Abstract
The pulmonary metastasis assay (PuMA) is an ex vivo lung explant and closed cell culture system that permits researchers to study the biology of lung colonization in osteosarcoma (OS) by fluorescence microscopy. This article provides a detailed description of the protocol, and discusses examples of obtaining image data on metastatic growth using widefield or confocal fluorescence microscopy platforms. The flexibility of the PuMA model permits researchers to study not only the growth of OS cells in the lung microenvironment, but also to assess the effects of anti-metastatic therapeutics over time. Confocal microscopy allows for unprecedented, high-resolution imaging of OS cell interactions with the lung parenchyma. Moreover, when the PuMA model is combined with fluorescent dyes or fluorescent protein genetic reporters, researchers can study the lung microenvironment, cellular and subcellular structures, gene function, and promoter activity in metastatic OS cells. The PuMA model provides a new tool for osteosarcoma researchers to discover new metastasis biology and assess the activity of novel anti-metastatic, targeted therapies.
Keywords: This Month in JoVE, Issue 133, Osteosarcoma, sarcoma, metastasis, lung, microscopy
Introduction
Improved outcomes for pediatric patients with metastatic osteosarcoma (OS) still remains a critical unmet clinical need 1. This underscores the importance of developing new molecularly-targeted therapies. Conventional chemotherapeutics that target tumor cell proliferation have not proven to be effective in treating metastatic disease, and thus novel strategies must target the metastatic process itself 2. The current article discusses the practical aspects of a relatively new type of ex vivo lung metastasis model, the pulmonary metastasis assay (PuMA) developed by Mendoza and colleagues3, which provides a useful tool in discovering new molecular drivers in lung metastasis progression in OS 4,5. Before proceeding, however, it would be prudent to briefly touch upon several current models of metastasis, and how the PuMA model offers several advantages over conventional in vitro assays.
Most experimental models used to study metastasis comprise of in vitro and in vivo systems that recapitulate either a specific step or several steps of the metastatic cascade. These steps include: 1) tumor cells migrating away from the primary tumor, 2) intravasation into nearby vessels (blood or lymphatic) and transit within circulation, 3) arrest at the secondary site, 4) extravasation and survival at the secondary site, 5) formation of micrometastases, and 6) growth into vascularized metastases (Figure 1). In vitro models of metastasis can include 2-dimensional (2D) migration and 3-dimensional (3D) Matrigel invasion assays which are reviewed in detail elsewhere 6. For in vivo models, the two commonly used model systems include: 1) the spontaneous metastasis model is where a tumor cells are orthotopically injected into a specific tissue type to form a local tumor which spontaneously sheds metastatic cells to distant sites; 2) the experimental metastasis model is where tumor cells are injected into the blood vessel upstream of the target organ. For example, a tail vein injection of tumor cells results in the development lung metastases5,7,8. Other experimental metastasis models include injection of tumor cells into the spleen or mesenteric vein which results in the development of liver metastases9,10. Practical considerations of these in vivo models are discussed in detail by Welch 11. Another in vivo model used to study metastasis in pediatric sarcomas is the renal kidney subcapsular tumor implantation model which results in local tumor formation and spontaneous metastasis to the lungs 12,13. A more technically demanding technique such as intravital videomicroscopy can directly visualize, in real-time, interactions between metastatic cancer cells and the microvasculature of a metastatic site (ie. lung or liver) as described by MacDonald14 and Entenberg15, or cancer cell extravasation in the chorioallantoic membrane as described by Kim 16.
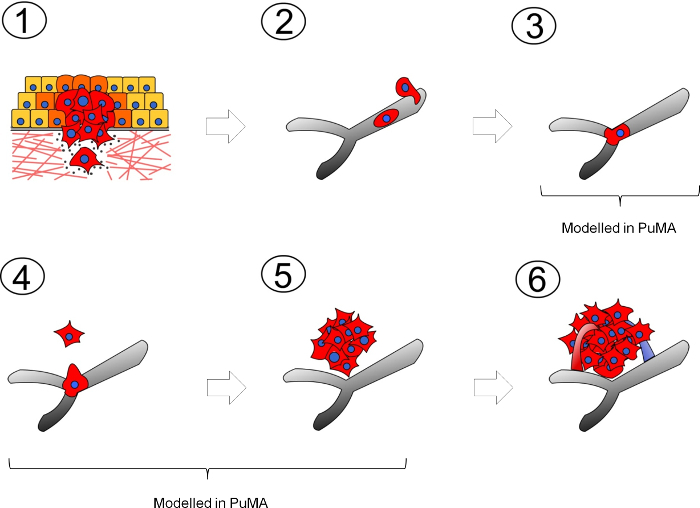
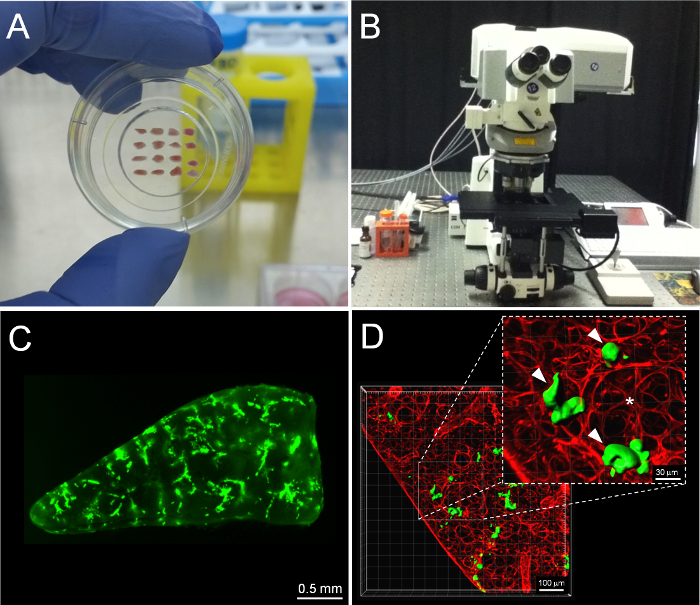
The PuMA model is an ex vivo, lung tissue explant, closed culture system where the growth of fluorescent tumor cells can be longitudinally observed via fluorescence microscopy over a period of a month (see Figure 2A). This model recapitulates the initial stages of lung colonization (steps 3 to 5) in the metastatic cascade. Some major advantages of the PuMA model over conventional in vitro models are: 1) it provides an opportunity to longitudinally measure metastatic cancer cell growth in a 3D microenvironment that retains many features of the lung microenvironment in vivo 3; 2) PuMA allows the researcher to assess whether the knockdown of a candidate gene or drug treatment has anti-metastatic activity in the context of a 3D lung microenvironment; 3) the PuMA model is flexible with many types of fluorescence microscopy platforms (Figure 2B) such as widefield fluorescence microscopy or laser-scanning confocal microscopy, examples of each are shown in Figure 2C & D, respectively. This article will discuss how to use the PuMA model to obtain longitudinal imaging data on the metastatic growth of enhanced green fluorescent protein (eGFP)-expressing, human high and low metastatic osteosarcoma cells (MNNG and HOS cells, respectively) using low-magnification widefield fluorescence. Examples of imaging a fluorescent dye which labels the lung parenchyma, and a red-fluorescent protein genetic reporter which labels mitochondria in OS cells in the PuMA model using confocal laser-scanning microscopy are also discussed.
