Abstract
The giant panda feeds almost exclusively on bamboo, a diet highly enriched in lignin and cellulose, but is characterized by a digestive tract similar to carnivores. It is still large unknown if and how the giant panda gut microbiota contributes to lignin and cellulose degradation. Here we show the giant pandas’ gut microbiota does not significantly contribute to cellulose and lignin degradation. We found that no operational taxonomic unit had a nearest neighbor identified as a cellulolytic species or strain with a significant higher abundance in juvenile than cubs, a very low abundance of putative lignin and cellulose genes existed in part of analyzing samples but a significant higher abundance of genes involved in starch and hemicellulose degradation in juveniles than cubs. Moreover, a significant lower abundance of putative cellulolytic genes and a significant higher abundance of putative α-amylase and hemicellulase gene families were present in giant pandas than in omnivores or herbivores.
Introduction
The giant panda (Ailuropoda melanoleuca) is an endemic flagship species in China. It is well known that the giant panda belong to the Arctoidea (a superfamily of Carnivora) [1]. Unlike other species in Arctoidea, >99% of the food consumed by the giant panda consists of highly fibrous plant—bamboo. In order to process and utilize highly lignified bamboo, the giant panda exhibits several specialized morphological adaptations, such as panda’s thumb, large skull, wide flaring zygomatic arches, a prominent sagittal crest, and flat cheek teeth with elaborate crown patterns [2, 3]. However, the giant panda’s gastrointestinal (GI) tract is largely characteristic of carnivores [2]. The giant panda’s cecum is absent and its large intestine may not have any function for digesta fermentation [4] and its genome lacks homologs of the enzymes needed for cellulose degradation [5]. It has thus been hypothesized that the degradation of cellulose, a key component of its bamboo diet, should be dependent on its gut microbiota.
Some researchers have found some cellulose-degrading bacteria in the feces from giant pandas (such as, [6]). Based on metagenomic technology to detect bacteria diversity in gut microbiome of the giant panda, Zhu et al. [7] detected 13 operational taxonomic units closely related to Clostridium groups I and XIVa, both of which contain taxa known to digest cellulose, and putative genes coding for two cellulose-digesting enzymes. In contrast, other studies suggested that the giant panda gut microbiome does not have significant cellulose degradation potential [4, 8, 9], and their overall microbiome composition converged with those of carnivorous and omnivorous bears and not with herbivores [9]. The giant panda has a very low daily energy expenditure, which can partially explain why the giant panda can survive and adapt to the low-quality bamboo with low-energy digestive efficiency [10]. The functional role of the giant panda gut microbiota, particularly in cellulose degradation, is still largely unknown.
Diet is one key factor influencing the composition of GI tract microbial communities [11–13]. Thus, if the giant panda has the capacity to break-down cellulose, its gut microbiota should adapt to the dietary change from breast milk to bamboo during growth. Thus, the abundance of putative cellulolytic genes and/or bacteria should increase from birth to juveniles. Here, we perform the first large scale metagenome sequencing study of giant panda gut microbiota following animals from nursing to juvenile stages switching from milk to a bamboo diet.
Materials and methods
Samples
To investigate the microbiota development of the giant panda from birth to juvenile, 14 captive-born giant pandas housed in the Chengdu Research Base of Giant Panda Breeding and born in 2008 or 2011 were enrolled in this study (Supplementary Table S1) following the method pipeline (Supplementary Figure S1–S2). Following their diet and birth days, giant pandas’ feces were divided into four groups: S1 (panda breast milk as diet and <2 months old), S2 (between 3 and 12 months old and no bamboo found in their feces, commercial milk as dietary supplement), S3 (>6 months old and bamboo stems or leaves as diet), and S4 (>6 months old and bamboo shoots as diet) (Supplementary Table S1). The ingestion and health status of each giant panda were monitored daily by veterinarians and giant panda keepers and no samples were included in our study if giant pandas were fed antibiotics in the 2 months prior to sampling. Fresh fecal samples were collected immediately after defecation and were snap frozen on dry ice and shipped to the laboratory on dry ice, and were stored at −80 °C for no > 6 months before DNA extractions.
