Abstract
The heterogeneity of exosomal populations has hindered our understanding of their biogenesis, molecular composition, biodistribution, and functions. By employing asymmetric-flow field-flow fractionation (AF4), we identified two exosome subpopulations (large exosome vesicles, Exo-L, 90-120 nm; small exosome vesicles, Exo-S, 60-80 nm) and discovered an abundant population of non-membranous nanoparticles termed “exomeres” (~35 nm). Exomere proteomic profiling revealed an enrichment in metabolic enzymes and hypoxia, microtubule and coagulation proteins and specific pathways, such as glycolysis and mTOR signaling. Exo-S and Exo-L contained proteins involved in endosomal function and secretion pathways, and mitotic spindle and IL-2/STAT5 signaling pathways, respectively. Exo-S, Exo-L, and exomeres each had unique N-glycosylation, protein, lipid, and DNA and RNA profiles and biophysical properties. These three nanoparticle subsets demonstrated diverse organ biodistribution patterns, suggesting distinct biological functions. This study demonstrates that AF4 can serve as an improved analytical tool for isolating and addressing the complexities of heterogeneous nanoparticle subpopulations.
Introduction
Exosomes are nanosized extracellular membrane vesicles of endosomal origin secreted by most cell types, including cancer cells1-3. Proteins, genetic material (e.g., mRNAs, miRNAs, lnRNAs, DNA), metabolites and lipids, are selectively recruited and packaged into exosomes, which horizontally transfer their cargo to recipient cells, thereby acting as vehicles of intercellular communication in both physiological and pathological conditions4-7. Harnesing this knowledge, translational researchers have focused on developing exosome-based diagnostic/prognostic biomarkers and therapeutic strategies.
Although our understanding of the biology, function and translational potential of exosomes is rapidly expanding, the heterogeneous nature of nanovesicles and technical limitations in efficiently separating exosomal subpopulations have hindered the characterization of their molecular composition and biogenesis. The state-of-the-art technology, asymmetric-flow field-flow fractionation (AF4)8, exhibits unique capability to separate nanoparticles and has been widely utilized to characterize nanoparticles and polymers in the pharmaceutical industry and to examine various biological macromolecules, protein complexes and viruses8, 9, but rarely tested for extracellular vesicle (EV) analysis10-14. Using AF4, nanoparticles are separated based on their density and hydrodynamic properties by two perpendicular flows, i.e., the forward laminar channel flow and the variable crossflow.
Here, we established and optimized AF4 parameters and protocols, followed by rigorous biophysical and molecular characterization of small EV fractions isolated from numerous cancer and normal cells. Through our modified AF4 protocols, we identified a distinct nanoparticle we term “exomere” as well as two exosome subpopulations that demonstrate distinct biophysical and molecular properties.
Results
Identification of a distinct nanoparticle population and subsets of exosomes
We first fractionated B16-F10 melanoma-derived sEVs by AF4 (see Methods). A linear separation of the sEV mixture was achieved based on the hydrodynamic radius (black dots, Y axis) along the time course (X axis) (Fig. 1a). The online QELS monitor for real-time dynamic light scattering (DLS) measurement (red trace) determined the hydrodynamic radius of particles. UV absorbance (blue trace) measured protein concentration and abundance of particles at specific time points for corresponding particle sizes. Particles with a 35-150 nm diameter were successfully separated by AF4 (Fig. 1a). We identified five peaks (P1-P5), corresponding to the time and particle size, at which most abundant particles were detected. P1 represented the void peak, a mixture of all types of nanoparticles. P5 was composed of individual or aggregated particles and protein aggregates with much larger sizes, which are outside the separation range of the current AF4 protocol, and eluted when crossflow dropped to zero (Supplementary Fig. 1a). The hydrodynamic diameters of peaks P2, P3 and P4 were 47 nm, 62 nm and 101 nm, respectively. To infer the hydrodynamic radius, correlation functions were fitted to single exponentials (Fig.1b, representative P3 fraction graph).
Figure 1.
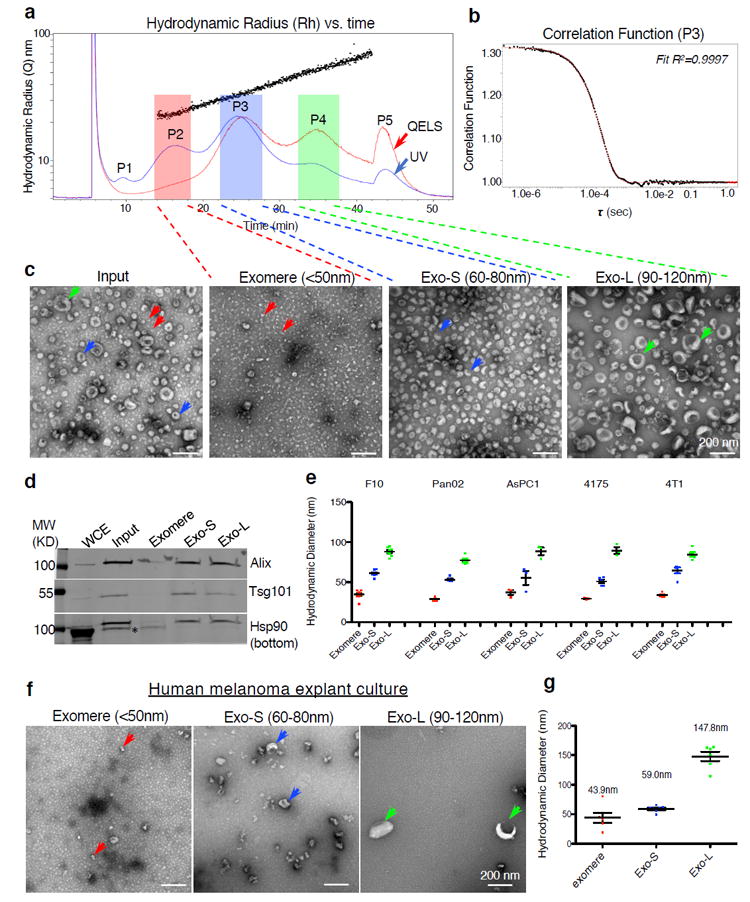
Identification, via AF4 and EM imaging analysis, of exomeres and two distinct subpopulations of exosomes released by tumor cells. (a) A representative AF4 fractionation profile of B16-F10-derived exosomes. x- axis, time (min); y-axis (scale) and black dots, hydrodynamic radius (nm); red and blue lines illustrate the QELS (DLS) intensity and UV absorbance (shown on a relative scale), respectively. P1-P5 marks the peaks detected based on UV absorbance. Fractions were pooled for exomeres (hydrodynamic diameter <50 nm,); Exo-S (60-80 nm); and Exo-L (90-120 nm). (b) Representative correlation function at peak 3 (P3), t = 25.1 min. For (a) and (b) the experiment was repeated independently 50 times with similar results. (c) TEM imaging analysis of exosome input mixture (pre-fractionation) and fractionated exomeres, Exo-S and Exo-L subpopulations. Arrows point to exomeres (red), Exo-S (blue) and Exo-L (green). Scale bar, 200 nm. This experiment was repeated 7 times independently with similar results. (d) Western blotting analysis of exosomal marker proteins in fractionated samples. 100 μg of whole cell extract (WCE) and 10 μg of exosome and exomere mixture input and each subset were analyzed. This experiment was done once. (e) Measurement of hydrodynamic diameters of exomeres, Exo-S and Exo-L derived from representative cell lines (i.e. B16-F10 (F10), AsPC-1, Pan02, MDA-MB-4175 (4175) and 4T1) in the batch mode using Zetasizer after pooling fractions collected for each subset of nanoparticles from an individual AF4 fractionation. Data are presented as mean±SEM (standard error of the mean), in the order of exomere, Exo-S and Exo-L: B16-F10 (n=10, 9, and 8 independent measurements, respectively); Pan02 (n=11, 6, 11); AsPC-1 (n=5, 5, 5); 4175 (n=3, 5, 3); 4T1 (n=5, 5, 5)) (f) TEM imaging analysis of fractions collected from explant culture of fresh human melanoma tissue. Scale bar, 200 nm. This experiment was performed with two independent specimens with similar results. (g) Batch mode measurement of hydrodynamic diameters of fractions shown in (f). Data are presented as mean±SEM (exomeres and Exo-L, n=6; Exo-S, n=7 independent measurements). Statistical source data are provided in Supplementary Table 8, and unprocessed blots are provided in Supplementary Figure 7.
Individual fractions were measured using Nanosight Tracking Analysis (NTA), validating consistent particle size for each fraction between 60 nm and 140 nm (Supplementary Fig. 1b). DLS combined with AF4 showed a broader dynamic range than NTA for those particles with a smaller (~70 nm) or larger (~160 nm) particle size (Supplementary Fig. 1c). Moreover, NTA of each individual fraction in the range of 60-160 nm revealed a monomodal profile with a peak of ~77 nm (Supplementary Fig. 1d).
