Abstract
Two low-molecular-weight proteins have been purified from Brassica napus pollen and a gene corresponding to one of them has been isolated. The gene encodes an 8.6-kD protein with two EF-hand calcium-binding motifs and is a member of a small gene family in B. napus. The protein is part of a family of pollen allergens recently identified in several evolutionarily distant dicot and monocot plants. Homologs have been detected in Arabidopsis, from which one gene has been cloned in this study, and in snapdragon (Antirrhinum majus), but not in tobacco (Nicotiana tabacum). Expression of the gene in B. napus was limited to male tissues and occurred during the pollen-maturation phase of anther development. Both the B. napus and Arabidopsis proteins interact with calcium, and the potential for a calcium-dependent conformational change was demonstrated. Given this affinity for calcium, the cloned genes were termed BPC1 and APC1 (B. napus and Arabidopsis pollen calcium-binding protein 1, respectively). Immunolocalization studies demonstrated that BPC1 is found in the cytosol of mature pollen. However, upon pollen hydration and germination, there is some apparent leakage of the protein to the pollen wall. BPC1 is also concentrated on or near the surface of the elongating pollen tube. The essential nature of calcium in pollen physiology, combined with the properties of BPC1 and its high evolutionary conservation suggests that this protein plays an important role in pollination by functioning as a calcium-sensitive signal molecule.
Pollination is a complex process involving an intricate series of physiological changes in the pollen and interactions between the pollen grain and stigma, which culminates in the successful fertilization of the egg (for review, see Dumas et al., 1994; Elleman and Dickinson, 1994). The pollen grain lands on the stigma in a desiccated, metabolically inactive state. In Brassica, a conduit is formed between the pollen grain and a stigmatic papillary cell to facilitate reciprocal transfer of materials essential for pollen recognition and germination. In a compatible interaction, pollen hydration occurs and a metabolically active state is resumed in the pollen grain. This leads to pollen tube development, emergence and directional growth, and penetration of the stigma. Initiation and sustenance of metabolic activities in the pollen involves passage of nutrients and signaling molecules from the stigma. Calcium is of key significance in this regard.
The general importance of calcium in pollen biology has been well established. Over 30 years ago, calcium was recognized as an essential constituent of in vitro pollen-germination media and as a potential chemoattractant guiding pollen tube growth (Kwack and Brewbaker, 1961; Mascarenhas and Machlis, 1964). Its physiological requirement during pollen-pistil interactions has been inferred from observation of active uptake of 45Ca2+ by pollen grains directly from the stigma (Bednarska, 1991). Calcium is likely released from the papillary cell by a regulated Ca2+-ATPase (Bednarska, 1991). Manipulation of intracellular concentrations of calcium in the pollen has been achieved using various chemicals known to affect calcium homeostasis (Picton and Steer, 1985; Bednarska, 1989; Heslop-Harrison and Heslop-Harrison, 1992; Pierson et al., 1994; Malhó and Trewavas, 1996). From these studies, the establishment and maintenance of a precise calcium gradient was determined to be essential for pollen germination, pollen tube elongation, and directional growth. This gradient is established in the pollen grain at the point of tube emergence before germination (Bednarska, 1989). The elongating tube then maintains a descending concentration gradient from the tip by the action of calcium channels and Ca2+-ATPase in the plasmalemma and mitochondria (Picton and Steer, 1985; Pierson et al., 1994; Malhó and Trewavas, 1996).
By interacting directly with the cytoskeletal apparatus (Picton and Steer, 1985; Pierson et al., 1994) or via a calmodulin intermediate (Hausser et al., 1984), calcium regulates cytoplasmic flow, vesicle fusion, and the function of cytoskeleton elements required for tube emergence and growth (Picton and Steer, 1985; Pierson et al., 1994; Malhó and Trewavas, 1996). Any modification of the calcium gradient can therefore inhibit germination and pollen tube elongation. Indeed, such modulation may act as a mechanism to facilitate the self-incompatibility response in some plant species (Singh et al., 1989; Franklin-Tong et al., 1993).
Many calcium-mediated effects have been well characterized in other systems (Carafoli, 1987; Heizman and Hunziker, 1991; Ikura, 1996; Niki et al., 1996). In these cases, the physiological effects of calcium are often mediated indirectly by signal proteins. By binding calcium, these proteins undergo conformational changes that enable interaction with and modulation of effector proteins that cause the physiological changes linked to the calcium stimulus. Many of these calcium-signal proteins are well characterized and have been subgrouped based on structural and biochemical features (Heizman and Hunziker, 1991; Nakayama and Kretsinger, 1994; Niki et al., 1996). The majority are intracellular constituents, but some members may be secreted and have intercellular actions (Zimmer et al., 1995). Several target proteins for these calcium-binding proteins have been identified, enabling biochemical definition of many of the physiological effects associated with calcium (Nakayama and Kretsinger, 1994; Zimmer et al., 1995; Niki et al., 1996).
The results of the present study reveal a potential link between the physiological effects of calcium on pollen and a possible mechanism for manifestation of these effects. A low-Mr cytosolic calcium-binding protein has been characterized from B. napus pollen extracts and is termed BPC1 (B. napus pollen calcium-binding protein 1). In an independent study, this protein and a related Brassica pollen protein were identified by their strong immunoreactivity to IgE from a human subject allergic to Brassica pollen (Toriyama et al., 1995). Immunolocalization demonstrates that BPC1 is located at the surface of the pollen tube, an area of calcium flux during pollen germination and tube elongation (Polito, 1983; Pierson et al., 1994; Malhó and Trewavas, 1996). Biochemical analysis suggests the protein can undergo a calcium-dependent conformational change; therefore, BPC1 may link calcium flux with physiological responses in pollen. This linkage is highly conserved among diverse species. In addition to the Brassica proteins, highly related pollen allergens have been identified in Betula verrucosa (Engel et al., 1997; Twardosz et al., 1997), Olea europaea (Batanero et al., 1997), Artemisia vulgaris (Engel et al., 1997), and the monocots Cynodon dactylon (Suphioglu et al., 1997), Phleum pratens, and Lilium longiflorum (Engel et al., 1997; Twardosz et al., 1997). The present study also describes cloning of APC1, a homolog from Arabidopsis, and demonstrates the presence of related sequences in Antirrhinum but not in tobacco (Nicotiana tabacum). Considering the biochemical and genetic characteristics of BPC1, physiological activity of this protein family likely involves the transduction of calcium signals associated with pollen germination and tube elongation.
MATERIALS AND METHODS
Plant Materials
Brassica napus subsp. oleifera W1 (Goring et al., 1992), tobacco (Nicotiana tabacum), and Arabidopsis ecotype Lansberg erecta were cultivated in a growth room or chamber with a light regime of 16 h of light/8 h of dark at 18°C. Anthers were collected from W1 flowers daily at anthesis, placed into microcentrifuge tubes, and allowed to dry on the benchtop to facilitate anther dehiscence. Pollen release was promoted by agitating the collected anthers using a vortex mixer. With this procedure pollen would pack at the bottom of the tube and the anther material would collect at the top, thereby permitting its removal. Microspores were isolated from flower buds according to the method of Coventry et al. (1988) and stored at −80°C. Whole flower buds, anthers, and stigmas were also stored at −80°C until RNA extraction.
Pollen-Protein Extraction and Analysis
Soluble pollen proteins were isolated following the method of Knox et al. (1975). Pollen was immersed in a solution of 50 mm Tris-HCl, pH 7.5, 1 mm CaCl2, and 5% (w/v) mannitol. Proteins were then separated by two-dimensional gel electrophoresis using a pH 3.0 to 10.0 gradient (Bio-lyte, Bio-Rad) in the first dimension and a 15% SDS-PAGE gel in the second dimension (Ausubel et al., 1988). The separated proteins were then either stained with Coomassie Blue R-250 or electroblotted (Ausubel et al., 1988) onto Immobilon P membranes (Millipore). After transfer, proteins were visualized using Ponceau S stain (Ausubel et al., 1988) and isolated by excising corresponding areas of the membrane.
