivo-jetPEI (Polyplus Transfection, Illkirch, France) three times per week for 3 weeks. At the end of the study, all mice were euthanized, and lungs were dissected, weighed, and imaged ex vivo for GFP signals. Lung tissues were further fixed with 10% formalin and subjected to hematoxylin and eosin (H&E) staining for histopathological evaluation in the Histology Facility at Roswell Park Cancer Institute (Buffalo, NY). H&E-staining images (100×) were captured using an Olympus camera (DP25) and CellSens software (Olympus, Center Valley, PA).
Induction of Cytokine Release
Female BALB/c mice at 5 to 6 weeks of age (Jackson Laboratory) were administered intravenously via tail vein with 30 μg of in vivo-jetPEI-formulated control tRNA, nCAR/miR-34a-5p, or nCAR/miR-124-3p. Separate groups of animals without any treatment or treated intravenously with 20 μg of lipopolysaccharide (LPS) were used as controls. Blood was collected 1 hour after administration, and serum was isolated for the quantification of cytokine IL-6 and TNFα levels using mouse enzyme-linked immunosorbent assay kits (Thermo Fisher Scientific).
Statistical Analysis
Data are presented as mean ± S.D. Statistics analysis was performed using unpaired Student’s t test, one-way analysis of variance, or two-way analysis of variance (GraphPad Prism). P < 0.05 was considered to be statistically significant.
Results
Identification of Pre-miR-34a G138U/139ΔG Derivative for ncRNA Carrier
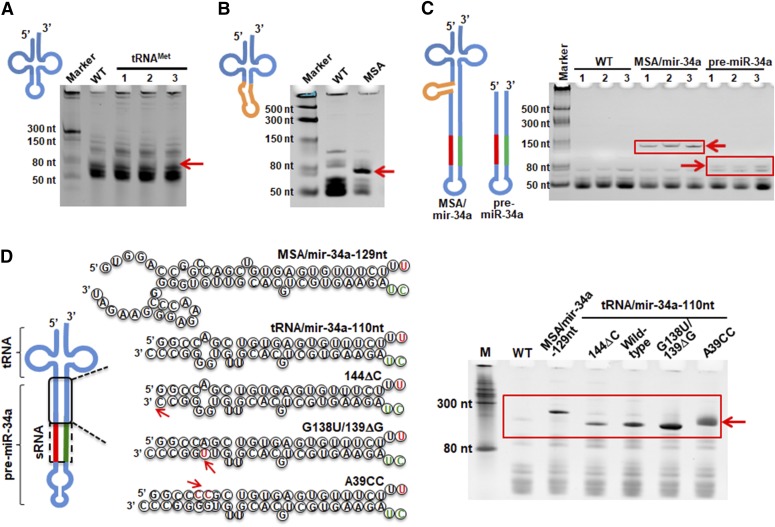
To enable heterogeneous expression of recombinant ncRNAs in bacteria, we sought to assess the foundational basis for chimeric ncRNA design. We first found that tRNAMet standalone showed no obvious expression in E. coli (Fig. 1A). In sharp contrast, tRNAMet fused Sephadex aptamer (MSA) was overexpressed, indicating the importance of Sephadex aptamer for accumulation of the chimera (Fig. 1B). Furthermore, human pre-miR-34a itself was noticeably expressed in bacteria (≤2% of total RNAs) and hybrid MSA/mir-34a showed an improved accumulation (10%–20% of total RNAs) (Fig. 1C), demonstrating the stabilities of pre-miR-34a and chimeric ncRNA for the success of heterogeneous expression.
Fig. 1.
Identification of a new ncRNA carrier (nCAR) toward higher-level expression of recombinant RNAs. tRNA standalone showed no expression in HST08 E. coli (A), whereas the fusion of a Sephadex aptamer (highlighted in orange) led to an overexpression of resulting MSA (B). (C) Hybrid MSA/mir-34a was overexpressed in E. coli and pre-miR-34a itself showed relatively lower-level expression. (D) The pre-miR-34a was refined toward a more stable structure (base changes dictated by red arrows), and the Sephadex aptamer was removed. Among these constructs, the tRNA/pre-miR-34a-G138U/139ΔG showed the highest level of heterogeneous expression (∼50% of total RNAs) and, therefore, was chosen as a carrier for ncRNA bioengineering.
Therefore, we aimed at refining pre-miR-34a to achieve a more stable and higher level of expression and assessed the dependence on Sephadex aptamer. Our approach was to use pre-miR-34a (110 nt) and rationally modify small bulges and kinks to yield more stable stems (Fig. 1D), which might lead to greater degree of RNA stability and accumulation in the host. Among five pre-miR-34a derivatives evaluated experimentally, we found that construct G138U/139ΔG showed the highest level of expression (>50% of total RNAs), which was also independent of Sephadex aptamer (Fig. 1D). Supporting this observation, the positional entropy of pre-miR-34a G138U/139ΔG was 4.6 kcal/mol lower than its wild-type counterpart (tRNA/mir-34a-110nt). In addition, the increased expression of tRNA-fused pre-miR-34a G138U/139ΔG derivative, as compared with MSA/pre-miR-34a (Chen et al., 2015), was shown in both HST08 and Top10 common E. coli stains (Supplemental Fig. 1). As a result, the pre-miR-34a G138U/139ΔG derivative was selected to fuse to a tRNA (e.g., tRNAMet) as ncRNA carrier (nCAR) for bioengineering RNAi agents.
nCAR Permits a Remarkable High-Level Production of Target ncRNAs at a High Success Rate
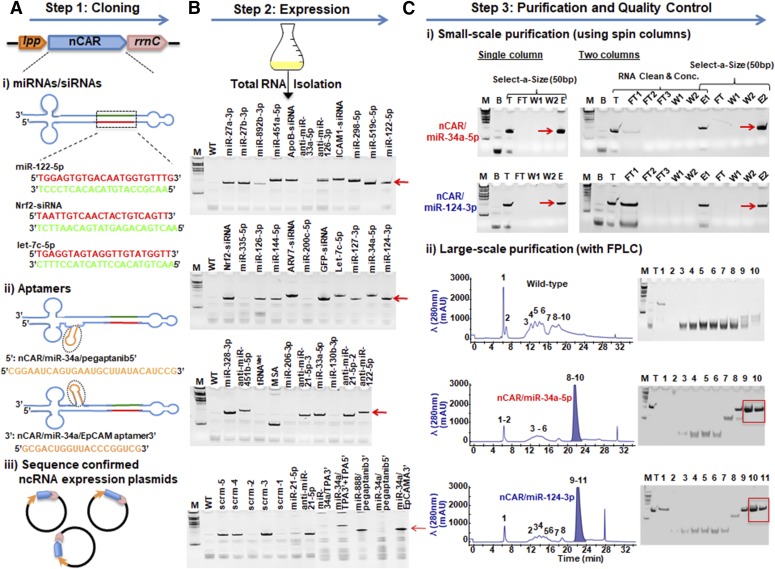
The nCAR was applied for the assembly of target RNAi agents, where small RNAs (sRNAs) such as miRNAs, siRNAs, and aptamers substituted miR-34a sequences or were directly added to designated locations (Fig. 2A). Considering the 5′ counting rule (Park et al., 2011), a total of 42 nCAR/sRNA agents were designed (Supplemental Tables 1 and 2). The coding sequences of individual target bioengineered ncRNA agents (BERA) were thus cloned into a target vector (Supplemental Fig. 2). Following transformation and fermentation, we assessed the expression of target BERAs by urea-PAGE analysis of total RNAs isolated from E. coli. Excitingly, we found that 33 BERAs were successfully expressed at a remarkably high level (40%–80% of total RNAs; Fig. 2B), which gave a success rate of 80% (33 out of 42 target ncRNAs). The results demonstrate the robustness of nCAR for an improved higher level, heterogeneous expression of target RNAi agents.
