Abstract
The P8 stem-loop region of the trnL intron, which is known to be hypervariable in size with multiple repeat motifs and created difficulties in alignment, is always excluded in phylogenetic as well as barcode analyses. This region was investigated for species discrimination in 98 taxa of orchids belonging to the tribe Vandeae using in silico mapping of restriction site polymorphism. The length of the P8 regions varied from 200 nucleotides in Aerides rosea to 669 nucleotides in Dendrophylax sallei. Forty two taxa had unique lengths, while as many as eight shared a common length of 521 nucleotides. Of the 35 restriction endonucleases producing digestions in the P8 regions, three, viz., AgsI, ApoI and TspDTI turned out to have recognition sites across all the 98 taxa being studied. When their restriction data were combined, 92 taxa could be discriminated leaving three taxon pairs. However, Acampe papillosa and Aeranthes arachnites despite having similar restriction sites differed in their P8 lengths. This is the first report on thorough investigation of the P8 region of trnL intron for search of species specific restriction sites and hence its use as a potential plant DNA barcode.
Introduction
For the past few decades there has been a hunt for a short DNA segment which can be used as a universal marker, popularly termed as DNA Barcode, for identification of faunal and floral species inhabiting this planet. For animal identification the mitochondrial gene cytochrome c oxidase subunit 1 (COX1) has proved successful [1] however, there is still difficulty in fixing a universal barcode for plants because of a more complex genetic background. Various nuclear and plastid coding and non-coding loci, viz., ITS1, ITS2, accD, matK, ndhJ, rpoB, rpoC1, ycf5, atpF-H, psbK-I, rbcL, rbcLa, trnH-psbA, trnL-F, etc. have been tested for validation either singly or in combination of two or more loci [2–4].
The chloroplast trnL-F non-coding region includes the trnL(UAA) intron ranging from 350 to 600 base pairs (bp) and the intergeneric spacer between trnL(UAA) 3′ exon and the trnF(GAA) gene [5,6]. The trnL(UAA) intron interrupts the anticodon loop of the tRNALeu, which is encoded in the large single copy region of the plastid genome. In the chloroplast DNA, trnL is the only Group I intron region having conserved secondary structure [7,8] with alternation of conserved and variable regions [9]. They are capable of catalyzing their own splicing from the flanking exons. The secondary structure of trnL intron contains regions of complimentary sequences that form nine stem-loop structures (P1-P9) [10]. Within these stems there are four regions (P, Q, R and S) conserved in primary sequences among all group I introns [11] and they are known as the catalytic core [12]. The P8 stem-loop region of the trnL intron is known to be hypervariable in size with multiple repeat motifs [12,13] and created difficulties in alignment. Therefore, this region is always excluded in phylogenetic as well as barcode analyses [14–17].
Orchidaceae comprises of 850 genera and 20,000 species which are arranged in five subfamilies, 22 tribes and 70 sub-tribes [18]. The tribe Vandeae consists of five sub-tribes, 139 genera with 2600 species of monopodial epiphytes distributed in tropical America, tropical and southern Africa, tropical and sub-tropical Asia, eastern Australia and Tasmania, much of the tropical Pacific south to New Zealand and east to Tahiti [19]. Considering the hypervariability to be an intrinsic property of the P8 region of trnL(UAA) intron, the present investigation was undertaken to find out whether the locus had any potential in discrimination of closely related species belonging to the tribe Vandeae based on in silico restriction site polymorphism analysis.
Materials and methods
Gene sequences
One hundred and twenty five sequences of tRNALeu (trnL) gene for orchids belonging to tribe Vandeae were downloaded from GenBank. These included ten sequences (GU185926, GU185928, GU185931, GU185932, GU185933, GU185934, GU185935, GU185938, GU185939 and GU185940) generated and submitted by the first author. Methods followed for DNA extraction and PCR amplification were given by Kishor and Sunitibala [20]. Sequencing was done using 3730 DNA Analyzer (Applied Biosystems, Warrington, UK) available at DNA Sequencing Facility, University of Delhi, South Campus. The primer pair ‘c’ (3′CGAAATCGGTAGACGCTACG5′) and ‘d’ (3′GGGGATAGAGGGACTTGAAC5′) [5] were used for amplification as well as sequencing.
In silico analysis of restriction site polymorphism for species discrimination
The trnL intron borders were delimited by identifying the binding sites of the primers ‘c’ and ‘d’. We looked for the P8 region by delimiting the borders following the method of Borsch et al. [13] and by considering the secondary structures of trnL of Campylopus flexuous [21] and Nymphaea odorata [13]. The correctness of the borders was again verified by considering the secondary structures of the P8 regions drawn for Aerides odorata, A. sukauensis and A. krabiensis [16].
In silico restriction mapping was done using the online software RestrictionMapper Version 3 (http://restrictionmapper.org/). Selection was made to include all commercially available restriction enzymes and find the base pair position where each enzyme cut the trnL P8 region for all the taxa. The program was set to allow providing maximum cuts with a minimum site length of 5 nucleotides. As many as 35 restriction endonucleases were found to have recognition sites in the P8 regions; however, only four, viz., AgsI, ApoI, TspDTI and VspI (five and six base cutters) were selected for the analysis because each of them had their recognition sites in the region across all the taxa.
The in silico mapped restrictions produced by the four enzymes were scored by presence (1) or absence (0). Further analyses were done using the computer package NTSYS [22]. Similarity matrix was generated using the program Qualitative in NTSYS 2.20e package. This matrix was subjected to the unweighted pair group method with arithmetical averages (UPGMA). Cluster analysis was performed on the similarity matrix with the SAHN program using UPGMA and the dendrograms were generated with the TREE program. Analysis was done for each enzyme as well as for combination of two, three and four of them.
Results
Length variation of trnL P8 region
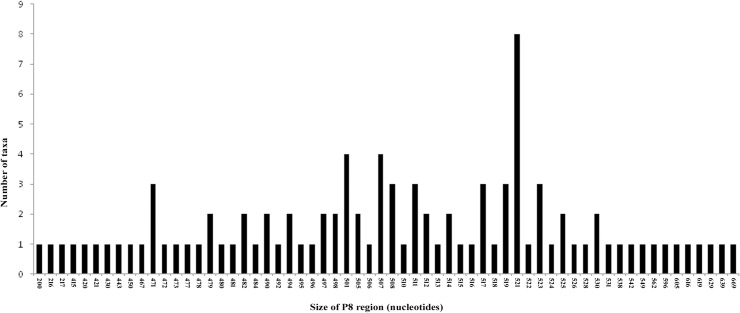
The downloaded trnL sequences for the 125 accessions contained many with incomplete sequences. After establishing the borders, only 98 taxa with complete sequence of the P8 region could be selected for the analysis. The length of the P8 regions varied from 200 nucleotides (Aerides rosea) to 669 nucleotides (Dendrophylax sallei) (Fig 1, Table 1). Forty two taxa had unique lengths, while as many as eight shared a common length of 521 nucleotides (Aeranthes arachnites, Aeranthes grandiflora, Angraecum conchiferum, Angraecum dives, Angraecum leonis, Bonniera appendiculata, Phalaenopsis amboinensis, and Phalaenopsis modesta). Within the genus Phalaenopsis, which is represented by 23 species in the present study, length variation of the P8 regions ranged from 494 (P. tetraspis) to 538 nucleotides (P. lindenii); while in eight taxa of Vanda, it ranged from 471 (V. testaceae) to 562 nucleotides (V. grifithii).
Fig 1. Length variation of the trnL(UAA) intron P8 regions of the 98 taxa of orchids versus number of taxa sharing common lengths.