For determining the amino acid sequence, proteins were subjected to in situ digestion with the endoproteinases Lys-C, KC, or AspN (Wako BioProducts, Richmond, VA). Digestion of excised spots was for 24 h at 37°C in 0.1 m Tris-HCl, pH 8.5, containing 40% (v/v) acetonitrile. The resulting peptides were isolated using a Hypersil ODS column (2.1 × 100 mm; Hewlett-Packard) employing a linear gradient of 8% to 80% acetonitrile in 0.1% (v/v) trifluoracetic acid over 40 min with a flow rate of 100 mL/min. Automated Edman degradations were performed on a protein-sequencing system (model G1005A, Hewlett-Packard). The amino acid sequences of the peptides derived from BPC1 are as follows: peptide I, ISASELE, and peptide II, IDTDGDGNISFQEFTEFASAN. The peptide sequences from PP-A are: peptide I, ISATELGDALK; peptide II, DFASANRG; peptide III, DVAKRM; and peptide IV, DG ISYNN (where the space represents an undetermined amino acid).
Gene Isolation
Partial cDNA clones encoding BPC1 were isolated by the 3′-RACE-PCR method as described previously (Frohman, 1990) using a thermocycler (Perkin-Elmer). Total RNA isolated from 1- to 4-mm W1 flower buds was used as a template. Two nested oligonucleotide primers were synthesized (University of Guelph DNA/Protein Analysis Facility) based on peptide sequence and accounting for degeneracy of the genetic code. Reverse transcription was performed as outlined previously (Frohman, 1990) with 1 μg of RNA, 200 units of Moloney murine leukemia virus reverse transcriptase (GIBCO-BRL), and the dT17-adapter primer (Goring et al., 1992). PCR amplification was catalyzed by Taq polymerase (Boehringer Mannheim). The initial PCR reaction was performed using OL175–65 (ATA/C/T GAC/T ACC/I GAC/T GGC/I GAC/T GGC/I AA, where I represents inosine) and the RACE-adapter primer (Goring et al., 1992) for five cycles of 30 s at 94°C, 60 s at 37°C, and 60 s at 72°C, followed by a further 30 cycles of 30 s at 94°C, 45 s at 40°C, and 60 s at 72°C. The ramp time was 2.5 min and 1 min between the annealing and extension periods of the two respective cycle series. A second PCR reaction was performed using the initial reaction products as a template for the further 3′ nested primer OL175-01 (TTC/T CAA/C GAA/G TTC/T ACC/I GAA/G TT) and the RACE-adapter primer. Reaction conditions were as outlined above. An approximately 250-bp product resulted, which was purified using a 1.5% low-melting-point agarose gel. This was cloned into the EcoRV site of pBluescript SK− (Stratagene) using Escherichia coli DH10B as the host and following standard techniques (Ausubel et al., 1988). The DNA sequence was determined using Sequenase II (United States Biochemical) and standard methods (Ausubel et al., 1988).
With the information derived from the 3′-RACE clones, two nested oligonucleotides were synthesized to enable cloning of the rest of the ORF using 5′-RACE-PCR, as described previously (Frohman, 1990). Reverse transcription of RNA from 4-mm flower buds was primed using OL176-92 (GA TGG CAA ATA CTA CCA TTC) as described above. After the reaction, excess primer was removed using a Microcon-30 (Amicon, Beverly, MA) column. Polyadenylation of the cDNA was catalyzed by terminal transferase (GIBCO-BRL) as described above. Serial dilutions of the cDNA were amplified using the dT-17 adapter primer, the RACE-adapter primer, and the internal nested primer OL176-91 (AA ACT TTG GCA ACA TCC TTC). Reaction conditions included an initial five cycles of 30 s at 94°C, 60 s at 37°C, a ramp time of 30 s, and 72°C for 60 s, followed by five cycles of 30 s at 94°C, 30 s at 51°C, and 60 s at 72°C. A final 25 cycles of 30 s at 94°C, 30 s at 60°C, and 60 s at 72°C concluded the reaction. The reaction products were separated on a 1.5% low-melting-point agarose gel, and isolated subfractions were used as a template in a subsequent series of PCR reactions primed by OL176-91 and the RACE-adapter primer. This yielded an approximately 320-bp fragment, which was cloned into the EcoRV site of pBluescript SK−. Correct clones were identified by colony hybridization using a 3′-RACE clone (pKR92–5) as a radiolabeled probe following standard procedures (Ausubel et al., 1988).
B. napus genomic clones were isolated from a W1 genomic DNA library prepared in Lambda Fix II (Stratagene) and screened with a 3′-RACE clone (pKR92-5). An Arabidopsis clone was isolated from a genomic library of the Columbia ecotype (obtained from the Arabidopsis Biological Resource Center, Ohio State University, Columbus) by screening with a B. napus cDNA clone encoding its entire ORF (pKR94-3). Standard procedures were followed for library screening (Ausubel et al., 1988). An approximately 2.5-kb SstI-XhoI fragment was isolated from one of the B. napus lambda clones and subcloned into pBluescript SK− (pKR108). The Arabidopsis lambda clone released an approximately 3.9-kb PstI-BamHI fragment, which was also subcloned into pBluescript SK− (pKR140). The DNA sequence in both orientations for the portion of these clones encoding the ORF was determined (data not shown).
DNA and RNA Analysis
Genomic DNA from B. napus, Arabidopsis, and tobacco was extracted as described previously (Ausubel et al., 1988). Antirrhinum DNA was kindly supplied by Dr. E. Coen (John Innes Centre, Norwich, UK). Total RNA was extracted from flower buds, anthers, and leaves as described previously (Ausubel et al., 1988). DNA and RNA blots were performed following standard methods (Ausubel et al., 1988), with conditions adjusted for high- (95% complementarity) or low- (65% complementarity) stringency wash conditions. Approximately 15 μg of genomic DNA from B. napus W1, Antirrhinum, and tobacco, or approximately 1 μg of Arabidopsis DNA was digested with restriction enzymes (GIBCO-BRL) and separated on a 0.8% agarose gel before blotting to Hybond N+ membranes (Amersham). RNA blots involved separation of 10 μg of RNA on a 1.2% formaldehyde gel before transfer to Biodyne B membrane (Pall Specialty Materials, Port Washington, NY).
Recombinant Protein Production and Purification
Recombinant BPC1 (rBPC1) and APC1 (rAPC1) were produced by cloning the modified ORFs into the E. coli expression vectors pGEX2T[128/129] (kindly provided by Dr. M.A. Blanar, University of California, San Francisco) and pT7-7 (Studier and Moffatt, 1986), respectively. pGEX-2T is similar to the vector pAR(ΔRI) (Blanar and Rutter, 1992) and enables production of rBPC1 as a fusion to GST, the FLAG peptide, and the heart-muscle kinase-recognition domain. Cleavage of fusion proteins with thrombin results in approximately 2 kD of additional amino acids at the N terminus of the recombinant protein. To generate the rBPC1 expression construct (pKR129), an EcoRI site was inserted at the 5′ end of the BPC1 ORF using a cDNA clone as a template for the primers OL177-80 (CG GAA TTC GCT GAT GCT GAG CAC GAA) and OL176-91 in a PCR reaction (5 cycles of 30 s at 94°C, 30 s at 51°C, and 60 s at 72°C, followed by 15 cycles of 30 s at 94°C, 30 s at 60°C, and 60 s at 72°C). The approximately 150-bp product was used to reconstruct a modified BPC1 ORF with its start codon removed by the newly introduced EcoRI site. The rAPC1 expression construct pKR195 was generated by placing a NdeI site integral with the APC1 ORF start codon and a ClaI site 3′ of the stop codon using the genomic APC1 clone as template and the primers OL9075 (GG AAT TCC AT ATG GCT GAT G CA ACG GAG) and OL9074 (C CAT CGA TT TAG AAA ATT TTG GCA ACA TCC) in a PCR reaction (20 cycles of 30 s at 94°C, 30 s at 60°C, and 60 s at 72°C). The 260-bp product was cloned into the NdeI and ClaI sites of pT7-7 (Studier and Moffatt, 1986). Fidelity of the modified ORF was confirmed by DNA sequencing in both orientations. E. coli BL21 (DE3) [pLysS] (Novagen, Madison, WI) was transformed with each construct.
