Abstract
A calcium-pectate-binding anionic isoperoxidase (APRX) from zucchini (Cucurbita pepo) was purified and subjected to N-terminal amino acid microsequencing. The cDNA encoding this enzyme was obtained by reverse transcriptase polymerase chain reaction from a cDNA library. It encoded a mature protein of 309 amino acids exhibiting all of the sequence characteristics of a plant peroxidase. Despite the presence of a C-terminal propeptide, APRX was found in the apoplast. APRX protein and mRNA were found in the root, hypocotyls, and cotyledons. In situ hybridization showed that the APRX-encoding gene was expressed in many different tissues. The strongest expression was observed in root epidermis and in some cells of the stele, in differentiating tracheary elements of hypocotyl, in the lower and upper epidermis, in the palisade parenchyma of cotyledons, and in lateral and adventitious root primordia. In the hypocotyl hook there was an asymmetric expression, with the inner part containing more transcripts than the outer part. Treatment with 2,3,5-triiodobenzoic acid reduced the expression of the APRX-encoding gene in the lower part of the hypocotyl. Our observations suggest that APRX could be involved in lignin formation and that the transcription of its gene was related to auxin level.
Plant peroxidases (EC 1.11.1.7) exist in numerous molecular forms. For example, more than 50 different sequences encoding peroxidase have been identified in Arabidopsis. These enzymes are mainly located in cell walls and vacuoles (Mäder, 1992) and catalyze the reduction of hydrogen peroxide into water using electrons from various donor molecules. This redox activity allows them to oxidize many substances, such as polyphenols, flavonoids (Gaspar et al., 1982), and alkaloids (Blom et al., 1991), or to promote the oxidative cross-linking of cell wall polymers (Fry, 1986). Their substrate specificity is generally considered to be low, except toward some electron donors such as scopoletin (Reigh et al., 1973) and extensin (Brownleader et al., 1995), which are oxidized by specific isoforms. In some cases, they can also oxidize molecules such as IAA (Gazaryan et al., 1996) through catalytic mechanisms that differ from the classical peroxidative cycle. They have also been shown to produce hydrogen peroxide in the presence of reducers such as NADH (Elstner and Heupel, 1976) and Cys (Pichorner et al., 1992).
The wide spectrum of biochemical reactions that peroxidases are able to catalyze and the great number of molecular isoforms explain the difficulty encountered in the study of plant peroxidases. Studies combining the techniques of molecular biology and biochemistry are necessary to get an insight into their precise function in plants. It is also essential to identify the mechanisms that determine their microlocalization in cells, particularly within the extracellular matrix network. Most of the known peroxidase mRNAs encode a signal peptide that targets the neosynthesized protein to the secretory pathway. Some sequences also encode a C-terminal extension thought to direct the protein to the vacuole (Welinder, 1992; Neuhaus et al., 1994).
All peroxidase molecules can diffuse randomly throughout the cell wall. Once they have been released out of the cell by exocytosis, it is possible that some of them are targeted to specific microdomains where they fulfill their catalytic function. This implies that certain peroxidase isoforms exhibit an affinity for particular polymers of the extracellular matrix acting as docking structures. Such a specific interaction has been observed between isoperoxidases from zucchini (Cucurbita pepo) (Penel and Greppin, 1994) and horseradish (Penel et al., 1996) and the pectins in their calcium-induced conformation. In zucchini one anionic and three cationic isoperoxidases have been shown to exhibit this particular binding property (Penel and Greppin, 1996), which has been shown to be dependent on the presence of cationic amino acid residues exposed at the surface of the protein (Penel and Greppin, 1996). In situ binding experiments done on hypocotyl cross-sections have demonstrated that the APRX binds in a calcium-dependent manner to the cell wall of several tissues, with the strongest affinity being exhibited by the cell walls of the epidermis (Penel et al., 1996).
Its particular binding property makes this APRX especially interesting for a molecular study. In this work we have purified and microsequenced APRX, cloned its mRNA, and sequenced the resulting full-length cDNA. To obtain information on the location of this isoperoxidase in zucchini seedlings, we have detected its transcripts by in situ hybridization on sections made from roots, hypocotyls, and cotyledons. The observed expression patterns suggest a role for this peroxidase isoform in xylem differentiation and relationships with auxin.
MATERIALS AND METHODS
Plant Material
Zucchini (Cucurbita pepo L. cv Black Beauty) seedlings were grown on moist, absorbent paper at 25°C in darkness. Four-day-old seedlings were harvested for protein and RNA extractions and for in situ hybridization.
Adventitious root formation was studied using 6-cm-tall etiolated seedlings whose root systems had been cut off. The seedlings were placed in beakers with their cut ends in nutrient solution (1% Sinesol, Iriland, Geneva, Switzerland) with or without 10−4 m TIBA and placed in the light at 20°C. Under these conditions the first adventitious roots appeared 4 d after root excision. Two-centimeter segments were taken each day at the bottom of the hypocotyls for RNA extraction.
Purification and Microsequencing of APRX
APRX was extracted and purified as described by Penel and Greppin (1996). The purified protein was separated by two-dimensional PAGE according to the method of O'Farrell (1978) and electroblotted onto a PVDF membrane (Bio-Rad). The electroblotted protein was incubated with pyroglutamate aminopeptidase according to the method of LeGendre et al. (1993) to remove the pyroglutamyl group from the blocked N terminus. The N-terminal sequence was determined by automated Edman degradation using a DNA sequencer (model 373A, Applied Biosystems, Foster City, CA).
cDNA Amplification, Cloning, and Sequencing of APRX cDNA
Total RNA was isolated from etiolated zucchini hypocotyls with a phenol-SDS procedure (Dean et al., 1985). Poly(A+) RNAs were isolated with oligo(dT)-cellulose (Redi-Col, Pharmacia). The cDNA library was synthesized by RT-PCR with a cDNA amplification kit (Marathon, CLONTECH, Basel, Switzerland), which added a specific sequence (adaptor) to the 5′ and 3′ ends of each cDNA.