Fig. 2.
Three-step strategy to bioengineer ncRNA agents using nCAR. (A) In step 1, the ncRNAs of interest are cloned into the target vector (Supplemental Fig. S2), where the miR-34a duplex (red/green) is replaced by target sRNAs (e.g., miRNA, siRNA or antisense RNA, RNA aptamers, etc.) of interest. (B) In step 2, the verified plasmid is transformed into E. coli and total RNAs are isolated postfermentation for urea-PAGE analysis of target BERA expression. Among 42 target ncRNAs, 33 showed remarkable high-level expression (40%–80% of total RNAs). Total RNAs from untransformed wild-type bacteria (WT) are used for comparison. (C) Finally, in step 3, ncRNAs are isolated either on small scale using spin columns or large scale using fast protein liquid chromatography (FPLC) methods to offer micrograms or milligrams of BERAs, respectively. B, blank nontransformed E. coli; E, eluate; FT, flow through; T, total RNA; W1-2, washes. Fractions 1–11 were collected at various times during FPLC isolation. RNA purity was verified by high-performance liquid chromatography (HPLC) analysis (Supplemental Fig. S3), and both methods could offer >98% pure, ready-to-use BERAs.
On-Demand Small- and Large-Scale Purification of Bioengineered ncRNAs
We next sought for methods to isolate target BERAs from total RNAs. While the utility of the nCAR platform is purposed for large-scale production of milligrams of BERAs, we did examine the practicality of purifying microgram quantities that may be used for initial or high-throughput screening. Since the nCAR system generally offers BERAs (e.g., nCAR/miR-34a-5p and nCAR/miR-124-3p, etc.) around a length of 180 nt, we assessed whether target BERAs could be isolated with commercially available Select-a-Size DNA Clean & Concentrator (Fig. 2C) based on similar physicochemical properties of nucleic acids. The 50-bp selection was able to produce >88% pure BERA (determined by HPLC method) with a yield of 15–20 μg/ml culture (Supplemental Table 3). When used in combination with the RNA Clean & Concentrator spin columns that remove large RNAs (Fig. 2C), the purity of isolated BERAs was over 97% (Supplemental Table 3). Although the two-column method unsurprisingly offered a lower yield than the single column, overall yield (∼14 μg/ml culture or 30%–40% of total RNAs) was still tremendously high, owing to the remarkable high-level expression.
To achieve large-scale purification, we established a new anion exchange FPLC method. Up to 15 mg of total RNAs were loaded onto an Enrich-Q 10 × 100 column and separated by a single-run, salt-gradient elution (Fig. 2C). The majority of FPLC-isolated BERAs displayed a high degree of homogeneity (>98%; Table 1), as determined by HPLC method (Supplemental Fig. 3). Given such enhanced expression and purification yield, we readily obtained up to 10 mg of target BERAs in a single run and 4–20 mg of pure BERAs from 1 l bacterial fermentation (Table 1). In addition, endotoxin was flushed through this process and the endotoxin activities of final BERAs were minimal (e.g., nCAR/miR-34a-5p: 0.85 EU/μg RNA; nCAR/miR-124-3p: 0.1 EU/μg RNA), which are much lower than the reported levels (2000 EU/μg DNA) to induce cytotoxicity (Butash et al., 2000) and safe for mouse studies by considering the recommended limit of 5 EU/kg of body weight/h for humans (Malyala and Singh, 2008) and the difference in doses between mice and humans (Copeland et al., 2005).
Selective Release of Target miRNAs from Bioengineered ncRNAs in Human Cells Rewrites miRNome, Dependent (miR-34a-5p), or Independent (miR-124-3p) on Endoribonuclease Dicer
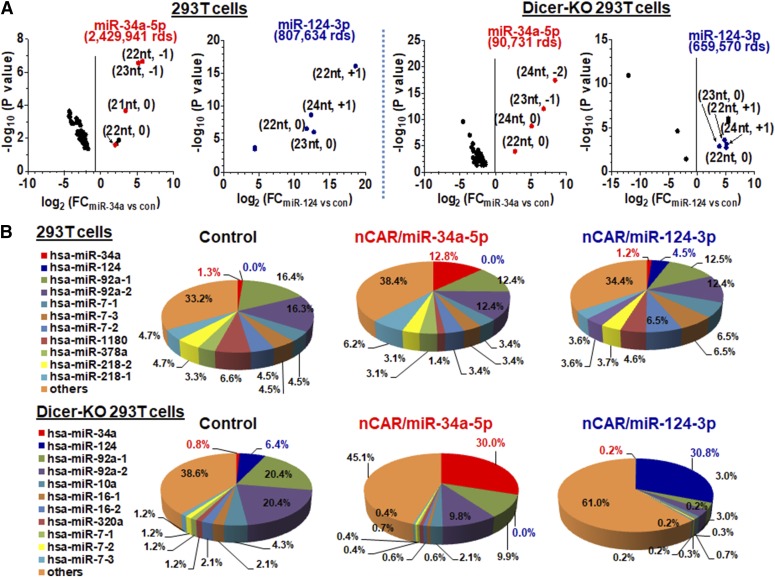
To examine whether target miRNAs can be specifically generated from BERAs in human cells and whether this process is Dicer dependent, we conducted a small RNA sequencing study on human 293T cells and Dicer-knockout (Dicer-KO) counterparts (Bogerd et al., 2014) treated with nCAR/miR-34a-5p, nCAR/miR-124-3p, and control RNA. The result showed that nCAR/miR-34a-5p was selectively processed to miR-34a-5p as its dominant isoform (22-nt; starting site 0) in 293T cells, as well as to lower reads of other isoforms (Fig. 3A). However, miR-34a-5p levels were 27-fold lower in Dicer-KO 293T cells than wild-type 293T cells, demonstrating a critical role for Dicer in the production of miR-34a-5p from nCAR/miR-34a-5p. Surprisingly, nCAR/miR-124-3p was mainly processed to a 23-nt isoform starting at position 0 in wild-type 293T cells, whereas a 22-nt species from position +1 (1-nt to the right) in Dicer-KO cells. Since miR-124-3p naturally exists as 20 nt, additional nucleotides carried over from pre-miR-34a at the 3′ end of the carrier indicates a unique cleavage of nCAR/miR-124-3p to offer miR-124-3p. Most interestingly, comparable levels of miR-124-3p were generated from nCAR/miR-124-3p in wild-type and Dicer-KO 293T cells (Fig. 3A), indicating the independence of Dicer for the production of miR-124-3p.