Transmission electron microscopy (TEM) with negative staining of AF4 input and representative fractions across the full dynamic range revealed three populations of particles (P2, P3, P4; Fig. 1a) with distinct morphology and size (Fig. 1c). P2 represented a distinct population of nanoparticles not previously described, which were smaller than 50 nm (~35nm) and clearly lacked an external membrane structure (Fig. 1c); we therefore named these structures “exomeres”. The other two nanoparticle subpopulations we refer to as small exosomes (Exo-S; 60-80nm [P3]) and large exosomes (Exo-L; 90-120nm [P4]) (Fig. 1c). All three particle types were readily detected in the input TEM image (Fig. 1c). Western blot analysis confirmed exosome markers Tsg101 and Alix for Exo-S and Exo-L, and heat shock protein 90 (Hsp90) for exomeres (Fig. 1d). The sizes of each particle type measured in batch mode showed consistent results (Fig. 1e).
In summary, a single run of AF4 can efficiently discern exomeres and two distinct exosome subpopulations in a robust and highly reproducible manner (Supplementary Fig. 1e, f). Freeze-thawing of samples led to inconsequential differences (Supplementary Fig. 1g). However, changes in culture conditions led to differences in relative abundance of each particle type (Supplementary Fig. 1h-i).
Importantly, only a minor peak eluted in the time range similar to exomeres in a blank media control compared to CM of B16-F10 and MDA-MB-4175 when processed in parallel (Supplementary Fig. 1j, k), thereby confirming that exomeres are indeed actively secreted by cultured cells and not mere aggregates present in media.
Using AF4, we detected distinct particles with diameters corresponding to exomeres and Exo-S/L in more than 20 cell lines analyzed (Supplementary Table 1, Supplementary Fig. 2a), findings confirmed by TEM analysis of pooled fractions from selected cell lines (Supplementary Fig. 2b). Based on UV absorbance and TEM analysis, all cells secreted higher amounts of exomeres relative to Exo-S/L, except for B16-F10 and B16-F1 where Exo-S were relatively more abundant (Supplementary Fig. 2a and Fig. 1a, b). Measurement of the hydrodynamic diameter of each of these particles using Zetasizer showed sizes similar to the B16-F10 preparations (Fig. 1e).
We also detected exomeres and Exo-S/L in AF4-fractionated sEVs from CM of human melanoma tumor explants by TEM (Figure 1f, arrows; Supplementary Fig. 3a). Exomere and Exo-S size, measured in batch mode using Zetasizer, was comparable to results from tumor cell lines (Fig. 1g). AF4 profiling and TEM imaging analysis showed that the normal mouse tissue explants (mammary fat pad and lung) also secreted exomeres, and Exo-S/L nanoparticles (Supplementary Fig. 3b).
Biophysical characterization of exomeres and exosome subpopulations
Given the structural differences between exomeres and Exo-S/L, we examined their biophysical properties, such as zeta potential and stiffness. Measuring zeta potential, an average surface charge, using Zetasizer, revealed all particles were negatively charged, with exomeres being the weakest negatively charged (-2.7 mV to -9.7 mV); Exo-L, the strongest (-12.3 mV to -16.0 mV); and Exo-S, intermediate (-9.0 mV to -12.3 mV) (Figure 2a).
Figure 2.
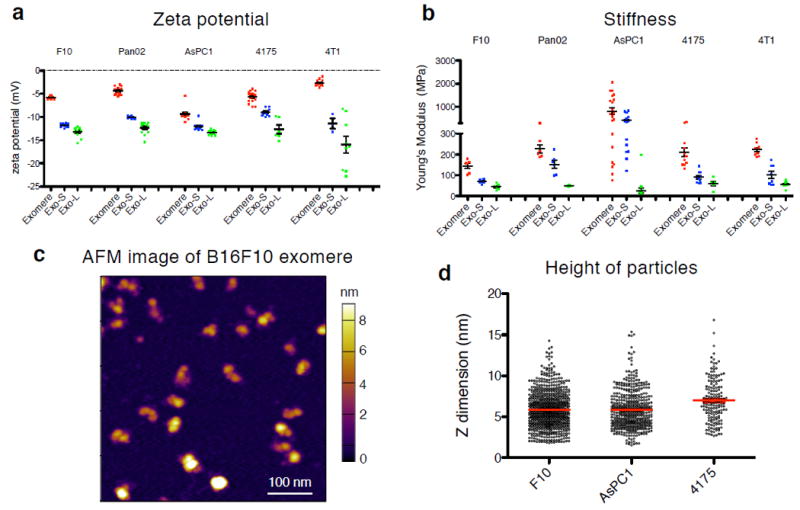
Characterization of physical and mechanical properties of exomeres and exosome subpopulations. Zeta potential (a) and stiffness (b) of exomeres and exosome subpopulations derived from various cancer cells were measured using Zetasizer and AFM indentation, respectively. Young’s modulus was used to express particle stiffness. At least 3 and 5 replicates for each group of particles was measured for zeta potential and stiffness, respectively. Data are presented as mean±SEM. For (a), in the order of exomere, Exo-S and Exo-L: B16-F10 (n=8, 10, and 12 independent measurements, respectively); Pan02 (n=13, 11, 13); AsPC-1 (n=12, 12, 12); 4175 (n=17, 9, 6); 4T1 (n=13, 3, 9); for (b), B16-F10 (n=6, 6, 6 particles measured); Pan02 (n=6, 6, 6); AsPC-1 (n=21, 19, 16); 4175 (n=11, 10, 5); 4T1 (n=9, 8, 9)). (c) Representative AFM image of exomeres derived from B16F10. This experiment was repeated with samples derived from 3 different cell lines with similar results. (d) AFM imaging analysis of the height (z-dimension) of exomeres derived from B16F10 (n=754 particles analyzed), AsPC1 (n=475) and MDA-MB-4175 (n=160). Mean±SEM is depicted. Statistical source data are provided in Supplementary Table 8.
For particle stiffness, atomic force microscopy (AFM) was performed in solution (see Methods). Exomeres demonstrated the highest stiffness (145~816 mPa) and Exo-L the lowest (26~73 mPa), with Exo-S stiffness being intermediate (70~420 mPa).
AFM analysis of exomeres derived from B16-F10, MDA-MB-4175, and AsPC-1 cell lines demonstrated exomere structural heterogeneity and average exomere heights of 5.9 nm, 7.0 nm and 5.8 nm, respectively (Fig. 2c, d).
Collectively, these findings demonstrate the diverse biophysical properties exhibited by exomeres versus distinct exosome subpopulations. How size, charge, and mechanical properties influence the differential stability, trafficking and uptake of the nanoparticles in vivo requires further investigation15, 16.
Distinct proteomic content and cellular functions among exomeres and exosome subpopulations
To characterize the molecular composition of exomeres and distinct exosome subpopulations, we conducted proteomic profiling of nanoparticles derived from B16-F10, Pan02, 4T1, AsPC-1, MDA-MB-4175 cells using label-free mass spectrometry. A range of 165-483 proteins were identified in exomeres, 433-1004 proteins in Exo-S, and 247-1127 proteins in Exo-L. Moreover, unique proteins were detected in each nanoparticle subtype (Fig. 3a), suggesting exomeres are unique entities released by cells rather than debris or fragments of exosomes.
Figure 3.
