Abstract
Plant seeds are home to diverse microbial communities whose composition is determined by plant genotype, environment, and management practices. Plant domestication is now recognized as an important driver of plant-associated microbial diversity. To what extent and how domestication affects seed microbiomes is less well studied. Here we propose a ‘back-to-the-future’ approach to harness seed microbiomes of wild relatives of crop cultivars to save and re-instate missing beneficial seed microbes for improved plant tolerance to biotic and abiotic stress.
Domestication leaves footprints of microbial diversity
Plant domestication was a crucial step in human civilization by developing a full-fledged, reliable, and productive agriculture [1]. In the co-evolution of this relationship between plants and humans, also microorganisms were involved secretly. Although the potential of the plant microbiome for plant health and growth was already discovered by Lorenz Hiltner more than a century ago, recent multi-omics technologies have provided a much deeper insight into the taxonomic and functional diversity of microorganisms living inside and outside of plant tissue [2]. Alike humans and other eukaryotes, plants are now considered as co-evolved species assemblages or holobionts, consisting of bacterial, archaeal, fungal, and other eukaryotic species [3]. The microbiome plays a pivotal role in plant biology, performing key functions in germination, growth, health, stress protection, and phytochemistry [4]. It is therefore critical to understand the temporal dynamics, spatial organization and evolution of plant microbiomes. Plant domestication and breeding have led to highly productive cultivars but also to profound changes in the holobiont with significant compositional shifts in the microbiomes of cultivars of various crop species including rice, maize, wheat, and common bean [5, 6]. Intriguingly, the microbial footprint of domestication revealed similar changes for different crops with a shift from slow-growing bacterial phyla such as Bacteriodetes on wild crop relatives and ancient cultivars to fast-growing phyla such as Proteobacteria on modern crop cultivars [6–8]. This striking shift in microbiome composition has also been described for the human gut of lean and obese people [9], exemplifying that the link between the ‘domestication syndrome’ and microbiome changes described for plants is also apparent for domesticated animals including man.
Seeds transmit the footprint of domestication
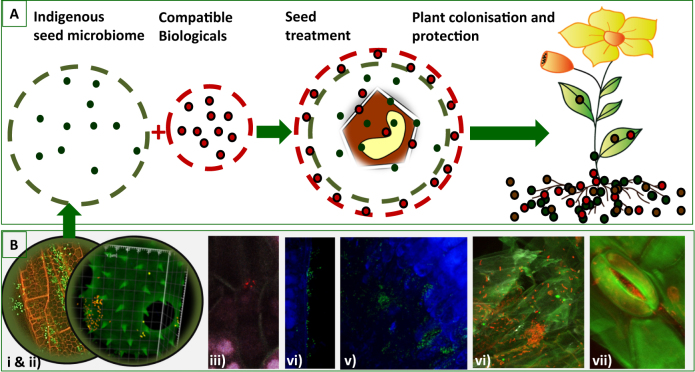
Key factors driving the observed shifts in microbiome composition identified to date include intrinsic host genotypic traits, life style and external environmental conditions [2]. For plants, it was unclear whether vertical transmission of microbes contributed to changes in microbial community composition across plant genotypes and whether such vertical transmission influences plant performance. Seeds are small embryonic plants enclosed in a seed coat that govern reproduction of angiosperm and gymnosperm plants. Healthy seeds were typically considered as ‘sterile’ and appropriate criteria for seed quality and purity, with a focus on transmissible plant pathogens, have been established at international and national level (e.g., International Seed Federation (ISF), International Seed Health Initiatives (ISHIs)). Hence, a variety of phytosanitary measures, including mechanical, thermal, physical, biological, and chemical treatments for seed cleaning have been developed over the past decades. However, recent omics-based analyses show that (i) plant seeds also contain beneficial, plant genotype-specific microbes, and (ii) these microbes can be vertically transmitted from one plant generation to the next [7, 10–12]. Therefore, the surface sterilization procedures are not fully effective in removing microorganisms located in the endosperm or close to the embryo. Microbes may enter seeds maternally via the carbohydrate transport route from the leaves to the seed’s outer coat, paternally via pollen grains or environmentally via penetration of the nectarthodes or the stigma style of the flowers. In addition to the vertical transmission via the parents, members of the plant microbiota are transmitted horizontally from the surrounding environment [12] or, for several wild plant species, possibly via passage through the gut of birds or other animals. Inside seeds, microbes have been localized in the seed coat but can also be found on the cotyledon as well as on the root hypocotyl embryo after seed germination (Fig. 1).
Fig. 1.