DNA extraction and data processing
DNA was isolated by bead-beating procedure described by Xue et al. [9] with the Qiagen QIAamp DNA Stool Mini Kit (Qiagen, Germany) according to the protocol for isolation of DNA for pathogen detection with slight improvements. For fecal samples without bamboo, ~1 g of each sample was resuspended in 1.2 ml of lysis buffer of the kit and transferred to a 2-ml screw-cap tube containing 0.3 g silica bead mixtures (0.2 g, 0.1 mm, and 0.1 g 1.0 mm silica beads) with a bead beater (BioSpec, Bartlesville, OK) set on high for 2 min. After agitation by the bead beater, DNA was extracted by following the manufacturer’s instructions. For samples with bamboo, a pretreatment protocol prior to DNA extraction according to the procedure described previously by Xue et al. [9] was added. Then, the cell suspension obtained from the pretreatment was used to extract DNA same as the samples without bamboos. The DNA concentrations of each sample were adjusted to 50 ng/μl for subsequent 16 S rRNA genes, first internal transcribed spacer (ITS-1), and metagenomic DNA libraries. Details on the 16 S, ITS-1, and metagenome data processing are available in the Supplementary Information data.
Comparison of carbohydrate-active genes between giant pandas and other mammalian metagenomes
To maximize the breadth of possible comparisons of carbohydrate-active genes, our data of S3 and S4 were combined with previously published mammalian metagenome data [11, 14] available on the MG-RAST database [15]. To minimize the potential effect of differences in sequence size among the different data sets, short reads in our study were mapped to the contig sequences of the pan-metagenome, then the mapping contigs were cut into 300 bp bins starting from the first mapping base for each single short read as a mimic data set. Then these data sets of S3 and S4 samples with other mammalian microbiome data were used to annotate against the CAZy database following previously published methods [11]. The Pearson’s correlation coefficient index between the mimic data set and corresponding paired-end data set was 0.75–0.802, which showed that our mimic data could represent the total data set.
Amylase copy number analysis in giant pandas host genomes
Primers for qPCR were designed to be specific to pancreatic amylase based on the giant panda, polar bear, cat, cheetah, and tiger reference genome sequences to compare copy numbers of amylase genes in Felidae (strict carnivore), Ursidae (omnivore), and the giant panda. In addition, primers for qPCR targeting the housekeeping gene TP53 were designed based on the five reference genome sequences and were used to adjust for DNA dilution quantity variation of different samples. DNA from blood samples of 41 giant pandas, 2 black bears, 2 brown bears, 22 tigers, 3 lions, 2 leopards was used as templates for qPCR. Experiments were performed and analyzed as described by Perry et al. [16]. All reactions were run in triplicate and normalized by comparisons with TP53. In addition, the PCR products from five giant pandas and five tigers were sequenced with the Sanger method.
Statistical analysis
The R packages Stats and Vegan were used to perform statistical analysis. Spearman rank correlation coefficient was used to assess the correlation between alpha diversity, taxonomic relative abundance, or genes relative abundance with increasing age of each panda. Significant differences in alpha diversity and taxonomy or relative abundance of genes between groups were tested using the Wilcoxon rank-sum test. ANOSIM and MRPP analyses were performed with the Vegan package in R software (version 2.15.3).
The random forest model has been shown to be a suitable model for 16 S rRNA and metagenomic data in R (R package “Random Forest”, ntree = 1000) with default parameters [17, 18]. Variable importance by mean decrease in accuracy was calculated for the random forest models using the full set of OTUs or gene data [19].
