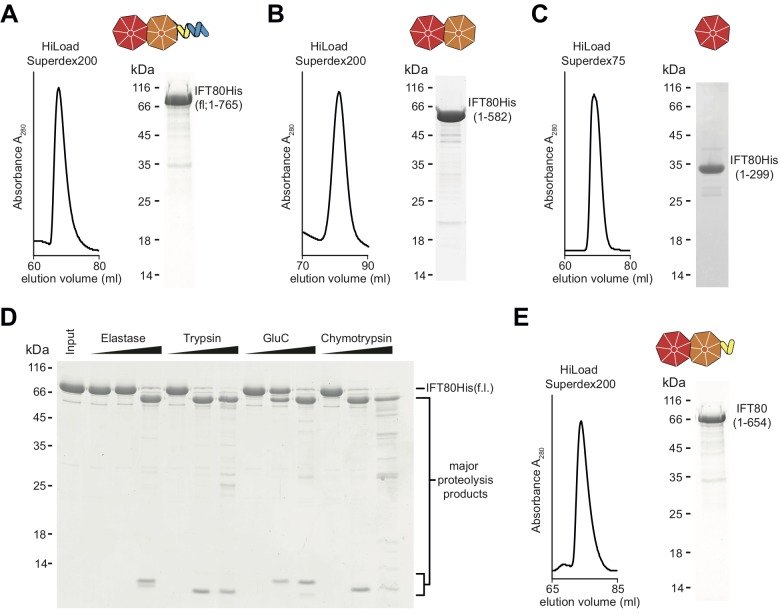
Figure 1. Purification of CrIFT80 constructs.
(A–C) SEC profiles (left) and corresponding Coomassie-stained SDS-PAGE pictures of CrIFT80 fragments designed based on domain prediction. A schematic representation of the proteins is shown above the gel picture for full-length CrIFT80 (residues 1–765) (A), CrIFT80 BP1-BP2 (residues 1–582) (B), and CrIFT80 BP1 (residues 1–299) (C). (D) Coomassie-stained SDS-PAGE gel showing the limited proteolysis results for full-length CrIFT80 treated with the proteases Elastase, Trypsin, GluC, and Chymotrypsin. The positions of the full-length protein as well as for the major proteolysis products are indicated. All proteases lead to the cleavage of the protein into stable fragments of similar sizes. (E) Large-scale proteolysis of CrIFT80 with the protease GluC followed by SEC (profile on the left) led to the identification by mass-spectrometry of a fragment encompassing residues 1–654. The Coomassie-stained SDS-PAGE gel on the right shows the purity of this product, and the schematic representation on top shows that it is composed of BP1 and BP2 followed by a short stretch of alpha-solenoid structure.