Abstract
Cytochrome P450 oxidoreductase (POR) is the primary electron donor in eukaryotic cytochrome P450 (CYP) containing systems. A wealth of ensemble biophysical studies of Cytochrome P450 oxidoreductase (POR) has reported a binary model of the conformational equilibrium directing its catalytic efficiency and biomolecular recognition. In this study, full length POR from the crop plant Sorghum bicolor was site-specifically labeled with Cy3 (donor) and Cy5 (acceptor) fluorophores and reconstituted in nanodiscs. Our single molecule fluorescence resonance energy transfer (smFRET) burst analyses of POR allowed the direct observation and quantification of at least three dominant conformational sub-populations, their distribution and occupancies. Moreover, the state occupancies were remodeled significantly by ionic strength and the nature of reconstitution environment, i.e. phospholipid bilayers (nanodiscs) composed of different lipid head group charges vs. detergent micelles. The existence of conformational heterogeneity in POR may mediate selective activation of multiple downstream electron acceptors and association in complexes in the ER membrane.
Introduction
Cytochrome P450s (CYPs) are multifunctional heme-containing enzymes, which catalyze some of nature’s most complex and versatile chemical reactions1,2. In eukaryotes, they are anchored to the ER membrane and activated by their co-localized redox partner the NADPH-dependent cytochrome P450 oxidoreductase (POR). POR is a diflavin protein3 binding one molecule each of the FAD and FMN cofactors in distinct globular domains connected by a flexible hinge region4. Crystallographic structures have provided static snapshots of its modular architecture in a “compact”/closed conformation4 that facilitates inter-flavin (FAD →FMN) electron transfer and “open”/extended conformation, anticipated to mediate electron donation to downstream electron acceptors5. Several extended crystallographic structures have been characterized using deletion6 and cross-linked mutants7 of the N-terminal truncated POR and recently in its complex with heme oxygenase8. SAXS and NMR-based investigations9 in consensus with the crystal structure of the yeast-human chimeric POR10 have provided evidence of an alternate extended conformation. Conformational oscillation between compact and extended conformations plays a pivotal role in vectorial electron transfer of several redox-active biological systems11. Over the last decade, a wide array of ensemble biophysical studies have provided complementary evidences for concerted domain motions of POR12 and a highly dynamic equilibrium of its two conformers, which is regulated by redox potential9,13 and ionic strength14,15. Additionally, ELDOR studies of the disemiquinoid form of POR16 (FADH· FMNH·) and recent SAXS data15 have suggested the existence of a continuum of numerous conformations rather than discrete forms and envisioned a multidimensional energy landscape. The conformational plasticity of POR has been intricately associated with its catalytic function and biomolecular recognition towards a broad range of P450s17 some of which may be integrated with POR in dynamic metabolons18–20. Nevertheless, a comprehensive understanding pertaining to the number, abundance and relative distribution of POR conformational spectrum is yet to be attained.
Ensemble biophysical techniques obscure the heterogeneities of a complex system by delivering an average value. Direct observations of the dynamics of POR can be achieved by single molecule spectroscopy, which has emerged as a powerful tool to reveal heterogeneities in systems that remain elusive in bulk studies21–24. This enables quantification of the abundance of unsynchronized, transient conformational sub-states. Single molecule fluorescence resonance energy transfer (smFRET) is a technique, which acts as a ratiometric ruler to probe distance based molecular fluctuations such as conformational dynamics, protein folding or unfolding25. A smFRET-based study of the structurally related diflavin enzyme nitric oxide synthase (nNOSr) recently demonstrated that it adopts multiple open conformations26. As the conformational equilibria of the diflavin reductases vary substantially27 and kinetic analyses of electron flux in POR reveals different rate-determining steps in plant and mammalian enzymes28, it is intriguing to probe the structural diversity of POR.
The conformational dynamics of membrane-bound enzymes as a function of their reconstitution environment have also received much attention. Molecular dynamics simulation studies of rhodopsin29 and the integral outer membrane protein A bacterial (OmpA)30 protein illustrated increased fluctuation freedom in detergent micelles compared to phospholipid bilayers. However, selection of an appropriate model system for membrane proteins amenable for in vitro biophysical investigation remains a challenge. Classically, detergents have been utilized for solubilization of membrane proteins, though posing the caveat of being unilamellar and often disrupting protein structural and functional integrity31. Nanodiscs, discoidal patches of phospholipid bilayers encapsulated by an amphipathic scaffold protein (MSP), are monodisperse entities whose size and physico-chemical properties can be precisely manipulated32,33. It is noteworthy that the majority of existing information on POR conformational equilibrium is derived from either detergent solubilized or soluble versions lacking the N-terminal membrane anchor34. We have previously demonstrated that POR function and stability strongly depends on the presence of the membrane environment35. Neutron reflectivity data have also revealed a difference in the net conformational equilibrium value of full-length POR36 in ‘nanodiscs’ as compared to its truncated soluble form9.
In this report, a smFRET37 based approach was utilized to directly observe and quantify the conformational sampling of POR. A full length, double cysteine variant of Sorghum bicolor POR (N181C/A552C) solubilized in detergent micelles (DM) was site specifically labeled with the fluorophores Cy3 (donor) and Cy5 (acceptor) by maleimide chemistry. The dual labeled POR was reconstituted into nanodiscs, which contained phospholipids with 25% negative (ND1) and net neutral (ND2) head group charges. smFRET experiments of DM, ND1 and ND2 were carried out under freely diffusing conditions by confocal microscopy. In addition, the effect of ionic strength on the conformational sampling was investigated.