Protocol
All animal protocols from which imaging data were obtained were performed with approval of the Animal Care and Use Committee of the National Cancer Institute, National Institutes of Health. All animal protocols discussed and portrayed in the article video have been approved by the University of British Columbia Animal Care Committee.
1. Preparation of tumor cells for injection and materials for the PuMA model
NOTE: The amount of solutions and cells will be enough for 1 mouse. Scale up as necessary if more mice are used in the study. For media recipes, refer to Table 1 and Table 2.
Pre-warm 5 mL of A-Media in a 15 mL conical tube in a 37 oC water bath.
Melt the 1.2% low melting agarose solution (in sterile water) using the lab microwave.
Transfer 5 mL of the molten agarose into a 15 mL conical tube, and keep warm in a 37 oC water bath. Ensure the melted agarose is at 37 oC and liquid prior to the lung insufflation step.
Pre-chill 30 mL of cell-culture grade PBS supplemented with 1X pen/strep in an ice bucket.
In one well of a 6-well plate, pre-soak a gelatin sponge in 1.5 mL of B-media. The sponge will be the support anchor for the lung slices.
Ensure the OS cells are about 70-90% confluent on the day of the procedure. Do not use cells that are over-confluent or if the media is orange to yellow since the cells are usually stressed and have diminished viability at this point. For the osteosarcoma cell lines, MNNG and HOS, 5 x 105 in a volume of 100 μL will be used to inject into the tail vein of 1 mouse. Upscale accordingly if more mice are used for the study.
Harvest the tumor cells using 0.25% trypsin-EDTA (3 mL for a T75 flask or 2 mL in a 10 cm culture dish). When cells are beginning to lift off the plate, neutralize the trypsin-EDTA with complete media. Spin down cells and rinse once with cell-culture grade PBS. Resuspend pellet in 5 mL of PBS.
Perform a cell count by standard methods. It is best to make more cell suspension than what is actually needed since loss of cell suspension often occurs during needle draw-up and failed injection attempts.
Perform a Trypan-Blue Exclusion Assay on a sample of the cell suspension to assess the viability of the cells. Proceed only if the cell show 90% viability or higher.
Inject 5 x 105 cells in a volume of 100 μL. To prepare an excess of 2X of this amount, 1 x 106 cells should be spun down and resuspended in 0.2 mL of HBSS.
Place cell suspension in an ice bucket while preparing the mice for injection.
2. Tail Vein Injection and Lung Insufflation
NOTE: The amount of solutions and cells in this section will be enough for 1 mouse. Scale up as necessary if more mice are used in the study. For a list of the equipment, materials and surgical instruments used in the following steps, refer to Table 3 and Table 4.
Warm a female mouse (age 6-8 weeks) under a heating lamp for 5 min in order to make their tail vein more visibly apparent. For murine K7M2 or K12 cells, use Balb/c mice; for human MG63.3, MG63, MNNG, and HOS cells, use severe combined immunodeficiency mice.
Make sure the cell suspension is uniform by gently shaking the tube. Carefully draw up the cell suspension into the 1 mL syringe without the needle. Cap the syringe with a 27 gauge needle. Make sure the needle bevel is on the same side as the volume markings on the needle.
Place the mouse in the restrainer, and swab the tail with an alcohol wipe.
Proceed to perform a tail vein injection and inject the 100 μL of the cell suspension. 5 min after the injection, place the mouse in a CO2 chamber and begin the euthanasia standard operating procedure (as outline by your institutional animal care committee). Cervical dislocation should not be used as a means of euthanasia since the procedure will damage the trachea.
Once the mouse is euthanized, begin preparation of the laminar flow hood for insufflation of the mouse lung with the agarose/A-media solution. Within the laminar flow hood, set up a work area with your sterile pad. Onto this pad you will place your sterilized instruments, IV catheter, IV extension set, and gravity perfusion apparatus (see Supplemental Figure 1).
Place the mouse in dorsal recumbancy. With sterile small scissors, carefully dissect out the sternum to expose the chest cavity. Take care to not puncture the lung since an agarose/A-media solution will be used to insufflate the lung.
When dissecting out the sternum, dissect past the thoracic inlet on both sides the trachea. Expose the trachea by dissecting away the surrounding soft tissue.
Cannulate the trachea with a 20 gauge IV catheter. Loosely tie a surgical knot using sterile catgut suture around the cannulated trachea.
Attach an IV extension set from the catheterized trachea to the 10 mL syringe of the gravity perfusion apparatus.
Combine the pre-warmed 37 oC agarose (5 mL) and A-media (5 mL) into a 1:1 mixture. Pour the liquid agarose/A-media solution into the 10 mL syringe of the gravity perfusion device. Prior to insufflation of lungs with agarose/A-media solution, ensure that the entire length of the extension set has been primed with agarose/A-media, thereby negating inadvertent insufflation of lung samples with air.
Fill the lung the agarose/A-media solution until the lung is fully insufflated.
Once the lung is fully insufflated, remove the cannula and firmly tie off the surgical knot to prevent leakage of the agarase/A-media solution through the trachea.
Proceed to dissect out the pluck (trachea, heart, and lung) from the chest cavity. Take care to not puncture or damage the surface of the lung.
Place the pluck (in no particular orientation) in the pre-chilled 30 mL of PBS supplemented with 1X pen/strep, and allow the agarose/A-media to solidify for 20 min.
Using fine scissors and tweezers, cut small pieces of the lung (3 mm x 1.5 mm) as shown in Figure 1A. Smaller lung slices can be easily imaged using a 2.5X objective. Multiple slices can be cut per experimental condition, typically 4-10 slices per group. NOTE: For each experimental group, place the lung slices into a separate well (6-well plate) containing a 2 x 2 cm gelatin sponge pre-soaked in B-media. The amount of media per well should be 1.5 mL. Change the media every 2-3 days. For drug studies, the frequency of changing the media/drug is user determined.
3. Widefield Fluorescence Imaging of Lung Slices and Analysis
NOTE: For widefield fluorescence imaging, smaller slices are cut (3 mm x 1.5 mm x 1 mm) in order to fit the lung section into 1 image using a 2.5X objective.