For PCR amplification of the cDNA encoding APRX, two degenerate oligonucleotide primers were synthesized (Microsynth, Balgach, Switzerland) whose nucleotide sequences were designed to be complementary to the coding strand for the peptide sequences FYDQTCP and TCPRLPNIV present in the N-terminal sequence of APRX. The sequence of primer 1 (5′-TTYTAYGAYCARACITGYCC-3′; where I is inosine, R is A/G, and Y is C/T) corresponded to the sense orientation of FYDQTCP; the sequence of primer 2 (5′-ACITGYCCIMGNYTICCIA AYATHGT-3′; where I and Y are as above, and H is A/T/C, M is A/C, and N is A/C/G/T) corresponded to the sense orientation of TCPRLPNIV. The first PCR amplification was performed with 0.2 μm primer 1 and with 0.2 μm AP1 primer (5′-CCATCCTAATACGACTCAC TATAGGGC-3′ situated in the adaptor added with the cDNA amplification kit) using the cDNA polymerase (Advantage, CLONTECH). The PCR procedure started with 1 min of denaturation at 94°C and was carried on with 30 cycles of 30 s of denaturation at 94°C, 30 s of annealing at 54°C, and 3 min of extension at 68°C. A nested PCR was performed with 0.2 μm primer 2 and 0.16 μm primer 3 (5′-TTCTAGAATTCAGCGGCCGC[T]30N1N-3′, where N1 is A/C/G and N is theA/C/G/T contained in the adaptor of the cDNA amplification kit) using the same PCR cycles as above, except that the annealing temperature was higher (64°C) and a final step at 68°C for 4 min was added. The nucleotide sequence of the 5′-untranslated leader of APRX was obtained by a 5′-rapid amplification of cDNA ends (RACE) PCR procedure with the same cDNA amplification kit using primer 4 (5′-CGATGCCGGGAGCATCCTCTAGC-3′) and primer AP1.
The PCR products were cloned into vector (pGEM-T Easy, Promega), and the resulting ligation product was transformed into Escherichia coli strain XL-1-Blue (Stratagene) according to the method of Sambrook et al. (1989). The dideoxy method was used for sequencing the double-stranded DNA using T7 Sequenase (version 2.0, Amersham).
RNA and DNA Extraction and Hybridization
Total RNA was extracted from 4-d-old etiolated seedlings (Dean et al., 1985) and DNA from 2-week-old plant leaves according to the method of Rogers and Bendich (1994). Northern and Southern analyses were performed according to the method of Sambrook et al. (1989). After blotting onto membranes (Hybond-N, Amersham), RNA and DNA were hybridized using as a probe the clone obtained by RT-PCR labeled with DNA- or RNA-labeling kits (DIG, Boehringer Mannheim). For control of RNA loading, membranes were rehybridized with a tobacco cDNA probe complementary to 18S rRNA.
In Situ Hybridization
In situ hybridization was performed following the procedure described below, which was modified from that of Komminoth (1996). A 300-bp untranslated 3′-end APRX-specific probe was obtained from the RT-PCR clone with PstI. This 300-bp clone was linearized with PstI to produce the antisense probe, and with EcoRI to produce the sense probe. Digoxigenin-labeled sense and antisense specific probes were synthesized according to the instructions of manufacturer. For the preparation of paraffin sections, pieces from different parts of seedlings were fixed with 3% formaldehyde and 0.25% glutaraldehyde in 100 mm phosphate buffer, pH 7.2, at 4°C overnight. After fixation, the samples were rinsed with the same phosphate buffer, dehydrated in a graded-alcohol series, and embedded in paraffin. Sections (10 μm) were attached on poly-l-Lys-coated slides (Polysine, Menzel-Glaser, Germany).
The sections were deparaffinized by soaking the slides in 100% xylene followed by several washes in xylene:ethanol (50:50, v/v), rehydrated by transferring them through a graded ethanol series (90%, 70%, 50%, and 25%), and briefly rinsed with DEPC-treated water. The sections were then incubated for 30 min at 37°C with 1 μg/mL proteinase K in 100 mm Tris/HCl, pH 7.5, containing 5 mm EDTA. The proteinase K was removed by rinsing three times with DEPC-treated water. The sections were then treated for 10 min at room temperature with 100 mm triethanolamine, pH 8.0, containing 0.25% acetic anhydride, followed by 5 min in 2× SSC, and twice for 5 min in DEPC-treated water. Finally, the sections were dehydrated through an ethanol series ranging from 25% to 100% ethanol, dried, and prehybridized for 2 h at 44°C in 4× SSC, 50% formamide, 1× Denhardt's solution, 5% dextran sulfate, 1 μg/mL salmon-sperm DNA, and 0.25 μg/mL yeast tRNA, and hybridized overnight in the same buffer containing 10 ng/μL sense or antisense probes.
After hybridization the slides were washed at 50°C with 2× SSC twice for 45 min, with 1× SSC for 45 min, and with 0.5× SSC for 15 min. Subsequently, an RNaseA treatment (0.5 μg/mL in 500 mm NaCl, 10 mm Tris/HCl, pH 7.5, and 1 mm EDTA) was performed at room temperature for 30 min followed by a 15-min rinse in 0.5× SSC, then with HIS 1 buffer (100 mm Tris/HCl, pH 7.5, and 150 mm NaCl). After blocking for 1 h in 2% BSA (w/v), 0.3% Triton X-100 in 100 mm Tris/HCl, pH 7.5, and 150 mm NaCl, the sections were incubated with antidigoxygenin antibody-alkaline phosphatase conjugate (dilution 1:500, Boehringer Mannheim) at room temperature for 1 h, and washed two times in HIS 1 buffer for 10 min. They were then stained with nitroblue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate according to the instructions of the manufacturer. The reaction was stopped by placing the slides into 10 mm Tris-HCl, pH 7.5, containing 1 mm EDTA. For each tissue, sections were incubated with an antisense probe to detect the APRX transcripts and with a sense probe as a control.
Separation of Isoperoxidases by IEF
Soluble proteins were extracted from roots, hypocotyls, and cotyledons by grinding in 20 mm Hepes, pH 7.0, containing 1 mm EGTA (1 mL for each 1 g fresh weight). The extract was filtrated and centrifuged for 10 min at 10,000g. Proteins were assayed with Coomassie Blue reagent (Spector, 1978). Extracellular proteins were obtained by the following procedure. Segments of 1 cm taken throughout hypocotyls were placed in 20 mm Hepes, pH 7.0, containing 5 mm EGTA or 2 mm CaCl2 and subjected to four 1-min periods of vacuum. The segments were then collected, dried on absorbing paper, and centrifuged for 5 min at 1,000g in a swingout rotor to collect the intercellular fluid, which was used directly for IEF separation performed as described previously (Penel and Greppin, 1996). Peroxidase bands were visualized with o-dianisidine/hydrogen peroxide.