Experimental
Materials
All chemicals were of analytical grade and purchased from Sigma-Aldrich (Denmark) unless otherwise stated. 2′5′-ADP Sepharose for affinity purification, size exclusion chromatography (SEC) column Superdex 200 HR 10/30 for high-resolution preparative separation, Cy3 and Cy5 maleimide fluorophores and PD-10 desalting columns were purchased from GE Healthcare Life Sciences, Brøndby, Denmark. Phospholipids: 1,2-dilauroyl-sn-glycero-3-phosphocholine (DLPC) and 1,2-dilauroyl-sn-glycero-3-phosphorylglycerol sodium salt (DLPG) were purchased from Avanti Polar Lipids (Alabaster, USA). CriterionTGX Stain-Free (12%) precast gels and Bio-Beads SM-2 were obtained from Bio-Rad.
Construct design
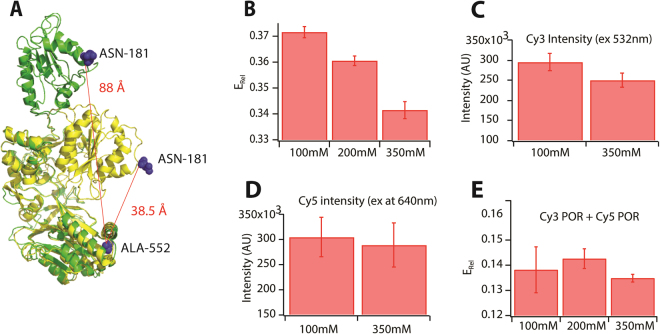
As the crystal structure of Sorghum bicolor POR is unavailable, homology modelling of Sorghum bicolor POR isoform 2b (GenBank: EES13592.1) was performed using the SWISS MODEL automated online server38. The compact conformation of SbPOR2b was based on POR from Rattus norvegicus as template (PDB ID: 1AMO_A4) while the open conformation was based on the yeast-human chimeric POR (PDB ID: 3FJO10), which displayed a sequence identity to SbPOR2b of 39% and 36%, respectively. Based on the 3D model structures, a pair of fully solvent accessible residues close to the FAD and FMN domains, but not directly involved in binding the flavin co-enzymes (N181 and A552 near the N- and C-termini, respectively) were selected for substitution with cysteine residues for fluorophore labeling. These residues would undergo major distance fluctuations during electron transfer with Cα-Cα distances of 38.5 Å to 88 Å (as calculated by the measurement wizard tool of PyMol software) in the compact and open conformations, respectively (Fig. 1A).
Figure 1.
(A) An overlay of the compact and extended conformations POR based on homology models. The calculated Cα-Cα inter-residue distances between N181 and A552 are shown. (B) Average ERel dependence on ionic strength. Increasing ionic strength shifts the average conformational equilibrium to low FRET states. Control experiments showing minimal effect of salt concentration on the fluorescence properties of Cy3 (C), Cy5 (D) or the mixture Cy3 and Cy5 (E).
Protein preparation and fluorophore labeling
The native surface exposed cysteine C536 was substituted to serine. The full-length, codon optimized POR gene containing specific cysteine mutations N181C/C536S/A552C was cloned into pET-52(b) expression vector (Genscript, USA). The full length POR mutant C536S/N181C/A552C was expressed in the E. coli BL21DE3 strain and purified using 2′5′-ADP Sepharose affinity chromatography as described previously36. The purified proteins were flash frozen in liquid nitrogen and stored at −80 °C until use. Fluorophore labeling of POR with an equimolar mixture of Cy3 and Cy5 mono-functional maleimides was carried according to manufacturer’s protocol (GE Healthcare). The free fluorophores were separated from the labeled proteins by gel filtration on PD-10 columns. Singly labeled POR with either Cy3 or Cy5 was prepared similarly for control experiments.
Cytochrome-c activity assay
The dual labeled POR enzyme (0.045 µM) in buffer containing Tris 50 mM (pH 7.3) and 100 mM NaCl was incubated with 50 µM cytochrome c in a 1 mL cuvette for 2–3 min. After recording a baseline for 1 min, the reaction was initiated by addition of 1 mM NADPH and the amount of cytochrome c reduced was monitored by the time-dependent increase in absorbance at 550 nm on a Perkin-Elmer Lambda 650 UV/Vis spectrophotometer (εCyt-c = 21.2 mM−1 cm−1). The slope of the curve in the linear region was used to determine the catalytic activity39.
Ensemble FRET
Dual labeled POR in micelles was diluted to a concentration of 15 nM in standard buffer containing 50 mM Tris (pH 7.3), 100 mM NaCl and 0.1% Triton-X 100. A spectrofluorometer (Fluoromax, Horiba) was used to excite the sample at 532 nm (slit width = 5 nm) while the emission spectrum was recorded between 550–720 nm. Control experiments were carried out in parallel with single labeled Cy3 and Cy5 POR. For ionic strength experiments, POR was dissolved in buffer containing 50 mM Tris (pH 7.3) and 0.1% Triton-X 100 while the concentration of NaCl was varied from 100 mM to 350 mM. Fluorescence spectra were acquired as described above.
Nanodisc reconstitution
POR reconstitution in nanodiscs was carried out following our recently published protocol40. In brief, the dual labeled POR solubilized in detergent was reconstituted into nanodiscs (NDs) containing the membrane scaffolding protein (MSP1E3D1) and either DLPC or a mixture of DLPC: DLPG (75/25 mol %) phospholipids. The molar ratios of MSP: Lipid: Protein was 1:120:0.1, which offers at least five-times molar excess of nanodiscs to POR enzyme. The incubation mixture also included 1 μM FMN and 1 μM FAD to replenish the enzyme with its co-factors. Detergent removal was facilitated by incubation with Bio-Beads for 4 h, which was followed by purification of nanodiscs by size exclusion chromatography (SEC) on a preparative HPLC (Shimadzu) equipped with a Superdex 200 Increase 10/300 GL (bed volume: 24 mL, flow rate: 0.5 mL/min). Cy3 POR and Cy5 POR were reconstituted into nanodiscs in a similar fashion.
