Abstract
Osteoclasts are unique bone-resorbing cells that differentiate from the monocyte/macrophage lineage of bone marrow. Dysfunction of osteoclasts may result in a series of bone metabolic diseases, including osteoporosis. To develop pharmaceutical targets for the prevention of pathological bone mass loss, the mechanisms by which osteoclasts differentiate from precursors must be understood. The ability to isolate and culture a large number of osteoclasts in vitro is critical in order to determine the role of specific genes in osteoclast differentiation. Inactivation of the mammalian/mechanistic target of rapamycin complex 1 (TORC1) in osteoclasts can decrease osteoclast number and increase bone mass; however, the underlying mechanisms require further study. In the present study, a RANKL-based protocol to isolate and culture osteoclasts from mouse bone marrow and to study the influence of mTORC1 inactivation on osteoclast formation is described. This protocol successfully resulted in a large number of giant osteoclasts, typically within one week. Deletion of Raptor impaired osteoclast formation and decreased the activity of secretory tartrate-resistant acid phosphatase, indicating that mTORC1 is critical for osteoclast formation.
Keywords: Developmental Biology, Issue 133, Osteoclast, mTORC1, Raptor, bone remolding, cell biology, RANKL, M-CSF
Introduction
Bone is an ever-changing organ and is remodeled by osteoblasts and osteoclasts throughout life. Osteoclasts are responsible for mineralized matrix resorption and osteoblasts synthesize and secrete new bone matrices1. The balance between bone resorption and bone formation is crucial for bone health including maintenance of bone mass and response to stimulation and injury. If this balance is disrupted, a series of bone metabolic diseases may occur, including osteoporosis and periodontal diseases. In these diseases, bone mass loss resulting from osteoclastic bone resorption exceeds the bone forming capacity of osteoblasts2,3. Thus, in order to develop pharmaceutical targets to treat skeletal disorders such as osteoporosis, it is critical to understand the generation and biology of osteoclasts4.
Osteoclasts are unique giant multinucleated cells located at or near the bone surface, and belong to the monocyte/macrophage family1. Ibbotson K. J. et al. reported a method to generate osteoclast-like cells in vitro with medium containing 1,25-dihydroxy-vitamin D35. The identification of macrophage-colony stimulating factor (M-CSF) and receptor activator for nuclear factor-κ B ligand (RANKL) as essential factors of osteoclast formation has dramatically increased the efficiency of osteoclastogenesis in vitro1,6,7. The ability to culture osteoclasts in vitro has improved our understanding of the generation and regulation of osteoclasts.
The mammalian/mechanistic target of rapamycin (mTOR) functions in two structurally and functionally distinct complexes, namely mTORC1 and mTORC28,9. The two multi-protein complexes are distinct from each other due to their different components and downstream substrates. mTORC1 contains the unique regulatory-associated protein of mTOR (Raptor), while mTORC2 contains the rapamycin-insensitive companion of mTOR (Rictor)9. mTORC1 can integrate and transmit important signals to regulate cell growth, proliferation and differentiation. Recently, we demonstrated that mTORC1 plays a key role in the network of catabolic bone resorption by deletion of Raptor to inactivate mTORC1 in osteoclasts10. However, the underlying mechanisms require further study. In the present study, a RANKL-based osteoclastogenic method was used to generate osteoclasts from bone marrow-derived macrophages (BMMs) of wild-type (WT) and RapCtsk mice, and to study the influence of mTORC1 inactivation on osteoclast formation.
Protocol
All procedures relating to the animals were performed according to the protocol approved by the Stanford Administrative Panel on Laboratory Animal Care (APLAC) and were approved by the Animal Care and Use Committee of the Shanghai Institute of Biochemistry and Cell Biology.
1. Preparation
Generate osteoclast specific Raptor deletion mice (Raptorfl/fl; Ctsk-cre, hereafter RapCtsk) by mating Raptorfl/fl mice with Ctsk-cre mice. Use the Raptorfl/fl mice as the WT control in this study.
Have a container with ice to keep isolated bone.
- Prepare culture medium.
- Prepare α-MEM culture medium by supplementing minimum essential medium alpha (α-MEM) with 1x glutamine, penicillin-streptomycin, and 10% fetal calf serum.
- Prepare bone marrow macrophage induction medium consisting of 50 mL of α-MEM medium and M-CSF at 20 ng/mL.