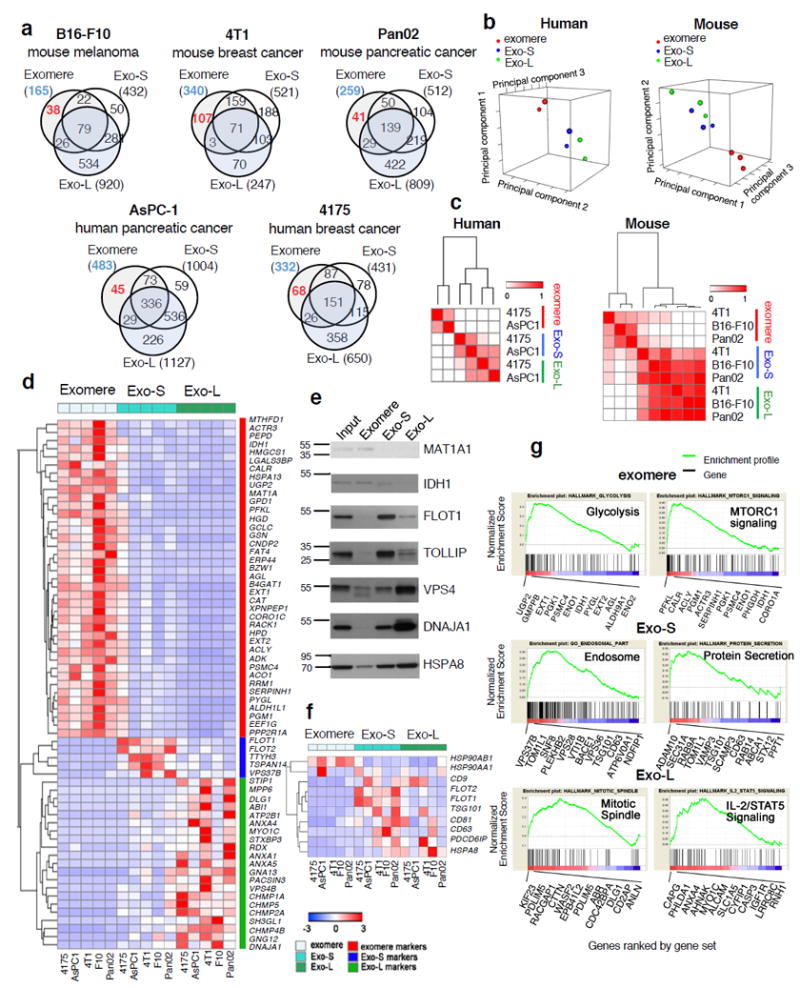
Proteomic profiling of exomeres and exosome subpopulations derived from various cancer cells. (a) Venn diagram of proteins identified in each subset of particles. (b) Principal component analysis and (c) Consensus clustering analysis of normalized proteomic mass spectrometry datasets from human (MDA-MB-4175 and AsPC1) and mouse (B16F10, 4T1, and Pan02) cell lines. (d) Heat map illustration of unique proteins specifically associated with exomeres, Exo-S and Exo-L. Scale shown is intensity (area) subtracted by mean and divided by row standard deviation (i.e. Δ (area-mean)/SD). (e) Western blot analysis of representative signature proteins in fractionated samples. An equal amount (10 μg) of exosome and exomere input mixture and each subset were analyzed. This experiment was done once. (f) Heat map illustration of the relative abundance of conventional exosome markers in exomeres, Exo-S and Exo-L. Scale shown is intensity (area) subtracted by mean and divided by row standard deviation (i.e. Δ (area-mean)/SD). (g) Identification of top candidate gene sets enriched in exomere, Exo-S and Exo-L populations by gene set enrichment analysis (GSEA). Proteins in each subset of nanoparticles are ranked by GSEA based on their differential expression level. Whether a pre-specified pathway is significantly overrepresented toward the top or bottom of the ranked gene list in each subset of nanoparticle is evaluated using the normalized enrichment score (the green line). Black vertical lines mark the positions where the members of a particular pathway appear in the ranked list of genes. Proteins that contributed most to the enrichment score are listed below the plot. For all proteomic analysis (b-d, f-g), a total of 30 samples (3 nanoparticle subtypes derived from 5 different cell lines; and two independent biological replicates for each nanoparticle sample) were subjected to statistical analysis. Unprocessed blots are provided in Supplementary Figure 7.
Examination of the subcellular localization annotation of proteins revealed the specific enrichment of Exo-S/L in membrane-associated proteins, which were relatively depleted in exomeres (Supplementary Table 3), consistent with our structural studies identifying Exo-S/L as membrane-encapsulated particles and exomeres as non-encapsulated particles. ESCRT- and Snare-related proteins, involved in vesicle budding, membrane fusion and exosome biogenesis17, 18, were identified within Exo-S/L. In particular, proteins associated with endosomes, multivesicular bodies, vacuoles, and phagocytic vesicles were enriched in Exo-S. Plasma membrane, cell-cell contact/junction, late-endosome, and trans-Golgi network proteins were enriched in Exo-L. Notably, proteins associated with extracellular matrix and space, proteasome accessory complex, endoplasmic reticulum, mitochondrion, and microtubule/cytoskeleton were packaged in exomeres. These findings imply possible fundamental differences in exomeres, Exo-S, and Exo-L biogenesis.
Principal component analysis (PCA) demonstrated closer correlation of protein expression for Exo-S and Exo-L compared to exomeres from the same cell-type (Supplementary Fig. 4a). According to PCA and consensus clustering analysis, exomeres from different cell types exhibited a higher degree of similarity to each other than to Exo-S and Exo-L from the same cell type (Fig. 3b, c).
To identify the signature proteins in each particle subset, we performed statistical analysis on the expression levels of proteins identified in these datasets. We pinpointed 64 proteins for exomeres and 99 proteins for Exo-S/L (Supplementary Table 4), with a false discovery rate (FDR) <0.05, positive enrichment in each particle subset of interest, and detection frequency of >80% (i.e., a particular protein was positively enriched in at least 4/5 samples for each subtype of nanoparticles derived from 5 different cell lines). Remarkably, exomeres were significantly enriched in proteins involved in metabolism (see gene set enrichment analysis [GSEA] analysis below), including MAT1A, IDH1, GMPPB, UGP2, EXT1, and PFKL. The sialoglycoprotein galectin-3-binding protein (LGALS3BP) and key proteins controlling glycan-mediated protein folding control (CALR)19 and glycan processing (MAN2A1, HEXB, GANAB)20-22 are also enriched in exomeres, suggesting exomere cargo may mediate the targeting of recipient cells through specific glycan recognition and modulate glycosylation in recipient cells. Among proteins uniquely represented in Exo-S/L were annexins, ESCRT components (charged multivesicular body proteins/CHMPs, vacuolar protein-sorting proteins, HGS, Alix1/PDCD6IP, and Tsg101), Hsp40 (DnaJ) family proteins, signaling transducer G protein subunits, integrins, Rab proteins, and solute carrier family members. Members of key signaling pathways, such as JAK1, TGFBR2, and MET, were also enriched in Exo-S/L. To evaluate the unique markers of Exo-S and Exo-L subpopulations, we compared protein expression between these two sample sets and exomeres separately using t-test. A second set of filters (protein intensity/area >108 and fold change ≥5.0) was applied to the identified signature for exomeres and Exo-L, but not for Exo-S (Fig. 3d). Fewer signature proteins were identified for Exo-S compared to exomeres and Exo-L, most likely due to similarity of Exo-S to the other particles. Representative signature proteins identified by proteomics in each subset were validated by western blot analysis (Fig. 3e).
We further mined these proteomic datasets for conventional exosome markers, including flotillins, CD9, CD63, CD81, Alix1, Tsg101, HSC70 (HSPA8) and Hsp90 (Fig. 3f). Among the five cell lines, flotillins (FLOT1 and FLOT2) represented bona fide markers of Exo-S, while HSP90AB1 was preferentially associated with exomeres. Although CD9, CD63 and CD81 all demonstrated specific association with Exo-S/L subsets, they all showed a cell type- and particle-dependent preferential expression. Consistent with Kowal et al 23, combining CD63, CD9 or CD81 will be necessary to isolate/label exosomes.
We found numerous Rab proteins in Exo-S/L subsets, but few of them in exomeres (Supplementary Fig. 4b), suggesting critical roles of Rab proteins for Exo-S/L formation and trafficking, but not for exomere biogenesis.
Next, we examined the most abundant proteins in each subset of nanoparticles. Hemoglobin, histones, cytoskeleton proteins (actins and tubulins), peptidylprolyl isomerase A (PPIA) and HSP ranked as the most abundant top 50 proteins in all three nanoparticle subpopulations (Supplementary Table 4). Hsp40/DnaJ family (HSP70 co-chaperones) members were also found in the top 50 proteins for Exo-L. Interestingly, HSP90AB1 was preferentially packaged in exomeres, while HSP70 members (HSPA8, HSPA2 and HSPA5) were more abundant in Exo-S/L. Other proteins relatively enriched in exomeres included inter-alpha-trypsin inhibitor heavy chain family members (ITIH), gelsolin (GSN), talin 1 (TLN1), WD repeat domain 1 (WDR1), and proteins involved in metabolism, such as phosphoglycerate kinase 1 (PGK1), pyruvate kinase muscle (PKM), and enolase 1 (ENO1). Consistent with the analysis above, SDCBP, PDCD6IP/Alix, tetraspanins (CD9, CD63, CD81 and others), G protein family proteins and integrins were highly represented in both Exo-S and Exo-L. Tetraspanins were preferentially enriched in Exo-S while G proteins and integrins were more prominent in Exo-L. Eukaryotic translation elongation factor 1 alpha 1 (EEF1A1) was most often present in exomeres and Exo-L. Other proteins preferentially associated with Exo-S included immunoglobulin superfamily member 8 (IGSF8) and its paralog prostaglandin F2 receptor inhibitor (PTGFRN), milk fat globule-EGF factor 8 protein (MFGE8), and components of the ESCRT-I complex. Notably, annexins and S100 proteins were only represented in the top 50 proteins of Exo-L.
Furthermore, to exclude the possibility of lipoprotein contamination in exomeres, we examined proteins that are typically associated with purified lipoprotein particles (high-, low-, and very low-density lipoproteins, i.e., HDL, LDL, and VLDL) by proteomic MS analysis and then evaluated their presence in exomeres and Exo-S/L. Much fewer proteins were found in lipoproteins (Supplementary Table 5) and only some of these proteins were detected in exomeres and Exo-S/L, suggesting most nanoparticle proteins are distinct from lipoproteins. A rough estimation showed that the lipoprotein-associated proteins account for 0-8% of total nanoparticle proteins (Supplementary Fig. 4c). Moreover, EM analysis revealed that lipoprotein morphology/structure was clearly distinct from exomeres and Exo-S/L (Supplementary Fig. 4d). Taken together, these analyses ruled out the possibility that exomeres were mere lipoprotein contaminants.