Image acquisition on a widefield fluorescence microscope:
Lung slices are typically imaged at 0, 3, 7, and 14 days post-injection. In the sterile environment of the biological cabinet, carefully transfer the lung slices from the gelatin sponges to a sterile 35 mm glass-bottom round dish. Take care to dab off the excess fluid from the lung slice as the excess fluid will act as a mirror and reflect fluorescent light coming from the tumor cells.
Arrange the lung slices in a similar manner depicted in Figure 1A. Each column would represent a different experimental condition. Take care to not cross-contaminate the lungs with the tweezers. Rinse with 70% ethanol and dry before handling lung slices from another experimental group.
Optimize the imaging parameters (ie. gain, offset, exposure time, binning) that provides the best contrast between the fluorescent tumor cells and background lung tissue in the control (vehicle untreated) group. Use the same parameters from the control group to image the experimental groups. Save as tiff image format.
For a scale reference, take a digital picture of a micrometer at the same objective.
Since lung auto-fluorescence changes over time, the imaging parameters must be adjusted to the control lungs each imaging session. The imaging parameters for one session may not necessarily be optimal for subsequent imaging sessions.
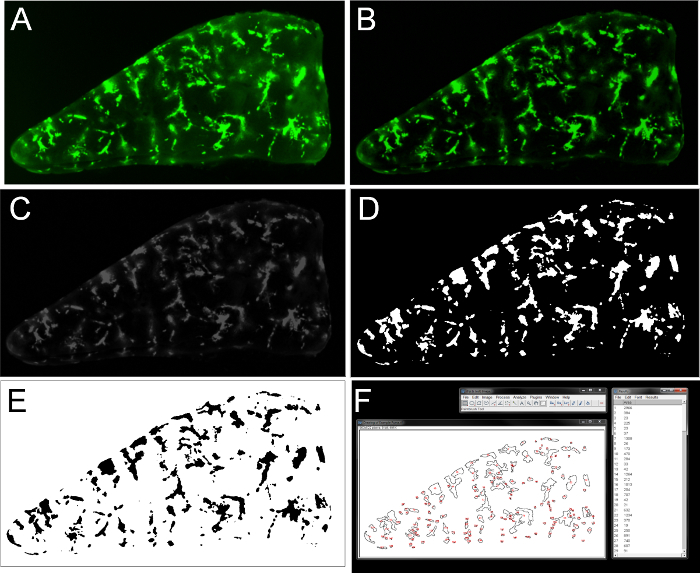
Image Analysis: NOTE: The following image processing steps are done with ImageJ 1.51h software package 17.
Open an image file in ImageJ (Figure 3A).
Subtract background: Process > Subtract Background > Rolling ball radius (start with 50 pixels), uncheck "Light Background" (Figure 3B).
Convert image to 8-bit format: Image > Type > 8-bit (Figure 3C).
Set units to pixels: Image > Enter "Pixels" in Unit of Length, enter 1 in "Pixel width", "Pixel Height", "Voxel Depth". Check "Global" box to apply to subsequent images.
Using the Polygon selection tool, outline the shape of the entire lung slice and determine the area (pixel2) of the total lung slice. This value will be used to calculate the percent tumor burden of the lung.
Threshold image: Image > Adjust > Threshold > highlight "Default" and "B&W" > use the slider to threshold the image such that the majority of tumor cells are accurately highlighted. Pressing "Apply" will result in a black and white image where the lung is all black, and the fluorescent lesions are white (Figure 3D). Pressing "Apply" a second time will invert the image where all fluorescent structures are now black shapes (Figure 3E).
Quantification of the number and shape of lesions: Analyze > Set Measurements > Check "Area". Uncheck all other boxes. Analyze Particles > Size (pixel2): 0-infinity represents the range of shapes that ImageJ will enumerate. Empirically determine the smallest lesion that is deemed to be a single tumor cell in Day 0 vehicle images. Use the area of this single tumor cell as the lower limit for the remaining images in the data set. For the work presented in this paper, 11 pixel2 is set as the lower limit. For the work presented in the video example, 24 pixel2 is set as the lower limit. Leave "Circularity" range as 0-1. "Show" can be set to "Nothing". Alternatively, if "Show" and "Outlines" is selected, a drawing of all outlined shapes is generated. Check "Display results" for a separate window of measurements to pop up. Optionally, having "Add to manager" box checked will save all the shapes quantified to a ROI manager, which can be saved for future reference. After pressing OK, a "Results" window will pop up containing all enumerated shapes and area measurements (Figure 3F).
Copy and paste the data from the "Results" window into a spreadsheet. Use the SUM mathematical function in Excel to sum all the areas of the metastatic lesions for that particular lung slice. To assess the percent lung tumor burden of lung slice, divide the sum of the areas of metastatic lesions by the total area of the lung slice. This parameter is also known as Area fraction (AA) which is described further by Underwood 18. Lung tumor burden = Sum of metastatic lesion areas/total area of lung slice
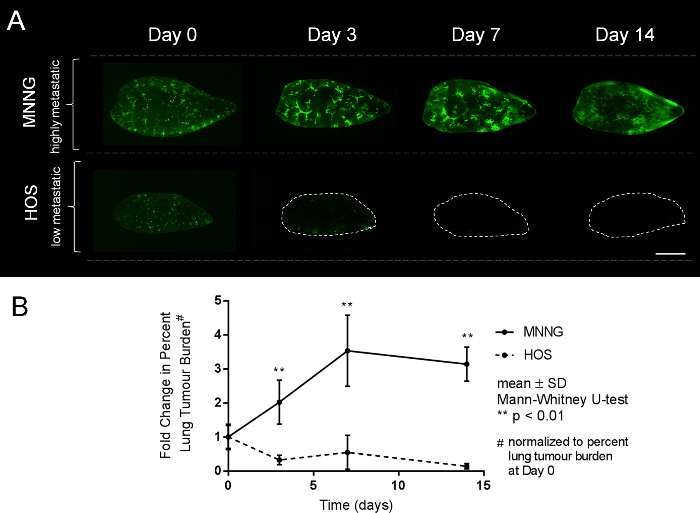
Calculate the lung tumor burden for the rest of the lung sections in control group and remaining groups. Plot the average lung tumor burden per group over 0, 3, 7 and 14 days. Representative widefield fluorescent pictures of high and low metastatic human OS cells growing in the PuMA model at progressive time points are shown (Figure 4A). A line graph showing the fold-change (normalized to Day 0) in percent metastatic tumor burden over time is shown (Figure 4B).
4. Confocal Fluorescence Imaging of the PuMA Model
NOTE: For confocal imaging, tissue processing is similar to that in the previous section except that larger lung slices are cut (complete transverse sections, 1-2 mm thick) to allow for more ROIs to be imaged.