For production of the fusion protein, 500-mL cultures of Luria-Bertani broth containing Glc (2%, w/v), carbenicillin (50 mg/mL), and chloramphenicol (34 mg/mL) were incubated at 37°C with vigorous shaking until the OD600 was 0.8 to 1.0. Expression was induced with the addition of 0.8 mm isopropylthio-β-galactoside, and cultures were incubated at 30°C with vigorous shaking for a further 4 h. For rBPC1, cells were harvested by centrifugation, washed with PBS, and lysed by the addition of 1% (w/v) Triton X-100. Viscosity of the suspension was reduced by the addition of DNase I (22 mg/mL) in combination with 10 mm MgCl2. Proteolysis was inhibited with PMSF (0.1 mg/mL). For rAPC1, cells were resuspended in buffer A (30 mm Tris-HCl, pH 7.5, 15% glycerol, and 2 mm β-mercaptoethanol) and disrupted using a French press.
rBPC1 was purified first as a fusion to GST using glutathione-Sepharose (Sigma). Following cleavage with thrombin (Sigma), as described previously (Ausubel et al., 1988), rBPC1 was purified to homogeneity by gel-filtration chromatography (data not shown; see “Calcium Interaction Assays” for details). rAPC1 was purified by loading the cell extract onto an anion-exchange column (2.5 × 15 cm; Fractogel EMD DEAE 650 (S), Merck, Darmstadt, Germany) at 3 mL/min. The column was washed extensively with buffer A, and bound proteins were eluted with a linear 0 to 600 mm NaCl gradient in buffer A. Fractions containing rAPC1 were identified by 16.5% Tris-Tricine SDS-PAGE analysis. Pooled fractions were purified further by the addition of ammonium sulfate to 60% saturation. Proteins that remained soluble were concentrated using a CentriPrep 3S column (Amicon) and then applied to a Superdex 75 gel-filtration column (1.6 × 60 cm, Pharmacia) at 0.5 mL/min in buffer A plus 150 mm NaCl. Fractions containing pure rAPC1, as judged by SDS-PAGE analysis, were pooled.
Calcium Interaction Assays
Calcium interactions with BPC1 and APC1 were investigated using the recombinant proteins. For mobility-shift assays, the conditions were as described previously (Sistrunk et al., 1994). Protein was mixed with 10 mm EGTA, 10 mm CaCl2 plus 2 mm EGTA, or 10 mm MgCl2 plus 2 mm EGTA prior to electrophoresis on native-PAGE gels (Ausubel et al., 1988). Gel filtration was on a 1- × 60-cm column packed with Sephacryl S-100H media (Pharmacia). Running buffers (pH 7.4) contained 20 mm Tris-HCl with either 10 mm EGTA or 10 mm CaCl2 plus 2 mm EGTA. Ionic strength was modified by the addition of 60 or 200 mm NaCl. Buffer supply at a rate of 0.25 mL/min was controlled with a protein-purification system (model 650E, Waters). Protein elution was monitored at 254 nm with a detector (model 441, Waters), and column fractions were analyzed by SDS-PAGE. The column was standardized using proteins of known molecular mass (thyroglobin, 670 kD; γ-globin, 158 kD; BSA, 66 kD; ovalbumin, 44 kD; carbonic anhydrase, 29 kD; myoglobin, 17 kD; Cyt c, 12.4 kD; and vitamin B-12, 1.4 kD; all from Sigma or Bio-Rad).
Development of Polyclonal Antibodies rAPC1 and Immunotechniques
rAPC1 was purified to homogeneity as described above and used to generate polyclonal antibodies in Balb/c mice. Fifty micrograms of rAPC1 in PBS was mixed with adjuvant (TiterMax, CytRx, Norcross, GA) at a 1:1 ratio and injected into mice subcutaneously. Two weeks later, a second injection was made subcutaneously using the same adjuvant. After an additional 2 weeks, a final intraperitoneal injection was made without adjuvant. The serum was collected 3 d after the final injection. Immunoblots with anti-APC1 serum were by standard procedures (Ausubel et al., 1988) using Immobilon-PSQ membrane (Millipore) and goat anti-mouse IgG-horseradish peroxidase conjugate (Sigma) with detection by enhanced chemiluminescence reagents (Amersham).
Whole, developing flowers between the 2-mm stage and anthesis, pistils of mature flowers, and germinated pollen grains were fixed in 3% glutaraldehyde, 2% formaldehyde, and 150 mm Suc in 25 mm potassium phosphate buffer (pH 7.2) for 12 h at 4°C. For germinated pollen, pollen from mature anthers of 30 to 50 B. napus flowers was dispersed directly into pollen-germination buffer (1 mm KNO3, 1.65 mm CaCl2, 0.16 mm H3BO3, 1 mm Tris base, and 20% [w/v] Suc; Roberts et al., 1983) onto a silanized glass plate that was then placed on blotter paper saturated with water in a sealed Petri plate and incubated at 25°C for 16 h. After fixation, all plant tissues were rinsed in distilled water, dehydrated stepwise to 100% ethanol, embedded in London White resin (London Resin Co., London, UK), and sectioned 1 μm thick. Solution changes for germinated pollen were facilitated by centrifugation at 600g for 4 min just prior to the change.
Immunolocalization of BPC1 was initiated by pretreating sections with 0.1 n HCl and then rinsing with 1.5× TBS (30 mm Tris-HCl and 225 mm NaCl, pH 7.3) with TBSTG buffer (0.2% [w/v] Tween 20 and 0.2% [w/v] Gly). Blocking was performed using 10% (v/v) rabbit serum (Sigma) in the same solution. Sections were incubated in mouse anti-rAPC1 antiserum at a dilution of 1:100 in TBSTG with 10% rabbit serum for 1 h at 37°C. Following thorough rinsing in TBSTG, the sections were incubated for 30 min at 37°C with affinity-purified rabbit-anti-mouse IgG antibodies (Sigma) tagged with 7 nm colloidal gold diluted in TBSTG with 10% rabbit serum. After thorough rinsing in TBSTG, TBS, and distilled water, the sections were briefly incubated in 1% aqueous glutaraldehyde. Enhancement of bound colloidal gold was accomplished by following the manufacturer's instructions for a western-blotting grade gold-enhancement kit (Bio-Rad), with the addition of 0.5% (w/v) gelatin to the enhancement solutions. Micrography was performed using a contrast microscope (Jenalumar, Zeiss-Jena).
RESULTS
Cloning and Characterization of BPC1 and APC1
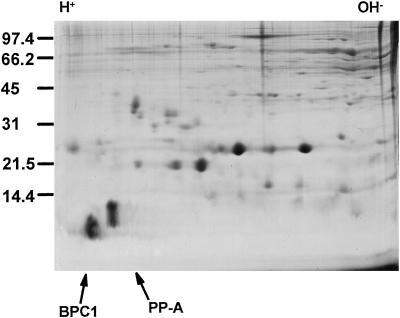
The spectrum of pollen proteins from the B. napus subsp. oleifera W1 line (Goring et al., 1992) released upon pollen hydration in isotonic buffer was characterized by two-dimensional gel electrophoresis, as illustrated in Figure 1. Comparison of this self-incompatible line with Westar, a near-isogenic, self-compatible line, revealed no obvious differences in pollen proteins when Coomassie-Blue-stained gels were examined (data not shown). Two relatively abundant, acidic, low-molecular-mass proteins can be observed. These were chosen for further investigation and called BPC1 and PP-A (pollen protein A). Although both purified proteins were blocked at their amino termini, a partial amino acid sequence for each was determined; internal peptides analyzed included two from BPC1 and four from PP-A (see Methods for the sequence). Given the more favorable amino acid sequence from BPC1 for developing oligonucleotides with low redundancy, it was chosen for further investigation.