Results
Microbial diversity development from birth to juvenile
After quality filtering and assembly, 2,739,883 16 S rRNA gene sequences from 332 fecal samples and 3,604,489 ITS-1 sequences from 97 samples (Supplementary Table S1) were grouped into 1161 and 1006 OTUs, respectively. Across all 16 S rRNA samples, 98.3% of the total sequences were assigned into five phyla (total 10 phyla, Supplementary Figure S3). Firmicutes and Proteobacteria were the main phyla, which was consistent with other studies (e.g., [7, 9]). Bahl et al. [20] reported that freezing could result in decreasing the abundance of Bacteroidetes. Although Bacteroidetes had a low abundance with average 0.25% ( ± 1.6%) in our study, this was in line with previous studies on the giant panda gut microbiota (e.g. [4, 7–9]). Moreover, snap freezing of samples in our study should also preserve the integrity of the microbiota, as shown by Fouhy et al. [21]. However, we cannot exclude that the low abundance of Bacteroidetes may (at least partly) be owing to lysis during storage. The top 20 families included 94.4% of the total sequences and > 85% of the bacterial sequences affiliate to Enterobacteriaceae, Streptococcaceae, Lactobacillaceae, Clostridiaceae, Campylobacteraceae and Veillonellaceae for each group (Supplementary Figure S4). From birth (S1) to juvenile stages (S3/S4), the relative abundances of Firmicutes increased and Proteobacteria decreased significantly (P < 0.00001, T-tests; Supplementary Figure S3). The abundances of bacterial phyla were relatively constant in groups S3/4: this constancy among samples collected over > 12 months is an indication that the giant panda cub gut microbiome has reached a stable state, which suggests that the 1-year-old giant panda gut microbiome has many of the functional attributes of the adult microbiome.
For 16 S rRNA, pronounced shifts in abundant lineages around the second and 12th month seemed to follow dietary changes (Fig. 1a–c; Supplementary Figures S3–S5). During the first 2 months, bacterial communities were largely composed of Enterobacteriaceae, Streptococcaceae, Lactobacillaceae, and Campylobacteraceae (Supplementary Figure S4). Then, Enterobacteriaceae and Streptococcaceae decreased and Lactobacillaceae and Clostridiaceae increased between 3rd and 12th months (Supplementary Figure S4). In addition, Campylobacteraceae was relatively scarce in > 4 month old pandas and Veillonellaceae was relatively abundant between the 3rd and 12th months and scarce in other months (Supplementary Figure S4). After month 12, Clostridiaceae and Streptococcaceae reached highest abundances and Enterobacteriaceae and Lactobacillaceae lowest abundance (Supplementary Figure S4). The non-parametric Spearman rank correlation showed that relative abundance of Streptococcaceae and Clostridiaceae had a positive relationship with ages from birth to juvenile and Enterobacteriaceae was on the contrary (Supplementary Figure S4). At the genus level, eight taxa represented each > 1% of the total sequences across all samples. Among these genera, Escherichia/Shigella and Streptococcus were present in all samples and Clostridium was the only genus having a positive correlation with age (Supplementary Figure S4). Compared with the two other groups, S1 and S2 displayed significantly lower α-diversity (Fig. 1a and Supplementary Figures S5, P < 0.01) and Shannon diversity indices were similar to each other in every group (Supplementary Figure S5).
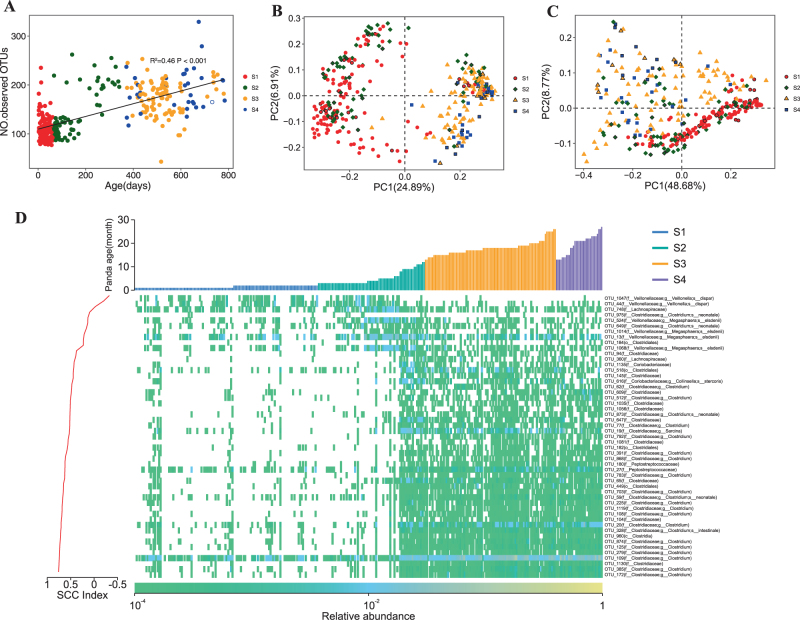
Fig. 1.