The possible contamination of exomeres with other types of protein complexes with high molecular weights was also ruled out when exomere proteins were surveyed for subunits of known complexes (the comprehensive resource of mammalian protein complexes (http://mips.helmholtz-muenchen.de/corum/#; 02.07.2017 CORUM current release)). The co-existence of multiple subunits of protein complexes of similar size to exomeres were not detected (Supplementary Table 5) except for 10 out of 59 subunits of Parvulin-associated pre-rRNP complex in 4T1 exomeres, 17 subunits of ribosomes in AsPC-1 exomeres, and 7 out of 16 subunits of Kinase maturation complex 1 in MDA-MB-4175 exomeres. However, these proteins account for only 1.8%, 2.1% and 1.8% of total exomere proteins in each case, respectively, suggesting their contribution diminishes the purity of exomeres by ~2%.
To gain insight into the function of these particle subsets, we conducted GSEA utilizing gene ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) and hallmark databases (Supplementary Table 6). Strikingly, GSEA demonstrated that exomere-specific proteins were selectively enriched in metabolic processes, including carbohydrate metabolism, protein synthesis, and small-molecules. At least 36 of the top 50 “GO-biological processes” pathways identified metabolic processes associated with exomeres in contrast to no metabolic processes associated with Exo-S/L (Supplementary Table 6). Genes encoding proteins involved in hypoxia, microtubule and coagulation were identified in exomeres (Supplementary Fig. 4e). Exo-S were enriched in membrane vesicle biogenesis and transport, protein secretion and receptor signaling gene sets. For Exo-L, enriched gene sets included mitotic spindle, IL-2/Stat5 signaling, multi-organism organelle organization, and G-protein signaling. Profiles of top rank gene sets enriched in exomeres (glycolysis and mTORC1 signaling), Exo-S (endosome and protein secretion) and Exo-L (mitotic spindle and IL-2/Stat5 signaling) are displayed in Figure 3g.
Collectively, these bioinformatic analyses of the proteomic content of each particle subset revealed the predominant link between exomere-associated proteins and metabolism and the link between Exo-S/L-associated proteins and multiple signaling transduction pathways, including biogenesis-related ESCRT complexes.
Distinct N-glycan profiles of exomeres and exosome subpopulations
Aberrant glycosylation is involved in pathological processes, including cancer24. Here, we aimed to determine the N-glycan profiles of each particle subset in three cell lines by conducting lectin blotting analysis (Fig. 4a) and glycomic mass spectrometry.
Figure 4.
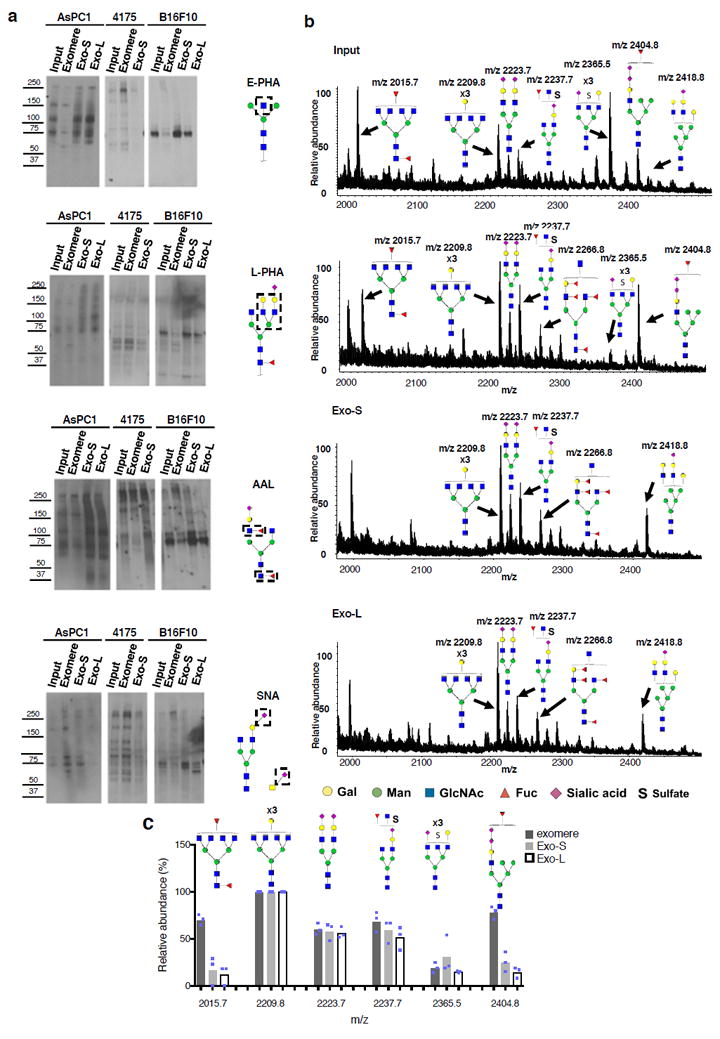
Characterization of N-glycosylation of proteins associated with exomere, Exo-S and Exo-L. (a) Lectin blotting analysis of N-glycan profile of proteins associated with exomeres versus exosome subpopulations Exo-S and Exo-L. Phaseolus vulgaris erythroagglutinin (E-PHA) and Phaseolus vulgaris leucoagglutinin (L-PHA) recognize bisected and branched N-glycans, respectively. Aleuria aurantia lectin (AAL) recognizes Fucα6GlcNAc and Fucα3GlcNAc. Sambucus nigra lectin (SNA) recognizes α-2,6-linked sialic acid. All experiments were repeated independently twice with similar results except for AAL and E-PHA blotting for B16-F10 and 4175 which were done once. (b) Mass spectrometric analysis of N-glycans of glycoproteins present in exomeres, Exo-S and Exo–L subsets of B16F10. One representative experiment of two biologically independent replicates is shown. (c) Comparison of the relative abundance of the top six most abundant N-glycan structures among exomere, Exo-S and Exo-L of B16F10. The assignments (m/z) [charge; neutral exchange] for MALDI-MS and nanoLC-ESI-MS/MS are the following: (2015.8 [-H; 0]; 1007.4a [-2H; 0]), (2209.8 [-H; 0]; 1104.4a [-2H; 0]), (2237.7b [-H; Na-H]; 732.57a [-3H; 0]), (2365.5b [-H;4K-4H]; 783.9a [-3H; 0] and 1182.4a,b [-2H; 4K-4H]), and (2404.8b [-H; 2K-2H]; 1201.9b [-2H; 2K-2H]). Data shown were quantified and normalized to the most abundant structure in the sample. Results are represented as average of three independent analytical measurements of one representative experiment. Statistical source data are provided in Supplementary Table 8, and unprocessed blots are provided in Supplementary Figure 7. Note: aThe product ion spectra for this species did not allow a complete structural assignment. bAssignments admit neutral exchanges of protons with cations in sialoglycans, including the presence of potassium and sodium.
E-PHA recognizing bisected N-glycans detected a major band at approximately 75 kDa in both Exo-S and Exo-L of B16-F10 and AsPC-1, with faint detection in exomeres across the three cell lines and Exo-S of MDA-MB-4175. E-PHA detected a high molecular-weight glycoprotein (240 kDa) in MDA-MB-4175 exomeres and a high molecular weight glycoprotein (150 kDa) in AsPC-1 and MDA-MB-4175 exomeres. L-PHA recognizing branched N-glycans detected a predominant band at 75 kDa in both Exo-S and Exo-L of B16F-10 and AsPC-1. Multiple bands ranging from 50 to 70 kDa were also detected in all exomeres (especially MDA-MB-4175). Using AAL, analysis of structures related to fucosylation (fucose linked α -1,6) to GlcNAc or fucose linked (α -1,3) to GlcNAc related structures revealed two abundant glycoproteins between 70 and 100 kDa in both Exo-S and Exo-L of B16-F10 and AsPC-1. Exomeres across all three cell lines and Exo-S of MDA-MB-4175 displayed strong fucosylation on high molecular-weight glycoproteins (200 –280 kDa). SNA, recognizing α-2,6-linked sialic acid, detected the presence of high molecular-weight α-2,6-sialylated glycoproteins (200- 250 kDa) in all exomeres. Moreover, a low molecular-weight protein (~60 kDa) displaying α-2,6-linked sialic acid modification was uniquely detected in Exo-L (but not Exo-S) from B16-F10. For AsPC-1, exomeres were the major carriers of sialylated glycoproteins, while these sialylated structures were almost absent in Exo-L. Lectin-binding profiles did not overlap with the most abundant proteins in the SDS-PAGE gel, indicating the specificity of lectin recognition independently of protein abundance (Supplementary Fig. 5a). Therefore, Exo-S and Exo-L versus exomeres display distinct N-glycosylation patterns. Notably, exomere and Exo-S/L-associated N-glycan profiles vary by cell type. Future studies will address the identity of these glycoproteins via glycoproteomic approaches.