Labelling of lung parenchyma with DAR4M:
Immerse lung slices in 10 μμM of DAR4M (in HBSS) for 45 min at 37 °C. DAR4M labels reactive nitrogen species within the cells of the lung.
After 45 min of incubation, rinse the lung slices with fresh HBSS.
Place the lung slice in a 35 mm glass-bottom round dish, and image on the confocal microscope.
The imaging parameters for the example images shown in Figure 5 are listed in Table 5.
Acquire a Z-stack for a region of interest. Use the suggested Z slice thickness for Nyquist sampling. A representative movie of a 3D stack showing green fluorescent OS cells in the DAR4M-labeled lung tissue is provided in Movie 1 and Supplemental Movie 1.
For the example confocal images shown in Figure 5, the imaging session was terminal. If longitudinal imaging is required, the use of a 2-photon or multi-photon equipped confocal LSM microscope for imaging is advised since there is less photodamage to living tissues19,20. Imaging mito-RFP expressing MG63 cells in the PuMA model:
Perform the PuMA protocol as outline in Step 1 and 2 with using MG63 cells expressing the mito-RFP construct.
Place the lung slice in a 35 mm glass-bottom round dish, and image on the confocal microscope.
The imaging parameters for the example images shown in Figure 6 are listed in Table 6. A representative movie of a 3D stack showing green fluorescent OS cells with red fluorescent mitochondria is provided in Movie 2 and Supplemental Movie 2.
Representative Results
Low-magnification widefield fluorescence microscopy
For widefield fluorescence microscopy of PuMA lung slices, representative images and quantification data are shown in Figure 2C, and Figure 4A and B. The metastatic propensities for high and low metastatic cell lines are visually apparent over progressive time points. MNNG cells can efficiently colonize the lung tissue whereas HOS cells cannot grow at all. From image analysis, quantitative data can be obtained to assess growth over time as shown in the graph of Figure 4B. Acquisition at low magnification (2.5X) is preferred in order to capture the entire lung slice in one image. This method permits batch imaging of a large number PuMA slices.
High-magnification confocal fluorescence microscopy
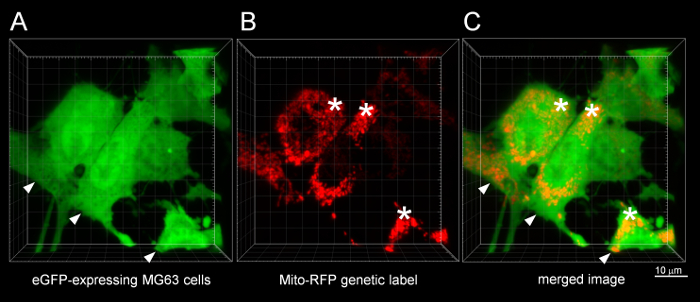
For confocal microscopy of PuMA lung slices, representative images are shown in Figure 5. Individual eGFP-expressing MG63 cells can be seen in Figure 5A in relation to the lung parenchyma, which is labeled by the DAR4M fluorescent dye in Figure 5B. A merged image is shown in Figure 5C, and a zoomedimage of a fluorescent tumor cell in a vessel is shown in Figure 5D. 3D movies of Figure 5C and 5D are shown in Movie 1 and Supplemental Movie 1, respectively. Confocal imaging of subcellular structures such as the mitochondria are shown inFigure 6. Cytosolic eGFP-expression permits the outline tumor cells in Figure 6A, and the mitochondria labeled with RFP are shown in Figure 6B. A merged image is seen in Figure 6C. 3D movies of Figure 6C are shown in Movie 2 and Supplemental Movie 2. Imaging subcellular structures such as the mitochondria can be combined with other fluorescent labels to study organelle biology in metastatic OS cells. When adding more labels, ensure that the laser lines and emission filters are compatible with the particular fluorophore/fluorescent protein of interest. Avoid imaging fluorophores/fluorescent proteins whose excitation and emission spectra closely overlap.
Figure 1: Diagram illustrating the metastatic cascade. The metastatic cascade can be broken down into several, rate-limiting steps. (1) Metastatic tumor cells leaving the primary tumor site and invades into the local parenchyma; (2) tumor cells intravasating into blood/lymphatic vessels and transit within the circulation; (3) tumor cell arrest at the secondary site; (4) tumor cell extravasation out from the microvasculature and survival in the secondary site; (5) growth into micrometastases; (6) growth into vascularized overt metastatic tumors. Please click here to view a larger version of this figure.
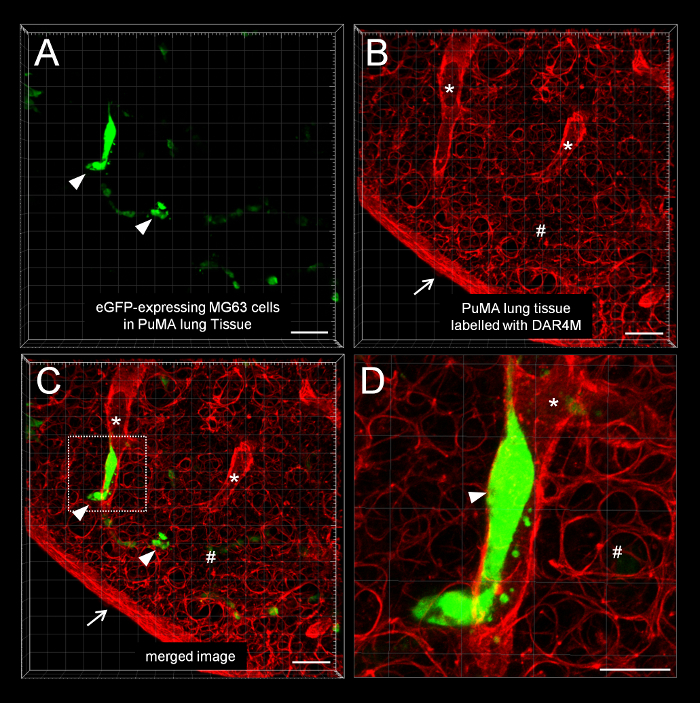
Figure 2: Imaging PuMA lung slices using widefield fluorescence and confocal fluorescence microscopy platforms. (A) Example PuMA slices are shown in a glass-bottom 35 mm dish. (B) confocal microscope equipment. (C) Example low-magnification image of a PuMA slice with eGFP-expression OS cells. Scalebar = 0.5 mm. (D) Example confocal image of a subregion of a PuMA slice. Scalebar = 100 μm. (*) Lung parenchyma is labeled red by DAR4M (see inset) and (∇) point to eGFP-expressing MG63 cells. Scalebar = 30 μm. Please click here to view a larger version of this figure.
Figure 3. Processing a low-magnification image of a PuMA slice using ImageJ. (A) Original PuMA image. (B) Subtraction of background. (C) Conversion to an 8-bit image. (D) Thresholding the image to highlight fluorescent tumor cells. (E) Inversion of image. (F) Enumeration of discrete shapes and quantification of shape areas. Please click here to view a larger version of this figure.