- Prepare osteoclast induction medium consisting of M-CSF and RANKL at 20 ng/mL, respectively, in 50 mL of α-MEM medium.
- Prepare the following buffers before TRAP staining.
- Prepare fix solution consisting of 6.5 mL of acetone, 2.5 mL of citrate solution and 0.8 mL of 37% (vol/vol) formaldehyde solution.
- Prepare TRAP staining solution.
- Prewarm deionized water to 37 °C.
- Add 100 µL of fast garnet GBC base solution and 100 µL of sodium nitrite solution into a 1.5-mL microtube and mix by gentle inversion for 30 s. Leave the mixture to stand for 2 min.
- Prewarm 9 mL of deionized water to 37 °C. Add 200 µL of diazotized fast garnet GBC base solution from step 1.4.2.2, 100 µL of naphthol AS-BI phosphate solution, 400 µL of acetate solution, and 200 µL of tartrate solution.
- Keep the TRAP staining mixture solution in a water bath at 37 °C.
- Prepare the following buffer before TRAP activity quantification of culture medium.
- Prepare tartrate buffer (50 mL): 2 mL of 0.33 M L-tartaric acid, 48 mL of 0.33 M sodium tartrate dibasic dehydrate. Adjust the pH value to 4.9.
- Prepare 4-Nitrophenyl phosphate disodium (pNPP) buffer (200 mL): 1.502 g of glycine, 41 mg of MgCl2, 27.2 mg of ZnCl2, and 180 mL of deionized water. Adjust the pH value to 10.4 with 3 M NaOH and the volume to 200 mL with deionized water.
- Prepare pNPP buffer with phosphatase substrate (for one 96-well plate): 49.37 mg of phosphatase substrate and 175 µL of pNPP buffer of step 1.5.2.
- Prepare substrate buffer (9 mL/96-well plate): 6.24 mL of deionized water, 360 μL of acetate solution, and 2.4 mL of tartrate buffer. Mix by vortex and preheat at 37 °C before use.
- Add 90 μL of pNPP buffer with phosphatase substrate(1.5.3) to 9ml of preheated substrate buffer(1.5.4) to make a substrate mixture and vortex for 10 s.
2. Dissection (Day -1)
Euthanize 3 one-month-old female WT and RapCtsk mice by CO2 separately. Perform CO2 inhalation with appropriate equipment by trained personnel.
Immerse 6 mice in a beaker with 100 mL of 75% ethanol (vol/vol) for 5 min to prevent bacterial contamination and then place on a dissection board in a supine position. Make an incision at the distal of the tibia vertically and dissect the skin along the hind limb with ophthalmic scissors.
Cut the articular ligament of the hip joint with scissors and dislocate the hind limbs from the trunk.
Cut the articular ligament of the knee joint and carefully disassociate the tibia and femur from the knee joint. Gently remove the soft tissue with scissors.
Place all bones of one genotype in of one mouse in each well of a six-well-plate with 2 mL of α-MEM on ice. NOTE: To preserve cell viability, the bone should be kept in these conditions for less than 1 h.
3. Isolation (Day -1)
Fill one well of a six-well plate with 75% ethanol (3 mL) and the other 5 wells with α-MEM medium (3 mL).
Put all bones of one genotype in the ethanol wash for 15 s and wash bones 5 times with α-MEM medium for 10 s each time.
Cut off the epiphyses with the scissor and insert a 0.45-mm syringe needle into the bone cavity and flush marrow out with α-MEM medium into a 50-mL centrifuge tube.
Flush the bone cavity using the same method from the other end of the bone. Repeat at least twice to wash the bone cavity thoroughly until the bone is pale.
Centrifuge the bone marrow to obtain a cell pellet at 800 x g and 4 °C for 5 min. Aspirate the supernatant.
Add 3 mL of red blood cell lysis buffer into the centrifuge tube and pipette gently to resuspend the bone marrow. Keep the centrifuge tube on ice for 8 min to lyse red blood cells.
Add 6 mL of α-MEM medium into the centrifuge tube to stop cell lysis. Centrifuge at 500 x g and 4 °C for 10 min and aspirate the supernatant.
Resuspend in 3 mL of α-MEM culture medium and place the cells into a six-well plate (generally one mouse per well).
Incubate at 37 °C in a 5% CO2 incubator overnight.