16S rRNA gene-based comparison of gut microbiota structure of giant pandas from birth to juvenile. a Relationship between alpha diversity and increasing age. Principal coordinate analysis (PCoA) using unweighted b and weighted c UniFrac distances of 16 S rRNA data. Each point corresponds to a community colored by collection date and diet (Table S1) and the black outer ring indicates metagenomic survey samples. d Heatmap of the 51 OTU-level phylotypes identified as key variables for differentiation between S1 and S3/S4 gut microbiota structure of the giant pandas. The OTUs are arranged according to their Spearman correlation coefficient (SCC) index between the relative abundance of OTUs and age of giant pandas. Each column represents one sample
Microbial community membership and structure showed significantly higher intra-individual variations between groups than that within each group. Fecal samples collected early in the time series harbored microbial communities more similar to each other than to samples collected later, and vice versa (P < 0.01; PERMANOVA with Monte Carlo). The giant pandas had a long time to shift their diet from panda milk to bamboo and this time was denoted by S2. During this time, supplementary milk and bamboo leaves/stem and/or shoots were provided, but no bamboo could be found in giant panda’s stools. The α- and β-diversity of S2 was overlapping with that of S1 or S3/4 (Fig. 1b–c, Supplementary Figure S5). In addition, the diversity of S3 (bamboo leaves/stem as diet) were not significantly different from that of S4 (bamboo shoots as diet) (Fig. 1b–c).
Some species of Clostridia were reported to be distantly related to known cellulose degraders [7, 9]. In our samples, we detected 196 OTUs affiliating to the class Clostridia (Supplementary Figure S6). Random Forest analysis identified 51 OTUs whose relative abundance reliably discriminated S1 and S3/4 samples (Fig. 1d; Supplementary Table S2). A total of 20 and 31 of these OTUs had a negative or positive correlation between their relative abundance and age, respectively, and none of these OTUs was closely associated with putative cellulolytic lineages (Fig. 1d and Supplementary Figure S6). In addition, six OTUs clustered with Cellulosilyticum lentocellum and Cellulosilyticum ruminicola who both can degrade cellulosic materials [22] and two with Clostridium cellulosi, a cellulolytic thermophile [23] (Supplementary Figure S6). Xue et al. [9] also found some OTUs closely related to the above three species. Zhu et al. [7] found some OTUs closely related to Clostridium cluster I and XIVa, which contains organisms known to digest cellulose. In this study, 10 OTUs affiliated to Clostridium cluster I and six to Clostridium cluster XIVa (Supplementary Figure S6). In addition, two OTUs were assigned into the cluster including Fibrobacter succinogenes, Ruminococcus flavefaciens, and Ruminococcus albus (Supplementary Figure S6) who are the main species involved in fiber digestion in rumen ecosystem [24]. In addition, 61 OTUs belonged to Bacteroidetes including potential cellulolytic organisms. These aforementioned OTUs showed no significant abundance difference between juveniles and cubs, which suggests that these OTUs may be not important for cellulose digestion in giant pandas.
Across all ITS samples, Ascomycota (on average: 63.35% ± 23.76%) and Basidiomycota (on average: 14.47% ± 15.27%) were the main fungal phyla and < 1% of the sequences were assigned into Blastocladiomycota, Chytridiomycota, and Zygomycota (Supplementary Figure S7). In addition, on average 22.08% (SD = 21.57%) of sequences belonged to unclassified phyla (Supplementary Figure S7). The dominant fungal classes were Eurotiomycetes, Sordariomycetes, Saccharomycetes, Tremellomycetes, and Dothideomycetes, which represented > 72% of the fungal sequences (Supplementary Figure S7). Out of the 419 fungal genera found in giant panda feces, some fungi have potential cellulolytic activity, including Aspergillus, Bjerkandera, Ganoderma, Humicola, Penicillium, Postia, Trametes, and Trichoderma, which have not been found in the GI tract of ruminants [25, 26]. Among these fungal genera with potential cellulolytic activity, only Aspergillus had moderately high abundance (on average: 2.82%) and was found in most samples (91 out of 97 samples); however, with no abundance difference between juveniles and cubs. A total of 1006 fungal OTUs were found using the ITS data and the numbers of OTUs increased with age (Supplementary Figure S8) and 253 OTUs were shared among the four groups. No OTUs belonged to the cellulolytic fungus Perenniporia, which was found in giant pandas by Tun et al. [27]. We did not find that fungal composition below phylum level had a positive correlation with age or to be enriched in one of the four groups (Supplementary Figures S9–S10), which may suggest that fungi are less important than bacteria for the gut microbiome development of giant pandas.