Figure 4. Low-magnification serial images showing the growth of highly and low metastatic human OS cells in the PuMA model over time. (A) Serial images of highly metastatic MNNG cells and low metastatic HOS cells are shown at 0, 3, 7, and 14 days post-injection. MNNG cells are able to grow better in the lung microenvironment in contrast to the HOS cells. Scalebar = 1 mm. (B) Fold-change in percent metastatic tumor burden for MNNG and HOS cells over time are shown as line graphs. Please click here to view a larger version of this figure.
Figure 5: Confocal microscopy of a DAR4M-labeled PuMA lung slice. A representative confocal image of eGFP-expressing MG63 cells in PuMA lung tissue labeled with DAR4M. (A) Green channel showing eGFP-expressing MG63 cells (∇). (B) Red channel showing the DAR4M dye binding to reactive nitrogen species in lung parenchyma cells. The dye highlights the lung parenchyma (#), and structures resembling a blood vessel (*). The edge of the PuMA lung section is denoted (↓). (C) Merged green and red image. Scalebar = 100 μm. (D) A zoomed image of the dashed white box (from C) shows a tumor cell in a vessel. Scalebar = 50 μm. Please click here to view a larger version of this figure.
Figure 6: Confocal microscopy of mitochondria in osteosarcoma cells in the PuMA model. A representative confocal image of mitochondria in eGFP-expressing MG63 cells. (A) Green channel showing eGFP-expressing MG63 cells (∇). (B) Red channel showing the RFP localized to mitochondria (*). (C) Merged green and red image. Scalebar = 10 μm. Please click here to view a larger version of this figure.
| Culture Media | Recipe |
| Complete Media (500mL) | •440 mL DMEM |
| •50 mL FBS | |
| •5 mL 10X Pen/strep solution (10000U/mL) | |
| •5 mL L-Glutamine (200 mM) | |
| A-Media (250mL) | •177.58 mL sterile water |
| •50 mL 10X M-199 media | |
| •15 mL 7.5% sodium bicarbonate solution | |
| •2.25 mL of 50 mM hydrocortizone | |
| •125 mL 0.4mg/ml retinol acetate (in sterile water) | |
| •5 mL of 10X Pen/strep solution (10000U/mL) | |
| •50 mL of 10mg/ml bovine insulin | |
| B-media (500ml) | •427.6 mL sterile water |
| •50 mL 10X M-199 media | |
| •15 mL 7.5% sodium bicarbonate solution | |
| •2.25 mL of 50 mM hydrocortizone | |
| •125 mL 0.4mg/ml retinol acetate (in sterile water) | |
| •5 mL of 10X Pen/strep solution (10000U/mL) | |
| •50 mL of 10mg/ml bovine insulin |
Table 1. Recipes for Complete Media, A-media, B-media. Recipes for cell culture and media for the PuMA assay are listed.
| Cell Culture Reagents | Description | Catalogue No. | Company |
| MNNG-HOS | highly metastatic OS cell line | CRL-1547 | ATCC |
| HOS | poorly metastatic OS cell line | CRL-1543 | ATCC |
| MG63.3 | highly metastatic OS cell line | N/A | Amy LeBlanc Laboratory (NCI) |
| MG63 | poorly metastatic OS cell line | CRL-1427 | ATCC |
| 10X M199 media | Base media for A-media and B-media | 11825015 | Thermofisher |
| Distilled Water (sterilized) | Component of A-media & | 15230-147 | Thermofisher |
| B-media | |||
| 7.5% sodium bicarbonate solution | Component of A-media & | 25080094 | Thermofisher |
| B-media | |||
| Hydrocortizone | Component of A-media & | H6909 | Sigma-Alrich |
| B-media | |||
| Retinol acetate-water soluable | Component of A-media & B-media | R0635-5MG | Sigma-Alrich |
| Penicillin/Streptomycin 10X concentrated (10000 U/ml) solution | Component of A-media & | 15140122 | Thermofisher |
| B-media, complete media | |||
| Bovine insulin solution (10mg/ml) | Component of A-media & | I0516-5ML | Sigma-Alrich |
| B-media | |||
| DMEM, high glucose | Base media of Complete Media | 11965092 | Thermofisher |
| L-Glutamine (200 mM) | Component of Complete Media | 25030081 | Thermofisher |
| Fetal Bovine Serum | Component of Complete Media | 16000044 | Thermofisher |
| Dulbecco’s Phosphate Buffered Saline | Used in cell culture | 14190144 | Thermofisher |
| Hank’s Buffered Salts Solution, no calcium, no magnesium, no phenol red | Used to resuspend cell pellet prior to injection | 14175095 | Thermofisher |
| Trypsin-EDTA (0.25%), phenol red | Used in cell culture | 25200114 | Thermofisher |
| DAR4M | Used to label lung parenchyma | ALX-620-069-M001 | Enzo |
Table 2. Cell culture reagents for A-media, B-media, and complete media. Components for media recipes, reagents, and buffers are listed with catalogue number and company name.
| Materials | Description | Catalogue No. | Company |
| Zeiss 710 Confocal LSM | Upright LSM confocal microscope | N/A | Zeiss |
| Zeiss 780 Confocal LSM | Inverted LSM confocal microscope | N/A | Zeiss |
| SCID mice | NOD.CB17-Prkdcscid/NcrCrl, female, age 6-8 weeks | N/A | Charles River |
| GelFoam | Used as a support for lung tissue sections | 59-9863 | Harvard Apparatus |
| SeaPlaque Agarose | Used during insufflation of the lung | 50100 | Lonza |
| 1 ml syringe with 27 gauge needle | Used for tail vein injection | Fisherscientific | |
| 14-826-87 | |||
| 10 ml syringe | Used for insufflation of the lung | 309604 | BD |
| 20 gauge catheter | Used during insufflation of the lung | SR-OX2032CA | Terumo |
| Abbott IV extension set (30", Sterile) | Used during insufflation of the lung | 8342 | Medisca |
| Alcohol swabs | For wiping tail vein before injection | 326895 | BD |
| Sterile surgical gloves | Asceptic handing of mouse lungs | Varies with size | Fisherscientific |
| 30 cm ruler | Used for insufflation of the lung | Staples | |
| Support stand for ruler | Used for insufflation of the lung | HS29022A | Pipette.com |
| 35 mm glass-bottomed culture dish | Used during imaging of lung slices | 81158 | Ibidi |
| Absorbent Underpads with Waterproof Moisture Barrier | Used to line the sterile work area in the biological hood | 56617-014 | VWR |
| Catgut Plain Absorbable Suture | Used to tie off cannulated trachea | N/A | Braun |
Table 3. Materials for PuMA. Materials required for the PuMA protocol are listed with catalogue number and company name.