Figure 1.
Two-dimensional PAGE analysis of B. napus W1 pollen proteins. PP-A and BPC1 are indicated with arrows. Positions of molecular-mass markers (in kilodaltons) and orientation of the IEF dimension are indicated.
Oligonucleotides were synthesized based on the amino acid sequence for BPC1, taking into account the degeneracy of the genetic code. These were used to isolate partial cDNAs via 3′-RACE-PCR using RNA from 1- to 4-mm flower buds as the template. An authentic PCR product could only be obtained from 4-mm bud RNA (data not shown). Sequence analysis of five independently cloned PCR products confirmed the presence of amino acid sequences from BPC1-derived peptides. Oligonucleotides were synthesized based on sequence information from the 3′-RACE clones for use in isolating the complete ORF using 5′-RACE-PCR. One of the 3′-RACE clones was used to screen a genomic library of B. napus W1. A positive λ-phage clone was partially sequenced in both orientations to confirm the sequence of the ORF derived from various PCR clones. Comparison of the cDNA sequence with the genomic sequence demonstrated that no introns are present in the BPC1 gene (data not shown). RNA-blot analysis demonstrated that accumulation of BPC1 transcript is developmentally regulated and only detectable in anthers and microspores (data not shown), which is in accordance with the results of Toriyama et al. (1995).
Using the BPC1 ORF as a probe, genomic clones of APC1, an Arabidopsis homolog, were isolated. A restriction fragment corresponding to the region encoding the ORF was subcloned and sequenced in both directions. As found for BPC1, no introns interrupt the APC1 ORF. RT-PCR using RNA isolated from Arabidopsis inflorescences demonstrated that APC1 is expressed in this plant (data not shown).
BPC1 and APC1 encode proteins of 79 and 83 amino acids, respectively, and share over 80% identity (Fig. 2). Comparison of the proteins with the database demonstrated that BPC1 is identical to a B. napus pollen allergen that was reported while this work was in progress (Toriyama et al., 1995), whereas APC1 is a novel protein. Both proteins are members of a growing family of highly conserved pollen proteins found in a variety of plant species (Fig. 2). In addition, the partial amino acid sequence obtained for PP-A indicates that it also is a member of this protein family. Analysis of APC1 and BPC1 to identify structural motifs revealed two potential calcium-binding domains known as EF hands (Heizmann and Hunziker, 1991; Nakayama and Kretsinger, 1994). These domains are conserved within the protein family (Fig. 2). No apparent signal peptide exists among the proteins, as indicated by the absence of an N-terminal hydrophobic sequence, although a short hydrophobic region does occur at the C terminus. Therefore, even though BPC1 was isolated with a method expected to enrich extracellular pollen wall proteins (Knox et al., 1975), there is no evidence from the sequence that it is extracellular.
Figure 2.
Comparison of APC1 and BPC1 with other calcium-binding proteins. Alignment of the sequence was performed using Clustal (scoring matrix, PAM250; Thompson et al., 1994) and evaluated with PROTOMAT (Henikoff et al., 1995). Identical amino acids are indicated by colons, and alignment was optimized by insertion of dashes. Unknown amino acids are represented by spaces. The potential site of N-linked glycosylation is underlined. Positions of EF-hand motifs are shown in bold. The fit of the APC1 and BPC1 sequence to EF-hand consensus was evaluated with Blocks Searcher (Henikoff and Henikoff, 1994). Gene names are shown on the left and the number of amino acid residues per line and per protein are shown on the right. Bra r II, Accession no. D63154; Bra r EST, accession no. L47857; BET v 4, accession no. X87153 or Y12560; Cyn d 7, accession no. U35683; C.d. B4, accession no. A28050; and C.d. B2, accession no. A28046. The partial amino acid sequence determined from purified B. napus W1 PP-A was manually aligned with other sequences.
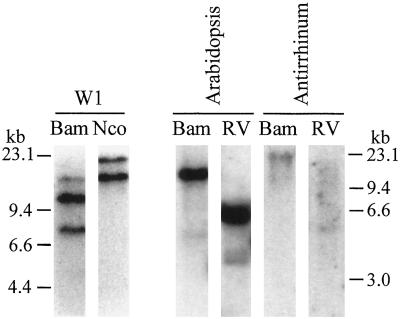
Using the cloned ORF of BPC1 as a probe, DNA-blot analysis of B. napus, Arabidopsis, Antirrhinum, and tobacco was performed. The W1 genome had at least two hybridizing regions when the blot was washed at high stringency (Fig. 3). Given the amphidiploid nature of B. napus, these bands likely correspond to two copies of BPC1. Indeed, two genomic clones encoding BPC1 but having different regulatory regions have been isolated (K. Rozwadowski and S.J. Rothstein, unpublished results). The amphidiploid genome of B. napus and the occurrence of at least two forms of BPC1-like proteins (Toriyama et al., 1995) could also account for at least two additional bands observed at low stringency (data not shown). Similar results were observed during analysis of other B. napus lines, which also demonstrated that BPC1 has no linkage to marker genes associated with the self-incompatibility response in B. napus (K. Rozwadowski and S.J. Rothstein, unpublished results). Hybridizing sequences were also detectable in Arabidopsis and snapdragon (Fig. 3), but not tobacco (data not shown), under low-stringency conditions. In Arabidopsis, these sequences corresponded to a prominent band and weak signal, suggesting that two related genes are present, whereas in Antirrhinum only weak signals were detectable, making clear interpretation of copy number difficult.
Figure 3.
Distribution of BPC1-like genes in B. napus W1, Arabidopsis, and Antirrhinum. DNA blots were probed with BPC1 cDNA. Washes were at high stringency for the B. napus W1 sample and at low stringency for the remaining samples. Genomic DNA was digested with BamHI (Bam), NcoI (Nco), or EcoRV (RV). Migration positions of DNA molecular-mass markers are indicated.
Biochemical Characterization of BPC1 and APC1
The protein sequence derived from the cloned genes indicated that both BPC1 and APC1 possess two potential calcium-binding domains. The effect of calcium on these proteins was evaluated by nondenaturing PAGE and/or gel-filtration chromatography. An abundant supply (several milligrams per liter of culture) of rBPC1 and rAPC1 was obtained by expression of the genes in E. coli BL21 (DE3) [pLysS]. rBPC1 was prepared as a fusion to GST using the vector pGEX-2T-[128/129], whereas rAPC1 was expressed in its native form using pT7-7. Both proteins remained soluble and undegraded in E. coli (data not shown) and were purified from E. coli lysates using a combination of glutathione-Sepharose affinity and/or other chromatographic procedures.
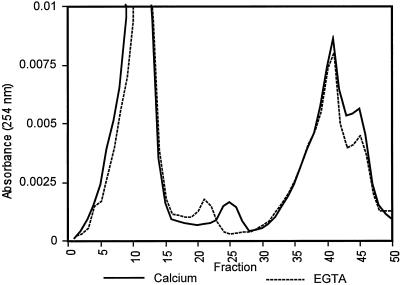
Interaction between calcium and rBPC1 and rAPC1 was initially detected as a calcium-dependent shift in electrophoretic mobility during nondenaturing PAGE (data not shown). This did not occur in the presence of MgCl2 or EGTA. Mobility of rBPC1 through a gel-filtration column was also affected by the presence of CaCl2 (Fig. 4; Table I). The GST and rBPC1 portions of the thrombin-cleaved fusion protein were separated by gel filtration in the presence of calcium or EGTA, and three peaks were resolved (Fig. 4). Analysis of column fractions by SDS-PAGE demonstrated that the first peak represented GST and the second rBPC1 (data not shown). The third peak was likely glutathione, which was added in excess to remove the fusion protein from glutathione-Sepharose during purification from E. coli lysate. The presence of calcium caused a shift in the elution volume of rBPC1 compared with that observed in the presence of EGTA (Fig. 4). This effect was not observed for GST or glutathione.