4. Plating (Day 0)
Transfer the supernatant to a 15 mL centrifuge tube to collect the unattached cells the next morning.
Centrifuge at 800 x g and 4 °C for 5 min and aspirate the supernatant.
Resuspend in 4 mL of α-MEM culture medium.
Take 20 µL of the cell solution and mix with 20 µL of Trypan Blue. Add 10 µL of the mixture to the counting chamber and obtain the cell count per mL of solution.
Add an appropriate volume of α-MEM culture medium to obtain a cell solution of 500,000 cells/mL.
Add 500 µL and 50 µL of bone marrow macrophage induction medium into each well of a 24-well plate and 96-well plate, respectively.
Add 500 µL and 50 µL of cell solution into each well of a 24-well plate and 96-well plate, respectively.
Incubate at 37 °C in a 5% CO2 incubator for 3 days.
5. Differentiation (Days 3-7)
Three days after plating, collect 50 µL medium from each well of the 96-well-plate and freeze at -20 °C, and then aspirate the residual medium.
Add 1 mL and 100 µL osteoclast induction medium into each well of a 24-well plate and a 96-well plate, respectively.
Incubate at 37 °C for 2 days.
Collect 50 µL of medium from each well and aspirate the residual medium.
Add osteoclast induction medium into each well.
Incubate at 37 °C for 1 day. NOTE: At this time point, large, multinucleated osteoclasts should be seen under an inverted microscope (Figure 2A).
If osteoclasts are not seen, change the osteoclast induction medium and observe daily until osteoclasts are seen.
Collect 50 µL of medium from each well of the 96-well plate after osteoclasts have formed.
6. Tartrate Resistant Acid Phosphatase (TRAP) Staining
NOTE: Mature osteoclasts may be present after 6-7 days of in vitro culture (step 4).
Aspirate medium and wash gently 3 times with 1x PBS.
Fix the cells in each well of the 96-well plate with 100 µL of fix solution for 30 s at room temperature.
After fixation, aspirate the fix solution and wash gently 3 times with deionized water prewarmed to 37 °C. Aspirate deionized water.
Add 100 µL of TRAP stain into each well of the 96-well plate and place the plate in an incubator at 37 °C for 30 min and shield from light.
After staining, aspirate the TRAP stain and gently wash 3 times with deionized water.
Image osteoclasts using an inverted microscope at 40X magnification.
7. TRAP Activity Quantification of Culture Medium
Collect 30 µL culture supernatant from each well of the 96-well plate from step 5.1, 5.4 and 5.8; Use 30 µL of non-cultured osteoclast induction medium as the negative control. Place the samples in a new 96-well plate.
Add 90 μL of substrate mixture(1.5.5) into each well of the 96-well plate.
Place the plate in an incubator at 37 °C for 1 h and shield from light.
Stop the reaction by adding 50 µL of 3 M NaOH into each well of the 96-well plate.
Measure theabsorbance at the wavelength of 405 nm.
8. Western Blotting
NOTE: After 6-7 days of in vitro culture in the 24-well plate, mature osteoclasts should be visible.
Aspirate the medium.
Wash gently with pre-cooled 1x PBS 3 times.
Aspirate PBS.
Add 100 µL of 1x SDS lysis buffer containing protease inhibitor cocktail.
Heat lysates at 105 °C for 10 min and centrifuge at 12,000 × g and 4 °C for 10 min. Collect the supernatants containing proteins.
Separate the lysates containing 30 µg of protein by 10% SDS-PAGE11 and then transfer to a polyvinylidene fluoride (PVDF) membrane.
Block the membranes with 5% nonfat milk for 30 min.
Incubate the membranes with anti-Raptor, anti-P-S6, anti-S6, anti-β-actin at 1:1,000 dilution in 5% nonfat milk at 4 °C overnight.
Wash the membranes with Tris Buffered Saline with Tween-20 (TBST) for 10 min at room temperature.
Repeat step 8.9 twice.
Incubate the membranes with horseradish peroxidase (HRP)-conjugated anti-mouse or rabbit IgG antibodies at 1:5,000 dilution in 5% nonfat milk for 1 h at room temperature.
Wash the membranes 3 times with TBST for 10 min at room temperature.
For chemiluminescent detection, add western chemiluminescent HRP substrate on the membranes and incubate for 5 min at room temperature. Drain the excess substrate and expose the blots to X-ray film.