We then aimed to identify profiles of the glycan structures enriched in each particle subset by MALDI-TOF mass spectrometry (MS). Two independent, semi-quantitative MS analyses were conducted on B16-F10-derived exomeres and Exo-S/L (Fig. 4b). Figure 4c depicts the quantification of the top six most abundant N-glycan structures detected in one of the representative experiments. We observed the ubiquitous expression of certain complex N-glycans in all subsets, corresponding to peaks at m/z 2209.8, 2223.7, 2237.7 and 2365.5. Specifically, a complex N-glycan at m/z 2015.7 and a hybrid N-glycan at m/z 2404.8 were enriched in exomeres. Moreover, four of these six N-glycans contained sialic acid, and three of six were fucosylated. Similarly, the ions m/z 2015.7 and 2404.8 were enriched in exomeres from MDA-MB-4175 (Supplementary Fig. 5b, d). The ion m/z 2404.8 was slightly enriched in AsPC1 exomeres, but the ion at m/z 2015.7 was not detected in AsPC-1 samples (Supplementary Fig. 5b and c). Instead, the ion at m/z 2012.7 was strongly detected in AsPC1 exomeres and Exo-S. Two other ions, at m/z 2117.7 and 2389.9, demonstrating Exo-S enrichment, were detected in AsPC-1 only (Supplementary Fig. 5b-d).
High-resolution MS analysis allowed further structural characterization of certain N-glycans (Supplementary Fig. 5e-j). This was the case of extracted ion chromatogram m/z 1111.39 (2-) and 1007.38 (2-) (corresponding to m/z 2223.7 and 2015.7 in Figure 4c, respectively). In addition, the combination of CID-MS/MS de novo sequencing and PGC-LC relative retention times for extracted ion chromatogram m/z 1111.39 (2-) revealed that this N-glycan from exomeres contained both α2,3-linked and α2,6-linked sialic acids, whereas the glycan from Exo-S contained exclusively α2,3-linked sialic acids. The unique presence of m/z 1007.38 (2-) in exomeres was also further confirmed.
Taken together, our glycomics study demonstrated the prevalence of complex N-glycans in all particle subsets with relatively high levels of sialylation, consistent with previous findings of complex N-glycans and sialoglycoproteins in tumor microvesicles/exosomes25-27. Furthermore, our study revealed differences in N-glycan composition and structures among exomeres, Exo-S, and Exo-L.
Distinct lipid composition among exomeres and exosome subpopulations
To investigate the lipid composition of each subset of particles, we performed quantitative lipidomics on these nanoparticles derived from B16-F10, MDA-MB-4175 and AsPC-1. By lipid MS, we found that Exo-S and Exo-L contained more lipids than exomeres for all cell lines (Fig. 5a, >5x fold in all subpopulations, except for Exo-S of MDA-MB-4175 (>3x fold)).
Figure 5.
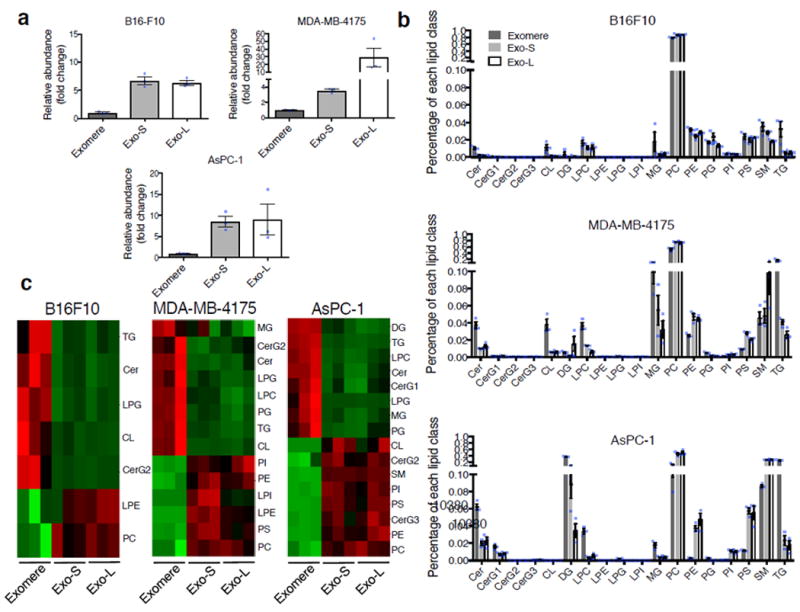
Characterization of lipid composition in exomeres and exosome subsets. (a) Comparison of total lipid content of each subset of nanoparticles derived from different cell lines. Total signal intensity of each sample after normalization to sample weight and internal standards was compared to that of exomeres from the same set of samples (expressed as fold change). Data are presented as mean±SEM (n=3 biologically independent samples). (b) Relative abundance of each lipid class present in each subset of nanoparticles from different cell lines. Data are presented as mean±SEM (n=3 biologically independent samples). For (a) and (b), statistical source data are provided in Supplementary Table 8. (c) Heat map illustration of lipid classes specifically associated with exomeres, Exo-S and Exo-L (ANOVA test, q<0.05). Statistical analysis was performed on a total of 9 samples for each cell line (3 different nanoparticle subtypes and 3 independent biological repeats for each nanoparticle sample). Abbreviation: Cer, ceramide; CerG1-3, glucosylceramides; CL, cardiolipin; DG, diglyceride; LPC, lysophosphatidylcholine; LPE, lysophosphatidylethanolamine; LPG, lysophosphatidylglycerol; LPI, lysophosphatidylinositol; MG, monoglyceride; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PG, phosphatidylglycerol; PI, phosphatidylinositol; PS, phosphatidylserine; SM, sphingomyelin; TG, triglyceride.
Eighteen lipid classes were commonly identified in all samples (Supplementary Table 7, Fig. 5b), and their relative frequency in each sample was compared. Phosphatidylcholine (PC) was the predominant lipid component in all subpopulations (46% ~ 89%) except for AsPC-1 exomeres (13%) (Fig. 5b), which contained higher levels of diglyceride (DG, 38%) and triglyceride (TG, 26%) instead. Other phospholipids, including phosphatidylethanolamine (PE) and phosphatidylserine (PS), accounted for 2-6% of total lipids in Exo-S/L across all cell lines (Fig. 5b). However, PE and PS levels were lower in exomeres from MDA-MB-4175 and AsPC-1, but similar to Exo-S/L in B16-F10 (Fig. 5b, c). Phosphatidylinositol (PI) levels were lower than other phospholipids but had a pattern of distribution across nanoparticle subsets similar to that of PE and PS (Fig. 5b, c). Sphingomyelin (SM) accounts for 2-10% of the total lipid in all samples except for AsPC-1 Exo-S/L, which contained a higher level of SM (28%, Fig. 5b, c). Cholesterol data were not collected in this study.
The relative levels of ceramide (Cer), TG and lysophosphatidylglycerol (LPG) varied significantly between exomeres and Exo-S/L across cell lines (ANOVA test, q<0.05). Additionally, simple glycosphingolipid CerG2 and mitochondrion-specific cardiolipin (CL) were more abundant in exomeres of B16-F10 and MDA-MB-4175 compared to exosome subsets. In contrast, CerG2 and CL were more abundant in Exo-S/L compared to exomeres isolated from AsPC-1 cells. Monoglyceride (MG), phosphatidylglycerol (PG) and lysophosphatidylcholine (LPC) were more abundant in exomeres than in Exo-S/L from MDA-MB-4175 and AsPC-1, but present at equal levels in all three B16-F10 nanoparticle subsets. Lastly, lysophosphatidylethanolamine (LPE) was detected at higher levels in Exo-S/L from B16-F10 and MDA-MB-4175, but not from AsPC-1. Thus, our study revealed cell type-dependent differences in the total lipid content and composition among distinct nanoparticle subsets.
Distinct nucleic acid content among exomeres and exosome subpopulations
Since we previously detected dsDNA in tumor-derived exosomes6, we determined the relative abundance of DNA in exomeres and Exo-S/L. DNA was detected in all three types of nanoparticles; however, relative abundance varied by cell-type (Fig. 6a). The relative amount of DNA was highest in exomeres derived from MDA-MB-4175 and in Exo-S from B16-F10 cells and AsPC-1. Bioanalyzer (Agilent) analysis revealed distinct size distribution of DNA associated with each subset of nanoparticles (Fig. 6b and Supplementary Fig. 6). Exomere DNA was relatively evenly distributed in a broad range of sizes between 100 bp and 10 kb with a slight enrichment around 2 kb in several cases. In contrast, a strong enrichment between 2 kb to 4 kb was detected for Exo-S/L DNA, and the peak size of Exo-L DNA was slightly larger than that of Exo-S DNA. This phenomenon may be due to the structural capacity and different biogenesis mechanisms of each particle subset.
Figure 6.