Table 1. Sequence lengths of trnL P8 regions of 98 orchid taxa along with the restriction sites for the endonucleases AgsI, ApoI, TspDTI and VspI.
| Taxa | GenBank Accession No. | Length of P8 region (nucleotides) |
Restriction endonucleases and their respective restriction sites (nucleotides) | |||
|---|---|---|---|---|---|---|
| Ags1 (TTSAA) | Apo1 (RAATTY) | TspDTI (ATGAA) | VspI (ATTAAT) | |||
| Acampe ochracea | DQ091438 | 421 | 259, 265, 293, 357, 400, 406 | 146, 180, 344, 380 | 27, 193, 289, 345, 361 | 121, 192 |
| Acampe papillosa | DQ091439 | 420 | 259, 265, 293, 357, 400 | 146, 180, 344, 380 | 27, 193, 289, 345, 361, 418 | 121, 192 |
| Aeranthes arachnites | DQ091536 | 521 | 259, 265, 293, 357, 400 | 146, 180, 344, 380 | 27, 193, 289, 345, 361, 418 | 121, 192 |
| Aeranthes grandiflora | DQ091537 | 521 | 238, 400, 428, 434, 452, 500 | 145, 238, 445 | 27, 263, 405, 463, 519 | 120, 301 |
| Aerides crassifolia | EF670402 | 492 | 195, 397, 403, 428, 471 | 146, 189, 196, 415, 451, 472 | 27, 201, 221, 226, 318, 374, 432 | 121, 263 |
| Aerides crispa | EF670407 | 484 | 189, 361, 389, 395, 412, 420, 463 | 140, 183, 190, 407, 443, 464 | 27, 195, 215, 310, 366, 424 | 115, 230, 241 |
| Aerides krabiensis | EF670404 | 217 | 196 | 155, 190, 197 | 27, 202 | 130 |
| Aerides multiflora | DQ194983 | 216 | 195 | 146, 189, 196 | 27, 190, 201 | 121 |
| Aerides odorata | EF670389 | 450 | 166, 327, 355, 361, 386, 429 | 125, 160, 167, 373, 409, 430 | 27, 172, 192, 197, 276, 332, 390 | 100, 221 |
| Aerides rosea | EF670405 | 200 | 138, 179 | 138, 173, 180 | 27, 174, 185 | 113 |
| Aerides sukauensis | EF670408 | 479 | 195, 356, 384, 390, 415, 458 | 189, 196, 402, 459 | 27, 201, 221, 305, 361, 419 | 121, 236 |
| Ancistrorhynchus cephalotes | EF670435 | 619 | 345, 526, 532, 598 | 145, 345, 544 | 27, 199, 370, 503 | 120, 603, 120, 603 |
| Angraecum calceolus | DQ091546 | 505 | 238, 420, 484 | 145, 238, 431 | 27, 263, 447 | 120, 294, 489 |
| Angraecum conchiferum | DQ091539 | 521 | 238, 428, 434, 500 | 145, 238, 446 | 27, 263, 405, 463 | 120, 301, 505 |
| Angraecum dives | DQ091547 | 521 | 238, 400, 428, 434, 500 | 145, 238, 446 | 27, 263, 405 | |
| Angraecum eichlerianum | AF506341 | 639 | 145, 238, 446 | 147, 359, 558 | 27, 194, 201, 384, 517, 575 | 122, 617, 623 |
| Angraecum florulentum | DQ091550 | 513 | 237, 392, 420, 426, 492 | 145, 237, 438 | 27, 262, 397, 455 | 120, 293, 497 |
| Angraecum leonis | DQ091551 | 521 | 238, 400, 428, 434, 500 | 145, 238, 446 | 27, 263, 405, 463 | 120, 301, 505 |
| Angraecum magdalenae | AF519973 | 616 | 334, 490, 524, 595 | 145, 254, 334, 536 | 27, 210, 359, 495, 553 | 120, 243, 391, 600 |
| Angraecum rutenbergianum | DQ091548 | 443 | 321, 349, 355, 421 | 145, 367 | 27, 326, 384 | 120, 222, 426 |
| Angraecum teres | DQ091552 | 542 | 145, 252, 266, 421, 449, 455, 521 | 266, 467 | 27, 291, 426, 484 | 120, 322, 526 |
| Arachnis labrosa | GU185926 | 478 | 195, 357, 385, 391, 416, 459 | 146, 189, 196, 403, 460 | 27, 201, 221, 306, 362, 420 | 121, 236, 250 |
| Ascocentrum curvifolium | EF670423 | 494 | 187, 371, 399, 405, 430, 473, 479 | 146, 180, 188, 417, 438, 453 | 27, 193, 213, 320, 376, 434 | 121, 228, 250, 265 |
| Beclardia macrostachya | DQ091497 | 523 | 259, 411, 439, 445, 502 | 145, 259, 457 | 27, 192, 198, 284, 416 | 120, 312, 507 |
| Bonniera appendiculata | DQ091541 | 521 | 238, 428, 434, 451, 500 | 145, 238, 446 | 27, 263, 405, 463 | 120, 301, 505 |
| Christensonia vietnamica | EF670413 | 497 | 200, 367, 395, 401, 426, 476, 482 | 146, 193, 201, 413, 434, 449 | 27, 206, 226, 316, 372, 43027, 206, 226, 316, 372, 430 | 121, 247 |
| Cleisostoma racemiferum | GU185928 | 525 | 231, 402, 430, 436, 461, 504, 510 | 146, 224, 232, 448, 484 | 27, 163, 214, 230, 237, 257, 351, 407, 465 | 121, 204, 273, 296 |
| Cryptopus elatus | DQ091450 | 523 | 240, 402, 430, 436, 453, 502 | 145, 240, 448 | 27, 265, 407, 465 | 120, 303, 507 |
| Cryptopus paniculatus | DQ091451 | 514 | 233, 393, 421, 427, 444, 493 | 145, 233, 439 | 27, 258, 398, 456 | 120, 498 |
| Dendrophylax sallei | AY147234 | 669 | 186, 379, 509, 582, 648 | 145, 379, 594 | 27, 192, 210, 272, 404, 553, 560, 611 | 120, 653 |
| Dimorphorchis rossii var. graciliscapa | EF670429 | 519 | 214, 396, 424, 430, 455, 498, 504 | 141, 208, 215, 278, 284, 442, 478 | 27, 220, 240, 345, 401, 459 | 116, 255, 269 |
| Holcoglossum kimballianum | EF670419 | 629 | 218, 506, 534, 540, 565, 608 | 146, 212, 219, 552, 588, 609 | 27, 224, 244, 455, 511, 569 | 121, 255, 262, 285, 292, 315, 322 |
| Holcoglossum subulifolium | EF670409 | 467 | 187, 344, 372, 378, 446 | 146, 181, 188, 390, 426, 447 | 27, 193, 213, 293, 349, 407 | 121, 224 |
| Jumellea maxillarioides | DQ091554 | 506 | 238, 408, 419, 485 | 145, 238, 431 | 27, 263, 392, 450 | 120, 286 |
| Jumellea sagittata | DQ091555 | 508 | 238, 387, 415, 421, 487 | 145, 238, 433 | 27, 263, 390, 448 | 120, 288, 492 |
| Lemurorchis madagascariensis | DQ091556 | 528 | 238, 407, 435, 441, 507 | 145, 238, 453 | 27, 412, 470 | 120, 512 |
| Listrostachys pertusa | DQ091509 | 524 | 238, 403, 431, 437, 454, 503 | 145, 238, 398, 449 | 27, 157, 263, 408, 466 | 120 |
| Luisia trichorrhiza | EF670428 | 605 | 213, 482, 510, 516, 541, 584 | 146, 207, 214, 376, 499, 528, 564, 585 | 219, 239, 243, 259, 263, 279, 387, 431, 487, 545 | 121, 303 |
| Luisia tristis | EF670426 | 596 | 219, 473, 501, 507, 532, 575 | 161, 213, 220, 490, 519, 576 | 225, 245, 249, 265, 269, 285, 374, 393, 422, 478, 536 | 136, 309 |
| Microcoelia stolzii | DQ091530 | 480 | 221, 359, 387, 393, 459 | 145, 221, 354, 405 | 27, 309, 364, 422 | 120 |