The seed microbiome. a showing the concept of compatible biologicals for crops and, b visualizing endophytes by in situ hybridization and confocal laser scanning microscopy (i) in the endosphere, (ii) on pollen, (iii) naturally occurring in seeds, (iv) after seed treatment within seed on the cotyledon, (v) after seed treatment within seed on the root hypocotyl embryo, (vi) in the rhizosphere and (vii) phyllosphere after seed treatment. Seeds were treated with Serratia plymuthica
The composition of seed microbiomes is diverse containing up to 9000 microbial species and up to two billion of bacterial cells [7, 10, 12]. Although a limited number of plants species has been investigated to date, seed microbiome composition appears to be plant species specific with an additional footprint of domestication [7, 13]. The majority of microbes residing inside seed tissue is alive and, after seed germination, colonize the rhizosphere when plants are grown under axenic conditions [7]. Under natural soil conditions, however, the carry-over of seed microbes to the rhizosphere and phylosphere is less well studied. While a substantial proportion of seed microbes were able to colonize the pumpkin rhizosphere [7], a study on endophytic bacteria of wild and modern maize varieties revealed a conserved core microbiota of maize seed endophytes, with at least one member (Enterobacter asburiae) successfully colonizing the rhizosphere [14]. These and a more recent study [13] also suggest a correlation between seed microbiome composition and protection against plant pathogens. If and how seed microbiomes affect plant germination, growth, development, quality, and (a)biotic stress protection is yet to be discovered. Hence, we propose to re-define the plant seed as a functional microbial entity and to go back-to-the-future to unravel microbiomes of seeds of wild relatives of modern crop cultivars.
Implications for sustainable agriculture
Understanding the specificity and efficiency of the transmission of microbes from seeds to plant roots, stems, and leaves will provide new exciting opportunities to exploit beneficial microbe-plant interactions in agriculture and horticulture. Microbiome analyses are a first step to gain a deeper understanding of genotype-dependent differences. This in turn should facilitate breeding of new cultivars that are better able to assemble a beneficial microbial community inside and outside seeds. Moreover, joining breeding and introducing beneficial microbes on or in seeds will allow a promising SynBiotic (combining prebiotic and probiotic treatment) approach for crop cultivation (Fig. 1). To date, however, only few examples are known of breeding programs that have considered microbiome composition and functions. Adam et al. [7] demonstrated for the Styrian oil pumpkin, a crop with a short breeding history, that microbiome engineering is indeed feasible. Several strategies have been proposed and tested to re-direct the seed and plant microbiome composition and activity [15]. For example, changing the quantity and quality of seed exudates or floral characteristics via plant breeding may be potential strategies to engineer the indigenous seed microbiome. Various Biotech companies are now embracing the plant microbiome for future applications and we expect that seed endophytes can serve as a potential source for new breeding targets. Currently, seed microbiome analyses have focused primarily on bacteria and to some extent on fungi, but there is a wealth of other (micro)organisms and consortia to be discovered for seed-based applications. Moreover, the concept of compatible seed biologicals (SynBiotics) offers the possibility to combine cosmopolitan seed cultivars with local biologicals. In this context, Pérez-Jaramillo et al. [16] suggested a ‘back to the roots’ approach that comprises the exploration of the microbiomes of wild relatives of crop plant species and their native habitats for the identification of beneficial microbes that may have been lost during the domestication process. This fundamental knowledge will allow us to re-instate the missing beneficial microbes from the wild relatives in seeds of modern crop species and to identify plant traits that can ultimately be used in dedicated microbiome engineering programs.
Saving seed-associated microbial biodiversity
Despite the potential of seed microbiomes for plant growth promotion and sustainable protection of crops against (a)biotic stress factors, the impact of current seed treatments (e.g., disinfection, (osmo)priming) on microbiome composition as well as the consequences of seed microbiome engineering for biosecurity need to be investigated. The international trade of seeds is immense, amounting >3.5 million tons per year (ISF). Seed companies operate world-wide with major seed production areas in the Northern and Southern hemisphere to secure seed production all year round. The centralized production and global trade of seeds may contribute to homogeneity of the plant microbiome at global scale but may also impact on soil microbial diversity and health. The essential roles of the plant microbiome for phenotypic and epigenetic plasticity as well as the evolution of plants, which was recently evidenced by Van der Heijden et al. [17], is therefore running the risk of being lost. Current seed treatments are primarily directed toward elimination of seed-borne microbial pathogens (e.g. International Rules for Seed Testing (ISTA Rules)) and synchronization of germination. We propose to evaluate the effects of these seed treatments on beneficial seed microbes or, in the future, on microbial consortia incorporated into seeds. To that end, also the impact of seed treatments on microbial diversity, a key factor controlling invasiveness of pathogens [18–20], needs to be evaluated. Therefore, international conservation strategies for seeds to preserve genetic diversity (e.g., Svalbard Global Seed Vault (Norway), the Millennium Seed Bank (UK)) should include conservation strategies for indigenous seed microbes. Saving seeds with their indigenous microbes offers an enormous potential to safe-guard, explore and exploit microbial diversity and their untapped metabolic potential for plant, human and environmental health [21, 22].