Figure 4.
Effect of calcium on chromatography of rBPC1. GST-rBPC1 fusion protein cleaved with thrombin was resolved by gel filtration in the presence of calcium (10 mm CaCl2, 2 mm EGTA, and 20 mm Tris-HCl, pH 7.4) or EGTA (10 mm EGTA and 20 mm Tris-HCl, pH 7.4).
Table I.
Effect of calcium and ionic strength on rBPC1 elution during gel filtration
| Buffer
|
rBPC1 Elution
|
|||||
|---|---|---|---|---|---|---|
| CaCl2 | EGTA | NaCl | Elution Volumea | MWb | MW/Mrc | MW EGTA/MW CaCl2 |
| mm | mL | kD | ||||
| + | − | 0 | 30.3 | 16.8 | 1.6 | — |
| − | + | 0 | 28.5 | 22.3 | 2.1 | 1.3 |
| + | − | 60 | 30.8 | 15.6 | 1.5 | — |
| − | + | 60 | 29 | 20.6 | 1.9 | 1.3 |
| + | − | 200 | 31.3 | 14.4 | 1.4 | — |
| − | + | 200 | 29.5 | 19.2 | 1.8 | 1.3 |
Elution volume for peak containing rBPC1 from 1- × 60-cm column of Sephacryl S-100 with flow rate of 0.25 mL/min.
Estimated molecular mass (MW) based on calibration with standard proteins (see Methods).
Actual Mr of rBPC1 is 10.6.
Comparison of the elution point of rBPC1 during gel filtration with those from known size standards enabled estimation of the molecular mass of the protein and an evaluation of the Stokes radius of rBPC1 in response to calcium. rBPC1 elution was affected by calcium and ionic strength (Table I). The ratio of estimated molecular mass, as determined by gel-filtration chromatography, to the actual molecular mass of rBPC1, based on amino acid composition, in the presence and absence of calcium or sodium chloride was determined. rBPC1 has a Mr of 10.6, an increase of 2 over the native protein, the result of additional amino acids present in the thrombin-cleaved recombinant protein. In the presence of calcium, rBPC1 eluted with an apparent molecular mass approximately 1.5 times that expected on the basis of its amino acid sequence. However, in the apo state created by the presence of EGTA, rBPC1 eluted at approximately twice its expected molecular mass. With increased ionic strength (up to 200 mm NaCl), a slight decrease in molecular mass was observed in the presence or absence of calcium. However, the effect of calcium on rBPC1 elution was independent of salt concentration, because the ratio of estimated molecular mass determined in the absence of calcium compared with that in its presence was constant (i.e. 1.3) at each salt concentration tested. This is consistent with a specific effect of calcium on the conformation of rBPC1.
Immunolocalization of BPC1
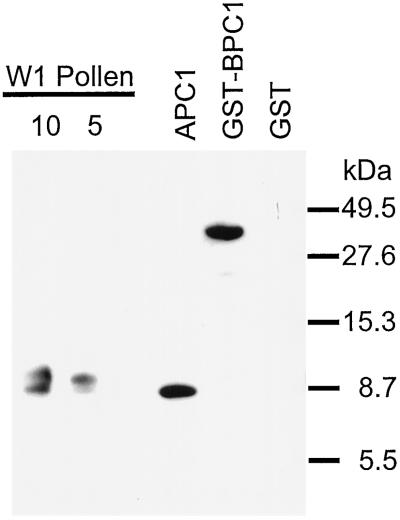
rAPC1 was purified to homogeneity (data not shown) and used to raise polyclonal antibodies in mice. Antisera from chickens and rabbits produced an unacceptable reactivity to a variety of pollen proteins (data not shown). Figure 5 illustrates the specificity of the antibody reacting with purified rAPC1, rBPC1, proteins of the expected size of BPC1, and the homologous but slightly larger Bra n II (Toriyama et al., 1995) in the pollen-protein extract from B. napus W1.
Figure 5.
Evaluation of anti-rAPC1 serum from mouse. Samples (in micrograms) of W1 pollen protein extract, purified rAPC1, and lysates of E. coli strains expressing GST-rBPC1 fusion protein or GST alone were probed with mouse anti-rAPC1 serum followed by goat-anti-mouse secondary antibody. Mouse preimmune serum or secondary antibody did not detect any proteins (data not shown). Positions of molecular-mass markers (in kilodaltons) are indicated.
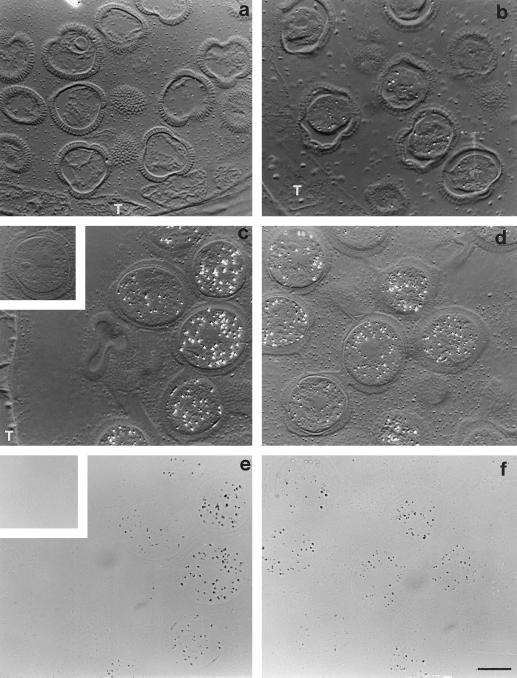
Immunocytochemistry was performed for cellular localization of BPC1-related proteins in floral tissues and pollen. As demonstrated in Figure 6, BPC1 accumulates in the latter stages of development of the pollen grains, in agreement with transcript accumulation (data not shown; Toriyama et al., 1995). The suspected cytosolic location of the protein, as deduced from sequence analysis, was confirmed. The protein was not detected in the tapetum. In the stigma, antibodies to BPC1 cross-reacted with proteins in parenchyma cells underlying the stigmatic surface and lying adjacent to the pistil vasculature (data not shown).
Figure 6.
Immunolocalization of BPC1 in the cytosol of developing pollen grains of B. napus W1. Sections of LR White embedded anthers were treated with mouse anti-rAPC1 followed by colloidal gold-tagged rabbit-anti mouse IgG and then silver enhanced. Silver grains, indicating the presence and location of BPC1, appear as bright refractive grains under differential interference contrast optics (a–d). a, Pollen grains from flower buds at the 4-mm stage; BPC1 protein was not detected at this stage. b, Pollen from flowers at the 6-mm stage; BPC1 protein is barely detectable, as illustrated by the sparse labeling in the four more prominent pollen grains in the micrograph. T, tapetum. c and d, Pollen from flowers at the 8-mm stage and the mature stage, respectively. There has been a marked increase in BPC1-labeling intensity. It is obvious that BPC1 is cytosolic, because neither pollen walls nor nuclei (c, grain center right; d, grains center and lower left) are labeled. e and f, Bright-field images of c and d, respectively, confirming the location of the label (silver grains appear as black grains) and demonstrating that the granular nature of the extracellular matrix seen in c and d is not due to labeling. The signal lies fully within the pollen grain walls and outside the nuclei. Inset in c and e show a grain from 8-mm stage flowers as a preimmune control. No labeling is evident in grains treated with preimmune mouse serum in place of anti-rAPC1. All micrographs are ×1000. Bar = 10 μm.