| Neofinetia falcata | EF670421 | 497 | 187, 374, 402, 408, 433, 476, 482 | 146, 181, 188, 391, 420, 456 | 27, 193, 213, 273, 323, 379, 437 | 121, 228, 237, 253 |
| Oeonia rosea | DQ091452 | 516 | 395, 423, 429, 495 | 145, 233, 441 | 27, 258, 400, 458 | 120, 296, 500 |
| Paraphalaenopsis labukensis | EF670425 | 507 | 384, 412, 418, 443 | 146, 401, 430, 466, 487 | 27, 210, 217, 222, 229, 314, 333, 389, 447 | 121, 250, 264 |
| Pelatantheria insectifera | GU185931 | 482 | 187, 383, 389, 406, 414, 461, 467 | 146, 180, 188, 401, 422, 437 | 27, 193, 213, 320, 382, 388, 418 | 121, 260 |
| Phalaenopsis amboinensis | AY273643 | 521 | 178, 398, 426, 432, 457, 500, 506 | 137, 172, 179, 444, 480 | 27, 184, 204, 211, 403, 461 | 112, 226, 234, 286 |
| Phalaenopsis bellina | AY273632 | 495 | 178, 372, 400, 406, 431, 474, 480 | 137, 172, 179, 418, 454 | 27, 184, 204, 377, 435 | 112, 219, 227, 246, 260 |
| Phalaenopsis celebensis | AY265799 | 507 | 187, 384, 412, 418, 443, 486, 492 | 146, 181, 188, 430 | 27, 193, 213, 389, 447 | 121, 228, 242, 262 |
| Phalaenopsis corningiana | AY273670 | 508 | 178, 385, 413, 419, 444, 487, 493 | 137, 172, 179, 431, 467 | 27, 184, 204, 390, 448 | 112, 219, 235, 249 |
| Phalaenopsis cornu-cervi | AY265751 | 501 | 378, 406, 412, 437, 480, 486 | 137, 172, 424 | 27, 184, 204, 383, 441 | 112, 219, 227, 242, 256 |
| Phalaenopsis deliciosa | DQ091444 | 415 | 292, 320, 326, 351, 394, 400 | 146, 181, 338, 374 | 27, 193, 297, 355 | 121 |
| Phalaenopsis inscriptiosinensis | AY273673 | 498 | 178, 375, 403, 409, 434, 477, 483, 676, 873, 901, 907, 932, 975, 981 | 137, 172, 179, 421, 457, 635, 670, 677, 919, 955 | 27, 184, 204, 380, 438, 525, 682, 702, 878, 936 | 112, 219, 241, 263, 610, 717, 739, 761 |
| Phalaenopsis lamelligera | AY273679 | 501 | 178, 378, 406, 412, 437, 480, 486 | 137, 172, 179, 424, 460 | 27, 184, 204, 321, 383, 441 | 112, 219, 227, 242, 256 |
| Phalaenopsis lindenii | AY273649 | 538 | 211, 415, 443, 449, 474, 517, 523 | 146, 205, 212, 461, 497 | 27, 217, 237, 420, 478 | 121, 252, 270, 284 |
| Phalaenopsis lueddemanniana | AY273640 | 517 | 394, 422, 428, 453, 496, 502 | 137, 172, 179, 440, 476 | 27, 184, 204, 399, 457 | 112, 219, 227, 246, 268 |
| Phalaenopsis maculata | AY273641 | 519 | 396, 424, 430, 455, 498, 504 | 147, 189, 197, 442, 478 | 27, 201, 222, 401, 459 | 122, 254, 268 |
| Phalaenopsis mannii | AF519969 | 523 | 185, 400, 428, 434, 459, 502, 508 | 137, 179, 186, 446, 482 | 27, 191, 211, 405, 463 | 112, 226, 234, 267, 288 |
| Phalaenopsis mariae | AY265770 | 519 | 396, 424, 430, 455, 498, 504 | 137, 172, 179, 442, 478 | 27, 184, 204, 231, 401, 459 | 112, 219, 227, 247, 261, 284 |
| Phalaenopsis micholitzii | AY273701 | 505 | 178, 382, 410, 416, 441, 484, 490 | 137, 172, 179, 428, 464 | 27, 184, 204, 387, 445 | 112, 219, 227, 246, 260 |
| Phalaenopsis modesta | AY265793 | 521 | 178, 398, 426, 432, 457, 500, 506 | 137, 172, 179, 444, 480 | 27, 184, 204, 403, 461 | 112, 236, 262, 272, 286 |
| Phalaenopsis pantherina | AY273666 | 501 | 178, 378, 406, 412, 437, 480, 486 | 137, 172, 179, 424, 460 | 27, 184, 204, 217, 321, 383, 441 | 112, 219, 242, 256 |
| Phalaenopsis philippinensis | AY273656 | 511 | 192, 388, 416, 422, 447, 490, 496 | 151, 186, 193, 434, 470 | 27, 198, 218, 393, 451 | 126, 245, 259 |
| Phalaenopsis pulchra | AY273639 | 517 | 394, 422, 428, 453, 496, 502 | 137, 179, 440, 476 | 27, 204, 399, 457 | 112, 218, 226, 245, 267, 281 |
| Phalaenopsis schilleriana | AY265781 | 511 | 192, 388, 416, 422, 447, 490, 496 | 151, 186, 193, 434, 470 | 27, 198, 218, 393, 451 | 126, 245, 259 |
| Phalaenopsis stuartiana | AY273654 | 507 | 384, 412, 418, 443, 486, 492 | 146, 181, 430, 466 | 27, 193, 213, 389, 447 | 121, 240, 254 |
| Phalaenopsis sumatrana | AY273695 | 508 | 178, 385, 413, 419, 444, 487, 493 | 137, 172, 179, 431, 467 | 27, 184, 204, 390, 448 | 112, 219, 235, 249 |
| Phalaenopsis tetraspis | AY265784 | 494 | 178, 371, 399, 405, 430, 473, 479 | 137, 172, 179, 417, 453 | 27, 184, 204, 376, 434 | 112, 219, 235, 249, 259 |
| Phalaenopsis violacea | AY273635 | 507 | 178, 384, 412, 418, 443, 486, 492 | 137, 172, 179, 430, 466 | 27, 184, 204, 389, 447 | 112, 219, 227, 246, 272 |
| Pomatocalpa diffusum | EF670432 | 471 | 187, 348, 376, 382, 399, 407, 450, 456 | 146, 180, 188, 394 | 213, 297, 353, 411 | 121, 228 |
| Pomatocalpa spicatum | EF670431 | 490 | 146, 187, 367, 395, 418, 426, 469 | 146, 180, 188, 413, 470 | 27, 193, 213, 316, 372, 430 | 121, 228, 247 |
| Rangaeris amaniensis | DQ091512 | 517 | 238, 424, 430, 496 | 145, 238, 442 | 27, 263, 401, 408, 459 | 120 |
| Rangaeris rhipsalisocia | DQ091511 | 515 | 145, 239, 427, 433, 494 | 239, 440 | 27, 185, 264, 404, 457 | 120 |
| Renanthera imschootiana | GU185932 | 472 | 147, 188, 349, 377, 383, 400, 408, 451, 457 | 147, 182, 189, 343, 395, 416, 431 | 27, 194, 214, 298, 354, 412 | 122, 229 |
| Renanthera matutina | AY273688 | 498 | 146, 375, 409, 434, 483 | 146, 181, 188, 421 | 27, 213, 324, 380 | 121, 228, 246, 256, 275, 448 |
| Renantherella histrionica | AY273692 | 496 | 146, 187, 386, 414, 420, 445 | 146, 181, 188, 380, 432, 468 | 27, 193, 208, 213, 335, 391, 449 | 121, 228, 241, 265 |
| Rhipidoglossum kamerunense | DQ091492 | 512 | 238, 418, 424, 491 | 145, 238, 437 | 27, 263, 269, 395, 454 | 120, 496 |
| Rhipidoglossum rutilum | DQ091494 | 522 | 238, 428, 434, 501 | 145, 238, 447 | 27, 263, 269, 405, 464 | 120, 506 |
| Rhipidoglossum subsimplex | DQ091496 | 512 | 238, 418, 424 | 145, 238, 437 | 27, 263, 269, 395, 454 | 120, 496 |