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
References
- 1.Purugganan MD, Fuller DQ. The nature of selection during plant domestication. Nature. 2009;457:843–8. doi: 10.1038/nature07895. [DOI] [PubMed] [Google Scholar]
- 2.Berg G, Rybakova D, Grube M, Köberl M. The plant microbiome explored: implications for experimental botany. Exp Bot. 2016;67:995–1002. doi: 10.1093/jxb/erv466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vandenkoornhuyse P, Quaiser A, Duhamel M, Le Van A, Dufresne A. The importance of the microbiome of the plant holobiont. New Phytol. 2015;206:1196–1206. doi: 10.1111/nph.13312. [DOI] [PubMed] [Google Scholar]
- 4.Mendes R, Raaijmakers JM. Cross-kingdom similarities in microbiome functions. ISME J. 2015;9:1905–7. doi: 10.1038/ismej.2015.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peiffer JA, Spor A, Koren O, Jin Z, Tringe SG, Dangl JL, et al. Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc Natl Acad Sci USA. 2013;110:6548–53. doi: 10.1073/pnas.1302837110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perez-Jaramillo et al. (2017) Linking rhizosphere microbiome composition of wild and domesticated Phaseolus vulgaris to genotypic and root phenotypic traits. ISME J. 2017;11:2244-2257. [DOI] [PMC free article] [PubMed]
- 7.Adam E, Bernhart M, Müller H, Winkler M, Berg G. (2016). The Cucurbita pepo seed microbiome: genotype-specific composition and implications for breeding. Plant Soil. 10.1007/s11104-016-3113-9
- 8.Germida J, Siciliano S. Taxonomic diversity of bacteria associated with the roots of modern, recent and ancient wheat cultivars. Biol Fertil Soils. 2001;33:410–15. doi: 10.1007/s003740100343. [DOI] [Google Scholar]
- 9.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–31. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 10.Johnston-Monje D, Lundberg DS, Lazarovits G, Reis VM, Raizada MN. Bacterial populations in juvenile maize rhizospheres originate from both seed and soil. Plant Soil. 2016;405:337–355. doi: 10.1007/s11104-016-2826-0. [DOI] [Google Scholar]
- 11.Nelson EB, (2017). The seed microbiome: Origins, interactions, and impacts. Plant Soil. 10.1007/s11104-017-3289-7.
- 12.Shade A, Jacques MA, Barret M. Ecological patterns of seed microbiome diversity, transmission, and assembly. Curr Opin Microbiol. 2017;37:15–22. doi: 10.1016/j.mib.2017.03.010. [DOI] [PubMed] [Google Scholar]
- 13.Rybakova D, Mancinelli R, Wirkstrom M, Birch-Jensen F, Postma J, et al. The seed microbiome: cultivar-dependent structure in oilseed rape affects the interaction with symbionts and pathogens. Microbiome. 2017;5:104. doi: 10.1186/s40168-017-0310-6.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnston-Monje D, Raizada MN. Conservation and diversity of seed associated endophytes in Zea across boundaries of evolution, ethnography and ecology. PLoS ONE. 2011;6:e20396. doi: 10.1371/journal.pone.0020396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mitter B, Pfaffenbichler N, Flavell R, Compant S, Antonielli L, Petric A, et al. A new approach to modify plant microbiomes and traits by introducing beneficial bacteria at flowering into progeny seeds. Front Microbiol. 2017;8:11. doi: 10.3389/fmicb.2017.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pérez-Jaramillo JE, Mendes R, Raaijmakers JM. Impact of plant domestication on rhizosphere microbiome assembly and functions. Plant Mol Biol. 2016;90:635–44. doi: 10.1007/s11103-015-0337-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van der Heijden MG, de Bruin S, Luckerhoff L, van Logtestijn RS, Schlaeppi K. A widespread plant-fungal-bacterial symbiosis promotes plant biodiversity, plant nutrition and seedling recruitment. ISME J. 2016;10:389–99. doi: 10.1038/ismej.2015.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Elsas JD, Chiurazzi M, Mallon CA, Elhottova D, Kristufek V, Salles JF. Microbial diversity determines the invasion of soil by a bacterial pathogen. Proc Natl Acad Sci USA. 2012;109:159–64. doi: 10.1073/pnas.1109326109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berg G, Köberl M, Rybakova D, Müller H, Grosch R, Smalla K. (2017). Plant microbial diversity is suggested as the key to future biocontrol and health trends. FEMS Microbiol Ecol. 93. doi: 10.1093/femsec/fix050. [DOI] [PubMed]
- 20.Wall DH, Nielsen UN, Six J. Soil biodiversity and human health. Nature. 2015;528:69–76. doi: 10.1038/nature15744. [DOI] [PubMed] [Google Scholar]
- 21.Gruber K. Agrobiodiversity: The living library. Nature. 2017;544:8–10. doi: 10.1038/544S8a. [DOI] [PubMed] [Google Scholar]
- 22.Raaijmakers JM, Mazzola M. Soil immune responses. Science. 2016;352:1392–93. doi: 10.1126/science.aaf3252. [DOI] [PubMed] [Google Scholar]