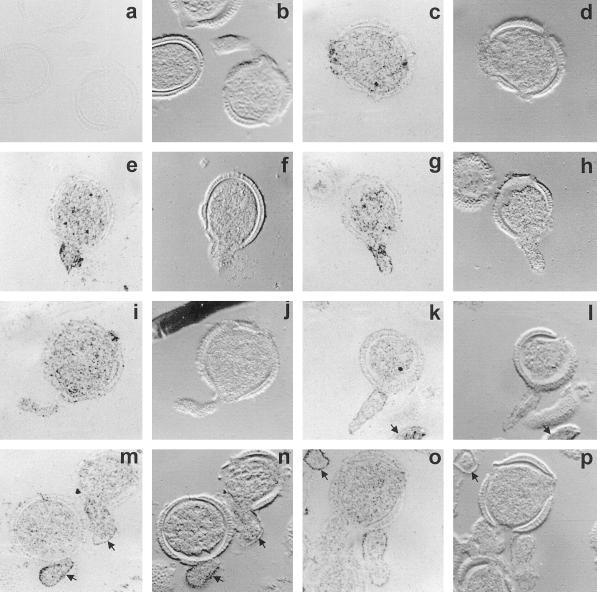
The majority of BPC1 remained cytosolic upon hydration and germination of the pollen. However, in contrast to the situation during pollen development (Fig. 6), some labeling was detected in the pollen walls after hydration and with germination (Fig. 7, c–p). This suggests that there may be some leakage of protein from the cytosol to the wall upon hydration and offers an explanation as to how BPC1 was initially isolated as an extracellular protein from hydrated grains. In addition, BPC1 presents itself on or near the surface of the extending pollen tubes (Fig. 7, e, g, k, m, and o).
Figure 7.
Immunolocalization of BPC1 in germinated pollen grains of B. napus W1. Sections of embedded germinated grains were treated with mouse anti-rAPC, followed by colloidal gold-tagged rabbit-anti-mouse IgG, and were then silver enhanced. Bright-field and corresponding differential interference contrast images are presented in each case for clarity. a and b, Preimmune control images. c to p, Immune images. Note that signals are not restricted to the cytosol, but some labeling is also seen in the pollen grain wall. Labeling is intense on or near the surface of the pollen tubes, as evidenced by intense edge labeling, seen in e to h and k to p. Lack of intensity of edge labeling of the tube in I and j is probably due to the grazing nature of the section. Arrows in k, l, o, and p indicate pollen tubes of grains lying above or below the plane of section. Arrows in m to p further indicate the edge labeling. All images are ×1000.
DISCUSSION
Structural and biochemical features of BPC1 and APC1 indicate that these proteins might serve as calcium-sensor signal molecules functioning during pollen germination and tube elongation. BPC1 has been characterized as an abundant, soluble, low-Mr protein present in B. napus pollen. Both BPC1 and APC1 possess two EF-hand domains that are responsible for calcium binding in a variety of proteins (Heizmann and Hunziker, 1991; Nakayama and Kretsinger, 1994; Travé et al., 1995). Functionality of these domains is implied by the conserved Asp and Glu residues at positions 10 and 21, respectively, and the essential Gly at position 15. These residues are involved in the coordination of calcium in the EF hand and in enabling the sharp bend required in the calcium-binding loop (Moncrief et al., 1990). rBPC1 and rAPC1 proteins were shown with an electrophoretic mobility shift assay to bind calcium, indicating that the EF-hand motifs were indeed functional. No interaction with magnesium was detected, in contrast to other proteins with EF-hands, which may have affinity for both calcium and magnesium (Nakayama and Kretsinger, 1994).
Interaction of rBPC1 with calcium was further examined using gel-filtration chromatography. A calcium-dependent shift in the elution point was observed, in which rBPC1 behaved as a smaller protein in the presence of calcium than it did in the apo state. Maintenance of these relative changes under various ionic strength conditions negates the possibility of chromatographic artifacts in the altered elution. Two possible types of structural changes could be responsible for the altered elution times. First, the apo- and calcium-bound states may have distinct conformations and Stokes radii that confer unique chromatographic behaviors to the protein depending upon calcium availability. In the presence of calcium, rBPC1 may assume a more compact conformation than in the apo state, enabling it to elute as a smaller protein. Conformational changes in response to calcium have been demonstrated in related proteins (Engel et al., 1997; Suphioglu et al., 1997; Twardosz et al., 1997). Second, it is possible that rBPC1 may interconvert between a monomeric and dimeric state in response to calcium. Calcium-dependent interactions between monomers of other proteins possessing EF hands have been demonstrated (Barger et al., 1992; Travé et al., 1995; Ikura, 1996). However, in general, calcium favors the dimerization of such proteins (Barger et al., 1992; Travé et al., 1995; Ikura, 1996), as opposed to its apparent effect on rBPC1.
BPC1 and APC1 are members of a highly conserved protein family identified in the pollen of a variety of plant species. In addition to the demonstration of homologous sequences in Antirrhinum described here, related proteins have been identified in Betula verrucosa (Engel et al., 1997; Twardosz et al., 1997), Olea europaea (Batanero et al., 1997), Artemisia vulgaris (Engel et al., 1997), Cynodon dactylon (Knox et al., 1992), Phleum pratense, and Lilium longiflorum (Engel et al., 1997; Twardosz et al., 1997), and multiple types were identified in Brassica rapa and B. napus (Toriyama et al., 1995; Lim et al., 1996) during the course of this study. The partial amino acid sequence from the second pollen protein purified in this study, PP-A, indicates that it, too, is a member of this protein family. These proteins share a very high degree of identity and a conserved pair of EF hands. Toriyama et al. (1995) classified BPC1-like proteins in Brassica into two groups with a composition of either 79 amino acids (group I), as found for BPC1, or 83 amino acids (group II), as found for APC1. Two immunoreactive proteins with slightly different molecular masses were detected in B. napus pollen with anti-APC1 serum, likely representing the two groups of proteins. The presence of multiple forms of the protein coincides with our DNA-blot data revealing a small gene family in B. napus, an amphidiploid derived from B. rapa and B. oleracea. Betula verrucosa and C. dactylon have also been shown to have at least two versions of the protein (Knox et al., 1992; Suphioglu et al., 1997; Twardosz et al., 1997). Two hybridizing DNA sequences were also detected in Arabidopsis, suggesting the presence of a second gene related to APC1. Differences of expression pattern between the two types of genes observed in Brassica may be physiologically important (Toriyama et al., 1995).
Immunolocalization studies indicated that BPC1 is distributed throughout the cytoplasm of developing and mature pollen. Following germination, however, BPC1 is apparently concentrated at or near the surface of emerging and elongating pollen tubes. This localization is suggestive of a role for BPC1 in pollen germination and tube growth and, in combination with the biochemical properties of BPC1, this function is likely to be related to calcium physiology. Calcium is known to play an essential role in pollen germination and pollen tube growth (Kwack and Brewbaker, 1961; Mascarenhas and Machlis, 1964; Pierson et al., 1994; Malhó and Trewavas, 1996).