Fig. 3.
Selective release of target miRNAs from nCAR/miRNAs in human cells, in a Dicer- dependent (miR-34a) or -independent (miR-124) manner, changes miRNome profiles. (A) Small RNA sequencing analyses showed that 1) nCAR/miR-34a-5p and nCAR/miR-124-3p were selectively processed to target miR-34a-5p and miR-124-3p isoforms (length, starting site/position), respectively, in human cells (fold of change (FC), nCAR/miR vs. control tRNA (con) treatment), and 2) the level of miR-34a-5p (reads or rds) produced from BERA was largely affected by Dicer status, whereas miR-124-3p production was Dicer independent. Individual read counts represent enriched miRNAs with FC >1.2 or <0.8 with differential significance by EdgeR analysis. (B) miR-34a-5p became the most abundant miRNA in both wild-type and Dicer-KO cells after transfection with nCAR-miR-34a-5p, which was in sharp contrast to a nominal portion in control cells. Similarly, miR-124-3p became the seventh-most and most abundant miRNA in wild-type and Dicer-KO cells, respectively, after transfection with nCAR-miR-124-3p.
The selective release of target miR-34a-5p from nCAR/miR-34a-5p in human cells led to a specific change of miRNome profiles, where miR-34a-5p became the most abundant miRNA in both wild-type 293T cells (>16 million reads of all miRNAs) and Dicer-KO cells (<0.5 million of reads) (Fig. 3B). Likewise, miR-124-3p became the seventh-most and most abundant miRNA in wild-type and Dicer-KO 293T cells, respectively, post treatment with nCAR-miR-124-3p. The increase in target miRNA levels among miRNome was further confirmed by qPCR analyses (Supplemental Fig. 4A). Although the fold of increase in miR-34a-5p levels was “surprisingly” higher in the Dicer-KO cells than wild-type 293T cells, this scenario is simply attributable to the low basal expression level of miR-34a-5p in Dicer-KO cells (Fig. 3A). Together, our results demonstrate that a large number of target miRNAs can be selectively generated from BERAs in human cells, in a Dicer-dependent (e.g., miR-34a-5p) or -independent (e.g., miR-124-3p) manner, which subsequently rewrites cellular miRNome profiles.
Bioengineered miRNAs Specifically Modulate the Transcriptome Profiles in Human Cells
To delineate the effects of BERA on miRNA target gene expression and assess its specificity, we processed the same set of RNA samples for mRNA sequencing study. The results showed that 112 genes were significantly downregulated and 193 upregulated in nCAR/miR-34a-treated 293T cells, as well as 260 genes downregulated and 290 upregulated in nCAR/miR-124-treated 293T cells (Fig. 4A). These downregulated genes include many well-documented targets for specific miRNA (e.g., AMER1, GAS1, and NECTIN1 for miR-34a; VAMP3, SNAI2, IQGAP1, TMEM109, and RHOG for miR-124) (Chang et al., 2007; Karginov et al., 2007; Chi et al., 2009; Kaller et al., 2011), as well as some genes that have not been reported before (e.g., BAG2 and BCL6B for miR-34a; NID1 and VIM for miR-124). The suppression of several transcripts was further verified by qPCR analyses (Supplemental Fig. 4B) with gene-selective primers (Supplemental Table 4), where housekeeping gene levels were not altered (Supplemental Fig. 4C).
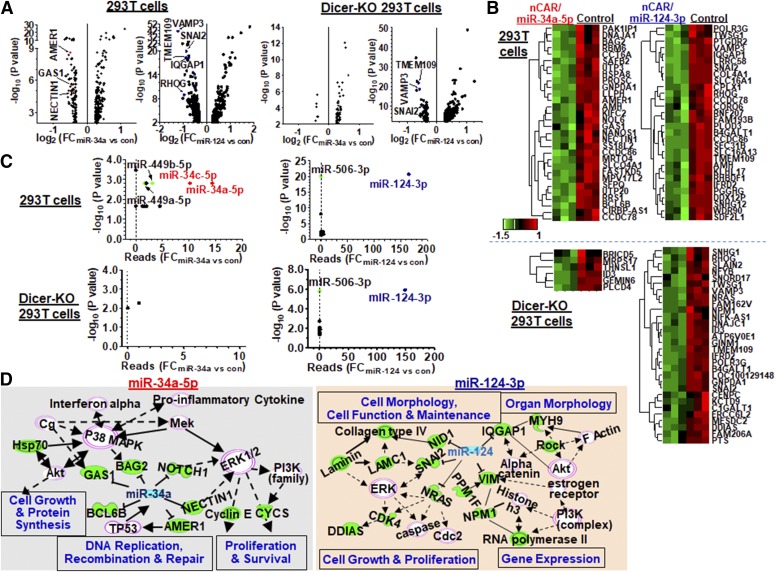
Fig. 4.
Specificity of nCAR/miRNA in the regulation of miRNA target gene expression. (A) Volcano plots of significantly-altered mRNAs (P < 0.01) in 293T cells transfected with nCAR/miR-34a-5p or miR-124-3p compared with control. Many transcripts were downregulated in 293T cells by nCAR/miR. In Dicer-KO cells, nCAR/miR-34a-5p showed minimal impact on the transcriptome (only six genes were significantly downregulated), whereas effects of nCAR/miR-124-3p retained. Several reported targets of miR-34a-5p (red) and miR-124-3p (blue) are designated with arrows. (B) Heatmap of the top 30 most downregulated genes in wild-type and Dicer-KO 293T cells treated with nCAR/miRNA compared with the control tRNA treatment. (C) Specificity of BERA in the regulation of miRNA target genes is supported by miRNA enrichment analyses, which readily identified miR-34/449 underlying nCAR/miR-34a-5p-downregulated mRNAs in 293T cells, whereas not in Dicer-KO cells. By contrast, miR-124/506 was enriched for nCAR/miR-124-3p-downregulated mRNAs in both wild-type and Dicer-KO 293T cells, which supports not only the specificity of BERA/miR-124-3p in the modulation of target gene expression but also its independence on Dicer. (D) Gene regulatory networks of major biologic pathways affected by miR-34a-5p and miR-124-3p, respectively, as identified by Ingenuity Pathway Analysis (IPA).