| Rhynchostylis gigantea | EF670411 | 479 | 195, 356, 384, 390, 407, 415, 458 | 146, 189, 196, 402, 438, 459 | 27, 73, 201, 221, 305, 361, 419 | 121, 236, 250 |
| Rhynchostylis retusa | EF670424 | 501 | 378, 406, 412, 429, 480 | 146, 424, 445, 460, 481 | 27, 201, 327, 383, 441 | 121, 226, 246, 265 |
| Sedirea japonica | EF670433 | 473 | 350, 378, 384, 401, 409, 452 | 146, 396, 432, 453 | 27, 193, 200, 355, 413 | 121, 222, 244 |
| Seidenfadenia mitrata | EF670414 | 490 | 200, 367, 395, 401, 426, 469, 475 | 146, 193, 201, 413, 449 | 27, 206, 226, 316, 372, 430 | 121, 247 |
| Smitinandia micrantha | EF670415 | 481 | 187, 319, 325, 353, 410, 417, 460, 466 | 146, 180, 188, 404, 440 | 27, 193, 213, 349, 405, 421 | 121, 228, 252 |
| Sobennikoffia humbertiana | DQ091558 | 514 | 238, 393, 421, 427, 493 | 145, 238, 439 | 27, 263, 398, 456 | 120, 295, 498 |
| Trichoglottis atropurpurea | DQ091440 | 430 | 330, 335, 341, 366, 409 | 146, 188, 302, 353, 410 | 27, 201, 256, 312, 370 | 121, 196, 200 |
| Trichoglottis bipunctata | EF670430 | 531 | 187, 436, 442, 459, 467, 510 | 146, 180, 188, 403, 454, 511 | 27, 193, 219, 356, 412, 471 | 121, 248, 279 |
| Tridactyle crassifolia | DQ091515 | 530 | 55, 145, 244, 437, 443, 509 | 244, 455 | 27, 269, 414, 472 | 120 |
| Tridactyle filifolia | DQ091516 | 530 | 145, 244, 437, 443, 509 | 244, 455 | 27, 269, 414, 472 | 120 |
| Tridactyle tanneri | DQ091520 | 518 | 239, 358, 425, 431, 497 | 146, 239, 443 | 27, 158, 264, 402, 460 | 121 |
| Vanda alpina | GU185933 | 526 | 187, 403, 431, 437, 462, 505, 511 | 146, 180, 188, 449, 485 | 27, 193, 213, 352, 408, 466 | 121, 228, 259, 277, 295 |
| Vanda coerulea | GU185935 | 549 | 208, 426, 454, 460, 485, 528, 534 | 146, 201, 209, 472, 508 | 27, 214, 234, 375, 431, 489 | 121, 257, 271, 275, 294, 311, 328 |
| Vanda flabellata | EF670410 | 477 | 187, 354, 382, 388, 413, 456, 462 | 146, 180, 187, 400, 436 | 27, 193, 213, 303, 359, 417 | 121, 234 |
| Vanda griffithii | GU185934 | 562 | 187, 444, 472, 478, 541, 547 | 146, 180, 188 | 27, 193, 213, 375, 393, 449, 503 | 121, 228, 245, 276, 302, 320, 338 |
| Vanda luzonica | AY273699 | 511 | 187, 387, 415, 421, 447, 490, 496 | 146, 180, 188, 434, 470 | 27, 193, 213, 336, 392, 451 | 121, 262 |
| Vanda motesiana* | GU185938 | 510 | 187, 387, 415, 421, 438, 446, 489, 495 | 180, 188, 433, 469 | 27, 193, 213, 336, 392, 450 | 121, 228, 248, 262 |
| Vanda stangeana# | GU185939 | 525 | 187, 402, 430, 436, 461, 504, 510 | 146, 180, 188, 448, 484 | 27, 193, 220, 230, 332, 350, 406, 465 | 121, 245, 277 |
| Vanda testacea |
GU185940 |
471 | 187, 348, 376, 382, 399, 407, 450, 456 | 180, 188, 394, 430 | 27, 193, 213, 297, 353, 411 | 121, 228, 242 |
| Vandopsis gigantea |
EF670417 |
482 | 187, 348, 376, 382, 407 | 146, 180, 188, 394, 430, 451, 462 | 27, 193, 213, 297, 353, 411 | 121, 228, 242 |
| Vandopsis lissochiloides | EF670418 | 471 | 187, 348, 376, 382, 407, 450 | 146, 180, 188, 394, 451 | 27, 193, 213, 297, 353, 411 | 121, 228, 242 |
R = A or G, S = C or G, Y = C or T; while submitting to GenBank the trnL sequences
Vanda motesiana* was entered as V. stangeana and
V. stangeana# entered as V. tessellate
In silico analysis of restriction site polymorphism
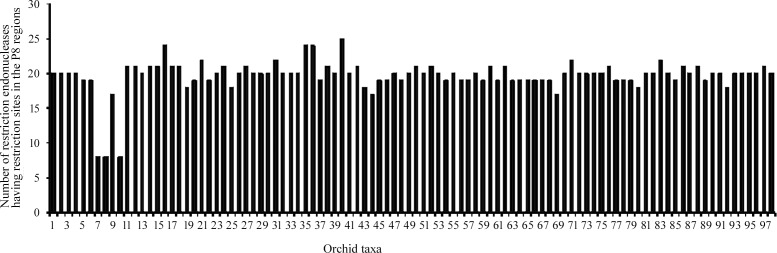
The result of the in silico restriction site analysis is presented in Table 1. Of all the commercially available restriction endonucleases being tried for the in silico restriction mapping using the online software ‘RestrictionMapper’, 35 endonucleases (AflII, AccI, AgsI, ApoI, AsuII, BciVI, BclI, BdaI, BglII, BsaAI, BsaBI, BsmI, CspCI, Eco57I, Eco57MI, EcoRI, EcoRV, HaeIV, MfeI, MboII, MslI, NdeI, PsiI, SmlI, SnaBI, Spe1, SspI, TfiI, TspDTI, TspGWI, TspRI, VspI, XbaI, XhoII and XmnI) were found to have recognition sites in the P8 region. Number of restriction endonucleases digesting the region varied from eight in Aerides krabiensis, A. odorata and A. rosea to 25 in Microcoelia stolzii (Fig 2). Again, from amongst the 35 restriction endonucleases, only three, viz., AgsI, ApoI and TspDTI turned out to have recognition sites across all the 98 taxa being studied; while VspI had in all excepting Angraecum dives. Hence, the above four were selected for subsequent analysis of the restriction site polymorphism.
Fig 2. Graph representing diversity of restriction endonucleases showing recognition sites in the P8 regions of the trnL(UAA) intron of the 98 taxa of orchids (1–98 represent names of 98 orchids in alphabetical order as presented in Table 1).
Species discrimination by single restriction endonuclease
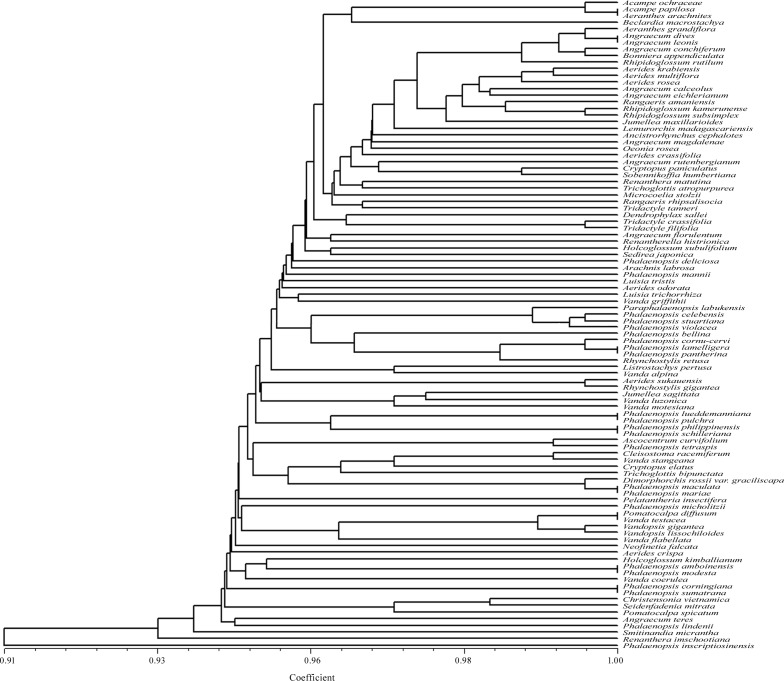
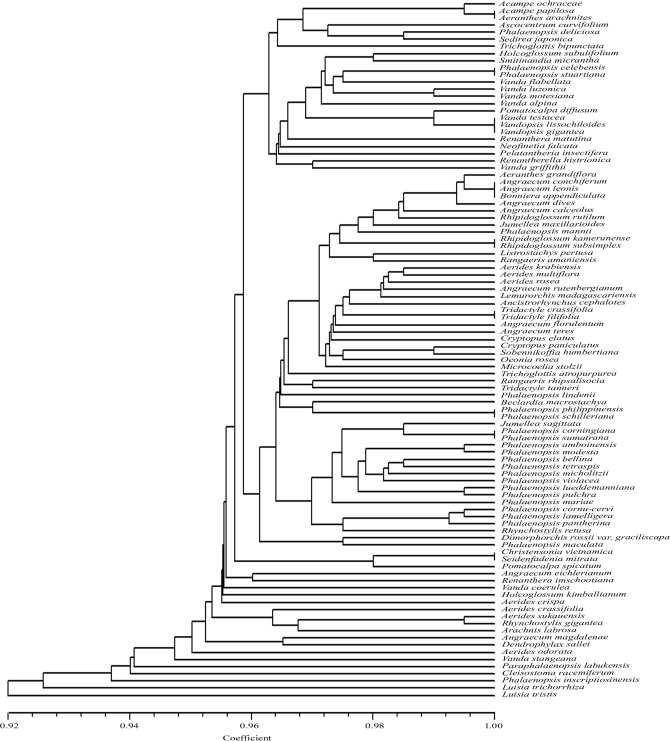
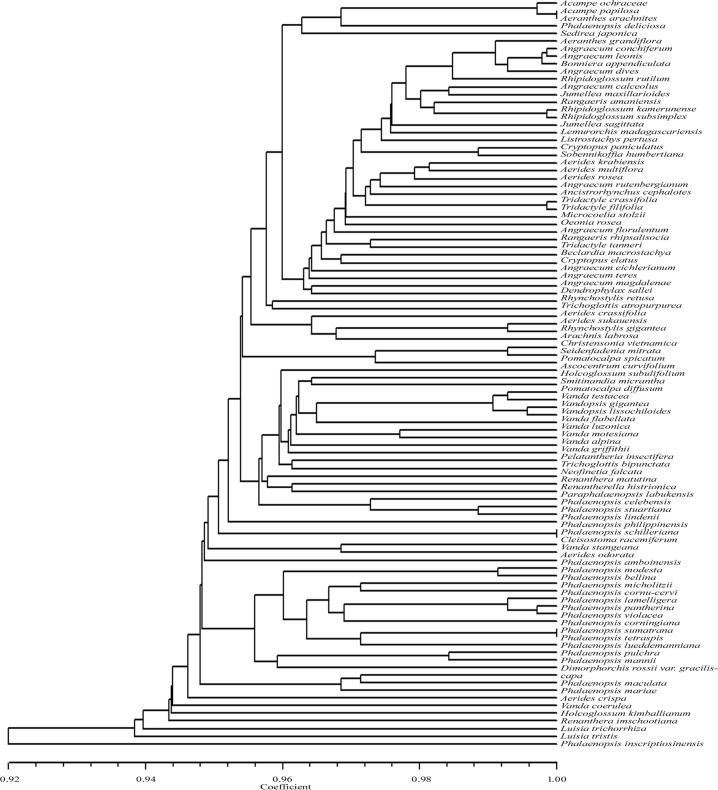
The restriction sites recognized by AgsI ranged from one (Aerides krabiensis and A. multiflora) to 14 (Phalaenopsis inscriptiosinensis) (Table 1). From the dendrogram constructed by the UPGMA method which showed similarity coefficient ranging from 0.91 to 1.00 (Fig 3), it was found that AgsI exhibited species specific restriction sites in 80 taxa; however, it could not provide discrimination for 18 taxa which included the taxon pairs Acampe papillosa and Aeranthes arachnites; Angraecum dives and Angraecum leonis; Phalaenopsis lamelligera and P. pantherina, P. lueddemanniana and P. pulchra; Phalaenopsis philippinensis and P. schilleriana; Phalaenopsis maculata and P. mariae; Phalaenopsis amboinensis and P. modesta; Phalaenopsis corningiana and P. sumatrana; Pomatocalpa diffusum and Vanda testacea. Each of these taxon pairs shared the same AgsI restriction sites.