In addition to its role within pollen, BPC1 may also function in pollen-stigma interactions. BPC1 was immunolocalized to the cytosol of pollen grains, which is in agreement with the absence of a conventional signal peptide in the deduced protein sequence. However, BPC1 was initially purified using a protein-extraction method to enrich for extracellular proteins by employing an isotonic buffer. Although leakage of some cytoplasmic constituents is to be expected, this leakage appeared to be limited to a subset of pollen proteins, as extraction in the presence of detergent produced a very distinct protein spectrum (K. Rozwadowski and S.J. Rothstein, unpublished results). Differential release of pollen cytosolic proteins in response to hydration has been observed previously (Vrtala et al., 1993). Furthermore, the possibility of release of some BPC1 protein during pollen germination in vivo cannot be excluded. Immunolocalization of some BPC1 in the pollen walls was observed after hydration and germination, suggesting that leakage of the protein from the cytosol to the wall occurred as the grains hydrated. In addition, BPC1-related proteins have been characterized as human allergens (Knox et al., 1992; Toriyama et al., 1995; Batenero et al., 1997; Engel et al., 1997; Suphioglu et al., 1997; Twardosz et al., 1997), as have the BET vIII and BET vI pollen proteins, which have 2- to 2.5-fold greater molecular masses than BPC1 (Vrtala et al., 1993; Seiberler et al., 1994). This allergenicity implies a pollen-surface location or release upon pollen hydration. Indeed, Bet vI has been shown to move to the pollen surface upon hydration and to be preferentially released, in contrast to nonallergens (Vrtala et al., 1993). Therefore, since BPC1-like proteins are smaller than Bet vI and Bet vIII, they may also be released to some extent during pollen hydration. Such release may have physiological significance, as a calcium-bound form of BPC1 could relay a signal to the stigma. Small calcium-binding proteins without conventional secretory signals in mammalian systems have been shown to have intercellular signaling functions (Zimmer et al., 1995).
Accumulation of the BPC1 transcript corresponded with accumulation of BPC1 protein, as indicated by RNA-blot and immunocytochemistry, respectively (data not shown; Toriyama et al., 1995). This suggests that protein accumulation is under transcriptional control. The high transcript level and gradual accumulation of protein correspond to the relative abundance of BPC1 observed by two-dimensional PAGE in a pollen-protein extract. BPC1 mRNA was detectable only in male floral tissues and its accumulation was initiated at the 4-mm stage of bud development (data not shown; Toriyama et al., 1995). At this point, the developing pollen enters the maturation phase and accumulates components required for rapid germination and tube growth upon landing on the stigma (Scott et al., 1991). Transcription of BPC1 declined just before anthesis when the pollen had matured, desiccated, and assumed a metabolically inactive state (Scott et al., 1991). Therefore, BPC1 appears to be a “late” gene associated with preparation for pollen germination.
The possible function of BPC1-like proteins as a signal molecule appears to rely on calcium binding. Microinjection of Bet v4 into pollen tubes has been demonstrated to alter cytoplasmic streaming and membrane depolarization (Engel et al., 1997). Mutant Bet v4 defective for calcium-binding did not induce these effects (Engel et al., 1997). Therefore, the calcium-dependent conformational changes of BPC1 may enable it to interact with and modulate effector proteins that manifest physiological responses to a calcium stimulus.
In conclusion, BPC1 is an abundant cytosolic low-Mr pollen protein highly conserved in a variety of dicot and monocot species. The protein is located at or near the pollen tube surface during emergence and elongation, binds calcium, and undergoes changes in conformation. Calcium-binding proteins may function as “buffer” or “trigger” proteins (Levine and Dalgarno, 1993; Zimmer et al., 1995; Ikura, 1996). In a buffering capacity, these proteins likely act by passively regulating the concentration of intracellular free calcium. However, trigger proteins bind calcium and then interact with and modulate the activities and function of other proteins. Considering the conformational changes of BPC1 exposed to calcium, and comparing this with what occurs in other systems, it likely functions in a trigger fashion. However, confirmation of the role of BPC1 will require additional study.
ACKNOWLEDGMENTS
We thank Nic Bate, Shoba Sivasankar, and Daphne Goring for useful discussions and reading of the manuscript. We also thank Jay Newsted for constructing the B. napus genomic library, Ulrike Schafer for assistance in preparing RNA samples, and Richard Stahl for assistance in preparing pollen for immunolocalization studies. pGEX-2T[128/129] was kindly provided by Dr. M.A. Blanar.
Abbreviations:
- GST
glutathione S-transferase
- ORF
open reading frame
- RACE
rapid amplification of cDNA ends
Footnotes
This work was supported by grants from the Natural Sciences and Engineering Research Council of Canada (NSERC) to S.J.R. K.L.R was supported by graduate scholarships from NSERC and by the Ontario Graduate Scholarship program.
LITERATURE CITED
- Ausubel FM, Brent R, Kingston RE, Moore DE, Seidman JG, Smith JA, Struhl K, eds (1988) Current Protocols in Molecular Biology. John Wiley & Sons, New York
- Barger SW, Wolchok SR, Van Eldik LJ. Disulfide-linked S100B dimers and signal transduction. Biochim Biophys Acta. 1992;1160:105–112. doi: 10.1016/0167-4838(92)90043-d. [DOI] [PubMed] [Google Scholar]
- Batanero E, Villalba M, Ledesma A, Puente X, Rodriguez R. Ole e 3, an olive-tree allergen, belongs to a widespread family of pollen proteins. Eur J Biochem. 1997;241:772–778. doi: 10.1111/j.1432-1033.1996.00772.x. [DOI] [PubMed] [Google Scholar]
- Bednarska E. The effect of exogenous Ca2+ ions on pollen grain germination and pollen tube growth. Sex Plant Reprod. 1989;2:53–58. [Google Scholar]
- Bednarska E. Calcium uptake from the stigma by germinating pollen in Primula officinalis L. and Ruscus aculeatus L. Sex Plant Reprod. 1991;4:36–38. [Google Scholar]
- Blanar MA, Rutter WJ. Interaction cloning: identification of a helix-loop-helix zipper protein that interacts with c-Fos. Science. 1992;256:1014–1018. doi: 10.1126/science.1589769. [DOI] [PubMed] [Google Scholar]
- Carafoli E. Intracellular calcium homeostasis. Annu Rev Biochem. 1987;56:395–433. doi: 10.1146/annurev.bi.56.070187.002143. [DOI] [PubMed] [Google Scholar]
- Coventry J, Kott L, Beversdorf WD (1988) Manual for Microspore Culture Technique. Department of Crop Science, University of Guelph, Ontario, Canada
- Dumas C, Gaude T, Heizmann P, Rougier M (1994) The cell biology of pollen development in B. napus.In EG Williams, AE Clarke, RB Knox, eds, Genetic Control of Self-Incompatibility and Reproductive Development in Flowering Plants. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 309–335
- Elleman CJ, Dickinson HG (1994) Pollen-stigma interaction during sporophytic self-incompatibility in B. napus oleracea.In EG Williams, AE Clarke, RB Knox, eds, Genetic Control of Self-Incompatibility and Reproductive Development in Flowering Plants. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 67–87
- Engel E, Richter RK, Obermeyer G, Briza P, Kungl AJ, Simon B, Auer M, Ebner C, Rheinberger HJ, Breitenbach M and others. Immunological and biological properties of Bet v 4, a novel birch pollen allergen with two EF-hand calcium-binding domains. J Biol Chem. 1997;272:28630–28637. doi: 10.1074/jbc.272.45.28630. [DOI] [PubMed] [Google Scholar]