RESULTS
APRX Purification and Localization
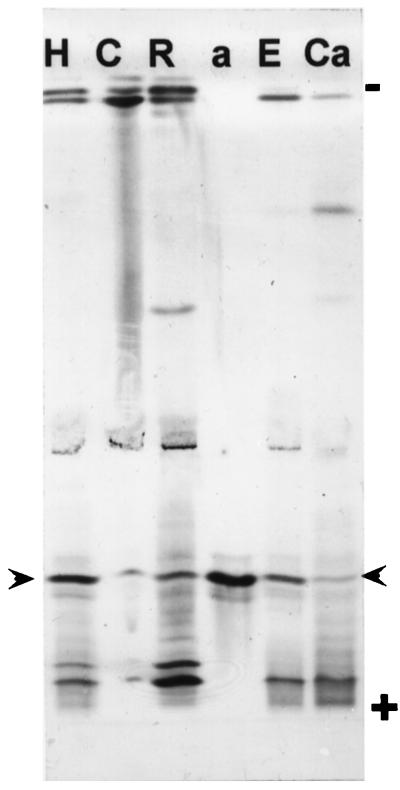
APRX was purified from whole etiolated seedlings as described previously (Penel and Greppin, 1996). The procedure yielded one major acidic peroxidase band (Fig. 1, lane a) that was found in extracts from roots, hypocotyls, and cotyledons. As cotyledons contain many proteins, a larger quantity of proteins had to be electrophoresed on the gel to detect APRX. In hypocotyls APRX could be released from extracellular spaces after vacuum infiltration, showing that it was an apoplastic isoperoxidase (Fig. 1). In addition, the amount of APRX that could be recovered by this technique was higher when vacuum infiltration was performed in the presence of EGTA, suggesting the calcium-dependent binding of this enzyme in the apoplast. A cationic isoperoxidase was found to exhibit the same calcium-dependent behavior, as shown previously (Penel and Greppin, 1994, 1996). In contrast, a moderately cationic band was recovered only when calcium was present in the infiltration buffer (Fig. 1).
Figure 1.
Peroxidase isoenzymes from various fractions of zucchini seedlings separated on acrylamide gel by IEF. Extracts are from hypocotyls (H) (10 μg of proteins); cotyledons (C) (100 μg); roots (R) (10 μg); purified APRX (a) (28 ng); and extracellular fluid obtained in the presence of 5 mm EGTA (E) or 2 mm CaCl2 (C). Arrowheads indicate the positions of APRX on the gel.
The N-terminal amino acid sequence of APRX was determined by subjecting purified enzyme to two-dimensional electrophoresis and transferring it onto a PVDF membrane. Microsequencing was then carried out after deblocking of the N terminus. The sequence obtained, TETFYDQTCPRLPNIVRQ, was 64% identical to a poplar (accession no. X97348) and a flax (accession no. L07554) peroxidase.
Isolation and Sequencing of the APRX cDNA Clone
The cDNA encoding APRX was obtained using a cDNA library prepared from etiolated zucchini hypocotyls. PCR reactions using the specific primers 1, 2, and 3 generated a 1,145-bp cDNA clone that encoded a plant peroxidase. To obtain the complete sequence, including the signal peptide and the 5′-untranslated sequences, we used the 5′-RACE-PCR procedure, with the specific primer 4 and primer AP1 from the cDNA amplification kit. The resulting 238-bp clone was sequenced and exhibited an overlap of 157 bp with the first 1,145-bp clone without any sequence discrepancy. APRX was therefore encoded by a 1,226-bp cDNA (Fig. 2). The open reading frame corresponded to a deduced protein of 309 amino acids with a signal peptide of 16 amino acids. It exhibited a calculated Mr of 33,894 and a pI of 5.1. The N terminus of the mature protein was blocked by a pyroglutamyl residue. Like other peroxidases, this protein contained two putative N-glycosylation sites (Asn-X-Thr/Ser). In addition, the cDNA contained a 5′-untranslated leader sequence of 6 bp and a 242-bp sequence in the 3′-untranslated region, without the animal consensus polyadenylation signal or a known plant polyadenylation signal. Southern analysis performed on zucchini leaf DNA digested with several restriction enzymes showed that APRX was encoded by a single gene (data not shown).
Figure 2.
The complete nucleotide and deduced amino acid sequences of APRX (accession no. Y17192). The signal peptide is shown in italics and the C-terminal propeptide is underlined. The four primers used to produce the full cDNA are shown. The two boxes designate the putative glycosylation sites. The stop codon is marked with an asterisk.
A comparison of APRX mature protein with the peroxidase sequences available in databases was made using the Gap program (Genetics Computer Group, Madison, WI). The best similarity scores were with two cucumber anionic peroxidases: 74% with accession no. M91373 and 88% with accession no. M91374 (Rasmussen et al., 1995) and 70% with tobacco anionic peroxidase (accession no. J02979; Lagrimini et al., 1987) (Fig. 3). Like other plant peroxidases, APRX contained eight Cys residues at positions 11, 44, 49, 90, 96, 174, 206, and 294, allowing the formation of four disulfide bridges (Kjærsgård et al., 1997). The amino acid sequence of mature APRX exhibited the conserved distal catalytic residues Arg-38 and His-42 and the proximal His-167 and Asp-241, which characterize plant peroxidases (Welinder, 1992).
Figure 3.
Mature peroxidase protein sequence comparison. The predicted protein sequences encoded by zucchini APRX clone (this study), two cucumber peroxidase cDNAs, Csprepera and Cspreper (Rasmussen et al., 1995), and a tobacco peroxidase cDNA, Ntpxdlf (Lagrimini et al., 1987), are shown. The alignment was created with the PileUp program and sequence homology was determined with the PrettyPlot program (both programs from Genetics Computer Group).
APRX Expression in Zucchini Seedlings
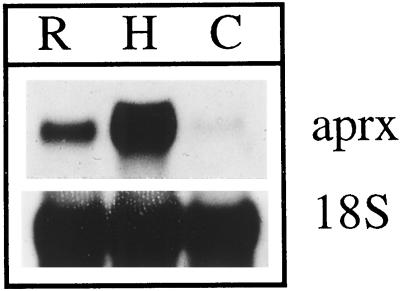
The level of expression of APRX was assessed in various organs of zucchini seedlings after RNA separation by electrophoresis and transfer onto a membrane. As shown in Figure 4, the mRNA encoding APRX was detected in hypocotyls, in roots, and, to a much lesser extent, in cotyledons.
Figure 4.
Northern-blot analysis using total RNA (10 μg) from root (R), hypocotyls (H), and cotyledons (C). APRX mRNA was detected with the 1145-bp cDNA as a specific probe. Detection of ribosomal 18S RNA was performed as a control.