Surface preparation
Glass slides were cleaned thoroughly35 by sonication in milli-Q water, 70% ethanol and 2% hellmanex and stored in methanol until use. Prior to experimental use, the glass slides were dried under nitrogen flow, plasma etched by a Plasma cleaner PDC 32 G and placed in custom-made teflon chambers. The slides were repeatedly washed with 80 µL of the standard buffer [50 mM Tris (pH 7.3), 100 mM NaCl].
Microscopy
FCS experiments on Cy3/Cy5 labeled POR were carried out in solution at nM concentrations. An inverted confocal microscope (Olympus IX71) was used along with an oil immersion objective HCX PL APO CS x 100 (NA 1.46) and two Avalanche Photodiode Detectors (APDs). Excitation at 543 nm was achieved by a diode laser (Thorlabs HGR020). The measurement was performed by focusing the laser 10 µm above the glass surface and the signals were collected in separate channels that were filtered with short-pass filters (Semrock) at above 650 nm for the donor channel and below 633 nm for the acceptor channel. The details of the microscopy setup used in this study and data acquisition are described in SI. (Supplementary methods section M1). All samples were measured at 100 mM and 400 mM NaCl concentrations and trajectories were collected for 300 sec.
Data treatment
The detailed explanation of parameters used for thresholding, FCS data binning, autocorrelation data fitting, EFRET calculation, and quantification of EFRET distribution using Bayesian Information Criterion (BIC) are presented in SI (Supplementary methods sections M2- M7).
Results
Preparation of dual labeled POR for FRET
The full-length POR containing site-specific mutations N181C/C536S/A552C was labeled with Cy3 (donor) and Cy5 (acceptor) maleimides. Dual labeled POR in detergent micelles were confirmed by SDS-PAGE by multichannel Cy3 and Cy5 detection (Supplementary Fig. S1). The absorption spectrum of dual labeled POR revealed equimolar labeling of Cy3 and Cy5 (Supplementary Fig. S2). Furthermore, dual labeled POR was functional (Supplementary Fig. S3), with a moderate loss (16%) in catalytic activity as compared to the unlabeled POR. This could be attributed to fluorophore labeling or loss of the loosely bound FMN cofactor during purification. Following, POR was reconstituted in nanodiscs and fractionated by size exclusion chromatography. Selected fractions were identified by absorbance at 280 nm (protein), 450 nm (flavins), 550 nm (Cy3) and 650 nm (Cy5) (Supplementary Fig. S4). The selected protein-loaded nanodisc fractions were combined and flash-frozen to be stored at −80 °C until use.
Ionic strength regulates conformational equilibrium of POR
The steady state fluorescence spectral analyses of dual-labeled POR in detergent micelles revealed a relative FRET efficiency of 37% (Fig. 1B). The effect of ionic strength on the POR conformational equilibrium was studied by incrementing the amount of NaCl in the buffer from 100 mM to 350 mM. This led to a 8% decrease from 0.37 to 0.34 in relative FRET efficiency related to an extension of the conformation towards a more open structure (Fig. 1B), which is in agreement with earlier findings41. Control experiments confirmed that addition of salt did not alter the fluorescence properties of Cy3, Cy5 or the equimolar mixture of Cy3 and Cy5 labeled POR (Fig. 1C–E).
Establishing a single molecule FRET platform to study conformational sampling
To directly observe and quantify the abundance of POR conformations we recorded smFRET by a parked beam confocal set-up. Experiments were performed using fluorescently labeled POR solubilized in detergent micelles (DM) or reconstituted in nanodiscs (NDs).
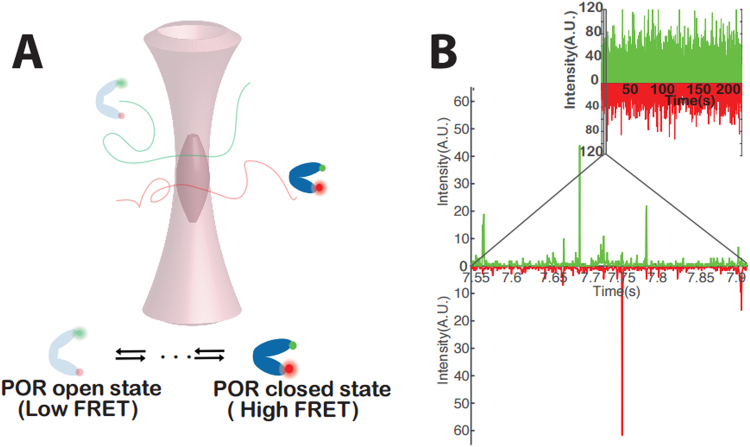
Our initial experiments were performed on POR in DM. Fluorescent bursts were collected from individual dual-labelled POR enzymes freely diffusing through the diffraction limited confocal volume (Fig. 2A). In typical trajectories, each coincident fluorescent burst in the Cy3 and Cy5 channels corresponds to an individual dual labeled POR enzyme diffusing through the confocal volume (Fig. 2B). We consequently evaluated the role of the reconstitution medium on the abundance and occupancy of POR conformational states. To do this, we recorded smFRET bursts of ND1 and ND2. To investigate the effect of ionic strength, a parallel set of smFRET experiments were conducted for DM, ND1 and ND2 in 400 mM NaCl.
Figure 2.
(A) A cartoon illustration of the experimental setup (B) Typical trajectory of detected photons recorded from ND reconstituted POR freely diffusing in solution. Coincident bursts correspond to an individual dual labeled POR traversing the diffraction-limited confocal volume. POR labeled with Cy3 or Cy5 only appear as single channel bursts. Inset shows an entire 250 s trace.