Fig 3. UPGMA dendrogram based on SAHN clustering of data from in silico restriction site polymorphism analysis of the trnL(UAA) intron P8 region using the restriction endonuclease AgsI among the 98 taxa of orchids.
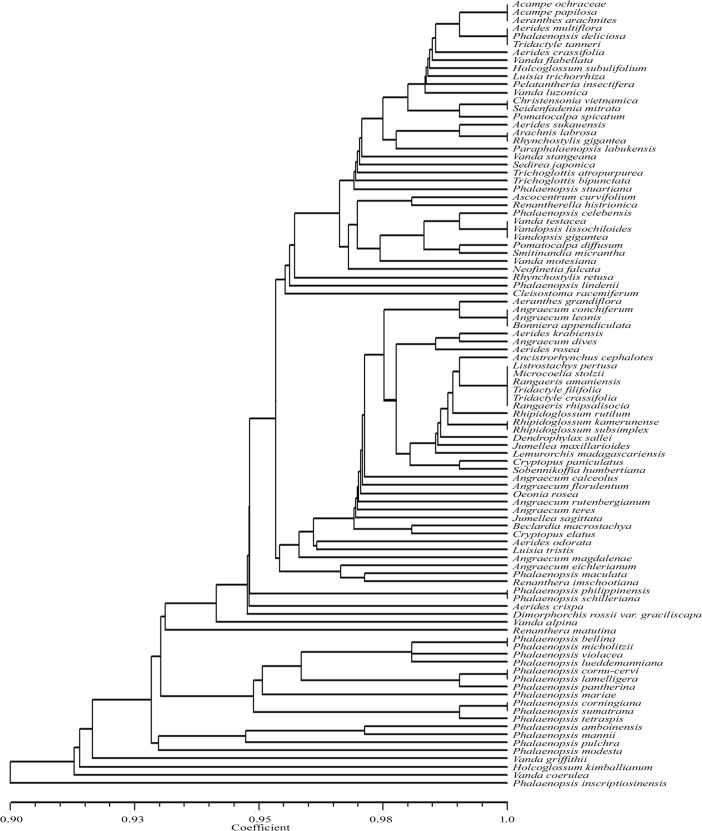
The ApoI recognized restriction sites ranged from two (in six taxa) to ten (Phalaenopsis inscriptiosinensis) (Table 1). As evidenced from the UPGMA dendrogram (Fig 4), of the 98 taxa analysed ApoI digestion discriminated 77 species, however; it could not show species specific restriction sites in 21 taxa. The following taxon pairs or groups shared the same restriction sites, Acampe ochraceae, Acampe papillosa and Aeranthes arachnites, Angraecum calceolus and Jumellea maxillarioides, Angraecum conchiferum, Angraecum dives, Angraecum leonis and Bonniera appendiculata, Rhipidoglossum kamerunense and Rhipidoglossum subsimplex, Tridactyle crassifolia and Tridactyle filifolia, Phalaenopsis amboinensis and P. modesta, Phalaenopsis lamelligera and P. pantherina, Phalaenopsis corningiana and P. sumatrana, Phalaenopsis philippinensis and P. schilleriana. This dendrogram showed similarity coefficient ranging from 0.92 to 1.00.
Fig 4. UPGMA dendrogram based on SAHN clustering of data from in silico restriction site polymorphism analysis of the trnL(UAA) intron P8 region using the restriction endonuclease ApoI among the 98 taxa of orchids.
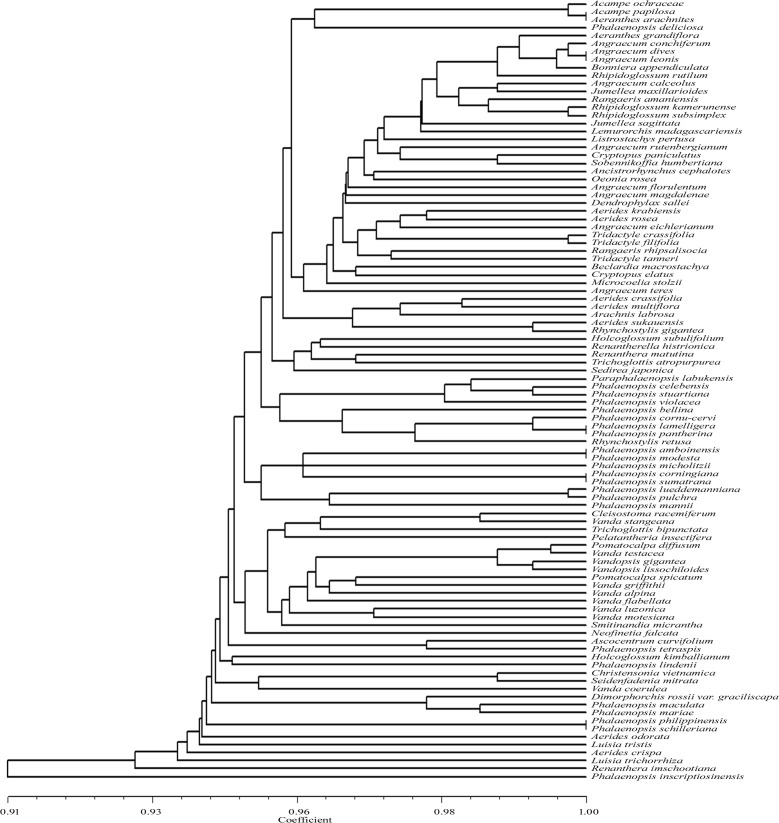
TspDTI exhibited species specific restriction patterns for 78 taxa and could not discriminate 20 taxa (Table 1, Fig 5). These twenty included the taxon pairs or groups Acampe papillosa and Aeranthes arachnites; Christensonia vietnamica and Seidenfadenia mitrata; Phalaenopsis celebensis and Phalaenopsis stuartiana; Phalaenopsis philippinensis and P. schilleriana; Phalaenopsis corningiana and P. sumatrana; Vanda testacea, Vandopsis lissochiloides and Vandopsis gigantean; Rhipidoglossum kamerunense and Rhipidoglossum subsimplex; Tridactyle crassifolia and Tridactyle filifolia. The UPGMA dendrogram showed similarity coefficient ranging from 0.92 to 1.00.