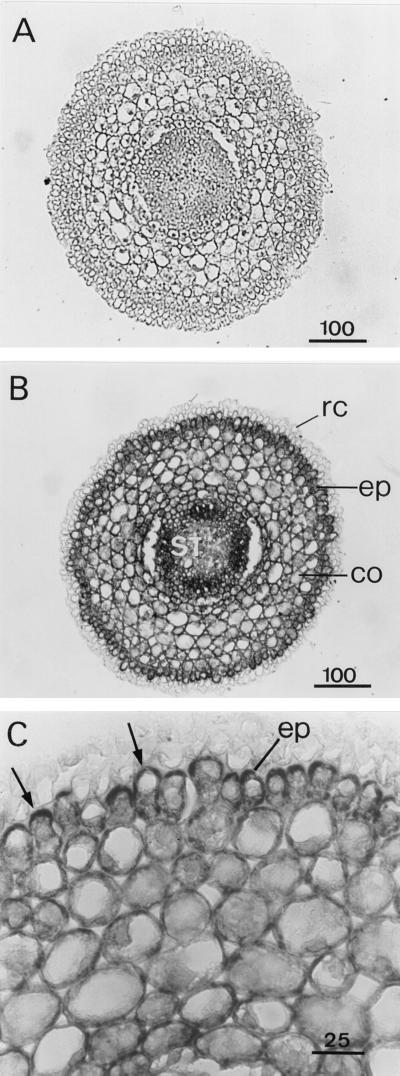
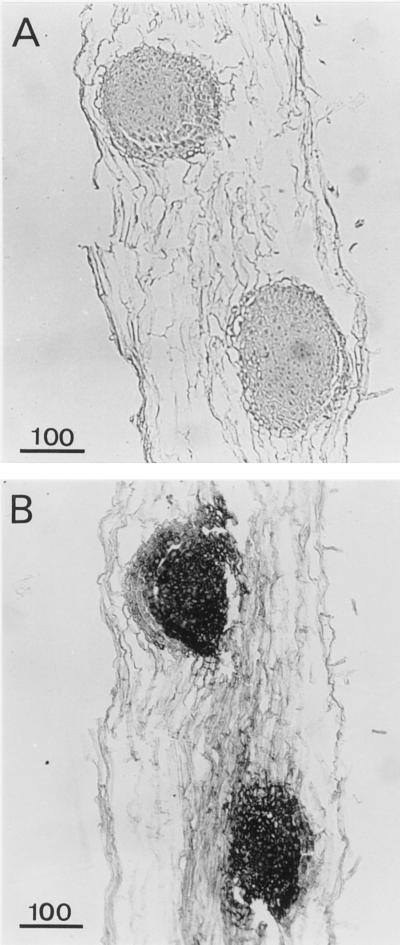
In situ hybridization was used to identify the tissues or cells expressing APRX using a 300-bp specific probe corresponding to the 3′-flanking untranslated region (Figs. 5–10). The mRNA encoding APRX was present in almost all cell types, but with great differences in the level of expression. Figure 5 shows root cross-sections taken 1 mm above the tip. At this level the differentiation of vascular tissues within the stele was not achieved. Only some protoxylem vessels could be distinguished. Control sections incubated with the sense probe did not show any staining (Fig. 5A) in any tissue observed in this work (Figs. 5–10). With the antisense probe the strongest staining was observed in the stele and in the epidermis (Fig. 5B). Epidermal cells exhibited an asymmetric distribution of transcripts that accumulated in the part of the cytoplasm facing the exterior of the root (Fig. 5C). This polarized distribution of APRX mRNA was not observed in other cell types. Lateral root primordia appeared in longitudinal sections made 2 or 3 mm above the tip (Fig. 6) and were heavily stained, with all cells containing a large amount of APRX-encoding mRNA.
Figure 5.
Detection of APRX mRNA by in situ hybridization of root cross-sections. Sections were hybridized with sense (A) or antisense (B and C) probe. co, Cortex; ep, epidermis; st, stele; rc, root cap. Arrows indicate the accumulation of transcripts in epidermal cells. Scale bars are in μm.
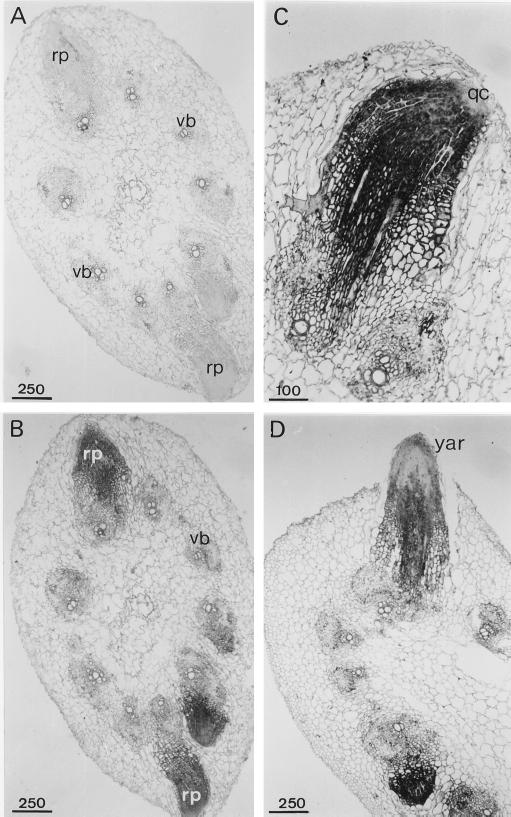
Figure 10.
Detection of APRX mRNA by in situ hybridization in cross-sections of hypocotyls. Shown are general views of adventitious root primordia in sections hybridized with sense (A) or antisense (B) probes, a young adventitious root (C), and an adventitious root emerging out of the hypocotyl (D) hybridized with the antisense probe. qc, Quiescent center; rp, root primordium; vb, vascular bundle; yar, young adventitious root. Scale bars are in μm.
Figure 6.
Detection of APRX mRNA by in situ hybridization in root longitudinal sections showing lateral root primordia. Sections were hybridized with sense (A) or antisense (B) probes. Scale bars are in μm.