Representative Results
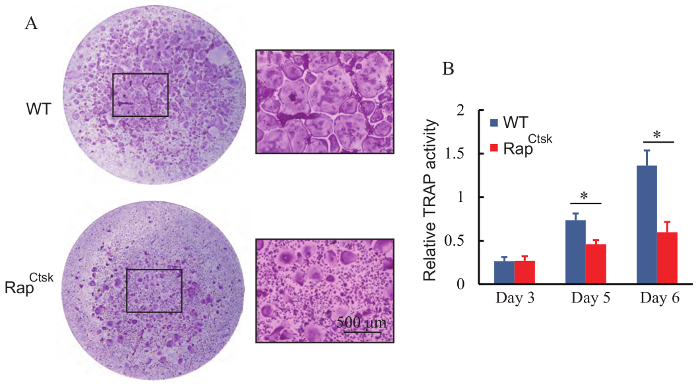
Using the present protocol, a large number of giant osteoclasts were seen on day 6; if giant osteoclasts are not seen, one more day of osteoclast differentiation may be needed (Figure 1). Successful osteoclast formation was confirmed by TRAP staining (Figure 2A). Osteoclasts were giant wine red/purple cells with more than 3 nuclei. More than 250 osteoclasts were obtained in each well of the 96-well plate in WT BMMs (Figure 4A).
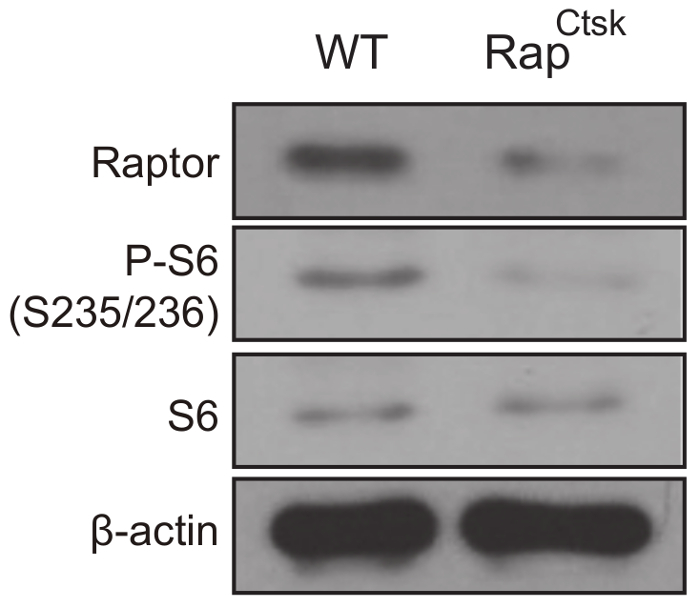
Deletion of Raptor was confirmed by western blotting analysis and the inactivation of mTORC1 was determined by a decrease in ribosomal protein S6 phosphorylation (P-S6), which is widely used as the readout for mTORC1 (Figure 3).
The number of TRAP-positive osteoclasts decreased in RapCtsk BMMs compared with the WT group (Figure 4A), which indicated that Raptor deficiency impaired osteoclast formation.
The activity of secretory TRAP is important for bone resorption, and TRAP activity in the culture medium was analyzed in the present study. As shown in Figure 4B, TRAP activity increased due to stimulation of RANKL in both WT and RapCtsk BMMs. In addition, TRAP activity decreased in RapCtskBMMs compared with the corresponding WT group (P <0.05).
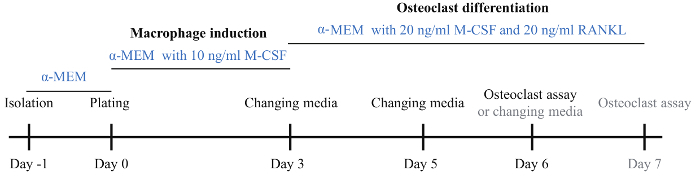
Figure 1: Schematic diagram of the osteoclast induction protocol. BMMs were isolated on day 1 and incubated with basic α-MEM medium overnight. On day 0, BMMs were collected and plated onto a 96-well plate and 24-well plate followed by incubation in α-MEM medium with 10 ng/mL M-CSF. On day 3, the medium was changed to α-MEM medium with 20 ng/mL M-CSF and 20 ng/mL RANKL for induction of osteoclast differentiation. On day 5, the osteoclast differentiation medium was changed. On day 6, a large number of osteoclasts were visible and osteoclast assays were performed. If osteoblasts were not visible, the osteoclast induction medium was changed and incubation continued for one more day. Please click here to view a larger version of this figure.