Fig 5. UPGMA dendrogram based on SAHN clustering of data from in silico restriction site polymorphism analysis of the trnL(UAA) intron P8 region using the restriction endonuclease TspDTI among the 98 taxa of orchids.
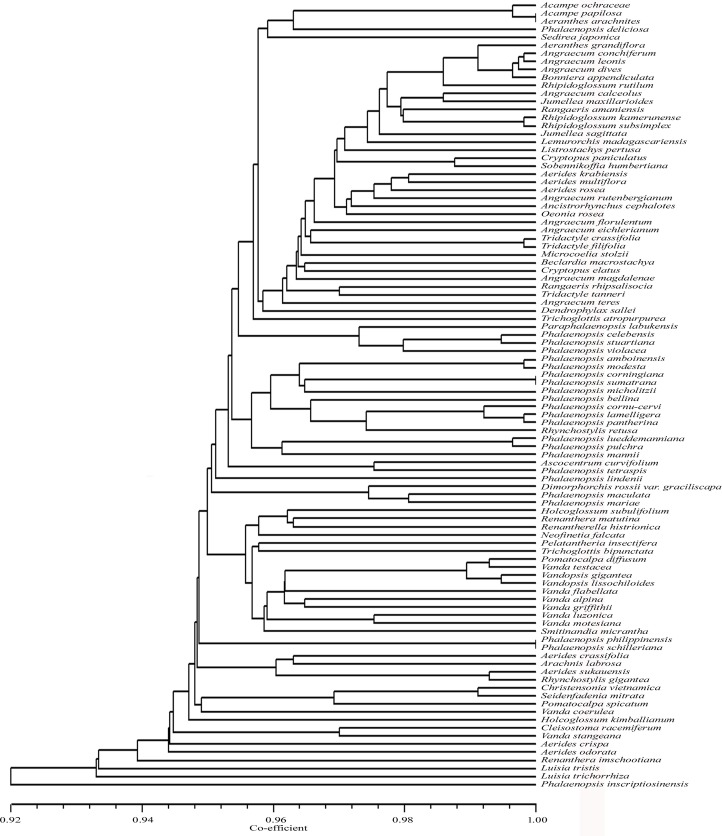
VspI also exhibited species specific restriction patterns for 66 taxa and could not discriminate 32 taxa (Table 1, Fig 6). These 32 include the taxon pairs or groups, Acampe ochraceae, Acampe papillosa and Aeranthes arachnites; Aerides multiflora, Phalaenopsis deliciosa and Tridactyle tanneri; Christensonia vietnamica and Seidenfadenia mitrata; Arachnis labrosa and Rhynchostylis gigantean; Vanda testacea, Vandopsis lissochiloides and Vandopsis gigantea; Angraecum conchiferum, Angraecum leonis and Bonniera appendiculat; Listrostachys pertusa, Microcoelia stolzii, Rangaeris amanuensis, Rangaeris rhipsalisocia, Tridactyle crassifolia and Tridactyle filifolia; Rhipidoglossum kamerunense and Rhipidoglossum subsimplex; Phalaenopsis philippinensis and P. schillerian; Phalaenopsis bellina and Phalaenopsis micholitzii; Phalaenopsis cornu-cervi and Phalaenopsis lamelligera; Phalaenopsis corningiana and P. sumatrana. The UPGMA dendrogram showed similarity coefficient ranging from 0.90 to 1.00.
Fig 6. UPGMA dendrogram based on SAHN clustering of data from the in silico restriction site polymorphism analysis of the trnL(UAA) intron P8 region using the restriction endonuclease VspI among the 98 taxa of orchids.
Species discrimination using combined data of restriction endonucleases
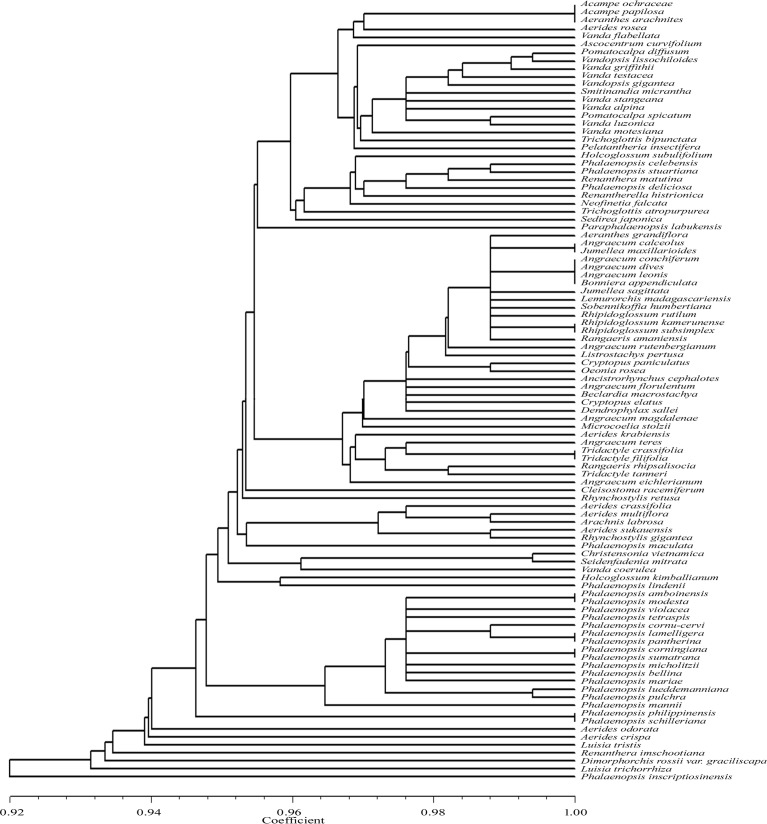
Analysis of the in silico restriction mapping using the combined data of AgsI and ApoI resulted in discrimination of 86 taxa (Fig 7) as against their individual resolutions of 80 and 77 taxa respectively. Those which had exactly the same restriction sites included six taxon pairs, Acampe papillosa and Aeranthes arachnites; Angraecum dives and Angraecum leonis; Phalaenopsis lamelligera and P. pantherina; Phalaenopsis amboinensis and P. modesta; Phalaenopsis corningiana and P. sumatrana; Phalaenopsis philippinensis and P. schilleriana. The UPGMA dendrogram showed similarity coefficient ranging from 0.91 to 1.00.
Fig 7. UPGMA dendrogram based on SAHN clustering of the combined data from in silico restriction site polymorphism analyses of the trnL(UAA) intron P8 region using the restriction endonuclease AgsI and ApoI among the 98 taxa of orchids.
Again, when ApoI, AgsI and TspDTI restriction data were combined, 92 taxa could be discriminated while the three taxon pairs which could not be discriminated from each other included Acampe papillosa and Aeranthes arachnites; Phalaenopsis corningiana and P. sumatrana; Phalaenopsis philippinensis and P. schilleriana (Table 1, Fig 8). The UPGMA dendrogram showed similarity coefficient ranging from 0.92 to 1.00.
Fig 8. UPGMA dendrogram based on SAHN clustering of the combined data from in silico restriction site polymorphism analyses of the trnL(UAA) intron P8 region using the restriction endonuclease AgsI, ApoI and TspDTI among the 98 taxa of orchids.
Combination of data of all the four enzymes ApoI, AgsI, TspDTI and VspI also gave the same result as that of the above three enzymes with no discrimination of the following three pairs Acampe papillosa and Aeranthes arachnites; Phalaenopsis corningiana and P. sumatrana; Phalaenopsis philippinensis and P. schilleriana (Table 1, Fig 9). The UPGMA dendrogram showed similarity coefficient ranging from 0.92 to 1.00.
Fig 9. UPGMA dendrogram based on SAHN clustering of the combined data from in silico restriction site polymorphism analyses of the trnL(UAA) intron P8 region using the restriction endonuclease AgsI, ApoI, TspDTI and VspI among the 98 taxa of orchids.