On the other hand, only six genes were significantly downregulated in Dicer-KO 293T cells by nCAR/miR-34a-5p (Fig. 4B), which have not been connected to miR-34a, again supporting the dependence on Dicer for miR-34a-5p production as well as the selectivity of nCAR/miR-34a-5p in the regulation of miR-34a target gene expression. In sharp contrast, nCAR/miR-124-3p significantly reduced the expression of 68 genes in Dicer-KO cells, among which many (e.g., VAMP3, SNAI2, and RHOG) are known miR-124 targets and show good overlap with targets identified in wild-type 293T cells (Fig. 4B). This is in agreement with the independence on Dicer in generating comparable high levels of miR-124-3p isoforms from nCAR/miR-124-3p in cells, whereas the major isoforms differed in length and position.
The selectivity of nCAR/miRNA in the regulation of miRNA target gene expression was further demonstrated by miRNA enrichment analyses. MiR-34a-5p was highly enriched from nCAR/miR-34a-5p-downregulated genes in 293T cells, whereas the same did not hold true for Dicer-KO cells (Fig. 4C), interweaving the specific effects on target gene expression due to Dicer-dependent excision of miR-34a-5p from nCAR/miR-34a-5p. Unsurprisingly, miR-449/34c within the same miR-34/449 family was also enriched from nCAR/miR-34a-5p-downregulated genes in 293T cells, supporting the similarity of their functions in gene regulation. However, miR-449/34c showed a low abundance (log2CPM < 6) and/or no significant (P > 0.05) change in its expression after BERA treatment and thus it was unlikely it had any contribution to the change of transcriptome. Interestingly, miR-124-3p was highly enriched for nCAR/miR-124-3p-downregulated genes in both wild-type and Dicer-KO 293T cells (Fig. 4C), which not only supports the selectivity of nCAR/miR-124-3p in the regulation of miR-124 target gene expression but also demonstrates the independence of miR-124-3p formation on Dicer. Likewise, miR-506 within the miR-124/506 family was enriched, although miR-506 was absent (0 reads) in all genotype and treatment cells. Additionally, we employed ingenuity pathway analysis to investigate the biologic pathways affected by BERA-downregulated genes. Gene regulation networks regulated by miR-34a were linked to cell growth, DNA replication, and cell proliferation and survival, whereas miR-124 was associated with cell morphology, maintenance, and gene expression (Fig. 4D). Taken together, these results demonstrate that nCAR/miRNAs displayed a high selectivity in gene regulation in human cells.
Biologic miRNAs Reduce the Protein Levels of Target Genes in Human Cells
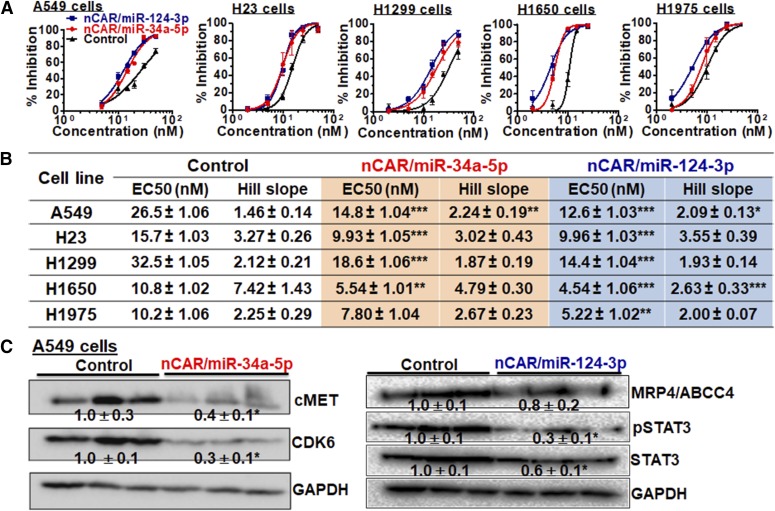
We further chose a few well-established targets for miR-34a (e.g., CDK6 and SIRT1) (Sun et al., 2008; Yamakuchi et al., 2008) and miR-124 (e.g., STAT3 and ABCC4/MRP4) (Hatziapostolou et al., 2011; Markova and Kroetz, 2014) to delineate the impact of bioengineered miRNAs on protein expression levels. Immunoblot analyses revealed a consistent suppression of CDK6 and SIRT1 protein levels by nCAR/miR-34a-5p in 293T cells (Supplemental Fig. 5A), as a result of high levels of released miR-34a-5p (Fig. 3A) However, in Dicer-KO cells, CDK6 protein levels were unchanged, whereas SIRT1 levels were steadily reduced by nCAR/miR-34a-5p, which could be due to a much lower level of miR-34a-5p produced in the absence of Dicer (Fig. 3A). This interesting observation might also indicate distinct sensitivities of different targets to the absolute amounts or extents of change of cellular miR-34a levels (Fig. 3A). Similarly, variable degrees of impact of nCAR/miR-124-3p on the protein levels of miR-124-3p targeted STAT3 and MRP4/ABCC4 (Supplemental Fig. 5B) were identified in wild-type and Dicer-KO 293T cells, which might be attributable to the difference in the most abundant miR-124-3p isoforms produced in those cells (Fig. 3A) with slightly altered seed sequences or nucleotides.
Bioengineered nCAR/miR-34a-5p and miR-124-3p Are Active in Suppressing Human Lung Carcinoma Cell Proliferation in Vitro
To further evaluate the utility of BERAs, we investigated the dose-dependent antiproliferation activities of two model BERAs, nCAR/miR-34a-5p and nCAR/miR-124-3p, against a variety of human lung carcinoma cells with different p53 and EGFR status, because lung cancer remains the most lethal cancer in the United States (Siegel et al., 2017) and restoration of miR-34a and miR-124 expression or function is effective to inhibit lung cancer cell growth (Wiggins et al., 2010; Kasinski and Slack, 2012; Cho et al., 2016; Yang et al., 2017). Compared with the control, both nCAR/miR-34a-5p and nCAR/miR-124-3p inhibited cell proliferation to a significantly greater degree (Fig. 5A), which was also demonstrated by the estimated EC50 and Hill Slope values (Supplemental Table 5). Interestingly, H1299 cells (mutant p53) were more sensitive to nCAR/miR-34a-5p than A549 (wild-type p53), under which the p53-miR-34a positive feedback loop confers an additional tumor suppressive effect when p53 is haploinsufficient (Okada et al., 2014). Similarly, H1650 and H1975 (mutant EGFR) cells carrying constitutively active EGFR were more sensitive to nCAR/miR-124-3p than A549 (wild-type EGFR). In addition, the suppression of A549 cell proliferation was associated with reduced protein levels of miR-34a targets (cMET and CDK6) and miR-124 targets (STAT3, pSTAT3, and MRP4/ABCC4) (Fig. 5B), supporting the presence of multiple targets/pathways for these miRNAs in reducing cancer cell proliferation.
Fig. 5.