Direct observation and quantification of conformational states by smFRET
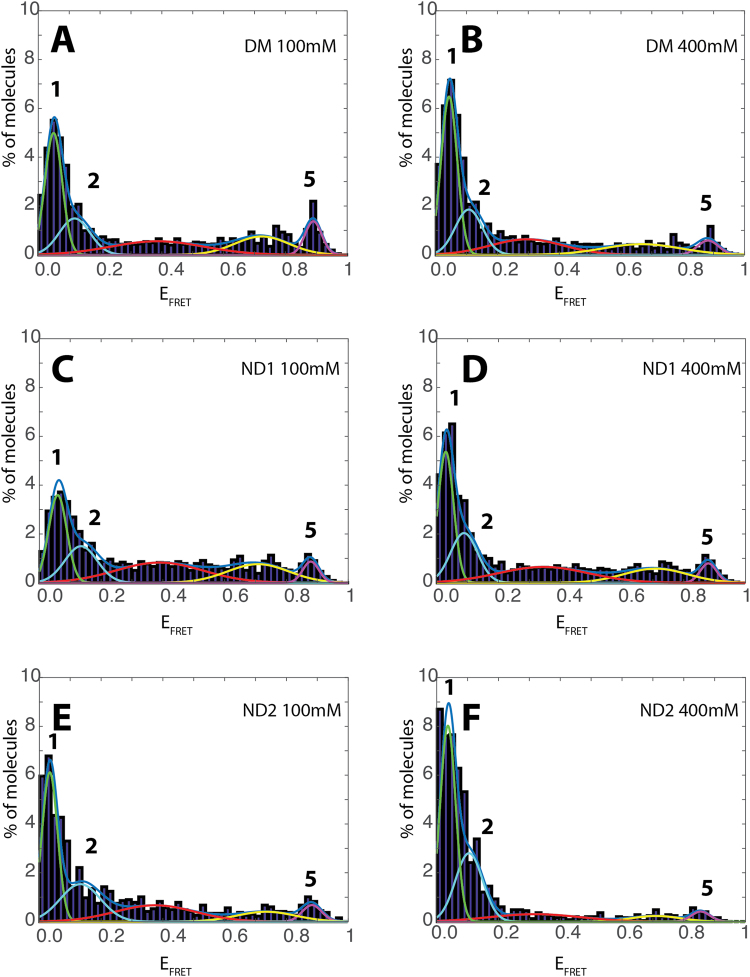
Several fluorescence trajectories were collected and binned at 300 μs and 600 μs, for detergent and nanodiscs reconstituted POR respectively. The bin time resolution was in accordance with the residence time of POR derived by autocorrelation curves (Supplementary Fig. S5, Supplementary Table 1). Particles below the threshold42 were discarded from the analysis. The signals obtained after threshold application were utilized to determine the FRET efficiency (EFRET) after correcting for donor bleed through, detector efficiency and acceptor cross excitation (Supplementary Fig. S6, method M1 and M6 for optimisation to avoid artefacts from dye photophysics). The histogram of EFRET for thousands of individual POR enzymes displayed a multimodal distribution indicating that POR adopts multiple conformations (Fig. 3A).
Figure 3.
(A,C and E) EFRET histograms of POR conformational sampling in detergent micelles (DM) and nanodiscs (ND1 and ND2) and (B,D and F) their dependence on increased ionic strength. The two “low FRET” peaks (1, 2) and high FRET peak (5) are indicated by line colors green, cyan and purple respectively.
To correctly evaluate the number and the position of the states we; 1) acquired data analysing a high number of molecules (typically between 104 and 7*104), 2) fitted an increasing number of additive single Gaussian distributions to non-binned data43,44 (Supplementary method M2). A Bayesian information criterion45 (BIC) was employed to find the number of distributions that best described our data (Supplementary Fig. S7). This revealed at least three dominating non-zero Gaussian peaks at EFRET 0.05 (peak 1), 0.12 (peak 2), 0.88 (peak 5) and two lower occupancy peaks (centered at ~0.4 and 0.6). The optimal fitting of data with five gaussians was also confirmed by Akaike information criterion (AIC) and the negative log likelihood in conjunction with Wilk’s theorem (Supplementary Table 2). Donor bleed through is also gaussian distributed and while accounted for by beta and gamma factor correction (Supplementary Fig. S6), some minor contribution might be observed in the low FRET states which might vary their ratios slightly, but not their existence (Supplementary Fig. S8). The EFRET values for peaks 1, 2 and 5 were utilized to calculate their inter-dye distances (Supplementary Table 3). The distances for peak 1 (86.6 Å) and peak 5 (38 Å) are in agreement with the Cα-Cα distances we expected from the homology models (i.e. 38.5 Å and 88 Å for the compact and extended conformations respectively, Fig. 1A). We note however that corrections for dye orientation may provide slightly different distances as compared to the homology model. Although we refrain ourselves from assigning defined conformational states based on single FRET derived distances, our single molecule data provide evidence for multiple conformations.
The FCS curves indicate the existence of low μs dynamics originating from POR and possibly from dye triplet state. Conformational dynamics occurring in time scales faster or comparable to the observation time46,47 may result in increased widths of the distributions as well as the observed plateau in intermediate EFRET values. Quantification of these dynamics falls out of the scope of this paper. The additional intermediate conformations could thus originate from unresolved conformational dynamics occurring in time scales faster or comparable to the observation time46,47. Additionally, the existence of two extended (Fig. 3A, peaks 1 and 2) and a compact conformation (Fig. 3A, peak 5) suggests a landscape of multiple conformations. The surprisingly narrow width of the high FRET state may indicate a very stable compact conformation as compared to more dynamic extended conformations. This is in agreement with recent SAXS and NMR studies, which suggests that POR exists in a dynamic equilibrium between a rigid closed state and a highly flexible open statecharacterized by a large ensemble of conformations15. Indeed the distance dependent broadening of the low FRET states is beyond dye photophysics48 (Supplementary Fig. S9). The two labeled POR domains (FAD and FMN) are connected via a flexible “hinge” domain. As this domain is highly flexible and partly disordered, this flexibility is expected to propagate to the POR domains introducing increased dynamics at greater distances of the extended conformations. These dynamics of the extended conformations would introduce the extra widening of the low EFRET distributions while the high EFRET distribution may corresponds to a well-defined compact conformation.