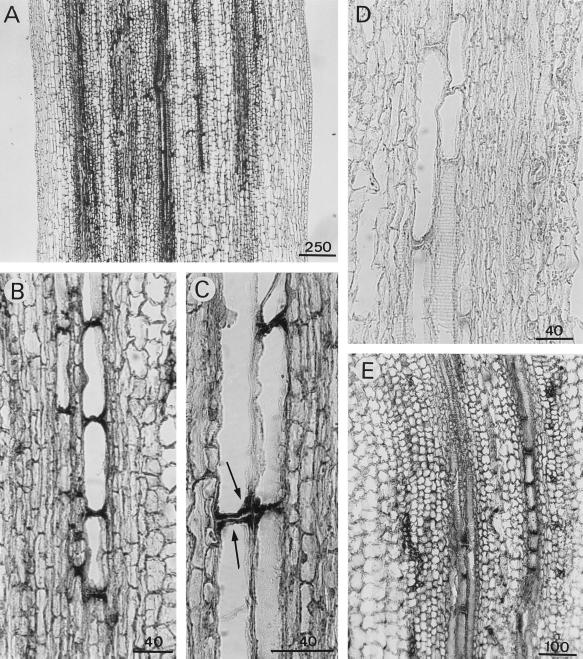
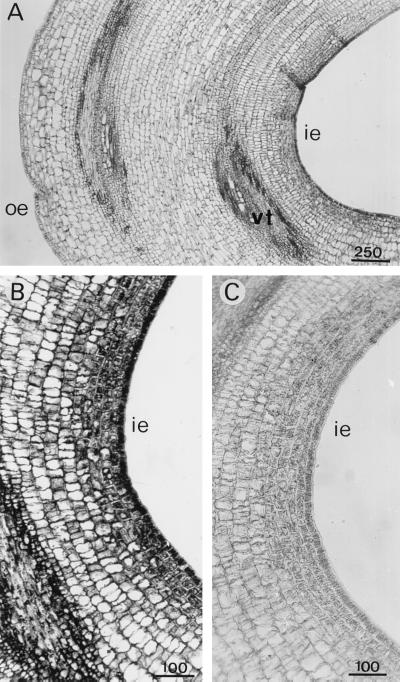
APRX expression varied in the various tissues of the hypocotyl. To study this expression, in situ hybridization was performed on longitudinal sections taken at different levels. APRX transcripts were much more abundant in the upper part of the hypocotyl than at its base. Some cells of the vascular bundles were particularly heavily stained throughout the hypocotyl, but mainly in the upper part, including the hypocotyl hook and the elongation zone (Fig. 7). They were frequently aligned one below the other (Fig. 7, B–D), which could mean that they correspond to the formation of vascular structures. Some large cells adjacent to the characteristic tracheary elements exhibited a strong accumulation of transcripts at their two extremities (Fig. 7C). These were identified as differentiating xylem elements because of their close association with typical xylem structures and because of their secondary wall thickenings. In the hook the distribution of transcripts was uneven (Fig. 8). Cells of the inner epidermis and cortical cells that were situated just below contained more transcripts than cells of the outer part. These latter cells were also much smaller than cells forming the outer epidermis.
Figure 7.
Detection of APRX mRNA by in situ hybridization in hypocotyl longitudinal sections taken 1 cm beneath the hook (A–D) or at the hook level (E). A, General view of a section hybridized with the antisense probe. B and C, Detailed views showing differentiating tracheary elements. D, Control section hybridized with the sense probe. Arrows in C indicate the accumulation of transcripts in differentiating xylem elements. Scale bars are in μm.
Figure 8.
Detection of APRX mRNA by in situ hybridization in longitudinal sections of the hypocotyl hook. Sections were hybridized with antisense (A and B) or sense (C) probes. ie, Inner epidermis; oe, outer epidermis; vt, vascular tissues. Scale bars are in μm.
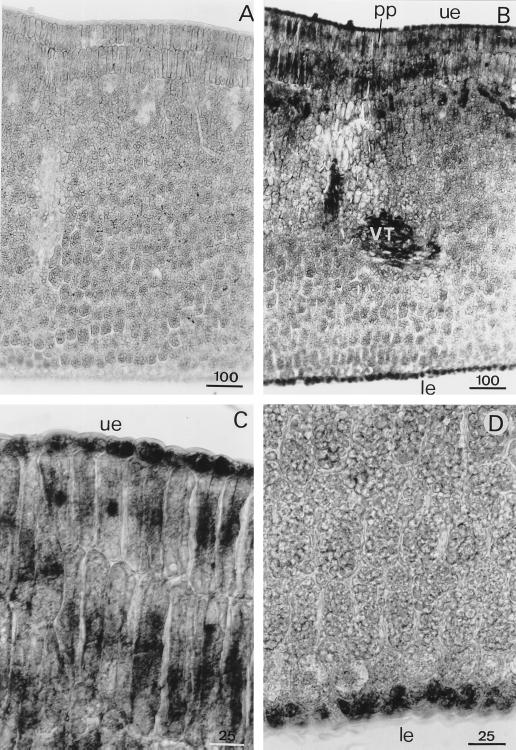
In cotyledons the presence of APRX transcripts was detected in lower and upper epidermis, in the two ranges of palisade parenchyma cells, and in the vascular system (Fig. 9). In some palisade cells, the nucleus was strongly labeled, indicating the presence of pre-mRNA-encoding APRX. Some cells of the vascular system exhibited strong APRX mRNA expression.
Figure 9.
Detection of APRX mRNA by in situ hybridization in transversal sections of cotyledons. Sections were hybridized with sense (A) or antisense (B) probes. Upper (C) and lower (D) parts of the cotyledon were hybridized with the antisense probe. le, Lower epidermis; pp, palisade parenchyma; ue, upper epidermis; VT, vascular tissues. Scale bars are in μm.
APRX mRNA Expression during Adventitious Root Formation
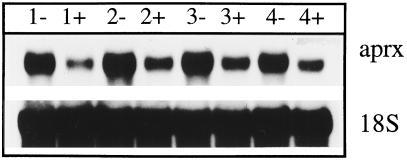
Root regeneration on zucchini hypocotyls occurred spontaneously 4 to 5 d after the ablation of their root system. Many root primordia appeared at the border of a vascular bundle or near the interfascicular cambium (Fig. 10). In situ hybridization revealed strong expression of APRX gene in the young root primordia, which contrasted with the slight staining of the surrounding hypocotyl cells (Fig. 10B). This massive expression decreased as the primordia became longer, which was particularly evident in roots emerging from the hypocotyl (Fig. 10D). Cells belonging to the quiescent center of a young root were not stained (Fig. 10C). Adventitious root formation on hypocotyls was prevented by the addition of 10−4 m TIBA, an anti-auxin substance known to inhibit root formation (Fujita and Syôno, 1996) (data not shown). Figure 11 shows that the relatively high level of APRX transcripts that could be detected in the lower part of hypocotyls by northern analysis was maintained during the days after root excision. In contrast, in hypocotyls maintained in the presence of TIBA, the level of transcripts that could be detected in the lower part of the hypocotyls was already substantially lower 1 d after the beginning of TIBA application.
Figure 11.
Detection of mRNA encoding APRX in the lower part of hypocotyl of zucchini seedlings 1 to 4 d after excision of their root system. Rootless seedlings were kept without (−) or with (+) 10−4 m TIBA. Northern analysis was performed with 10 μg of total RNA. The detection of ribosomal 18S RNA was performed as a control.