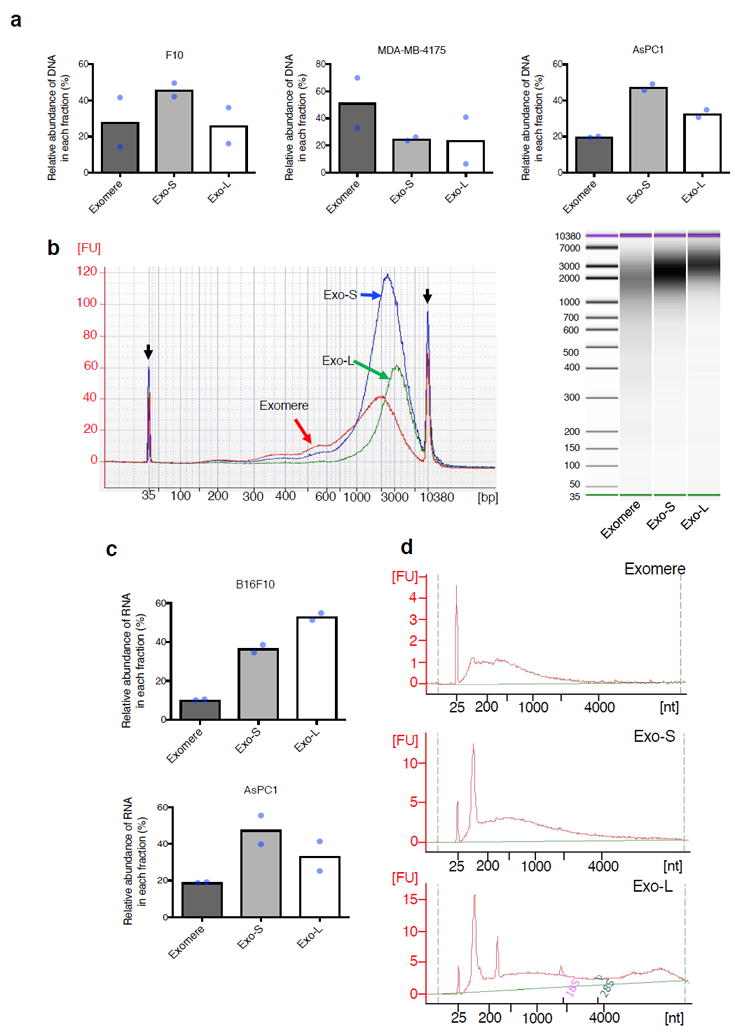
Characterization of nucleic acid association with exomere and exosome subsets. (a) Relative abundance of DNA associated with each subpopulation of particles from representative fractionations of B16F10, AsPC1 and MDA-MB-4175. (b) Agilent Bioanalyzer analysis of the size distribution of DNA associated with different subsets of particles. Data shown are the electropherograms (left) and electrophoresis images (right) from a representative of two independent experiments on AsPC1-derived particles. Black arrows, internal standards (35bp and 10380bp). Red line, exomeres; blue line, Exo-S; green line, Exo-L. (c) Relative abundance of total RNA associated with each subpopulation of particles from representative fractionations of B16F10 and AsPC1. (d) Size distribution of RNA isolated from different fractions of B16F10. Shown are representative profiles from one of two independent experiments. For (a) and (c), data shown are mean (n=2 biologically independent samples). Statistical source data are provided in Supplementary Table 8.
RNA was preferentially associated with Exo-S/L in both B16-F10 and AsPC-1 (Fig. 6c). RNA associated with exomeres and Exo-S showed a monomodal distribution (peak at 400nt and 500nt, respectively), whereas Exo-L RNA displayed a bimodal distribution (Fig. 6d) (additional peak >4000nt). Specifically, 18S and 28S rRNAs were detected at very low levels in Exo-L, barely detected in Exo-S and absent in exomeres compared to cellular RNA. A strong small RNA peak (corresponding to tRNAs, microRNAs and other small RNAs) was detected in Exo-S and Exo-L, but not in exomeres. Remarkably, a unique RNA peak of unknown identity, of ~315nt in size, was detected only in Exo-L.
Distinct organ biodistribution of exomeres and exosome subpopulations
Next, we investigated the organ biodistribution of B16-F10-derived nanoparticle subsets in naïve mice. Twenty-four hours post intravenous injection of near infrared dye (NIR)-labeled exomeres, Exo-S and Exo-L into mice, organs were collected and analyzed using the Odyssey imaging system (LI-COR Biosciences; Fig. 7). Interestingly, all nanoparticles were uptaken by hematopoietic organs, such as the liver (~84% of total signals), spleen (~14%) and bone marrow (~1.6%). The lungs (~0.23%), lymph nodes (~0.07%), and kidneys (~0.08%) showed less uptake of all nanoparticle subtypes. We did not detect particle uptake in the brain. Subsequently, the dynamic range of signal intensity in each organ was adjusted to compare the uptake of each subset of nanoparticles in the same organ (Fig. 7a). Punctuated distribution patterns of nanoparticles were detected specifically in the lung and lymph nodes. This is in contrast to the homogenous distribution pattern found for all nanoparticle subsets in the liver, spleen, and bone marrow. Importantly, although exomeres and Exo-S/L were predominantly uptaken in the liver, Exo-L displayed lymph node tropism. In addition, though not statistically significant, a trend of higher uptake of exomeres in the liver was observed. Quantification is shown in Fig. 7b. Distinct organ distributions indicate that nanoparticle subsets may be involved in different aspects of tumor progression and metastasis.
Figure 7.
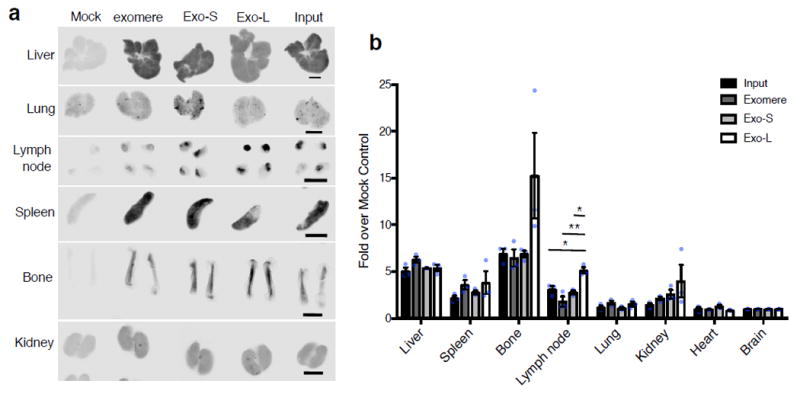
Organ biodistribution of B16F10-derived exomeres and exosome subpopulations in syngeneic naïve mice. (a) Whole organ imaging of NIR dye-labeled exomeres, Exo-S and Exo-L from a representative experiment using the Odyssey imaging system (LI-COR Biosciences; n=4 independent experiments). The dynamic range of signal intensity was adjusted for each organ so that the differences among these nanoparticle subsets can be easily recognized. Scale bar, 2.5 mm. (b) Quantification of the nanoparticle uptake in different organs in one representative experiment. This experiment was repeated independently 4 times with similar results. Signal intensity in each organ was acquired using the Image Studio (LI-COR Biosciences), and normalized to the brain from the same animal due to undetectable uptake of nanoparticles in this organ. Fold changes (y axis) were then calculated for each organ between the experimental group (i.e. input, exomere, Exo-S and Exo-L) versus the mock control. n=3 animals per group, results shown are mean ± SEM. Statistical significance determined using one way ANOVA (* p<0.05; ** p<0.01, unmarked, not significant). For lymph nodes, the p value for comparison between input versus Exo-L, exomere versus Exo-L and Exo-S versus Exo-L are 0.022, 0.001 and 0.01 respectively. Statistical source data are provided in Supplementary Table 8.
Discussion
Dissecting the heterogeneity of EV populations by differential ultracentrifugation, immuno-affinity capture, ultrafiltration and size-exclusion chromatography, polymer-based precipitation, and microfluidics28-33 in an attempt to separate nanoparticle populations has proven daunting. By employing state-of-the-art AF4 technology, we succeeded in separating two discernible exosome subpopulations, Exo-S and Exo-L, and identified a distinct nanoparticle, named exomere, which differs in size and content from other reported particles. Unlike labor-intensive and time-consuming gradient methods, AF4 is highly reproducible, fast, simple, label-free and gentle. Moreover, we were able to efficiently resolve the exosome subpopulations and exomeres in a single AF4 run with real-time measurements of various physical parameters of individual particles.
Our analyses revealed that exomeres were selectively enriched in proteins involved in metabolism, especially “glycolysis” and “mTORC1” metabolic pathways, suggesting their potential roles in influencing the metabolic program in target organ cells, as well as in proteins associated with coagulation (e.g., Factors VIII and X) and hypoxia. Our proteomic analysis also showed that exomeres were enriched in key proteins controlling glycan-mediated protein folding control (CALR)19 and glycan processing (MAN2A1, HEXB, GANAB)20-22, suggesting exomere cargo may modulate glycosylation in distant recipient cells. Subcellular localization analysis of exomere-enriched proteins revealed their specific association with ER, mitochondria and microtubules, demonstrating the potential roles of these proteins in exomere biogenesis and secretion.