Discussion
The P8 stem-loop region is reported to be the most length-variable region of the trnL intron which is due to presence of repeats of various sizes [21]. These repeats together with the enormous length and sequence variation hampered alignment in most of the plant groups being studied for phylogeny or barcoding and therefore, this region is often excluded from analysis of trnL intron sequences [14–16]. Due to this exclusion, the results inferred from analysis of the trnL intron sequences do not represent the exact information it should naturally infer. By virtue of its hypervariability, the P8 region should be having certain information which may be analysed and interpreted for applying in species level identification or barcoding, if not for phylogenetic inference. As there is difficulty in sequence alignment of the P8 regions, the only other method for analysis should be the restriction analysis. In silico restriction site polymorphism was opted over the conventional PCR-Restriction Fragment Length Polymorphism (PCR-RFLP) as there were already a large number of trnL sequences deposited to GenBank. Moreover, the PCR-RFLP may not show the true picture in case two fragments of equal lengths coming from different parts of the sequence cannot be distinguished by observing the gel picture. For plant species identification PCR-RFLP is seldom used, however, a few investigators employed it for identification of mangrove and mangrove associate species [23], fine roots of trees from the Alps [24], Cinnamomum spp. [25], upland grassland species from roots [26] and Vasconcellea species [27], Dendrobium orchids [28].
Sequence length analysis
The tribe Vandeae (Family: Orchidaceae) is very robust consisting of 2600 species of monopodial epiphytes under 139 genera. Some previous investigators had already worked on phylogeny of these orchids using trnL intron sequences [15–17]. As an objective of the present investigation we aimed to discriminate closely related species belonging to a genus or a subtribe or a tribe. While searching the GenBank database, 125 accessions of trnL intron sequence for taxa, under the tribe Vandeae, was available when we started the investigation and they were retrieved for analysis. Upon further investigation 98 out of the 125 sequences had complete P8 hypervariable regions, and thus were considered for the final analysis. Sequence length variation of the P8 regions could discriminate 92 out of the 98 taxa of orchids being investigated. It was observed that the length of the P8 regions of all the taxa being varied from 200 (Aerides rosea) to 669 nucleotides (Dendrophylax sallei). Dendrophylax sallei had 30 more nucleotides longer than Luisia curtisii (Orchidaceae) which was reported earlier to be the longest recorded angiosperm P8 (639 nucleotides) [16]. As evidenced from our finding, those taxa sharing a common length did not necessarily belong to the same genus and that there could be great differences in P8 lengths within the same genus, which is in conformity with that of Kocyan et al. [16]. This difference in P8 lengths might be due to slipped-strand mispairing (SSM) resulting into high repetition of A motifs [21, 29].
Restriction site polymorphism analysis
Our study represents the first ever in silico restriction analysis of the P8 region of trnL in an attempt to utilize the genetic information present in it for a meaningful interpretation in species identification of a certain group of angiospermic plants. From all the UPGMA dendrograms generated in this investigation it is evidenced that species belonging to one genus are clustered with those belonging to other genus or genera, or in the other sense they are haphazardly clustered owing to the hypervariable nature of the P8 regions. This showed that the present approach might not be applicable for phylogenetic inference of the taxa being investigated. However, since 95.9% of these taxa could be discriminated based on restriction site polymorphisms and sequence length data, this technique might be adopted for rapid species identification and hence as a plant DNA barcode.
So far there has not been much report on utilization of PCR-RFLP for identification of orchid species except for certain Thai Dendrobium orchids using rDNA-ITS and cpDNA regions [28]. Some investigators already showed the efficacy of double digestion or using two or more restriction endonucleases over single in generation of more polymorphic fragments in PCR-RFLP experiments. PCR-RFLP of the chloroplast trnS-psbC gene regions using a combination of two enzymes, HaeIII and MspI could successfully identify all the 119 accessions of millet into 7 species [30]. 579 grasses roots were distinguished to ten species using PCR-RFLP of trnL intron, with one or two enzyme digest [26]. Again, 16 taxa out of 30 tree species from the Alps were identified using PCR-RFLP with four restriction endonucleases TaqI, HinfI, RsaI and CfoI [24]. However, in our investigation, it was observed that as many as 35 restriction endonucleases had their recognition sites in the region. Combined data from analyses of the P8 regions with three restriction endonucleases ApoI, AgsI and TspDTI, could discriminate 92 of the 98 taxa based on species specific restriction sites. Hence, the advantage of screening a large number of restriction endonucleases is required for higher success rate in species discrimination of the plant specimens being investigated.
Aerides rosea, having the shortest P8 length of 200 nuclotides, showed to have recognition sites of at least eight enzymes, while Dendrophylax sallei despite having the longest P8 length (669 nucleotides) had recognition sites for only 20 enzymes. Microcoelia stolzii with a P8 length of 480 nucleotides had the maximum number (25) of restriction endonucleases cutting the region. Again, considering the number of recognition sites per restriction endonuclease for an individual taxon, Phalaenopsis inscriptiosinensis, with a P8 length of 498 nucleotides, had as many as 14 AgsI, 10 ApoI, 10 TspDTI and 8 VspI recognition sites. Hence, our result showed that the longest P8 length neither had the maximum number of restriction endonuclease recognizing it nor maximum number of recognition sites for an individual enzyme.
It was observed that there were 20 restriction endonucleases having recognition sites in the P8 regions of both Acampe papillosa and Aeranthes arachnites. These two taxa also exhibited the same number of restriction sites for all the twenty enzymes. The UPGMA trees drawn for each of the four enzymes as well as those for their combined data revealed them to have 100% genetic similarity. Hence, the only information to differentiate them as different species would be their P8 sequence lengths, 420 nucleotides for Acampe papilosa and 521 nucleotides for Aeranthes arachnites.
Phalaenopsis corningiana and P. sumatrana are treated as two separate species [31–33]. Our investigation revealed that both of them had the same P8 length (508 nucleotides) and identical sequence and hence could not be distinguished from each other as they had similar restriction sites for all the 19 restriction enzymes. Tsai [34] also could not separate the two taxa based on information derived from nrITS, IGS and atpB-rbcL sequences. The only morphological differences between them are in callus and marking patterns on the petals. Distribution of P. sumatrana is widespread ranging from Myanmar, Thailand, Vietnam, Indonesia, Malaysia and the Philippines; whereas P. corningiana is restricted to Borneo and Sarawak. Phalaenopsis philippinensis and P. schilleriana are also endemic to Philippines and they have distinct distinguishing morphological characters. From our analysis, it is observed that both possessed identical P8 sequences and hence identical restriction endonucleases and their restriction sites. Whether these two taxon pairs should be treated as natural hybrids or ecotypes may need evidences from other coding, non-coding sequences or protein markers.
It was suggested that for a gene region to be practical as a DNA barcode the following three criteria must be fulfilled: (i) contain significant species-level genetic variability and divergence, (ii) possess conserved flanking sites for developing universal PCR primers for wide taxonomic application, and (iii) have a short sequence length so as to facilitate current capabilities of DNA extraction and amplification [3]. From our result, it is learnt that P8 regions of the 98 taxa contained good amount of genetic variation either in sequence length or restriction sites of the enzymes being used. Those taxa which could not be discriminated might require understanding of their maternal origin, in case of natural hybridization; plastid DNA barcodes will fail in case of natural hybrids. Second, the primer pair (c and d) used to amplify trnL intron is well conserved from brayophytes to angiosperms [35]. The present study also employed this primer pair for both PCR amplification as well as sequencing. Using these primers, the whole trnL region could be amplified and sequenced and from it an intact P8 region be retrieved easily, which is an advantage. Third, sequence lengths of the P8 regions observed in the present investigation ranged from 200 to 669 nucleotides which were short enough to be considered as DNA barcodes. The only disadvantage of the P8 region lie in their inability to be aligned due to hypervariability, and hence cannot be used for further processing using standardized phylogenetic or barcoding techniques.
Conclusions
A technique for molecular identification using sequence length variation and in silico restriction site polymorphism analysis of the trnL intron P8 sequence was developed and utilized to discriminate 94 out of 98 taxa of orchids to the level of species. Investigations using this technique for species level discrimination across all the angiospermic families may be tried. The four restriction endonucleases ApoI, AgsI, TspDTI and VspI could be utilized for further analysis of the trnL intron P8 sequences of other uninvestigated orchid taxa either in silico or in PCR-RFLP. A plant DNA barcoding system using restriction site polymorphism of the trnL P8 region has not been suggested yet; and with this report there is high possibility of using this tool to establish a barcoding system.
Acknowledgments
This work was supported by the Council of Scientific and Industrial Research (CSIR), Government of India by granting Senior Research Associateship to Rajkumar Kishor (Grant No. SRA-8627A).
Data Availability
All relevant data are within the paper.
Funding Statement
This work was supported by the Council of Scientific and Industrial Research (CSIR), Government of India by granting Senior Research Associateship to Rajkumar Kishor (grant no. SRA-8627A).