Bioengineered miR-34a and miR-124 inhibit human lung cancer cell proliferation via the suppression of (proto-)oncogene expression. (A) Dose-response curves of nCAR/miR-34a-5p and nCAR-miR-124-3p in inhibiting human lung carcinoma A549, H23, H1299, H1650, and H1975 cell proliferation. Each nCAR/miR is significantly more effective than control RNA (P < 0.001, two-way ANOVA with Bonferroni post tests), except miR-34a-5p against H1975 cells. Values are mean ± S.D. (N = 3 per group). (B) Estimated EC50 and Hill Slope values. Data were fit to the normalized dose-response relationship with variable slope. Values are mean ± S.D. (N = 3 per group). *P < 0.05; **P < 0.01; ***P < 0.001, compared with corresponding control (one-way ANOVA). (C) Antiproliferative effects were associated with downregulation of miRNA target gene expression (cMET and CDK6 for miR-34a; STAT3, pSTAT3 and ABCC4/MRP4 for miR-124), as demonstrated by Western blots. *P < 0.05, Student’s t test.
Bioengineered nCAR/miR-34a-5p Significantly Reduces Metastatic Lung Xenograft Tumor Growth in Vivo
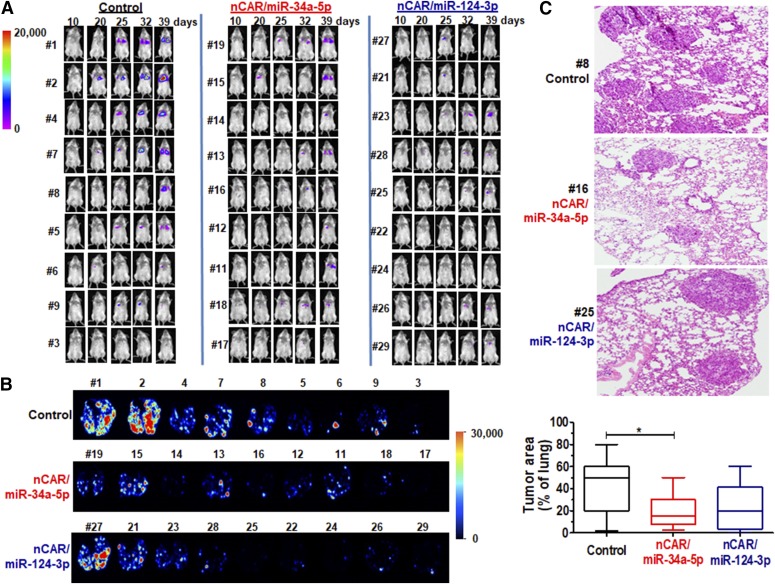
Lastly we determined the effectiveness of BERA miR-34a-5p and miR-124-3p in the suppression of lung tumor progression in vivo. Metastatic lung xenograft mouse models were established via tail vein injection of luciferase/GFP-expressing A549 cells and then treated intravenously with in vivo-jetPEI-formulated nCAR/miR-34a-5p, nCAR/miR-124-3p, or control RNA (30 μg, three times per week for 3 weeks). BERAs appeared to be well tolerated in mice, because, in addition to normal behaviors, all mice showed a steady increase in body weights that did not differ between treatments (Supplemental Fig. 6A). Tumor growth was monitored over the course of 6 weeks by bioluminescent imaging, and control mice showed stronger signals compared with mice treated with nCAR/miR-34a-5p and nCAR/miR-124-3p (Fig. 6A). At the end of the study, whole lung tissues were excised, weighed, and imaged ex vivo (Fig. 6B). As expected, tumors were obvious by visual inspection of lungs collected from mice that showed strong bioluminescent signals, which were indicated by more apparent ex vivo GFP signals. Lung tissues from mice treated with nCAR/miR-34a-5p were also found to be significantly lighter than the control group (Supplemental Fig. 6B). Furthermore, we performed histopathological analyses of all excised lung tissues to verify xenograft tumors and quantitatively determine the effectiveness of BERA therapy (Fig. 6C). In agreement with the observations from live animal and ex vivo lung images, we found that mice treated with nCAR/miR-34a-5p had significantly lower degrees of lung tumor nodules. These results demonstrate the effectiveness of biologic miR-34a-5p in the control of lung cancer progression in metastatic xenograft mouse models.
Fig. 6.
Application of bioengineered miRNAs to control lung xenograft tumor progression in vivo. (A) Effects of nCAR/miR-34a-5p, nCAR/miR-124-3p, and control RNA treatment on the progression of lung tumor progression in metastatic xenograft tumor mouse models (N = 9 per group), which were monitored through live animal bioluminescent imaging after the administration of D-luciferin. Bioluminescent images were taken on days 10, 20, 25, 32, 39 postinoculation of A549 cells and normalized to the same exposure time. (B) Local lung xenograft tumors were assessed via GFP imaging of ex vivo lung tissues at the end of the study. (C) Representative H&E-stained slides of lung tumors (100×). The tumor areas were thus quantified as percentages of corresponding lung areas, which were significantly (P < 0.05) lower for nCAR/miR-34a-5p treatment than the control.
Bioengineered ncRNAs Have Minimal Impact on Cytokine Release in Immunocompetent Mice in Vivo
Finally, we assessed possible immunogenicity of biologic ncRNAs in immunocompetent BALB/c mouse models by measuring the most sensitive cytokines IL-6 and TNFα around the peak time point (1 hour after treatment). Our data showed that LPS induced an immediate cytokine release syndrome in mice, as manifested by sharp increase in blood IL-6 and TNFα levels (Supplemental Fig. 6C). In contrast, compared with untreated mice, administration of in vivo-jetPEI-formulated nCAR/miR-34a-5p, nCAR/miR-124-3p and control RNA just led to insignificant increase of serum IL-6 TNFα levels that are remarkably and significantly lower than LPS treatment. The results indicate that biologic ncRNAs are tolerable in mice with minimal effects on cytokine release.
Discussion
We established a novel ncRNA bioengineering technology following the identification of a stable pre-miR-34a G138U/139ΔG derivative fused to tRNA molecule as a versatile carrier. This platform included the development of new complementary small- and large-scale purification methods. This approach offered a remarkable high-yield (40%–80% of total RNAs) and large-scale (4–20 mg from 1 l bacterial fermentation) production of target ncRNAs in E. coli, with a high success rate (80%; 33 ncRNAs out of 42). Using two bioengineered ncRNAs as examples, we further demonstrated a selective release of target miRNAs from nCAR/miRNAs and thus specific regulation of target genes, leading to altered miRNome and transcriptome profiles in human cells. In addition, we showed that the introduction of tumor suppressive miR-34a-5p and miR-124-3p with corresponding nCAR/miRNA prodrugs was proven to be effective for the control of human lung cancer cell proliferation in vitro and metastatic xenograft tumor progression in vivo. These results support the robustness of this new ncRNA bioengineering pipeline and broad applications of biologic ncRNA agents to basic research and experimental therapy.