| Materials | Description | Catalogue No. | Company |
| Micro Dissecting Scissors 3.5" Straight Sharp/Sharp | For cutting lung sections | RS-5910 | Roboz |
| 4” (10 cm) Long Serrated Straight Extra Delicate 0.5mm Tip | For manipulating/holding lung sections | RS-5132 | Roboz |
| 4” (10 cm) Long Serrated Slight Curve 0.8mm Tip | For manipulating/holding lung sections | RS5135 | Roboz |
| Thumb Dressing Forceps; Serrated; Delicate; 4.5" Length; 1.3 mm Tip Width | For general dissection | RS-8120 | Roboz |
| Thumb Dressing Forceps 4.5" Serrated 2.2 mm Tip Width | For general dissection | RS-8100 | Roboz |
| Extra Fine Micro Dissecting Scissors 3.5" Straight Sharp/Sharp, 20mm blade | For general dissection | RS-5880 | Roboz |
| Knapp Scissors; Straight; Sharp-Blunt; 27mm Blade Length; 4" Overall Length | For general dissection | RS-5960 | Roboz |
Table 4. Surgical instruments for PuMA. Various stainless steel instruments used in the protocol are listed with catalogue number and company name.
| Parameters | 488 laser | 561 laser |
| objective | Zeiss W N-Achroplan 10x/0.3 W (DIC) M27 | |
| power | 2% | 2% |
| Pixel dwell | 1.61 | 1.61 |
| Average | 0 | 0 |
| Zoom | 1 | 1 |
| Master gain | 820 | 814 |
| Digital gain | 1 | 0.51 |
| Digital offset | -4.5 | -9.24 |
| Pinhole | 51 | 57 |
| Tunable Filter | 496-553 | 570-618 |
Table 5. Parameters for confocal imaging of DAR4M-labeled PuMA sections. Specific image parameters used to obtained the confocal images in the current paper are provided in the table.
| Parameters | 488 laser | 561 laser |
| objective | Plan-Apochromat 63x/1.40 Oil DIC M27 | |
| power | 2% | 2% |
| Pixel dwell | 0.6 | 0.6 |
| Average | 2 (Line) | 2 (Line) |
| Zoom | 2.3 | 2.3 |
| Master gain | 449 | 390 |
| Digital gain | 1 | 1.23 |
| Digital offset | 0 | 0 |
| Pinhole | 136 | 136 |
| Tunable Filter | 493-550 | 566-703 |
Table 6. Parameters for confocal imaging of mitochondria MG63 cells in PuMA sections. Specific image parameters used to obtained the confocal images in the current paper are provided in the table.
Supplemental Figure 1: Gravity perfusion apparatus. The gravity perfusion apparatus is used to insufflate the lung with a liquid agarose/A-media solution at 20 cm of H20 hydrostatic pressure. The agarose/A-media solution is poured into the 10 mL syringe, which is attached by an IV extension set to the cannulated trachea (not shown). Please click here to download this figure.
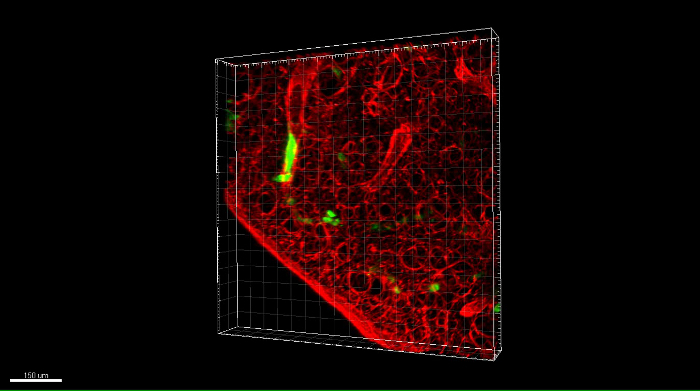 Movie 1: Rotation of a 3D confocal z-stack image of eGFP-expressing OS cells in a DAR4M-labeled PuMA lung slice. DAR4M (red channel) is used to highlight lung parenchyma and eGFP-expressing MG63 cells can also be seen (green channel). Scalebar = 100 μm. Please click here to download this move.
Movie 1: Rotation of a 3D confocal z-stack image of eGFP-expressing OS cells in a DAR4M-labeled PuMA lung slice. DAR4M (red channel) is used to highlight lung parenchyma and eGFP-expressing MG63 cells can also be seen (green channel). Scalebar = 100 μm. Please click here to download this move.
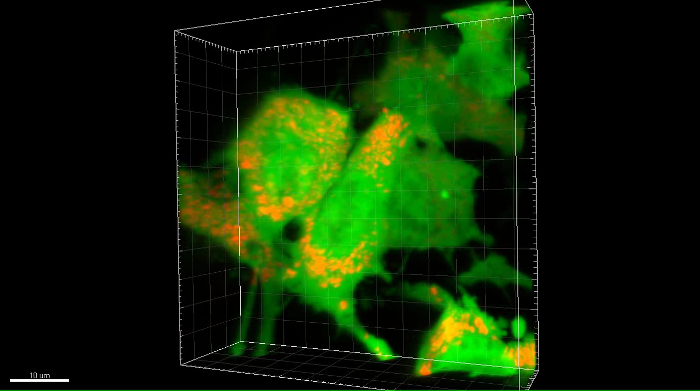 Movie 2: Rotation of a 3D confocal z-stack image of RFP-labeled mitochondria in eGFP-expressing OS cells in the PuMA model. Subcellular organelles such as mitochondria are shown labeled with RFP (red channel) in eGFP-expressing MG63 cells (green channel) in the PuMA model. Scalebar = 10 μm. Please click here to download this move.
Movie 2: Rotation of a 3D confocal z-stack image of RFP-labeled mitochondria in eGFP-expressing OS cells in the PuMA model. Subcellular organelles such as mitochondria are shown labeled with RFP (red channel) in eGFP-expressing MG63 cells (green channel) in the PuMA model. Scalebar = 10 μm. Please click here to download this move.
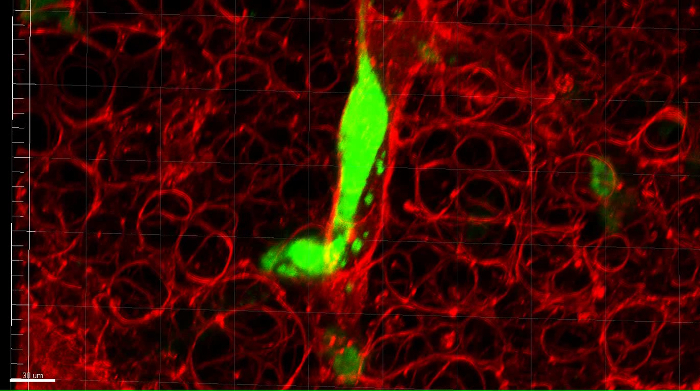 Supplemental Movie 1: Zoomed 3D movie of a metastatic OS cell in a vessel in the lung. A eGFP-expressing MG63 cell is shown in vessel within the lung microenvironment. Scalebar = 30 μM. Please click here to download this move.