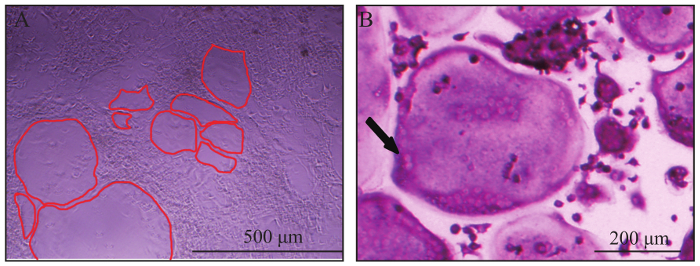
Figure 2: Representative view of osteoclasts. (A) Giant osteoclasts observed in bright field on day 6. The red line outlined the border of osteoclasts. (B) A typically giant, multinucleated, TRAP positive (wine red) osteoclast. The black arrow indicated two nuclei of the multinucleated osteoclast. Please click here to view a larger version of this figure.
Figure 3: mTORC1 inactivation in RapCtsk BMMs. Western blotting analysis of Raptor, P-S6 and S6 expression in WT and RapCtsk BMMs on day 6. Please click here to view a larger version of this figure.
Figure 4: Deletion of Raptor impaired osteoclast formation. (A) TRAP staining of BMMs on day 6. High magnification images of the squares in WT and RapCtsk groups are shown, respectively. There were less giant, TRAP positive osteoclasts formed in the RapCtsk BMMs in comparison with WT group. (B) TRAP activity of cultured WT and RapCtsk BMMs. Data represent means ± S.D.* P <0.05, n = 3. Please click here to view a larger version of this figure.
Discussion
The osteoclastogenic assay is the most widely used method to isolate and culture osteoclasts in vitro12,13. While several RANKL-based osteoclast inductions have been described13,14,15, the present study described a protocol with some modifications based on the previous methods.
In the previous study, BMMs were plating immediately after isolation14,15. We recommended that BMMs were cultured with basic medium overnight before plating to isolate BMMs from mesenchymal stem cells and fibroblasts which immediately adhere to the dish. M-CSF can promote the proliferation of monocytes to osteoclast precursors and stimulate osteoclast precursors to express RANK that binds RANKL and then induces the formation of mature osteoclasts6,16,17. Therefore, in our study, the BMMs were induced with 10 ng/mL M-CSF for 3 days before osteoclast induction, instead for osteoclast induction on the second day after plating15. The cell density is critical for osteoclast formation in vitro. It is recommended that 25,000 BMMs were seeded in each well of a 96-well plate following by 3 days incubation with 10 ng/mL M-CSF. When the cell confluence reached 80–90%, it is a suitable time for osteoclast induction of RANKL. Usually, mature osteoclasts form on day 6. The most common reason for few osteoclasts is suboptimal cell density, which can be the result of a low seeding density or low cell viability. BMMs are delicate primary cells and need to be treated gently. Keeping the bone on ice for a long time or resuspending vigorously decreases the viability of BMMs and subsequently impairs the formation of osteoclasts. Thus, the optimal seeding density of viable BMMs is critical for obtaining a large number of osteoclasts.
The activity of secretory TRAP is critical for osteoclastic bone resorption. In this study, we describe a method to quantitatively analyze secretory TRAP activity in the culture medium. The change of TRAP activity in the culture medium can result from changes of osteoclast number or secretory TRAP of osteoclast or a combination of both. Thus, this may be a useful method for analyzing osteoclast function in vitro together with the osteoclast resorption assay described previously18.
mTOR is an evolutionarily-conserved protein kinase that can regulate cell metabolism and differentiation9. In this study, we found that mTORC1 played an important role in bone resorption by regulating osteoclast formation. mTORC1 may be a potential pharmacological target for the treatment of bone metabolic disorders such as osteoporosis in the future.
In summary, our protocol for osteoclast induction successfully resulted in a large number of giant osteoclasts in vitro and our data suggest that mTORC1 is critical for osteoclast formation.
Disclosures
All authors state that they have no conflicts of interest.