Regulation of conformational sampling by ionic strength and membrane environment
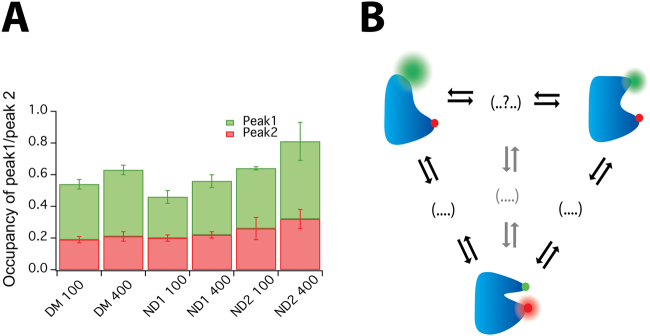
POR conformational equilibrium is regulated by ionic strength and membrane environment as previously determined by ensemble studies. Therefore, we evaluated the role of the reconstitution medium on the abundance and occupancy of the herein described POR conformational states. To do this, smFRET bursts of ND1 and ND2 were recorded and the data were binned at 600 µs based on the residence time in the confocal volume (Supplementary Fig. S5C). The positions and relative occupancies of the peaks did not change significantly upon moderate increasing in binning time (Supplementary Fig. 10A,B). In agreement with the data for POR in DM, BIC data analysis of ND1 and ND2 supported similar number of EFRET states and peak positions, albeit with a remodelled conformational distribution (Fig. 3C,E, supplementary methods M7). Both the EFRET and the state occupancy were found to strongly depend on the reconstitution medium. In order to rule out photo physical phenomenona of Cy3 when in close proximity to membranes or a protein49, we performed control experiments comparing Cy3 POR intensity distributions in detergent micelles and nanodiscs. Under our experimental conditions, we recorded a minute increase in the Cy3 intensity distribution in detergents as compared to nanodiscs (Supplementary Fig. S11). The relative occupancy of two extended conformations (sum of peaks 1 and 2) varied from POR reconstituted in DM (0.54 ± 0.03) to POR in NDs 0.47 ± 0.04 (13%) and 0.64 ± 0.12 (19%) for ND1 and ND2 respectively (Supplementary Table 4). More importantly, the relative ratio of occupancies for the two extended conformations was strongly dependent on the reconstitution medium and decreased from 1.8 for DM to 1.3 and 1.5 for ND1 and ND2 respectively (Fig. 4A) highlighting the strong effect of the reconstitution medium on membrane protein behaviour.
Figure 4.
(A) Ratio of the occupancies of peaks 1 and 2 for POR in detergent micelles (DM) and nanodiscs (ND1 and ND2) as a function of ionic strength (B) An illustrative model of conformational sampling in POR.
To investigate the effect of ionic strength, a parallel set of smFRET experiments were conducted in 400 mM NaCl. Control experiments revealed that addition of salt did not alter the residence time of POR in the confocal volume (Supplementary Fig. S5B,D) or the intensity of the donor or acceptor fluorophore (Supplementary Fig. S12). The Gaussian peak positions of the main peaks remained practically unchanged, however the occupancy of particular states varied significantly (Fig. 3B,D,F) indicating a regulatory mechanism enabling POR to redistribute the conformational landscape based on conformational selection50. For consistency, the data of ND2 were fit with 5 gaussians despite the practically complete shift towards the fully open states. The relative occupancy of two extended conformations (sum of peaks 1 and 2) increased 19% (from 0.54 ± 0.03 to 0.64 ± 0.02) for DM, 18% (from 0.47 ± 0.04 to 0.56 ± 0.03) for ND1 and 27% (from 0.64 ± 0.12 to 0.81 ± 0.16) for ND2 (Supplementary Table 4). This observed shift in equilibrium to extended states with increasing ionic strength is in agreement with previous reports proposing ionic strength as an efficient modulator of the POR conformational equilibrium and favouring open conformations15. Interestingly for all samples, this effect was mainly attributed to an increase in peak 1 corresponding to the most extended conformation of POR (Fig. 4A). This illustrates that the reconstitution medium affects the conformational landscape and its regulation by ionic strength.
Discussion
In conclusion, our smFRET measurements of full-length POR in solution allowed the first direct observation and quantification of at least three dominant FRET states that in agreement with earlier studies support the existence of multiple conformational states. Our data does not distinguish between the “swinging” and “rotating” domain motion models5 or a delicate mixture of the two. The recently published 2.3 Å resolution crystal structure of A. thaliana POR (ATR2)51 in the compact conformation implies that the edge-to-edge distance between FAD and FMN is 10.6 Å, which is much longer than that in rat or yeast POR. Furthermore, a “half-closed” state with the FMN domain rotated away from the rest of the protein suggests that POR may sample a broader diversity of conformational structures. The existence of two “extended” conformers could facilitate alternative transition pathways to the compact conformation leading to altered rates of electron flux. We have previously described that POR fluctuates between two functional states by single turnover measurements35 involving enzymatic conversion of the prefluorescent substrate resazurin to resorufin52. These states may be associated with the two alternative transitions pathways and pinpoints a central feature of enzyme chemistry defining the relationship between structure and function. We speculate that the existence of two or more transition pathways (Fig. 4B) may mediate selective activation of >50 downstream P450 partners within dynamic metabolons, which stoichiometric outnumber POR by a factor 10–2018,53. Importantly, environmental challenges and local lipid and solvent environment may act as additional regulatory cues biasing conformational sampling pathways and consequently POR selectivity18,19.
An increase of the ionic strength caused POR to redistribute its conformational equilibrium and populate more open conformations (Fig. 4A). The proposition that the conformational ensemble of proteins varies between detergent micelles and bilayers has been validated for several systems34,54,55.