Proteins unique to exosomes (Exo-L and Exo-S) versus exomeres were also identified. Multiple components of ESCRT complexes were specifically associated with Exo-S and Exo-L, but not observed within exomeres, suggesting a major role for ESCRT complexes in Exo-S/L but not exomere production. Other exosome-enriched proteins included Rab proteins, annexins, Hsp40 members, and proteins involved in multiple signaling transduction pathways, such as integrins, G-proteins, JAK1 and TGFBRs.
We found further differences between Exo-S and Exo-L protein cargo. Flotillin 1, flotillin 2, tweety family member 3, tetraspanin 14, and ESCRT-I subunit VPS37B were specifically enriched in Exo-S. In contrast, levels of such proteins as annexin A1/A4/A5, charged multivesicular body protein 1A/2A/4B/5, vacuolar protein sorting 4 homolog B, DnaJ heat shock protein family (Hsp40) member A1, and myosin IC were relatively higher in Exo-L. Interestingly, tissue factor, a well-studied exosome protein34, was enriched in Exo-L. It is thus plausible that exomeres and Exo-L cooperate to optimize the coagulation cascade in vivo.
Exo-S were predominantly enriched in proteins associated with endosomes, multivesicular bodies, vacuoles, and phagocytic vesicles, while Exo-L were specifically enriched in plasma membrane, cell-cell contact/junction, late-endosome, and trans Golgi network proteins. These data indicate that Exo-S are most likely bona fide/canonical exosomes (i.e., derived from intraluminal vesicles of endosomal compartments), whereas Exo-L may represent non-canonical exosomes or probably sEVs of different sub-cellular origin (i.e., plasma membrane budding).
Identifying specific exosome and exomere markers to better isolate and characterize these nanoparticles is critical to advancing our knowledge of EV biology. Since Flotillin 1 and 2 were specifically associated with Exo-S, these proteins may represent reliable markers of conventionally defined exosomes. Other previously reported exosome markers, including CD9, CD63, CD81, Tsg101 and Alix1, were present in Exo-S and/or Exo-L in a cell type-dependent manner, and therefore would have to be combined with size exclusion to distinguish exosome subpopulations. Notably, Hsp90-b, highly represented in exomeres, could be a potential exomere marker, whereas several Hsp70 family members, such as HSC70/HSPA8 could serve as possible markers for Exo-S/L subpopulations.
Our glycomic, lipidomic, and genomic studies also revealed additional distinct molecular signatures in exomeres and exosomes. Similar to the expression in metastatic tumor cells, exosome subsets were enriched with sialylated glycoproteins, supporting the role of these structures in exosome-mediated cellular recognition. One predominant sialoglycoprotein previously identified in exosomes25, 35, the galectin-3-binding protein (LGALS3BP), a modulator of cell communication and immune responses36, 37, was highly enriched in exomeres. This ligand could mediate the specific interaction of exomeres with target cells through proteins, such as collagens, fibronectin, nidogen, galectin-3 and integrin beta-138,39.
Interestingly, our lipidomics analyses revealed that exomeres contained fewer lipids compared to Exo-S and Exo-L. Phospholipids and SM, the major structural components of plasma lipid bilayer membrane40 ranked top in all nanoparticles examined. Such an observation is expected for Exo-S/L subsets due to their vesicular membrane structure, however, exomeres seem to lack external membrane structures. Yet, differences in several lipid classes distinguished exomeres from Exo-S and Exo-L. For instance, exomeres were found to contain higher levels of triglycerides and ceramides compared to exosome subpopulations and thus may serve to transport these metabolites to recipient cells. Our study further revealed that DNA packaging in exomeres and exosomes varied by tumor-type, while RNA was packaged in Exo-S and Exo-L independent of tumor classification.
Collectively, our findings demonstrate that proteins, glycans, lipids, and nucleic acids are selectively packaged in exomeres, Exo-S, and Exo-L, further supporting the idea that these are distinct nanoparticle subsets.
Our observation that nanoparticle subtypes have different organ biodistribution patterns suggests they mediate the pleiotropic effects of cancer. The punctate pattern of Exo-L uptake and its apparent tropism for lymph nodes implicate this nanoparticle in facilitating metastasis of disseminated tumor cells. Exomeres, along with exosomes, were uptaken by hematopoietic organs, including the liver, spleen and bone marrow. Interestingly, the predominant exomere uptake by the liver and the exomere enrichment in protein cargo involved in metabolism lead us to speculate that exomeres may specifically target the liver for metabolic reprogramming during tumor progression. Our data indicate that the size of nanoparticles, in addition to their specific cargo, may influence metastatic patterning and systemic effects of cancer.
Our identification of exomeres highlights the diversity of EVs and particles secreted by cells. Elucidating their biogenesis will be essential to unravel their roles in cellular and organ function. Target cells and the functional outcomes exerted by each nanoparticle subset in organs need to be further delineated to advance our understanding of the collective, systemic effects of nanoparticles in the metastasis process. Undoubtedly, these discoveries will open avenues for translational studies of EVs and particles in diagnostic, prognostic, and therapeutic applications.
Supplementary Material
Acknowledgments
We are grateful for the great technical support from Wyatt Technology and especially Dr. John Champagne. We thank the Genomics Resource Core facility (WCM) for their service of high quality. We thank Cyrus Ghajar and Joshua Weiss for feedback on the manuscript and members of the Lyden laboratory for helpful discussions. Our study was supported by National Cancer Institute (U01-CA169538, D.L. and M.S.B.), National Institutes of Health (R01-CA169416, D.L. and H.P.; R01-CA218513, D.L. and H.Z.), United States Department of Defense (W81XWH-13-10249, D.L.), W81XWH-13-1-0425 (D.L., J.B., B.A.G. and Y.K.), Sohn Conference Foundation (H.P. and H.Z.), the Children’s Cancer and Blood Foundation (H.P. and D.L.), The Manning Foundation (D.L.), The Hartwell Foundation (D.L.), The Nancy C. and Daniel P. Paduano Foundation (H.P. and D.L.), The Starr Cancer Consortium (H.P. and D.L.; D.L. and H.Z.), Pediatric Oncology Experimental Therapeutic Investigator Consortium (POETIC, D.L. and H.P.), James Paduano Foundation (D.L. and H.P.), National Institutes of Health/WCM CTSC (NIH/NCATS (UL1TR00457) (H.M. and H.Z.); NIH/NCATS (UL1TR002384) (D.L., H.M. and H.Z.);), Beth Tortolani Foundation (D.L. and J.B.), Charles and Marjorie Holloway Foundation (J.B.), Sussman Family Fund (J.B.), Lerner Foundation (J.B.), Breast Cancer Alliance (J.B.), Manhasset Women’s Coalition Against Breast Cancer (J.B.), National Institute on Minority Health and Health Disparities (NIMHD) of NIH (MD007599, H.M.), NIH/NCATS (UL1TR00457, H.M.). C.R., A.M., D.F., A.F., A.S. and H.O. acknowledge FEDER – Fundo Europeu de Desenvolvimento Regional funds through COMPETE 2020 – POCI, Portugal 2020 (NORTE-01-0145-FEDER-000029), and FCT – Fundação para a Ciência e a Tecnologia in the framework of the project “Institute for Research and Inovation in Health Sciences” (POCI-01-0145-FEDER-007274), and the FCT project POCI-01-0145-FEDER-016585 (PTDC/BBB-EBI/0567/2014). We acknowledge FCT for the grants to A.M. (SFRH/BPD/75871/2011) and A.F. (SFRH/BPD/111048/2015). D.F. acknowledges FCT (SFRH/BD/110636/2015), BiotechHealth PhD Programe (PD/0016/2012) and American Portuguese Biomedical Research Fund.
Footnotes
Author Contributions
H.Z. designed the experimental approach, performed the experimental work, analyzed the data, coordinated the project and wrote the manuscript. K.F., S.R., J.F. and H.M. performed zeta potential and stiffness measurement. C.R., D.F. A.M., J.A.F., A.S. and H.O. conducted the glycomics analysis. M.T.M. and H.M. performed and analyzed exosome mass spectrometry. Z.L. conducted and analyzed lipidomic mass spectrometry. H.S.K. conducted proteomic data analysis. H.C., L.B., A.B.M., M.N., A.P.M., P.S., W.B., H.W., A.M., G.G. and J.R.C. facilitated with exosome isolation/fractionation and other experimental work. J.P.J. and L.C performed electron microscopy. N.P., M.B. and K.M. performed atomic force microscopy. A.H., I.M. and C.K. contributed to manuscript reading, editing and providing feedback. P.G., A.M.C, J.B., R.S, H.P. and J.B. discussed the hypothesis and contributed to data interpretation and manuscript writing. M.S.B. contributed to studies involving human specimens. D.L. conceived the hypothesis, led the project, interpreted the data and wrote the manuscript.