- Franklin-Tong VE, Ride JP, Read ND, Trewavas AJ, Franklin CH. The self-incompatibility response in Papaver rhoeas is mediated by cytosolic free calcium. Plant J. 1993;4:163–277. [Google Scholar]
- Frohman MA. RACE: rapid amplification of cDNA ends. In: Innis M, Gelfand DH, Sninsky JJ, White TJ, editors. PCR Protocols: A Guide to Methods and Applications. San Diego, CA: Academic Press; 1990. pp. 28–38. [Google Scholar]
- Goring DR, Banks P, Beversdorf WD, Rothstein SJ. Use of the polymerase chain reaction to isolate an S-locus glycoprotein cDNA introgressed from Brassica campestris into B. napus ssp. oleifera. Mol Gen Genet. 1992;234:185–192. doi: 10.1007/BF00283838. [DOI] [PubMed] [Google Scholar]
- Hausser I, Herth W, Reiss HD. Calmodulin in tip-growing plant cells, visualized by fluorescing calmodulin-binding phenothiazines. Planta. 1984;162:33–39. doi: 10.1007/BF00397418. [DOI] [PubMed] [Google Scholar]
- Heizmann CW, Hunziker W. Intracellular calcium-binding proteins: more sites than insights. Trends Biochem Sci. 1991;16:98–103. doi: 10.1016/0968-0004(91)90041-s. [DOI] [PubMed] [Google Scholar]
- Henikoff S, Henikoff JG. Protein family classification based on searching a database of blocks. Genomics. 1994;19:97–107. doi: 10.1006/geno.1994.1018. [DOI] [PubMed] [Google Scholar]
- Henikoff S, Henikoff JG, Alford WJ, Pietrokovski S (1995) Automated construction and graphical presentation of protein blocks from unaligned sequences. Gene-COMBIS, Gene 163: GC17–26 [DOI] [PubMed]
- Heslop-Harrison J, Heslop-Harrison Y. Germination of monocolpate angiosperm pollen: effects of inhibitory factors and the Ca2+-channel blocker, nifedipine. Ann Bot. 1992;69:395–403. [Google Scholar]
- Ikura M. Calcium binding and conformational response in EF-hand proteins. Trends Biochem Sci. 1996;21:14–17. [PubMed] [Google Scholar]
- Knox RB, Heslop-Harrison J, Heslop-Harrison Y (1975) Pollen-wall proteins: localization and characterization of gametophytic and sporophytic fractions. In J Duckett, P Racey, eds, The Biology of the Male Gamete. Biol J Linn Soc 7 (Suppl): 177–187
- Knox RB, Singh MB, Smith PM (1992) Isolated protein allergens and antigenic fragments of Cynodon dactylon for treatment, prevention and diagnosis of allergic reactions to Bermuda grass pollen. Patent no. WO9216554-A
- Kwack BH, Brewbaker JL (1961) The essential role of calcium in pollen germination and the population effect (suppl). Plant Physiol 36: xv
- Levine BA, Dalgarno D. The dynamics and function of calcium-binding proteins. Biochim Biophys Acta. 1983;726:187–204. doi: 10.1016/0304-4173(83)90005-8. [DOI] [PubMed] [Google Scholar]
- Lim CO, Kim HY, Kim MG, Lee SI, Chung WS, Park SH, Hwang I, Cho MJ. Expressed sequence tags of Chinese cabbage flower bud cDNA. Plant Physiol. 1996;111:577–588. doi: 10.1104/pp.111.2.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhó R, Trewavas AJ. Localized apical increases of cytosolic free calcium control pollen tube orientation. Plant Cell. 1996;8:1935–1949. doi: 10.1105/tpc.8.11.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascarenhas JP, Machlis L. Chemotropic response of the pollen of Antirrhinum majus to calcium. Plant Physiol. 1964;39:70–77. doi: 10.1104/pp.39.1.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moncrief ND, Kretsinger RH, Goodman M. Evolution of EF-hand calcium-modulated proteins. I. Relationships based on amino acid sequences. J Mol Evol. 1990;30:522–562. doi: 10.1007/BF02101108. [DOI] [PubMed] [Google Scholar]
- Nakayama S, Kretsinger RH. Evolution of the EF-hand family of proteins. Annu Rev Biophys Biomol Struct. 1994;23:473–507. doi: 10.1146/annurev.bb.23.060194.002353. [DOI] [PubMed] [Google Scholar]
- Niki I, Yokokura H, Sudo T, Kato M, Hidaka H. Ca2+ signaling and intracellular Ca2+ binding proteins. J Biochem. 1996;120:685–698. doi: 10.1093/oxfordjournals.jbchem.a021466. [DOI] [PubMed] [Google Scholar]
- Picton JM, Steer MW. The effects of ruthenium red, lanthanum, fluorescein isothiocyanate and trifluoperazine on vesicle transport, vesicle fusion and tip extension in pollen tubes. Planta. 1985;163:20–26. doi: 10.1007/BF00395892. [DOI] [PubMed] [Google Scholar]
- Pierson ES, Miller DD, Callaham DA, Shipley AM, Rivers BA, Cresti M, Hepler P. Pollen tube growth is coupled to the extracellular calcium ion flux and the intracellular calcium gradient: effect of BAPTA-type buffers and hypertonic media. Plant Cell. 1994;6:1818–1828. doi: 10.1105/tpc.6.12.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polito VS. Membrane-associated calcium during pollen grain germination: a microfluorometric analysis. Protoplasma. 1983;117:226–232. [Google Scholar]
- Roberts IN, Gaude TC, Harrod G, Dickinson HG. Pollen-stigma interactions in Brassica oleracea: a new pollen germination medium and its use in elucidating the mechanism of self incompatibility. Theor Appl Genet. 1983;65:231–238. doi: 10.1007/BF00308074. [DOI] [PubMed] [Google Scholar]
- Scott R, Dagless E, Hodge R, Paul W, Soufleri I, Draper J. Patterns of gene expression in developing anthers of Brassica napus. Plant Mol Biol. 1991;17:195–207. doi: 10.1007/BF00039494. [DOI] [PubMed] [Google Scholar]
- Seiberler S, Scheiner O, Kraft D, Lonsdale D, Valenta R. Characterization of a birch pollen allergen, Bet v III, representing a novel class of Ca2+ binding proteins: specific expression in mature pollen and dependence of patients' IgE binding on protein-bound Ca2+ EMBO J. 1994;13:3481–3486. doi: 10.1002/j.1460-2075.1994.tb06654.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Perdue T, Paolill DJ., Jr Pollen-pistil interactions in Brassica oleracea: cell calcium in self and cross pollen grains. Protoplasma. 1989;151:57–61. [Google Scholar]
- Sistrunk MV, Antosiewicz DM, Purugganan MM, Braam J. Arabidopsis TCH3 encodes a novel Ca2+ binding protein and show environmentally induced and tissue-specific regulation. Plant Cell. 1994;6:1553–1565. doi: 10.1105/tpc.6.11.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier FW, Moffatt BA. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- Suphioglu C, Ferreira F, Knox RB. Molecular cloning and immunological characterisation of Cyn d 7, a novel calcium-binding allergen from Bermuda grass pollen. FEBS Lett. 1997;402:167–172. doi: 10.1016/s0014-5793(96)01520-7. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. ClustalW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toriyama K, Okada T, Watanabe M, Ide T, Ashida T, Xu H, Singh MB. A cDNA clone encoding and IgE-binding protein from Brassica anther has significant sequence similarity to Ca2+-binding proteins. Plant Mol Biol. 1995;29:1157–1165. doi: 10.1007/BF00020459. [DOI] [PubMed] [Google Scholar]
- Travé G, Lacombe PJ, Pfuhl M, Saraste M, Pastore A. Molecular mechanism of the calcium-induced conformational change in the spectrin EF-hands. EMBO J. 1995;14:4922–4931. doi: 10.1002/j.1460-2075.1995.tb00175.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twardosz A, Hayek B, Seibler S, Vangelista L, Elfman L, Gronlund H, Kraft D, Valenta R. Molecular characterization, expression in Escherichia coli, and epitope analysis of a two EF-hand calcium-binding birch pollen allergen, Bet v 4. Biochem Biophys Res Commun. 1997;239:197–204. doi: 10.1006/bbrc.1997.6860. [DOI] [PubMed] [Google Scholar]
- Vrtala S, Grote M, Duchene M, vanRee R, Kraft D, Scheiner O, Valenta R. Properties of tree and grass pollen allergens: reinvestigation of the linkage between solubility and allergenicity. Int Arch Allergy Immunol. 1993;102:160–169. doi: 10.1159/000236567. [DOI] [PubMed] [Google Scholar]
- Zimmer DB, Cornwall EH, Landar A, Song W. The S100 protein family: history, function, and expression. Brain Res Bull. 1995;37:417–429. doi: 10.1016/0361-9230(95)00040-2. [DOI] [PubMed] [Google Scholar]