DISCUSSION
The zucchini APRX purified and cloned in the present study is an apoplastic enzyme that was previously shown to have a strong affinity for pectin in the presence of calcium ions in cell walls both in vitro and in situ (Penel and Greppin, 1994, 1996; Penel et al., 1996). The larger amount of APRX that could be recovered from hypocotyl segments when EGTA was present in the buffer used for the vacuum infiltration (Fig. 1) confirmed the importance of calcium for the binding of this enzyme within the apoplast.
APRX is the first isoperoxidase from zucchini to be sequenced. The strongest homology it exhibits is with two APRXs from cucumber and one from tobacco. None of the numerous Arabidopsis peroxidase sequences that are known exhibits a homology higher than 58%. The Mr calculated from the deduced amino acid sequence was 33,894, whereas it was previously estimated to be approximately 41,400 by SDS-PAGE (Penel and Greppin, 1994). The difference could be explained by the presence of carbohydrate chains, since it appears from the sequence analysis that APRX contains two putative glycosylation sites. There is also a difference between the calculated pI (5.1) and the pI that was determined experimentally (4.3) (Penel and Greppin, 1994). APRX sequence analysis also showed the presence of a C-terminal propeptide that is generally supposed to target the protein toward the vacuole (Johansson et al., 1992; Neuhaus et al., 1994; Omann and Tyson, 1996). In spite of that, APRX was found at least partly in apoplastic spaces (Fig. 1). Other proteins containing a C-terminal extension, such as α-mannosidase, chitinase, and glucanase, were also reported to have an extracellular localization, and the hypothesis was put forward that these enzymes were secreted into the medium via a pathway in which the proteins pass through the vacuolar compartment (Kunze et al., 1998).
The APRX gene appears to be expressed in all parts of zucchini seedlings. Roots, hypocotyls, and cotyledons contain the mRNA encoding the isoperoxidase and the isoperoxidase itself. Both are also present in leaves and other organs of light-grown plants (data not shown), probably indicating an essential role for this isoperoxidase. A similar wide expression was reported for some isoperoxidases (Kjærsgård et al., 1997; Teichmann et al., 1997; Klotz et al., 1998), whereas others exhibit an organ-specific expression or are synthesized only after a specific treatment (Morgens et al., 1990; Mohan et al., 1993; Rasmussen et al., 1995; Omann and Tyson, 1996). The northern analyses done in this work (Fig. 4) could suggest that hypocotyl cells contain a higher amount of transcripts than root and cotyledon cells. The detection of transcripts on sections does not completely confirm such a conclusion (Figs. 5–10). Actually, the three organs contain cells that express APRX quite strongly.
The data obtained by in situ hybridization showed that APRX expression is tissue-specific and developmentally regulated, as was reported for other peroxidases such as the anionic peroxidase of tobacco (Klotz et al., 1998). In the hypocotyl there is a gradient of expression, with the upper part being more active than the lower part. Root primordia also exhibited a strong expression. Therefore, like many other peroxidases, the presence of APRX must not be correlated with cell growth cessation (Gaspar et al., 1982; Zheng and Van Huystee, 1992; Lagrimini et al., 1997). In the hypocotyl hook, however, small cells from the inner zone contained more transcripts than large cells situated in the outer part (Fig. 8). As seen on longitudinal sections from upper hypocotyls (Fig. 7), the transcripts accumulated mainly in vascular bundles in files of cells that most likely correspond to differentiating vessels.
Elongated cells exhibiting secondary thickenings of their walls were also heavily labeled at their two extremities, indicating that APRX could be involved in lignin deposition during the formation of xylem vessels. Such a function has been repeatedly attributed to peroxidases and has been confirmed by cytochemical and biochemical studies (Catesson et al., 1986; Ros Barceló, 1995; Sato et al., 1995; Christensen et al., 1998) and by immunolocalization (Smith et al., 1994). It is shown here, for the first time to our knowledge, that one particular peroxidase gene is expressed in xylem cells at the beginning of their differentiation.
APRX transcripts were also abundant in the epidermis of cotyledons and in the inner part of the hypocotyl hook and roots. The reason for this presence in dermal tissues has not been elucidated. The epidermis was often shown to contain a strong peroxidase activity (Gaspar et al., 1982), which could be involved in the polymerization of phenolics to produce the lignin-like material found in cutin and suberin (Riley and Kolattukudy, 1975; Hendricks and van Loon, 1990).
Young lateral roots (Fig. 6) and adventitious root primordia (Fig. 10) also exhibited a strong expression in every cell. Peroxidases have often been used as marker enzymes in the rooting process (Gaspar et al., 1992). The characteristic evolution of peroxidases during rhizogenesis includes always an increase in the activity of APRXs during the late phase of this process, namely the appearance of root primordia (Gaspar et al., 1992). The function of peroxidase in these rapidly growing tissues remains to be discovered. APRX mRNA was still produced near the tips of fully developed roots and was expressed in almost every cell, but at different levels. The stronger expression was observed in the stele and in epidermal cells, mainly in the part of cytoplasm facing the root surface. One possible interpretation of this uneven distribution could be that the neo-synthesized APRX molecules are synthesized and secreted mainly or exclusively toward the root surface. In addition to the transport of proteins, the control of mRNA localization within the cytoplasm constitutes an important mechanism to orientate the distribution of newly synthesized proteins (Okita et al., 1998).
This work provides some indications about a possible correlation between the level of auxin and the rate of expression of the APRX gene. Both differentiating tracheary cells and cells forming young root primordia contain high amounts of transcripts. These two processes, vascular differentiation and root initiation and growth, are known to be promoted by auxin (Davies, 1987). In addition, treatment with the anti-auxin TIBA drastically lowered the level of transcripts that could be extracted from the bottom of rootless hypocotyls (Fig. 11). As an auxin transport inhibitor, TIBA acts most likely by preventing auxin accumulation in the lower part of hypocotyls (Depta et al., 1983), inhibiting adventitious root formation (Fujita and Syôno, 1996). This positive correlation with auxin could also explain the high amount of APRX transcripts found in hypocotyl hook inner epidermis (Fig. 8), which was shown to contain up to 10-fold more IAA than the outer epidermis (Schwark and Bopp, 1993).
According to the results presented here, the main sites of expression of the gene encoding APRX are the dermal tissues and xylem elements during their differentiation. This expression pattern suggests that APRX could be responsible for the formation of lignin or lignin-like material found in cutin and suberin, as was suggested for other APRXs (Kolattukudy, 1987; Hendricks and van Loon, 1990; Teichmann et al., 1997). APRX could use its pectin-binding property to associate the newly synthesized polymers to the cell wall. It has been reported that the phenolic domain of the cuticle or the suberized layers is probably attached to the cell wall through linkages with pectinaceous domains (Kolattukudy, 1987).