Competing Financial Interests
The authors have no competing financial interests.
References
- 1.Thery C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nature reviews Immunology. 2002;2:569–579. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- 2.S ELA, Mager I, Breakefield XO, Wood MJ. Extracellular vesicles: biology and emerging therapeutic opportunities. Nature reviews Drug discovery. 2013;12:347–357. doi: 10.1038/nrd3978. [DOI] [PubMed] [Google Scholar]
- 3.Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. The Journal of cell biology. 2013;200:373–383. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balaj L, et al. Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nature communications. 2011;2:180. doi: 10.1038/ncomms1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choi DS, Kim DK, Kim YK, Gho YS. Proteomics, transcriptomics and lipidomics of exosomes and ectosomes. Proteomics. 2013;13:1554–1571. doi: 10.1002/pmic.201200329. [DOI] [PubMed] [Google Scholar]
- 6.Thakur BK, et al. Double-stranded DNA in exosomes: a novel biomarker in cancer detection. Cell research. 2014;24:766–769. doi: 10.1038/cr.2014.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tetta C, Ghigo E, Silengo L, Deregibus MC, Camussi G. Extracellular vesicles as an emerging mechanism of cell-to-cell communication. Endocrine. 2013;44:11–19. doi: 10.1007/s12020-012-9839-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fraunhofer W, Winter G. The use of asymmetrical flow field-flow fractionation in pharmaceutics and biopharmaceutics. European journal of pharmaceutics and biopharmaceutics. 2004;58:369–383. doi: 10.1016/j.ejpb.2004.03.034. [DOI] [PubMed] [Google Scholar]
- 9.Yohannes G, Jussila M, Hartonen K, Riekkola ML. Asymmetrical flow field-flow fractionation technique for separation and characterization of biopolymers and bioparticles. Journal of chromatography A. 2011;1218:4104–4116. doi: 10.1016/j.chroma.2010.12.110. [DOI] [PubMed] [Google Scholar]
- 10.Oh S, et al. Miniaturized asymmetrical flow field-flow fractionation: application to biological vesicles. Journal of separation science. 2007;30:1082–1087. doi: 10.1002/jssc.200600394. [DOI] [PubMed] [Google Scholar]
- 11.Sitar S, et al. Size characterization and quantification of exosomes by asymmetrical-flow field-flow fractionation. Analytical chemistry. 2015;87:9225–9233. doi: 10.1021/acs.analchem.5b01636. [DOI] [PubMed] [Google Scholar]
- 12.Petersen KE, et al. A review of exosome separation techniques and characterization of B16-F10 mouse melanoma exosomes with AF4-UV-MALS-DLS-TEM. Analytical and bioanalytical chemistry. 2014;406:7855–7866. doi: 10.1007/s00216-014-8040-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ashby J, et al. Distribution profiling of circulating microRNAs in serum. Analytical chemistry. 2014;86:9343–9349. doi: 10.1021/ac5028929. [DOI] [PubMed] [Google Scholar]
- 14.Agarwal K, et al. Analysis of exosome release as a cellular response to MAPK pathway inhibition. Langmuir. 2015;31:5440–5448. doi: 10.1021/acs.langmuir.5b00095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beningo KA, Wang YL. Fc-receptor-mediated phagocytosis is regulated by mechanical properties of the target. Journal of cell science. 2002;115:849–856. doi: 10.1242/jcs.115.4.849. [DOI] [PubMed] [Google Scholar]
- 16.Key J, et al. Soft Discoidal Polymeric Nanoconstructs Resist Macrophage Uptake and Enhance Vascular Targeting in Tumors. ACS nano. 2015;9:11628–11641. doi: 10.1021/acsnano.5b04866. [DOI] [PubMed] [Google Scholar]
- 17.Colombo M, Raposo G, Thery C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol. 2014;30:255–289. doi: 10.1146/annurev-cellbio-101512-122326. [DOI] [PubMed] [Google Scholar]
- 18.Hessvik NP, Llorente A. Current knowledge on exosome biogenesis and release. Cell Mol Life Sci. 2017 doi: 10.1007/s00018-017-2595-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Molinari M, Helenius A. Chaperone selection during glycoprotein translocation into the endoplasmic reticulum. Science. 2000;288:331–333. doi: 10.1126/science.288.5464.331. [DOI] [PubMed] [Google Scholar]
- 20.Fukuda MN, Masri KA, Dell A, Luzzatto L, Moremen KW. Incomplete synthesis of N-glycans in congenital dyserythropoietic anemia type II caused by a defect in the gene encoding alpha-mannosidase II. Proc Natl Acad Sci U S A. 1990;87:7443–7447. doi: 10.1073/pnas.87.19.7443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang WH, et al. An intrinsic mechanism of secreted protein aging and turnover. Proc Natl Acad Sci U S A. 2015;112:13657–13662. doi: 10.1073/pnas.1515464112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martiniuk F, Ellenbogen A, Hirschhorn R. Identity of neutral alpha-glucosidase AB and the glycoprotein processing enzyme glucosidase II. Biochemical and genetic studies. The Journal of biological chemistry. 1985;260:1238–1242. [PubMed] [Google Scholar]
- 23.Kowal J, et al. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc Natl Acad Sci U S A. 2016;113:E968–977. doi: 10.1073/pnas.1521230113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pinho SS, Reis CA. Glycosylation in cancer: mechanisms and clinical implications. Nature reviews Cancer. 2015;15:540–555. doi: 10.1038/nrc3982. [DOI] [PubMed] [Google Scholar]
- 25.Escrevente C, et al. Sialoglycoproteins and N-glycans from secreted exosomes of ovarian carcinoma cells. PloS one. 2013;8:e78631. doi: 10.1371/journal.pone.0078631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Batista BS, Eng WS, Pilobello KT, Hendricks-Munoz KD, Mahal LK. Identification of a conserved glycan signature for microvesicles. Journal of proteome research. 2011;10:4624–4633. doi: 10.1021/pr200434y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saraswat M, et al. N-linked (N-) glycoproteomics of urinary exosomes. Molecular & cellular proteomics. 2015;14:263–276. doi: 10.1074/mcp.M114.040345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thery C, Amigorena S, Raposo G, Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Current protocols in cell biology. 2006;Chapter 3(Unit 3):22. doi: 10.1002/0471143030.cb0322s30. [DOI] [PubMed] [Google Scholar]
- 29.Merchant ML, et al. Microfiltration isolation of human urinary exosomes for characterization by MS. Proteomics Clinical applications. 2010;4:84–96. doi: 10.1002/prca.200800093. [DOI] [PubMed] [Google Scholar]
- 30.Lasser C, Eldh M, Lotvall J. Isolation and characterization of RNA-containing exosomes. Journal of visualized experiments. 2012:e3037. doi: 10.3791/3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen C, et al. Microfluidic isolation and transcriptome analysis of serum microvesicles. Lab on a chip. 2010;10:505–511. doi: 10.1039/b916199f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jorgensen M, et al. Extracellular Vesicle (EV) Array: microarray capturing of exosomes and other extracellular vesicles for multiplexed phenotyping. Journal of extracellular vesicles. 2013;2 doi: 10.3402/jev.v2i0.20920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tauro BJ, et al. Comparison of ultracentrifugation, density gradient separation, and immunoaffinity capture methods for isolating human colon cancer cell line LIM1863-derived exosomes. Methods. 2012;56:293–304. doi: 10.1016/j.ymeth.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 34.Gardiner C, et al. Extracellular vesicles, tissue factor, cancer and thrombosis - discussion themes of the ISEV 2014 Educational Day. Journal of extracellular vesicles. 2015;4:26901. doi: 10.3402/jev.v4.26901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liang Y, et al. Complex N-linked glycans serve as a determinant for exosome/microvesicle cargo recruitment. The Journal of biological chemistry. 2014;289:32526–32537. doi: 10.1074/jbc.M114.606269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.White MJ, Roife D, Gomer RH. Galectin-3 Binding Protein Secreted by Breast Cancer Cells Inhibits Monocyte-Derived Fibrocyte Differentiation. Journal of immunology. 2015;195:1858–1867. doi: 10.4049/jimmunol.1500365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laubli H, et al. Lectin galactoside-binding soluble 3 binding protein (LGALS3BP) is a tumor-associated immunomodulatory ligand for CD33-related Siglecs. The Journal of biological chemistry. 2014;289:33481–33491. doi: 10.1074/jbc.M114.593129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hellstern S, et al. Functional studies on recombinant domains of Mac-2-binding protein. The Journal of biological chemistry. 2002;277:15690–15696. doi: 10.1074/jbc.M200386200. [DOI] [PubMed] [Google Scholar]
- 39.Sasaki T, Brakebusch C, Engel J, Timpl R. Mac-2 binding protein is a cell-adhesive protein of the extracellular matrix which self-assembles into ring-like structures and binds beta1 integrins, collagens and fibronectin. The EMBO journal. 1998;17:1606–1613. doi: 10.1093/emboj/17.6.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Meer G, Voelker DR, Feigenson GW. Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