In sharp contrast to the studies on protein functions and therapeutics using recombinant proteins produced and folded within live cells rather than those made by peptide synthesis, current research on ncRNA macromolecules rely heavily on chemically synthesized ncRNA mimics containing extensive artificial modifications. Although chemical modifications may increase ncRNA stability and thus offer more favorable pharmacokinetic properties (e.g., longer half-life) and even higher potency, synthetic ncRNA agents are fundamentally different molecules that undoubtedly have distinct higher-order structures as well as altered chemical and biologic properties. Therefore, the relevance of chemoengineered ncRNAs to cellular ncRNAs needs reconsideration. In addition, synthetic ncRNA agents from different manufacturers vary largely in the type, site, and degree of artificial modifications, which create another layer of uncertainty. Conversely, ncRNAs produced by the bioengineering platform presented in this report resemble the biogenesis of natural ncRNAs in live cells and thus offer highly structured, stable macromolecules without or just with necessary posttranscriptional modifications (Li et al., 2015; Wang et al., 2015). Since recombinant ncRNAs are produced in live cells to tolerable levels, biologic ncRNAs unlikely trigger any severe immune response (Wang et al., 2015; Zhao et al., 2016). Most importantly, the present approach displays substantial advantages over existing recombinant RNA methods (Ponchon and Dardel, 2007; Ponchon et al., 2009; Huang et al., 2013; Chen et al., 2015), because it achieves a remarkable high-level expression of target ncRNA molecules (40%–80% of total RNAs) at a high success rate (80%). The resultant ncRNA molecules are also different from viral or nonviral vector/plasmid based ncRNA expression materials that are truly DNA reagents (Ho and Yu, 2016). Therefore, the nCAR-based technology represents a more practical and cost-effective endeavor that can be easily adapted by a general biomedical research laboratory for the production of ncRNA agents of interest, either in microgram or milligram quantities.
In addition to the confirmation of a selective release of target miRNAs from bioengineered nCAR/miRNA agents in human cells, our small RNA sequencing results show that intracellular miRNAs are present as various isoforms, consistent with other studies (Ebhardt et al., 2009; Llorens et al., 2013). This may affect the binding affinity of miRNAs to Ago proteins (Elkayam et al., 2012) or turnover in the cell (Chatterjee and Grosshans, 2009). In particular, nCAR/miR-34a-5p was predominately processed into mature miR-34a-5p as a 22-nt isoform, whereas other species (21- and 23-nt shifts in cleavage start site) were produced at much lower abundance. However, nCAR/miR-124-3p was predominantly processed to a 23-nt form in 293T cells, whereas the other 22-nt species shifted 1 nt to the right in Dicer-KO cells, whose levels were rather comparable levels. The lack of significant increase of other miRNAs also supports the selectivity in producing target miRNAs from nCAR/miRNAs, whereas the proportional decrease of some high-abundance miRNAs could be a result of sharp increase of miR-34a-5p. Moreover, the dependence (miR-34a-5p) and independence (miR-124-3p) on Dicer for processing the nCAR/miRNAs indicate the versatility of using nCAR/miRNAs to introduce particular miRNAs into human cells to selectively change intracellular miRNome. Although it is unknown whether genetic background in Dicer-KO cells remains unchanged or not, our RNA sequencing results support the presence of Dicer-independent factors and pathways for miRNA biogenesis that might be altered in Dicer-KO cells and/or induced by nCAR/miR-124-3p treatment.
Upon the introduction of target miRNA into human cells, bioengineered ncRNA was effective to modulate target gene expression, leading to a specific change of transcriptome profile. Particularly, nCAR/miR-34a-5p and nCAR/miR-124-3p downregulated many well-documented targets (Chang et al., 2007; Karginov et al., 2007; Chi et al., 2009; Kaller et al., 2011) in 293T cells, as well as others (BAG2 and BCL6B for miR-34a; NID1 and VIM for miR-124) not reported before, although synthetic miRNA reagents were not included for comparison in present study. Although those genes have not been experimentally identified or verified by others, many are tentative targets (e.g., BCL6B for miR-34a-5p; and NID1 and VIM for miR-124-3p) predicted by various algorithms such as miRanda (www.microrna.org), TargetScan (http://www.targetscan.org/), and miRWalk (http://zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk/micrornapredictedtarget.html). Some genes (e.g., BAG2) are not predicted targets whose changes could be consequent or spatially controlled effects of the changes of primary target gene expression, whereas we cannot rule out possible off-target effects. Nevertheless, the specificity of nCAR/miRNA in the modulation of target gene expression is further demonstrated by “unbiased” enrichment analyses, which identified specific miRNAs behind corresponding downregulated genes. A step toward understanding ncRNAs as biologic macromolecules may advance our knowledge of these regulators in their natural forms to enable the progression of new discoveries.
As the altered transcriptomes were redefined for multiple intercalating pathways underlying cell growth, proliferation, and survival, the antiproliferative activities of nCAR-carried miR-34a-5p and miR-124-3p were observed in multiple human lung carcinoma cell lines. Cells with mutant p53 or constitutively active EGFR backgrounds seem to be more sensitive to miRNA treatment. Since naked biologic RNAs are degradable by serum RNases, formulation is necessary for therapeutic applications (Wang et al., 2015; Jilek et al., 2017). Further studies with in vivo-jetPEI-formulated RNAs in metastatic lung xenograft mouse models not only support the effectiveness of bioengineered miRNA agents in vivo but also establish the feasibility of developing biologic ncRNAs as therapeutics. Although the sample size was relatively small and the nCAR/miR-124-3p treatment group was more variable, the suppression of xenograft tumor growth by nCAR/miR-34a-5p was identified as statistically significant from control treatment. Moreover, we demonstrate that biologic ncRNAs are well tolerated in immunocompetent mice, as indicated by minimal impact on cytokine release. Rather, the assessment of biologic ncRNAs for cancer therapy should be challenged by more comprehensive studies with larger sample sizes and different models.
Limited by the array of possible directions, downstream in depth studies were carried out for two of many bioengineered ncRNAs to exemplify their biologic and pharmacological actions in the present study, but warrant additional investigations for bioengineered siRNA and RNA aptamer agents. While we focused on establishing the robust ncRNA bioengineering platform and assessing miRNA replacement strategy (Bader et al., 2010; Rupaimoole and Slack, 2017) with biologic miR-34a-5p and miR-124-3p molecules for lung cancer therapy, the utilities of BERAs cannot be underestimated. As supported by current studies, ncRNA bioengineering technology and the resulting biologic ncRNA agents should have direct impact on basic biomedical research and development of ncRNA therapies, although current ncRNA carriers may not be extended for the production of long ncRNAs playing important roles in various diseases (Cech and Steitz, 2014; Liu et al., 2017) because longer RNAs are more susceptible for degradation by bacterial RNases (Li et al., 2014, 2015).