Our assay could be expanded to study the conformational repertoire of various missense mutants of POR in the functionally significant regions that lead to clinical symptoms in human beings56. Additionally, elucidating the conformational profiles of multiple POR isoforms found in plants57 would be of physiological relevance as compelling evidences reveal that class I PORs are constitutively expressed while class II includes those induced by stresses58. Integrating such heterogeneities may provide the basis for an improved understanding of P450-centric metabolic systems59, and the molecular mechanisms controlling encountered substrate channeling18,60,61.
Electronic supplementary material
Acknowledgements
Financial support from the VILLUM Foundation to the VILLUM Research Center for Plant Plasticity is acknowledged. K.B acknowledges financial support from the European Union’s FP7/2007–2013/ (REA grant agreement number 289217). B.L.M. was supported by ERC Advanced Grant (ERC-2012-ADG_20120314, Project No: 323034), N.S.H. was supported by the VILLUM young Investigator program (Project No: 10099) and Carlsberg foundation distinguished associate professor fellowship CF16–0797. T.L. is the recipient of a post-doctoral fellowship awarded by the VILLUM Foundation (Project No: 95–300–73023). We thank Marijonas Tutkus (Department of Molecular Compound Physics, Center for Physical Sciences and Technology, Saulėtekio Avenue 3, LT-10257 Vilnius, Lithuania) for help with data treatment and modeling.
Author Contributions
K.B. designed the POR mutants and performed the protein expression, purification and nanodisc reconstitution. N.S.H. designed the single molecule measurements conducted by D.L. under the supervision of E.N.H. The ensemble FRET assays were performed by S.E. T.C.P. provided help with statistical analyses. Project design and scientific advice was provided by B.L.M., N.S.H. and T.L. K.B., T.L., N.S.H. and B.L.M. wrote the paper with subsequent contributions from the other authors. All authors have approved submission of the final version of the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Krutika Bavishi and Darui Li contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-24922-x.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Birger Lindberg Møller, Email: blm@plen.ku.dk.
Nikos S. Hatzakis, Email: hatzakis@nano.ku.dk
Tomas Laursen, Email: tola@plen.ku.dk.
References
- 1.Denisov IG, Makris TM, Sligar SG, Schlichting I. Structure and chemistry of cytochrome P450. Chem. Rev. 2005;105:2253–2277. doi: 10.1021/cr0307143. [DOI] [PubMed] [Google Scholar]
- 2.Munro AW, Girvan HM, Mason AE, Dunford AJ, McLean KJ. What makes a P450 tick? Trends Biochem. Sci. 2013;38:140–150. doi: 10.1016/j.tibs.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 3.Murataliev MB, Feyereisen R, Walker FA. Electron transfer by diflavin reductases. Biochim. Biophys. Acta, Proteins Proteomics. 2004;1698:1–26. doi: 10.1016/j.bbapap.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 4.Wang M, et al. Three-dimensional structure of NADPH-cytochrome P450 reductase: prototype for FMN- and FAD-containing enzymes. Proc. Natl. Acad. Sci. USA. 1997;94:8411–8416. doi: 10.1073/pnas.94.16.8411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Laursen T, Jensen K, Moller BL. Conformational changes of the NADPH-dependent cytochrome P450 reductase in the course of electron transfer to cytochromes P450. Biochim. Biophys. Acta, Proteins Proteomics. 2011;1814:132–138. doi: 10.1016/j.bbapap.2010.07.003. [DOI] [PubMed] [Google Scholar]
- 6.Hamdane D, et al. Structure and function of an NADPH-cytochrome P450 oxidoreductase in an open conformation capable of reducing cytochrome P450. J. Biol. Chem. 2009;284:11374–11384. doi: 10.1074/jbc.M807868200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xia C, et al. Conformational changes of NADPH-cytochrome P450 oxidoreductase are essential for catalysis and cofactor binding. J. Biol. Chem. 2011;286:16246–16260. doi: 10.1074/jbc.M111.230532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sugishima M, et al. Structural basis for the electron transfer from an open form of NADPH-cytochrome P450 oxidoreductase to heme oxygenase. Proc. Natl. Acad. Sci. USA. 2014;111:2524–2529. doi: 10.1073/pnas.1322034111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellis J, et al. Domain motion in cytochrome P450 reductase: conformational equilibria revealed by NMR and small-angle x-ray scattering. J. Biol. Chem. 2009;284:36628–36637. doi: 10.1074/jbc.M109.054304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aigrain L, Pompon D, Moréra S, Truan G. Structure of the open conformation of a functional chimeric NADPH cytochrome P450 reductase. EMBO Rep. 2009;10:742–747. doi: 10.1038/embor.2009.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leys D, Scrutton NS. Electrical circuitry in biology: emerging principles from protein structure. Curr. Opin. Struct. Biol. 2004;14:642–647. doi: 10.1016/j.sbi.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 12.Aigrain L, Fatemi F, Frances O, Lescop E, Truan G. Dynamic control of electron transfers in diflavin reductases. Int. J. Mol. Sci. 2012;13:15012–15041. doi: 10.3390/ijms131115012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang WC, Ellis J, Moody PC, Raven EL, Roberts GC. Redox-linked domain movements in the catalytic cycle of cytochrome p450 reductase. Structure. 2013;21:1581–1589. doi: 10.1016/j.str.2013.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jenner M, et al. Detection of a Protein Conformational Equilibrium by Electrospray Ionisation-Ion Mobility-Mass Spectrometry. Angew. Chem. Int. Ed. 2011;50:8291–8294. doi: 10.1002/anie.201101077. [DOI] [PubMed] [Google Scholar]