References
- 1.Hebert PDN, Ratnasingham S, De Waard JR. Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related animals. Proc Roy Soc London, Ser B, Biol Sci. 2003; 270: S96–S99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chase MW, Cowan RS, Hollingsworth PM, van den Berg C, Madrinan S, Petersen G, et al. A proposal for a standardized protocol to barcode all land plants. Taxon 2007; 56: 295–299. [Google Scholar]
- 3.Kress W J, Erickson D L. DNA barcodes: Genes, genomics, and bioinformatics. Proc Natl Acad Sci U S A. 2008; 105: 2761–2762. doi: 10.1073/pnas.0800476105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hollingworth PM, Grahm SW, Little DP. Choosing and using a plant DNA Barcode. PLos One. 2011; 6(5)e19254:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taberlet P, Gielly L, Pautou G, Bouvet J. Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Mol Biol. 1911; 17: 1105–l109. [DOI] [PubMed] [Google Scholar]
- 6.Gielly L, Taberlet P. A phylogeny of European gentians inferred from chloroplast trnL(UAA) intron sequences. Bot J Linnean Soc.1996; 120: 57–75. [Google Scholar]
- 7.Davies RW, Waring RB, Ray JA, Brown TA, Scazzocchio C. Making ends meet—a model for RNA splicing in fungal mitochondria. Nature.1982; 300: 719–724. [DOI] [PubMed] [Google Scholar]
- 8.Michel F, Jacquier A, Dujon B. Comparison of fungal mitochondrial introns reveals extensive homologies in RNA secondary structure. Biochimie. 1982; 64: 867–88. [DOI] [PubMed] [Google Scholar]
- 9.Quandt D, Muller K, Stech M, Frahm JP, Frey W, Hilu KW, et al. Molecular evolution of the chloroplast trnL-F region in land plants. Monogr Syst Bot Missouri Botanic Garden. 2004; 98:13–37. [Google Scholar]
- 10.Gielly L, Taberlet P. The use of chloroplast DNA to resolve phylogenies: noncoding versus rbcL sequences. Mol Biol Evol. 1994; 11: 769–777. doi: 10.1093/oxfordjournals.molbev.a040157 [DOI] [PubMed] [Google Scholar]
- 11.Kushel M G, Strickland R, Palmer JD. An ancient group I intron shared by Eubacteria and chloroplasts. Science. 1990; 250: 1570–l 573. [DOI] [PubMed] [Google Scholar]
- 12.Cech TR, Damberg SH, Gutell RR. Representation of the secondary and tertiary structure of group I introns. Nat Struct Biol. 1994; 1: 273–280. [DOI] [PubMed] [Google Scholar]
- 13.Borsch T, Hilu KW, Quandt D, Wilde V, Neinhuis C, Barthlott W. Noncoding plastid trnT–trnF sequences reveal a well resolved phylogeny of basal angiosperms. J Evol Biol. 2003; 16: 558–576. [DOI] [PubMed] [Google Scholar]
- 14.Borsch T, Hilu KW, Wiersema JH, Lohne C, Barthlott W, Wilde V. Phylogeny of Nymphaea (Nymphaeaceae): Evidence from substitutions and microstructural changes in the chloroplast trnT-trnF region. Int J Plant Sci. 2007; 168: 639–671. [Google Scholar]
- 15.Carlsward BS, Whitten WR, Williams NH, Bytebier B. Molecular phylogenetics of Vandeae (Orchidaceae) and the evolution of leaflessness. Am J Bot. 2006; 93:770–786. doi: 10.3732/ajb.93.5.770 [DOI] [PubMed] [Google Scholar]
- 16.Kocyan A, de Vogel EF, Conti E, Gravendeel B. Molecular phylogeny of Aerides (Orchidaceae) based on one nuclear and two plastid markers: A step forward in understanding the evolution of the Aeridinae. Mol Phylogenet Evol.2008; 48: 422–44. doi: 10.1016/j.ympev.2008.02.017 [DOI] [PubMed] [Google Scholar]
- 17.Tsai CC, Chiang YC, Huang SC, Chen CH, Chou CH. Molecular phylogeny of Phalaenopsis Blume (Orchidaceae) on the basis of plastid and nuclear DNA. Plant Syst Evol. 2010; 288: 77–98. [Google Scholar]
- 18.Dressler RL. Phylogeny and classification of the orchid family Dioscorides Press, Portland, OR: 1993. [Google Scholar]
- 19.Schuiteman A. Dendrobieae and Aeridinae In: Pridgeon AM, Cribb PJ, Chase MW, Rasmussen FN, editors. Genera Orchidacearum: Volume 6: Epidendroideae, Part 3, Oxford University Press; 2014. [Google Scholar]
- 20.Kishor R, Sunitibala DH. Induction of multiple shoots in a monopodial orchid hybrid (Aerides vandarum Reichb.f x Vanda stangeana Reichb.f) using thidiazuron and analysis of their genetic stability. Plant Cell Tiss Organ Cult. 2009; 97: 121–129. [Google Scholar]
- 21.Quandt D, Stech M. Molecular evolution of the trnL(UAA) intron in bryophytes. Mol Phylogenet Evol 2005; 36: 429–443. doi: 10.1016/j.ympev.2005.03.014 [DOI] [PubMed] [Google Scholar]
- 22.Rolf FJ. NTSYS-PC Numerical Taxonomy and Multivariate Analysis System, Exter Software, Setauket, NY. 1993
- 23.Parani M, Lakshmi M, Ziegenhagen B, Fladung M, Senthilkumar P, Parida A. Molecular phylogeny of mangroves VII. PCR-RFLP of trnS-psbC and rbcL gene regions in 24 mangrove and mangrove-associate species. Theor Appl Genet. 2000; 100:454–460. [Google Scholar]
- 24.Brunner I, Brodbeck S, Buchler U, Sperisen C. Molecular identification of fine roots of trees from the Alps: reliable and fast DNA extraction and PCR-RFLP analyses of plastid DNA. Mol Ecol. 2001; 10:2079–2087. [DOI] [PubMed] [Google Scholar]
- 25.Kojoma M, Kurihara K, Yamada K, Sekita S, Satake M, Iida O. Genetic identification of cinnamon (Cinnamomum spp.) based on the trnL-trnF chloroplast DNA. Planta Medica. 2002; 68: 94–96. doi: 10.1055/s-2002-20051 [DOI] [PubMed] [Google Scholar]
- 26.Ridgway KP, Duck JM, Young JPW. Identification of roots from grass swards using PCR-RFLP and FFLP of the plastid trnL (UAA) intron. BMC Ecology. 2003; 3: 8 doi: 10.1186/1472-6785-3-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van Droogenbroeck B, Kyndt T, Maertens I, Romeijn-Peeters E, Scheldeman X, Romero-Motochi JP, et al. Phylogenetic analysis of the highland papayas (Vasconcellea) and allied genera (Caricaceae) using PCR-RFLP. Theor Appl Genet. 2004; 108:1473–1486. doi: 10.1007/s00122-003-1575-7 [DOI] [PubMed] [Google Scholar]
- 28.Peyachoknagula S, Mongkolsiriwatanab C, Wannapinpongd S, Huehnea PS, Srikulnatha K. Identification of native Dendrobium species in Thailand by PCR-RFLP of rDNA-ITS and chloroplast DNA. ScienceAsia. 2014; 40:113–120 [Google Scholar]
- 29.Levinson G, Gutman GA. Slipped-strand mispairing: a major mechanism for DNA sequence evolution. Mol Biol Evol. 1987; 4: 203–21. doi: 10.1093/oxfordjournals.molbev.a040442 [DOI] [PubMed] [Google Scholar]
- 30.Parani M, Rajesh K, Lakshmi M, Parducci L, Szmidt AE, Parida A. Species identification in seven small millet species using polymerase chain reaction–restriction fragment length polymorphism of trnS-psbC gene region. Genome 2001; 44: 495–499 [DOI] [PubMed] [Google Scholar]
- 31.Sweet HR. Revision of the genus Phalaenopsis, Part 2. Am Orchid Soc Bull. 1968; 37: 1089–1104. [Google Scholar]
- 32.Fowlie JA. A note on Phalaenopsis sumatrana and Phalaenopsis corningiana. Orchid Digest. 1969; 33: 301. [Google Scholar]
- 33.Christenson E A. Phalaenopsis Timber Press, Portland Oregon, 2001; pp 330. [Google Scholar]
- 34.Tsai CC. Molecular Phylogeny and Biogeography of Phalaenopsis Species In: Chen W-H, Chen HH editors. Orchid Biotechnology II. World Scientific, 2011. [Google Scholar]
- 35.Taberlet P, Coissac E, Pompanon F, Gielly L, Miquel C, Valentini A, et al. Power and limitations of the chloroplast trnL(UAA) intron for plant DNA barcoding. Nucleic Acid Res.2007; 35: e14 doi: 10.1093/nar/gkl938 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.