Acknowledgments
The authors thank Dr. Minghan Tong and S. Kato for kindly providing reagents and mice. We thank the members of the Zou lab for useful discussions. This work was supported in part by grants from 973 Program from the Chinese Ministry of Science and Technology (MOST) [2014CB964704 and 2015CB964503], Clinical Research Program of 9th People's Hospital, Shanghai Jiao Tong University School of Medicine. Thanks for the help of Core Facility for Cell Biology and Core Facility for Chemical Biology, CAS Center for Excellence in Molecular Cell Science, Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences.
References
- Boyle WJ, Simonet WS, Lacey DL. Osteoclast differentiation and activation. Nature. 2003;423(6937):337–342. doi: 10.1038/nature01658. [DOI] [PubMed] [Google Scholar]
- Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nature Genetics. 2003;33:245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- Feng X, McDonald JM. Disorders of bone remodeling. Annu Rev Pathol. 2011;6:121–145. doi: 10.1146/annurev-pathol-011110-130203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyce BF. Advances in osteoclast biology reveal potential new drug targets and new roles for osteoclasts. J Bone Miner Res. 2013;28(4):711–722. doi: 10.1002/jbmr.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibbotson KJ, Roodman GD, McManus LM, Mundy GR. Identification and characterization of osteoclast-like cells and their progenitors in cultures of feline marrow mononuclear cells. J Cell Biol. 1984;99(2):471–480. doi: 10.1083/jcb.99.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacey DL, et al. Osteoprotegerin ligand is a cytokine that regulates osteoclast differentiation and activation. Cell. 1998;93(2):165–176. doi: 10.1016/s0092-8674(00)81569-x. [DOI] [PubMed] [Google Scholar]
- Wong BR, et al. TRANCE is a novel ligand of the tumor necrosis factor receptor family that activates c-Jun N-terminal kinase in T cells. J Biol Chem. 1997;272(40):25190–25194. doi: 10.1074/jbc.272.40.25190. [DOI] [PubMed] [Google Scholar]
- Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 2011;12(1):21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaskar PT, Hay N. The two TORCs and Akt. Dev Cell. 2007;12(4):487–502. doi: 10.1016/j.devcel.2007.03.020. [DOI] [PubMed] [Google Scholar]
- Dai Q, et al. Inactivation of Regulatory-associated Protein of mTOR (Raptor)/Mammalian Target of Rapamycin Complex 1 (mTORC1) Signaling in Osteoclasts Increases Bone Mass by Inhibiting Osteoclast Differentiation in Mice. J Biol Chem. 2017;292(1):196–204. doi: 10.1074/jbc.M116.764761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. Cold spring harbor laboratory press; 1989. [Google Scholar]
- Weischenfeldt J, Porse B. Bone Marrow-Derived Macrophages (BMM): Isolation and Applications. CSH Protoc. 2008;2008 doi: 10.1101/pdb.prot5080. [DOI] [PubMed] [Google Scholar]
- Bradley EW, Oursler MJ. Osteoclast culture and resorption assays. Methods Mol Biol. 2008;455:19–35. doi: 10.1007/978-1-59745-104-8_2. [DOI] [PubMed] [Google Scholar]
- Tevlin R, et al. Osteoclast derivation from mouse bone marrow. J Vis Exp. 2014. p. e52056. [DOI] [PMC free article] [PubMed]
- Xing L, Boyce BF. RANKL-based osteoclastogenic assays from murine bone marrow cells. Methods Mol Biol. 2014;1130:307–313. doi: 10.1007/978-1-62703-989-5_23. [DOI] [PubMed] [Google Scholar]
- Hsu H, et al. Tumor necrosis factor receptor family member RANK mediates osteoclast differentiation and activation induced by osteoprotegerin ligand. Proc Natl Acad Sci U S A. 1999;96(7):3540–3545. doi: 10.1073/pnas.96.7.3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underwood JC. From where comes the osteoclast? J Pathol. 1984;144(4):225–226. doi: 10.1002/path.1711440402. [DOI] [PubMed] [Google Scholar]
- Wein MN, et al. Control of bone resorption in mice by Schnurri-3. Proc Natl Acad Sci U S A. 2012;109(21):8173–8178. doi: 10.1073/pnas.1205848109. [DOI] [PMC free article] [PubMed] [Google Scholar]