In summary, we established a novel ncRNA bioengineering technology that can be easily adapted for the production of ncRNA agents bearing sRNAs of interest. Our findings indicate that bioengineered ncRNAs represent unique biologic materials and can be an invaluable addition to current tools for broad biomedical research including the development of ncRNA therapeutics. Although we cannot exclude the possibility that more superior ncRNA carriers would be expanded, the principle of producing biologic ncRNA macromolecules for basic and translational research will remain. We speculate the capability of multiplexing biologic ncRNAs in the near future for identification of new ncRNA actions and potential therapeutics.
Acknowledgments
We thank Dr. Qiu-Xia Chen and Dr. Junjian Wang for technical supports and helpful discussion.
Abbreviations
- ABCC4
ATP binding cassette subfamily C member 4
- BERA
bioengineered RNA agent
- CDK6
cyclin-dependent kinase 6
- cMET
tyrosine-protein kinase Met
- DNB
DNA nanoball
- FPLC
fast protein liquid chromatography
- HPLC
high performance liquid chromatography
- KO
knockout
- LPS
lipopolysaccharide
- miRNA
microRNA
- nCAR
ncRNA carrier
- ncRNA
noncoding RNA
- RNAi
RNA interference
- RT
reverse transcription
- siRNA
small interfering RNA
- SIRT1
sirtuin 1
- sRNA
small RNA
- STAT3
signal transducer and activator of transcription 3
- TNF
tumor necrosis factor
- tRNA
transfer RNA
Authorship Contributions
Participated in research design: Ho, Yu.
Conducted experiments: Ho, Duan, Batra, Jilek, Tu, Qiu.
Contributed new reagents or analytic tools: Qiu, Hu, Wun, Lara, DeVere White, Chen, Yu.
Performed data analysis: Ho, Duan, Qiu, Hu, Yu.
Wrote or contributed to the writing of the manuscript: Ho, Qiu, Hu, Yu.
Footnotes
This work was supported in part by the National Institutes of Health [Grants R01GM113888 and U01CA175315 (to A.-M.Y.); P30CA093373 (to R.D.V.W. and P.N.L.)]. J.L.J. was supported by a National Institutes of General Medical Sciences-funded Pharmacology Training Program Grant [T32GM099608].
 This article has supplemental material available at jpet.aspetjournals.org.
This article has supplemental material available at jpet.aspetjournals.org.
References
- An J, Lai J, Lehman ML, Nelson CC. (2013) miRDeep*: an integrated application tool for miRNA identification from RNA sequencing data. Nucleic Acids Res 41:727–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader AG, Brown D, Winkler M. (2010) The promise of microRNA replacement therapy. Cancer Res 70:7027–7030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckert B, Masquida B. (2011) Synthesis of RNA by in vitro transcription. Methods Mol Biol 703:29–41. [DOI] [PubMed] [Google Scholar]
- Beg MS, Brenner AJ, Sachdev J, Borad M, Kang YK, Stoudemire J, Smith S, Bader AG, Kim S, Hong DS. (2017) Phase I study of MRX34, a liposomal miR-34a mimic, administered twice weekly in patients with advanced solid tumors. Invest New Drugs 35:180–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogerd HP, Whisnant AW, Kennedy EM, Flores O, Cullen BR. (2014) Derivation and characterization of Dicer- and microRNA-deficient human cells. RNA 20:923–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramsen JB, Kjems J. (2012) Development of therapeutic-grade small interfering RNAs by chemical engineering. Front Genet 3:154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butash KA, Natarajan P, Young A, Fox DK. (2000) Reexamination of the effect of endotoxin on cell proliferation and transfection efficiency. Biotechniques 29:610–614, 616, 618–619. [DOI] [PubMed] [Google Scholar]
- Cech TR, Steitz JA. (2014) The noncoding RNA revolution-trashing old rules to forge new ones. Cell 157:77–94. [DOI] [PubMed] [Google Scholar]
- Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, Feldmann G, Yamakuchi M, Ferlito M, Lowenstein CJ, et al. (2007) Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell 26:745–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee S, Grosshans H. (2009) Active turnover modulates mature microRNA activity in Caenorhabditis elegans. Nature 461:546–549. [DOI] [PubMed] [Google Scholar]
- Chen QX, Wang WP, Zeng S, Urayama S, Yu AM. (2015) A general approach to high-yield biosynthesis of chimeric RNAs bearing various types of functional small RNAs for broad applications. Nucleic Acids Res 43:3857–3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi SW, Zang JB, Mele A, Darnell RB. (2009) Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature 460:479–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho CY, Huang JS, Shiah SG, Chung SY, Lay JD, Yang YY, Lai GM, Cheng AL, Chen LT, Chuang SE. (2016) Negative feedback regulation of AXL by miR-34a modulates apoptosis in lung cancer cells. RNA 22:303–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland S, Warren HS, Lowry SF, Calvano SE, Remick D, the Inflammation and the Host Response to Injury Investigators, (2005) Acute inflammatory response to endotoxin in mice and humans. Clin Diagn Lab Immunol 12:60–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corey DR. (2007) Chemical modification: the key to clinical application of RNA interference? J Clin Invest 117:3615–3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebhardt HA, Tsang HH, Dai DC, Liu Y, Bostan B, Fahlman RP. (2009) Meta-analysis of small RNA-sequencing errors reveals ubiquitous post-transcriptional RNA modifications. Nucleic Acids Res 37:2461–2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkayam E, Kuhn CD, Tocilj A, Haase AD, Greene EM, Hannon GJ, Joshua-Tor L. (2012) The structure of human argonaute-2 in complex with miR-20a. Cell 150:100–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang PY, Gómez Ramos LM, Holguin SY, Hsiao C, Bowman JC, Yang HW, Williams LD. (2017) Functional RNAs: combined assembly and packaging in VLPs. Nucleic Acids Res 45:3519–3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatziapostolou M, Polytarchou C, Aggelidou E, Drakaki A, Poultsides GA, Jaeger SA, Ogata H, Karin M, Struhl K, Hadzopoulou-Cladaras M, et al. (2011) An HNF4α-miRNA inflammatory feedback circuit regulates hepatocellular oncogenesis. Cell 147:1233–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho PY, Yu AM. (2016) Bioengineering of noncoding RNAs for research agents and therapeutics. Wiley Interdiscip Rev RNA 7:186–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung V, Guenthner-Biller M, Bourquin C, Ablasser A, Schlee M, Uematsu S, Noronha A, Manoharan M, Akira S, de Fougerolles A, et al. (2005) Sequence-specific potent induction of IFN-alpha by short interfering RNA in plasmacytoid dendritic cells through TLR7. Nat Med 11:263–270. [DOI] [PubMed] [Google Scholar]