- 15.Frances O, et al. A Well-Balanced Preexisting Equilibrium Governs Electron Flux Efficiency of a Multidomain Diflavin Reductase. Biophys. J. 2015;108:1527–1536. doi: 10.1016/j.bpj.2015.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hay S, et al. Nature of the Energy Landscape for Gated Electron Transfer in a Dynamic Redox Protein. J. Am. Chem. Soc. 2010;132:9738–9745. doi: 10.1021/ja1016206. [DOI] [PubMed] [Google Scholar]
- 17.Hedison, T. M., Hay, S. & Scrutton, N. S. Real-time analysis of conformational control in electron transfer reactions of human cytochrome P450 reductase with cytochrome c. FEBS J., 10.1111/febs.13501 (2015). [DOI] [PMC free article] [PubMed]
- 18.Laursen T, et al. Characterization of a dynamic metabolon producing the defense compound dhurrin in sorghum. Science. 2016;354:890–893. doi: 10.1126/science.aag2347. [DOI] [PubMed] [Google Scholar]
- 19.Dastmalchi M, Facchini PJ. Plant metabolons assembled on demand. Science. 2016;354:829–830. doi: 10.1126/science.aal2948. [DOI] [PubMed] [Google Scholar]
- 20.Bassard J-E, Møller BL, Laursen T. Assembly of Dynamic P450-Mediated Metabolons—Order Versus Chaos. Curr. Mol. Biol. Rep. 2017;3:37–51. doi: 10.1007/s40610-017-0053-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li M, et al. Single Enzyme Experiments Reveal a Long-Lifetime Proton Leak State in a Heme-Copper Oxidase. J. Am. Chem. Soc. 2015;137:16055–16063. doi: 10.1021/jacs.5b08798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ha T. Single-molecule methods leap ahead. Nat. Methods. 2014;11:1015–1018. doi: 10.1038/nmeth.3107. [DOI] [PubMed] [Google Scholar]
- 23.Veshaguri S, et al. Direct observation of proton pumping by a eukaryotic P-type ATPase. Science. 2016;351:1469–1473. doi: 10.1126/science.aad6429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Velonia K, et al. Single-enzyme kinetics of CALB-catalyzed hydrolysis. Angew. Chem. Int. Ed. 2005;44:560–564. doi: 10.1002/anie.200460625. [DOI] [PubMed] [Google Scholar]
- 25.Bavishi K, Hatzakis NS. Shedding light on protein folding, structural and functional dynamics by single molecule studies. Molecules. 2014;19:19407–19434. doi: 10.3390/molecules191219407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He Y, Haque MM, Stuehr DJ, Lu HP. Single-molecule spectroscopy reveals how calmodulin activates NO synthase by controlling its conformational fluctuation dynamics. Proc. Natl. Acad. Sci. USA. 2015;112:11835–11840. doi: 10.1073/pnas.1508829112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haque MM, et al. Distinct conformational behaviors of four mammalian dual-flavin reductases (cytochrome P450 reductase, methionine synthase reductase, neuronal nitric oxide synthase, endothelial nitric oxide synthase) determine their unique catalytic profiles. FEBS J. 2014;281:5325–5340. doi: 10.1111/febs.13073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Whitelaw DA, Tonkin R, Meints CE, Wolthers KR. Kinetic analysis of electron flux in cytochrome P450 reductases reveals differences in rate-determining steps in plant and mammalian enzymes. Arch. Biochem. Biophys. 2015;584:107–115. doi: 10.1016/j.abb.2015.09.002. [DOI] [PubMed] [Google Scholar]
- 29.Kusnetzow AK, Altenbach C, Hubbell WL. Conformational states and dynamics of rhodopsin in micelles and bilayers. Biochemistry. 2006;45:5538–5550. doi: 10.1021/bi060101v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bond PJ, Sansom MS. Membrane protein dynamics versus environment: simulations of OmpA in a micelle and in a bilayer. J. Mol. Biol. 2003;329:1035–1053. doi: 10.1016/S0022-2836(03)00408-X. [DOI] [PubMed] [Google Scholar]
- 31.Popot JLA. nanodiscs, and fluorinated surfactants: three nonconventional approaches to studying membrane proteins in aqueous solutions. Annu. Rev. Biochem. 2010;79:737–775. doi: 10.1146/annurev.biochem.052208.114057. [DOI] [PubMed] [Google Scholar]
- 32.Denisov IG, Sligar SG. Nanodiscs in Membrane Biochemistry and Biophysics. Chem. Rev. 2017;117:4669–4713. doi: 10.1021/acs.chemrev.6b00690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Denisov IG, Grinkova YV, Lazarides AA, Sligar SG. Directed Self-Assembly of Monodisperse Phospholipid Bilayer Nanodiscs with Controlled Size. J. Am. Chem. Soc. 2004;126:3477–3487. doi: 10.1021/ja0393574. [DOI] [PubMed] [Google Scholar]
- 34.Pudney CR, Khara B, Johannissen LO, Scrutton NS. Coupled Motions Direct Electrons along Human Microsomal P450 Chains. PLoS Biol. 2011;9:e1001222. doi: 10.1371/journal.pbio.1001222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laursen T, et al. Single Molecule Activity Measurements of Cytochrome P450 Oxidoreductase Reveal the Existence of Two Discrete Functional States. ACS Chem. Biol. 2014;9:630–634. doi: 10.1021/cb400708v. [DOI] [PubMed] [Google Scholar]