Supplemental Movie 1: Zoomed 3D movie of a metastatic OS cell in a vessel in the lung. A eGFP-expressing MG63 cell is shown in vessel within the lung microenvironment. Scalebar = 30 μM. Please click here to download this move.
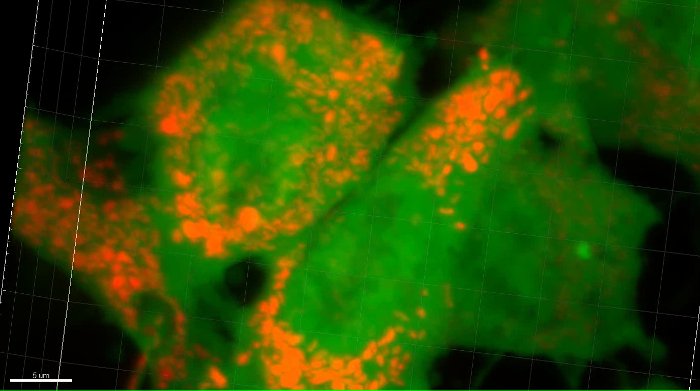 Supplemental Movie 2: Zoomed 3D movie of mitochondria in a metastatic OS cell in the lung. Mitochondria labeled with RFP are shown in MG63 cells in the lung microenvironment. Scalebar = 5 μM. Please click here to download this move.
Supplemental Movie 2: Zoomed 3D movie of mitochondria in a metastatic OS cell in the lung. Mitochondria labeled with RFP are shown in MG63 cells in the lung microenvironment. Scalebar = 5 μM. Please click here to download this move.
Discussion
The following technical article describes some practical aspects of the PuMA model in studying lung colonization in OS. Some critical steps in the protocol where researchers should take extra care include the following:
a) Cannulation of the trachea. The trachea can be easily damaged while dissecting the surrounding muscle and connective tissue. In addition, the needle of the catheter can easily be pushed through the trachea. Pay close attention to how the bevel of the needle enters the trachea when inserting the cannula.
b) Puncturing or damaging the surface of the lung. The lung can be easily punctured or damaged with dissecting scissors while removing the sternum and exposing the chest cavity. Be careful when dissecting out the pluck from the chest cavity. Puncturing or damaging the surface of the lung will cause the liquid agarose/A-media to leak out and the lung will be deflated.
c) Remove excess media from lung sections while imaging. Excess liquid media surrounding the lung section can cause a "mirror" effect when viewed under the microscope. The excess liquid can reflect fluorescent light from the tumor cells thereby exaggerating the fluorescence area in the image. Removal of excess media fluid will prevent this artifact in the image. Alternatively, the image artifact can be removed during post-processing of the images.
d) Photodamage of the living tissues. When using a mercury bulb or lasers of a 1-photon microscope, be sure to limit the time exposed to the light. The intensity/power percentage of the light source can also be reduced. Overexposure to the light source can cause photodamage to the lung tissue. Microscopes equipped with light emitting diodes (LED) or 2-photon/multi-photon microscopes would be ideal because they produce less photodamage during longitudinal imaging studies.
The PuMA model can be adapted and modified to study many aspects of the lung colonization process. Another variant of this ex vivo approach is described by van den Bijgaart and colleagues 21. For studies examining the effects of gene knock-down or anti-metastatic drug activity on tumor cell growth in the lung, scaling back the number of cells injected from 5 x 105 to 3 x 105 is advised since the effects of the intervention can be masked during the exponential growth of a larger cell innoculum. Several studies have used the PuMA model to study drivers of lung metastatic progression 4,5,8,22,23,24. Various fluorescent indicator dyes or fluorescent reporter genes can be used to label tumor cells to ascertain changes in cell physiology or gene expression 8 in the lung microenvironment.
One limitation of the PuMA model includes a limited number of compatible cell lines. The established cell lines compatible with this assay are listed by Mendoza and colleagues 3 . For high and low metastatic osteosarcoma cell lines, the follow pairs of clonally related cell lines have been found to grow and maintain their metastatic propensity in the PuMA model: human MG63.3 & MG63 cells, MNNG & 143B , and HOS cells, murine K7M2 and K12 cells. Researchers must empirically determine whether or not their cell lines can remain viable in B-media. Another limitation to consider is the limited length of time the lung tissues can be maintained in vitro. Lung tissue can retain its cellular components for 30 days, beyond that time and the cellular components of the lung begin to die off. Therefore studying the interactions between tumor cells and lung stromal cells should not exceed 30 days.
The PuMA model provides an unprecedented method to study how metastatic OS cells colonize lung tissue. The most striking aspect of the PuMA model comes from the fact that several low metastatic OS cell lines (HOS, MG63, K12), whose in vivo phenotypes have been characterized elsewhere 25, cannot grow in the PuMA model. In contrast, clonally related highly metastatic OS cells (MNNG, MG63.3, K7M2) have a greater propensity to colonize lung tissue in vivo25 and in the PuMA model. This suggests that the cellular and extracellular components of the lung microenvironment that prevent low metastatic OS cell lines from growing in vivo, are still maintained in the PuMA model despite the lack of blood flow. In other words, the lung microenvironment of the PuMA model still exerts a "selection pressure" that prevents low metastatic OS cells from growing in the lung. Indeed, studying high metastatic OS cells grow in PuMA has been useful in identifying new molecular drivers of the metastatic phenotype 4,5. Moreover, genetic down-modulation of said targets in highly metastatic cells was shown to diminish metastatic capacity in both the PuMA model and in vivo 5. For candidate anti-metastatic drug studies, the PuMA model can be used to determine which concentration range can reduce metastatic outgrowth in lung tissues, which in turn, can then be validated in vivo.
Future applications of this model should exploit the plethora of commercially available fluorescent dyes and reporter genes in order to delve deep into the basic biology of OS metastasis progression. For example, fluorescent dyes such as 2',7' -dichlorofluorescin diacetate or dihydroethidium can be used to assess the redox state of tumor cells in the PuMA model. Fluorescent reporter genes can be used to study organelle biology or assess promoter activity to determine which signaling pathways are activated in highly metastatic OS cells during the colonization process. Such approaches can be used to visualize whether a drug treatment has activity in tumor cells growing in the lung. Fluorescent microscopy platforms that produce less photodamage to tissues, such as LED-equipped microscopes or 2-photon/multiphoton confocal microscopes, can be used study how metastatic OS cells migrate and invade throughout lung tissues. Furthermore, adhesion interactions between tumor cells and lung stromal cells can be monitored with fluorescent reporter genes.