Cells of very young roots and of cotyledon palisade parenchyma also exhibit a strong expression. At this point it is difficult to put forward a hypothesis concerning the possible function of APRX in these cells. Both root primordia and palisade parenchyma are rather compact tissues, consisting of cells that seem to be tightly associated. APRX could have a role in the establishment of the close association existing between these cells at the cell wall level.
Many plant functions are commonly attributed to peroxidases, with most occurring in the apoplast. This study and other works cited herein demonstrate that each particular peroxidase isoenzyme exhibits a specific expression pattern within the plant. The detailed study of this expression, combined with a precise knowledge concerning both the catalytic characteristics of the isoperoxidase and the structural features that could determine its localization within the extracellular matrix, should allow for progress in the understanding of the real role of peroxidases in plants. APRX, which is preferentially expressed in particular tissues of zucchini seedlings and which binds specifically to pectins in the presence of calcium, is likely to provide a good model for this kind of study.
ACKNOWLEDGMENTS
We thank Evelyne Vasquez for her excellent technical assistance and Dr. S. Hamdi (University of Tours, France) for generously providing the ribosomal 18S cDNA clone.
Abbreviations:
- APRX
anionic isoperoxidase
- RT
reverse transcriptase
- TIBA
2,3,5-triiodobenzoic acid
Footnotes
This work was partly supported by the Federal Office of Education and Science (grant no. 93-0090). S.C. received a grant from the Agence Universitaire de la Francophonie.
LITERATURE CITED
- Blom TJM, Sierra M, van Vliet TB, Franke-van Dijk MEI, de Koning P, van Iren F, Verpoorte R, Libbenga KR. Uptake and accumulation of ajmalicine into isolated vacuoles of cultured cells of Catharanthus roseus (L.) G. Don. and its conversion into serpentine. Planta. 1991;183:170–177. doi: 10.1007/BF00197785. [DOI] [PubMed] [Google Scholar]
- Brownleader M, Ahmed N, Trevan M, Chaplin MF, Dey PM. Purification and partial characterization of tomato extensin peroxidase. Plant Physiol. 1995;109:1115–1123. doi: 10.1104/pp.109.3.1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catesson A-M, Imberty A, Goldberg R, Czaninski Y (1986) Nature, localization and specificity of peroxidases involved in lignification processes. In H Greppin, C Penel, T Gaspar, eds, Molecular and Physiological Aspects of Plant Peroxidases. University of Geneva, Switzerland, pp 189–198
- Christensen JH, Bauw G, Welinder KG, Van Montagu M, Boerjan W. Purification and characterization of peroxidases correlated with lignification in poplar xylem. Plant Physiol. 1998;118:125–135. doi: 10.1104/pp.118.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies PJ. Plant Hormones and their Role in Plant Growth and Development. Dordrecht, The Netherlands: Martinus Nijhoff Publishers; 1987. [Google Scholar]
- Dean C, Elzen P, Tamaki S, Duinsmuir P, Bedbrook J. Differential expression of the eight genes of the petunia ribulose biphosphate carboxylase small subunit multigene family. EMBO J. 1985;5:3055–3061. doi: 10.1002/j.1460-2075.1985.tb04045.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depta H, Eisele K-H, Hertel R. Specific inhibitors of auxin transport: action on tissue segments and in vitro binding to membranes from maize coleoptiles. Plant Sci Lett. 1983;31:181–192. [Google Scholar]
- Elstner EF, Heupel AL. Formation of hydrogen peroxide by isolated cell walls from horseradish (Armoracia lapathifolia Gilib.) Planta. 1976;130:175–180. doi: 10.1007/BF00384416. [DOI] [PubMed] [Google Scholar]
- Fry SC. Cross-linking of matrix polymers in the growing cell wall of angiosperms. Annu Rev Plant Physiol. 1986;37:165–186. [Google Scholar]
- Fujita H, Syôno K. Genetic analysis of the effects of polar auxin transport inhibitors on root growth in Arabidopsis thaliana. Plant Cell Physiol. 1996;37:1094–1101. doi: 10.1093/oxfordjournals.pcp.a029059. [DOI] [PubMed] [Google Scholar]
- Gaspar T, Kevers C, Hausman JF, Berthon JY, Ripetti V. Practical uses of peroxidase activity as a predictive marker of rooting performance of micropropagated shoots. Agronomie. 1992;12:757–765. [Google Scholar]
- Gaspar T, Penel C, Thorpe T, Greppin H (1982) Peroxidases 1970–1980. A Survey of Their Biochemical and Physiological Roles in Higher Plants. University of Geneva, Switzerland
- Gazaryan IG, Lagrimini LM, Ashby GA, Thorneley RNF. The mechanism of indole-3-acetic acid oxidation by plant peroxidases: anaerobic stopped-flow spectrophotometric studies on horseradish and tobacco peroxidases. Biochem J. 1996;313:841–847. doi: 10.1042/bj3130841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendricks T, van Loon LC. Petunia peroxidase a is localized in the epidermis of aerial plant organs. J Plant Physiol. 1990;136:519–520. [Google Scholar]
- Johansson A, Rasmussen SK, Harthill J, Welinder KG. cDNA, amino acid and carbohydrate sequences of barley seed-specific peroxidase BP1. Plant Mol Biol. 1992;18:1151–1156. doi: 10.1007/BF00047718. [DOI] [PubMed] [Google Scholar]
- Kjærsgård IVH, Jespersen HM, Rasmussen SK, Welinder KG. Sequence and RT-PCR expression analysis of two peroxidases from Arabidopsis thaliana belonging to a novel evolutionary branch of plant peroxidases. Plant Mol Biol. 1997;33:699–708. doi: 10.1023/a:1005707813801. [DOI] [PubMed] [Google Scholar]
- Klotz KL, Liu TTL, Liu L, Lagrimini M. Expression of the tobacco anionic peroxidase gene is tissue-specific and developmentally regulated. Plant Mol Biol. 1998;36:509–520. doi: 10.1023/a:1005939600344. [DOI] [PubMed] [Google Scholar]
- Kolattukudy PE (1987) Lipid-derived defensive polymers and waxes and their role in plant-microbe interaction. In PK Stumpf, EE Conn, eds, The Biochemistry of Plants, Vol 9. Academic Press, New York, pp 291–314
- Komminoth P (1996) Detection of mRNA in tissue sections using DIG-labeled RNA and oligonucleotide probes. In Nonradioactive in Situ Hybridization: Application Manual. Boehringer Mannheim, Mannheim, Germany, pp 126–135