- Huang L, Jin J, Deighan P, Kiner E, McReynolds L, Lieberman J. (2013) Efficient and specific gene knockdown by small interfering RNAs produced in bacteria. Nat Biotechnol 31:350–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jilek JL, Tian Y, Yu AM. (2017) Effects of microRNA-34a on the pharmacokinetics of cytochrome P450 probe drugs in mice. Drug Metab Dispos 45:512–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaller M, Liffers ST, Oeljeklaus S, Kuhlmann K, Röh S, Hoffmann R, Warscheid B, Hermeking H. (2011). Genome-wide characterization of miR-34a induced changes in protein and mRNA expression by a combined pulsed SILAC and microarray analysis. Mol Cell Proteomics 10:M111 010462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karginov FV, Conaco C, Xuan Z, Schmidt BH, Parker JS, Mandel G, Hannon GJ. (2007) A biochemical approach to identifying microRNA targets. Proc Natl Acad Sci USA 104:19291–19296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasinski AL, Slack FJ. (2012) miRNA-34 prevents cancer initiation and progression in a therapeutically resistant K-ras and p53-induced mouse model of lung adenocarcinoma. Cancer Res 72:5576–5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khvorova A, Watts JK. (2017) The chemical evolution of oligonucleotide therapies of clinical utility. Nat Biotechnol 35:238–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120:15–20. [DOI] [PubMed] [Google Scholar]
- Li B, Dewey CN. (2011) RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MM, Addepalli B, Tu MJ, Chen QX, Wang WP, Limbach PA, LaSalle JM, Zeng S, Huang M, Yu AM. (2015) Chimeric microRNA-1291 biosynthesized efficiently in Escherichia coli is effective to reduce target gene expression in human carcinoma cells and improve chemosensitivity. Drug Metab Dispos 43:1129–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MM, Wang WP, Wu WJ, Huang M, Yu AM. (2014) Rapid production of novel pre-microRNA agent hsa-mir-27b in Escherichia coli using recombinant RNA technology for functional studies in mammalian cells. Drug Metab Dispos 42:1791–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li PC, Tu MJ, Ho PY, Jilek JL, Duan Z, Zhang QY, Yu AX, Yu AM. (2018) Bioengineered NRF2-siRNA is effective to interfere with NRF2 pathways and improve chemosensitivity of human cancer cells. Drug Metab Dispos 46:2–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Yang Z, Wu J, Zhang L, Lee S, Shin DJ, Tran M, Wang L. (2017) Long noncoding RNA H19 interacts with polypyrimidine tract-binding protein 1 to reprogram hepatic lipid homeostasis. Hepatology DOI: 10.1002/hep.29654 [published ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorens F, Hummel M, Pantano L, Pastor X, Vivancos A, Castillo E, Mattlin H, Ferrer A, Ingham M, Noguera M, et al. (2013) Microarray and deep sequencing cross-platform analysis of the mirRNome and isomiR variation in response to epidermal growth factor. BMC Genomics 14:371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malyala P, Singh M. (2008) Endotoxin limits in formulations for preclinical research. J Pharm Sci 97:2041–2044. [DOI] [PubMed] [Google Scholar]
- Markova SM, Kroetz DL. (2014) ABCC4 is regulated by microRNA-124a and microRNA-506. Biochem Pharmacol 87:515–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada N, Lin CP, Ribeiro MC, Biton A, Lai G, He X, Bu P, Vogel H, Jablons DM, Keller AC, et al. (2014) A positive feedback between p53 and miR-34 miRNAs mediates tumor suppression. Genes Dev 28:438–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JE, Heo I, Tian Y, Simanshu DK, Chang H, Jee D, Patel DJ, Kim VN. (2011) Dicer recognizes the 5′ end of RNA for efficient and accurate processing. Nature 475:201–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira P, Pedro AQ, Queiroz JA, Figueiras AR, Sousa F. (2017) New insights for therapeutic recombinant human miRNAs heterologous production: Rhodovolum sulfidophilum vs Escherichia coli. Bioengineered 8:670–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira PA, Tomás JF, Queiroz JA, Figueiras AR, Sousa F. (2016) Recombinant pre-miR-29b for Alzheimer’s disease therapeutics. Sci Rep 6:19946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponchon L, Beauvais G, Nonin-Lecomte S, Dardel F. (2009) A generic protocol for the expression and purification of recombinant RNA in Escherichia coli using a tRNA scaffold. Nat Protoc 4:947–959. [DOI] [PubMed] [Google Scholar]
- Ponchon L, Dardel F. (2007) Recombinant RNA technology: the tRNA scaffold. Nat Methods 4:571–576. [DOI] [PubMed] [Google Scholar]
- Robbins M, Judge A, MacLachlan I. (2009) siRNA and innate immunity. Oligonucleotides 19:89–102. [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. (2010) edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupaimoole R, Slack FJ. (2017) MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov 16:203–222. [DOI] [PubMed] [Google Scholar]
- Siegel RL, Miller KD, Jemal A. (2017) Cancer statistics, 2017. CA Cancer J Clin 67:7–30. [DOI] [PubMed] [Google Scholar]
- Sun F, Fu H, Liu Q, Tie Y, Zhu J, Xing R, Sun Z, Zheng X. (2008) Downregulation of CCND1 and CDK6 by miR-34a induces cell cycle arrest. FEBS Lett 582:1564–1568. [DOI] [PubMed] [Google Scholar]
- Thyagarajan A, Shaban A, Sahu RP. (2018) MicroRNA-directed cancer therapies: implications in melanoma intervention. J Pharmacol Exp Ther 364:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WP, Ho PY, Chen QX, Addepalli B, Limbach PA, Li MM, Wu WJ, Jilek JL, Qiu JX, Zhang HJ, et al. (2015) Bioengineering novel chimeric microRNA-34a for prodrug cancer therapy: high-yield expression and purification, and structural and functional characterization. J Pharmacol Exp Ther 354:131–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiggins JF, Ruffino L, Kelnar K, Omotola M, Patrawala L, Brown D, Bader AG. (2010) Development of a lung cancer therapeutic based on the tumor suppressor microRNA-34. Cancer Res 70:5923–5930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamakuchi M, Ferlito M, Lowenstein CJ. (2008) miR-34a repression of SIRT1 regulates apoptosis. Proc Natl Acad Sci USA 105:13421–13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Wan L, Xiao C, Hu H, Wang L, Zhao J, Lei Z, Zhang HT. (2017) Inhibition of LHX2 by miR-124 suppresses cellular migration and invasion in non-small cell lung cancer. Oncol Lett 14:3429–3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Tu MJ, Wang WP, Qiu JX, Yu AX, Yu AM. (2016) Genetically engineered pre-microRNA-34a prodrug suppresses orthotopic osteosarcoma xenograft tumor growth via the induction of apoptosis and cell cycle arrest. Sci Rep 6:26611. [DOI] [PMC free article] [PubMed] [Google Scholar]