- 36.Wadsater M, et al. Monitoring shifts in the conformation equilibrium of the membrane protein cytochrome P450 reductase (POR) in nanodiscs. J. Biol. Chem. 2012;287:34596–34603. doi: 10.1074/jbc.M112.400085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kalinin S, et al. A toolkit and benchmark study for FRET-restrained high-precision structural modeling. Nat. Methods. 2012;9:1218–1225. doi: 10.1038/nmeth.2222. [DOI] [PubMed] [Google Scholar]
- 38.Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guengerich FP, Martin MV, Sohl CD, Cheng Q. Measurement of cytochrome P450 and NADPH-cytochrome P450 reductase. Nat. Protoc. 2009;4:1245–1251. doi: 10.1038/nprot.2009.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bavishi K, Laursen T, Martinez KL, Moller BL, Della Pia EA. Application of nanodisc technology for direct electrochemical investigation of plant cytochrome P450s and their NADPH P450 oxidoreductase. Sci. Rep. 2016;6:29459. doi: 10.1038/srep29459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang WC, Ellis J, Moody PCE, Raven EL, Roberts GCK. Redox-Linked Domain Movements in the Catalytic Cycle of Cytochrome P450 Reductase. Structure. 2013;21:1581–1589. doi: 10.1016/j.str.2013.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Deniz AA, et al. Ratiometric single-molecule studies of freely diffusing biomolecules. Annu. Rev. Phys. Chem. 2001;52:233–253. doi: 10.1146/annurev.physchem.52.1.233. [DOI] [PubMed] [Google Scholar]
- 43.Solomatin SV, Greenfeld M, Chu S, Herschlag D. Multiple native states reveal persistent ruggedness of an RNA folding landscape. Nature. 2010;463:681–687. doi: 10.1038/nature08717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Do, C. B. & Batzoglou, S. What is the expectation maximization algorithm? Nat. Biotechnol. 26, 897, 10.1038/nbt1406. [DOI] [PubMed]
- 45.McKinney SA, Joo C, Ha T. Analysis of Single-Molecule FRET Trajectories Using Hidden Markov Modeling. Biophys. J. 2006;91:1941–1951. doi: 10.1529/biophysj.106.082487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schuler B, Eaton WA. Protein folding studied by single-molecule FRET. Curr. Opin. Struct. Biol. 2008;18:16–26. doi: 10.1016/j.sbi.2007.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gopich, I. V. & Szabo, A. In Single-Molecule Biophysics 245–297 (John Wiley & Sons, Inc., 2011).
- 48.Kalinin S, Sisamakis E, Magennis SW, Felekyan S, Seidel CAM. On the Origin of Broadening of Single-Molecule FRET Efficiency Distributions beyond Shot Noise Limits. J. Phys. Chem. B. 2010;114:6197–6206. doi: 10.1021/jp100025v. [DOI] [PubMed] [Google Scholar]
- 49.Stennett EMS, Ciuba MA, Lin S, Levitus M. Demystifying PIFE: The Photophysics Behind the Protein-Induced Fluorescence Enhancement Phenomenon in Cy3. J. Phys. Chem. Lett. 2015;6:1819–1823. doi: 10.1021/acs.jpclett.5b00613. [DOI] [PubMed] [Google Scholar]
- 50.Hatzakis NS. Single molecule insights on conformational selection and induced fit mechanism. Biophys. Chem. 2014;186:46–54. doi: 10.1016/j.bpc.2013.11.003. [DOI] [PubMed] [Google Scholar]
- 51.Niu G, et al. Structure of the Arabidopsis thaliana NADPH-cytochrome P450 reductase 2 (ATR2) provides insight into its function. FEBS J. 2017;284:754–765. doi: 10.1111/febs.14017. [DOI] [PubMed] [Google Scholar]
- 52.Janssen KPF, et al. Single molecule methods for the study of catalysis: from enzymes to heterogeneous catalysts. Chem. Soc. Rev. 2014;43:990–1006. doi: 10.1039/C3CS60245A. [DOI] [PubMed] [Google Scholar]
- 53.Peterson JA, Ebel RE, O’Keeffe DH, Matsubara T, Estabrook RW. Temperature dependence of cytochrome P-450 reduction. A model for NADPH-cytochrome P-450 reductase:cytochrome P-450 interaction. J. Biol. Chem. 1976;251:4010–4016. [PubMed] [Google Scholar]
- 54.Zoghbi ME, Cooper RS, Altenberg GA. The lipid bilayer modulates the structure and function of an ATP-binding cassette exporter. J. Biol. Chem. 2016;291:4453–4461. doi: 10.1074/jbc.M115.698498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Akyuz N, et al. Transport domain unlocking sets the uptake rate of an aspartate transporter. Nature. 2015;518:68–73. doi: 10.1038/nature14158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fluck CE, Nicolo C, Pandey AV. Clinical, structural and functional implications of mutations and polymorphisms in human NADPH P450 oxidoreductase. Fundam. Clin. Pharmacol. 2007;21:399–410. doi: 10.1111/j.1472-8206.2007.00520.x. [DOI] [PubMed] [Google Scholar]
- 57.Andersen TB, Hansen NB, Laursen T, Weitzel C, Simonsen HT. Evolution of NADPH-cytochrome P450 oxidoreductases (POR) in Apiales - POR 1 is missing. Mol. Phylogenet. Evol. 2016;98:21–28. doi: 10.1016/j.ympev.2016.01.013. [DOI] [PubMed] [Google Scholar]
- 58.Parage C, et al. Class II Cytochrome P450 Reductase Governs the Biosynthesis of Alkaloids. Plant Physiol. 2016;172:1563–1577. doi: 10.1104/pp.16.00801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tripathi S, Li H, Poulos TL. Structural basis for effector control and redox partner recognition in cytochrome P450. Science. 2013;340:1227–1230. doi: 10.1126/science.1235797. [DOI] [PubMed] [Google Scholar]
- 60.Gleadow RM, Møller BL. Cyanogenic glycosides: synthesis, physiology, and phenotypic plasticity. Annu. Rev. Plant Biol. 2014;65:155–185. doi: 10.1146/annurev-arplant-050213-040027. [DOI] [PubMed] [Google Scholar]
- 61.Laursen T, Møller BL, Bassard JE. Plasticity of specialized metabolism as mediated by dynamic metabolons. Trends Plant Sci. 2015;20:20–32. doi: 10.1016/j.tplants.2014.11.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.