- Kunze I, Kunze G, Bröker M, Manteuffel R, Meins F, Müntz K. Evidence for secretion of vacuolar α-mannosidase, class I chitinase, and class I β-1,3-glucanase in suspension cultures of tobacco cells. Planta. 1998;205:92–99. doi: 10.1007/s004250050300. [DOI] [PubMed] [Google Scholar]
- Lagrimini LM, Burkhart W, Moyer M, Rothstein S. Molecular cloning of complementary DNA encoding the lignin-forming peroxidase from tobacco: molecular analysis and tissue-specific expression. Proc Natl Acad Sci USA. 1987;84:7542–7546. doi: 10.1073/pnas.84.21.7542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagrimini LM, Gingas V, Finger F, Rothstein S, Liu T-TY. Characterization of antisense transformed plants deficient in the tobacco anionic peroxidase. Plant Physiol. 1997;114:1187–1196. doi: 10.1104/pp.114.4.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeGendre N, Mansfield M, Weiss A, Matsudaira P (1993) Purification of proteins and peptides by SDS-PAGE. In P Matsudaira, ed, A Practical Guide to Protein and Peptide Purification for Microsequencing. Academic Press, New York, pp 71–101
- Mäder M (1992) Compartmentation of peroxidase isoenzymes in plant cells. In C Penel, T Gaspar, H Greppin, eds, Plant Peroxidases 1980–1990. University of Geneva, Switzerland, pp 37–46
- Mohan R, Vijayan P, Kolattukudy PE. Developmental and tissue-specific expression of a tomato anionic peroxidase (tap1) gene by a minimal promoter, with wound and pathogen induction by an additional 5′-flanking region. Plant Mol Biol. 1993;22:475–490. doi: 10.1007/BF00015977. [DOI] [PubMed] [Google Scholar]
- Morgens PH, Callahan AM, Dunn LJ, Abeles FB. Isolation and sequencing of cDNA clones encoding ethylene-induced putative peroxidases from cucumber cotyledons. Plant Mol Biol. 1990;14:715–725. doi: 10.1007/BF00016504. [DOI] [PubMed] [Google Scholar]
- Neuhaus J-M, Pietrzak M, Boller T. Mutation analysis of the C-terminal vacuolar targeting peptide of tobacco chitinase: low specificity of the sorting system, and gradual transition between intracellular retention and secretion into the extracellular space. Plant J. 1994;5:45–54. doi: 10.1046/j.1365-313x.1994.5010045.x. [DOI] [PubMed] [Google Scholar]
- O'Farrell PH. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1978;250:4007–4021. [PMC free article] [PubMed] [Google Scholar]
- Okita TW, Choi S-B, Ito H, Muench DG, Wu Y, Zhang F. Entry into the secretory system: the role of mRNA localization. J Exp Bot. 1998;49:1081–1090. [Google Scholar]
- Omann F, Tyson H. An anionic, stem-specific flax peroxidase cDNA with C-terminal motifs also found in a blue copper-type pea protein correlated with lignin deposition. Aust J Plant Physiol. 1996;23:773–789. [Google Scholar]
- Penel C, Crèvecoeur M, Greppin H (1996) The binding of peroxidases to pectins. In C Obinger, U Burner, R Ebermann, C Penel, H Greppin, eds, Plant Peroxidases: Biochemistry and Physiology. University of Geneva, Switzerland, pp 259–263
- Penel C, Greppin H. Binding of plant isoperoxidases to pectin in the presence of calcium. FEBS Lett. 1994;343:51–55. doi: 10.1016/0014-5793(94)80605-5. [DOI] [PubMed] [Google Scholar]
- Penel C, Greppin H. Pectin binding proteins: characterization of the binding and comparison with heparin. Plant Physiol Biochem. 1996;34:479–488. [Google Scholar]
- Pichorner H, Couperus A, Korori SAA, Ebermann R. Plant peroxidase has a thiol oxidase function. Phytochemistry. 1992;31:3371–3376. [Google Scholar]
- Rasmussen JB, Smith JA, Williams S, Burkhart W, Ward E, Sommerville SC, Ryals J, Rasmussen R. cDNA cloning and systemic expression of acidic peroxidases associated with systemic acquired resistance to disease in cucumber. Physiol Mol Plant Pathol. 1995;46:389–400. [Google Scholar]
- Reigh DL, Wender SH, Smith EC. Scopoletin: a substrate for an isoperoxidase from Nicotiana tabacum tissue culture W-38. Phytochemistry. 1973;12:1265–1268. [Google Scholar]
- Riley RG, Kolattukudy PE. Evidence for covalently attached p-coumaric acid and ferulic acid in cutins and suberins. Plant Physiol. 1975;56:650–654. doi: 10.1104/pp.56.5.650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers SO, Bendich AJ. Extraction of total cellular DNA from plants, algae and fungi. In: Gelvin SB, Schilperoort RA, editors. Plant Molecular Biology Manual. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1994. , Section D1, pp 1–8. [Google Scholar]
- Ros Barceló A. Peroxidase and not laccase is the enzyme responsible for cell wall lignification in the secondary thickening of xylem vessels in Lupinus. Protoplasma. 1995;186:41–44. [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sato Y, Sugiyama M, Komamine A, Fukuda H. Separation and characterization of the isoenzymes of wall-bound peroxidase from cultured Zinnia cells during tracheary element differentiation. Planta. 1995;196:141–147. [Google Scholar]
- Schwark A, Bopp M. Interaction of ethylene and auxin in the regulation of hook growth. II. The role for ethylene in different growing regions of the hypocotyl hook of Phaseolus vulgaris. J Plant Physiol. 1993;142:585–592. [Google Scholar]
- Smith CL, Rodgers MW, Zimmerlin A, Ferdinando D, Bolwell GP. Tissue and subcellular immunolocalisation of enzymes of lignin synthesis in differentiating and wounded hypocotyl tissue of French bean (Phaseolus vulgaris L.) Planta. 1994;192:155–164. [Google Scholar]
- Spector T. Refinement of the Coomassie blue method of protein quantitation: a simple and linear spectrophotometric assay for 0.5 to 50 μg of protein. Anal Biochem. 1978;86:142–146. doi: 10.1016/0003-2697(78)90327-5. [DOI] [PubMed] [Google Scholar]
- Teichmann T, Guan C, Kristoffersen P, Muster G, Tietz O, Palme K. Cloning and biochemical characterization of an anionic peroxidase from Zea mays. Eur J Biochem. 1997;247:826–832. doi: 10.1111/j.1432-1033.1997.00826.x. [DOI] [PubMed] [Google Scholar]
- Welinder KG (1992) Peroxidases: structure-function relationships. In C Penel, T Gaspar, H Greppin, eds, Plant Peroxidases 1980–1990. University of Geneva, Switzerland, pp 1–24
- Zheng X, Van Huystee RB. Peroxidase-regulated elongation of segments from peanut hypocotyls. Plant Sci. 1992;81:47–56. [Google Scholar]