To summarize, the current paper discusses the practical aspects of the PuMA model, first developed by Mendoza and colleagues 3. An example of using low-magnification, widefield fluorescence microscopy and high resolution confocal microscopy to assess the growth of high and low metastatic human OS cells is shown. Several critical steps of the protocol have been described. In addition, the advantages and limitations of the PuMA model have been discussed. Future applications of the PuMA model to study the lung colonization process have been put forward, and it is hoped that this model will have a wider use in the osteosarcoma research community.
Disclosures
The authors have nothing to disclose.
Acknowledgments
We would like to thank Dr. Arnulfo Mendoza who provided training in the PuMA technique. Additionally, we would like to acknowledge Drs. Chand Khanna, Susan Garfield (NCI/NIH), and Sam Aparicio (BC Cancer Agency) for providing use of their microscopes during the course of this study. This research was supported (in part) by the Intramural Research Program of the National Institutes of Health, Center for Cancer Research, Pediatric Oncology Branch. M.M.L. was supported by the National Institutes of Health Intramural Visiting Fellow Program (award 15335), and is currently supported by a Joan Parker Fellowship in Metastasis Research. P.H.S. is supported by British Columbia Cancer Foundation.
References
- Khanna C, et al. Toward a drug development path that targets metastatic progression in osteosarcoma. Clin Cancer Res. 2014;20(16):4200–4209. doi: 10.1158/1078-0432.CCR-13-2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steeg PS. Perspective: The right trials. Nature. 2012;485(7400):S58–S59. doi: 10.1038/485S58a. [DOI] [PubMed] [Google Scholar]
- Mendoza A, et al. Modeling metastasis biology and therapy in real time in the mouse lung. J Clin Invest. 2010;120(8):2979–2988. doi: 10.1172/JCI40252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SH, Ren L, Mendoza A, Eleswarapu A, Khanna C. Apoptosis resistance and PKC signaling: distinguishing features of high and low metastatic cells. Neoplasia. 2012;14(3):249–258. doi: 10.1593/neo.111498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lizardo MM, et al. Upregulation of Glucose-Regulated Protein 78 in Metastatic Cancer Cells Is Necessary for Lung Metastasis Progression. Neoplasia. 2016;18(11):699–710. doi: 10.1016/j.neo.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouliot N, Pearson HB, Burrows A. Metastatic Cancer: Clinical and Biological Perspectives. Austin, Texas: Landes Bioscience; 2012. Investigating Metastasis Using In Vitro Platforms. [Google Scholar]
- Cameron MD, et al. Temporal progression of metastasis in lung: cell survival, dormancy, and location dependence of metastatic inefficiency. Cancer Res. 2000;60(9):2541–2546. [PubMed] [Google Scholar]
- Morrow JJ, et al. mTOR inhibition mitigates enhanced mRNA translation associated with the metastatic phenotype of osteosarcoma cells in vivo. Clinical Cancer Research. 2016. [DOI] [PMC free article] [PubMed]
- Varghese HJ, et al. In vivo videomicroscopy reveals differential effects of the vascular-targeting agent ZD6126 and the anti-angiogenic agent ZD6474 on vascular function in a liver metastasis model. Angiogenesis. 2004;7(2):157–164. doi: 10.1007/s10456-004-1941-3. [DOI] [PubMed] [Google Scholar]
- Khanna C, Hunter K. Modeling metastasis in vivo. Carcinogenesis. 2005;26(3):513–523. doi: 10.1093/carcin/bgh261. [DOI] [PubMed] [Google Scholar]
- Welch DR. Technical considerations for studying cancer metastasis in vivo. Clin Exp Metastasis. 1997;15(3):272–306. doi: 10.1023/a:1018477516367. [DOI] [PubMed] [Google Scholar]
- Somasekharan SP, et al. YB-1 regulates stress granule formation and tumor progression by translationally activating G3BP1. J Cell Biol. 2015;208(7):913–929. doi: 10.1083/jcb.201411047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Naggar AM, et al. Translational Activation of HIF1alpha by YB-1 Promotes Sarcoma Metastasis. Cancer Cell. 2015;27(5):682–697. doi: 10.1016/j.ccell.2015.04.003. [DOI] [PubMed] [Google Scholar]
- MacDonald IC, Groom AC, Chambers AF. Cancer spread and micrometastasis development: quantitative approaches for in vivo models. Bioessays. 2002;24(10):885–893. doi: 10.1002/bies.10156. [DOI] [PubMed] [Google Scholar]
- Entenberg D, et al. A permanent window for the murine lung enables high-resolution imaging of cancer metastasis. Nat Methods. 2018;15(1):73–80. doi: 10.1038/nmeth.4511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, et al. Quantification of cancer cell extravasation in vivo. Nat Protoc. 2016;11(5):937–948. doi: 10.1038/nprot.2016.050. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9(7):671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underwood EE. Quantitative stereology. Addison-Wesley Pub. Co; 1970. [Google Scholar]
- Tanaka K, et al. In vivo optical imaging of cancer metastasis using multiphoton microscopy: a short review. Am J Transl Res. 2014;6(3):179–187. [PMC free article] [PubMed] [Google Scholar]
- Prouty AM, Wu J, Lin DT, Camacho P, Lechleiter JD. Multiphoton laser scanning microscopy as a tool for Xenopus oocyte research. Methods Mol Biol. 2006;322:87–101. doi: 10.1007/978-1-59745-000-3_7. [DOI] [PubMed] [Google Scholar]
- Bijgaart RJ, Kong N, Maynard C, Plaks V. Ex vivo Live Imaging of Lung Metastasis and Their Microenvironment. J Vis Exp. 2016. p. e53741. [DOI] [PMC free article] [PubMed]
- Guha M, et al. Mitochondrial retrograde signaling induces epithelial-mesenchymal transition and generates breast cancer stem cells. Oncogene. 2014;33(45):5238–5250. doi: 10.1038/onc.2013.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren L, Morrow JJ, et al. Positively selected enhancer elements endow osteosarcoma cells with metastatic competence . Nat Med. 2018. [DOI] [PMC free article] [PubMed]
- Ren L, et al. Metabolomics uncovers a link between inositol metabolism and osteosarcoma metastasis. Oncotarget. 2017;8(24):38541–38553. doi: 10.18632/oncotarget.15872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren L, et al. Characterization of the metastatic phenotype of a panel of established osteosarcoma cells. Oncotarget. 2015;6(30):29469–29481. doi: 10.18632/oncotarget.5177. [DOI] [PMC free article] [PubMed] [Google Scholar]