Metagenome development from birth to juvenile
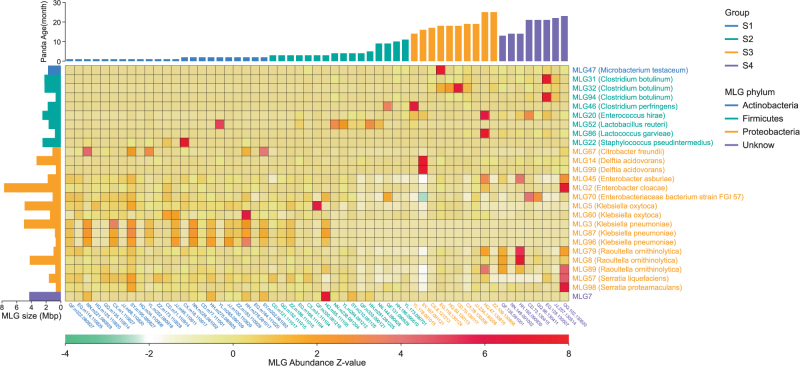
Based on the bacterial diversity and Unifrac distances, 57 samples (Fig. 1b–c; Supplementary Table S1) were selected for shotgun metagenome sequencing. In total, 299 gigabases (Gb) of paired-end read sequence data, with an average of 5.25 Gb (ranging from 1.5 to 9.7 Gb) for each sample (Supplementary Table S3), were obtained and a 552 Mbp Pan-metagenome including 461,872 contigs (Supplementary Tables S3–S4) was constructed. Then, Metagenome Linking Group (MLG), the group of metagenome sequences probably deriving from the same microbial genome, was assembled based on the Pan-metagenome. A total of 1406 MLGs ( > 10Kbp, Supplementary Table S5, Supplementary Figure S11) were obtained of which 811 MLGs were identified as Proteobacteria and 304 MLGs as Firmicutes. Out of the 1406 MLGs, 107 MLGs were > 0.5 Mbp with scaffold length totals in every MLG bin. The results of CheckM (version 1.0.7) using the 107 MLGs showed there were 58 MLGs with completeness% > = 50%. Out of the 58 MLGs, 30 MLGs showed completeness% >80% and contamination% < 25% and 9 MLGs received a very good evaluation result with completeness% > 97% and contamination% < 3% (Supplementary Table S6). We are thus confident that the MLGs obtained are of high quality. Twenty-six MLGs revealed significant abundance differences between different age stages (T-test, P < 0.01; Fig. 2, Supplementary Table S7). None of these 26 MLGs belonged to potential cellulolytic bacteria and most Firmicutes MLGs showed higher abundances at S3/S4, whereas most Proteobacteria MLGs were on the contrary (Fig. 2, Supplementary Table S7), which largely agrees with our results from 16 S data (Supplementary Figure S3).
Fig. 2.
Heatmap for metagenome linking groups (MLGs) with significant differences in different groups
From the Pan-metagenome of giant pandas, we predicted 816,364 genes with MetaGeneMark and 681,167 unique genes, which occupied 84% of the contigs. The number of genes increased from birth to juvenile (S1 to S3/4) and more genes were found in samples with bamboo shoots as diet (S4) than stem/leaves (S3) (Supplementary Figure S12). In total, 50.59% of the genes were classified into function database groups, 7.40% to carbohydrate-active enzymes (CAZy) level_2, 38.66% to eggNOG clusters, 25.60% to KEGG orthology, and 19.30% genes assigned to KEGG pathways, respectively. No significant difference of relative abundance among S1, S2, S3, and S4 were found using the functional annotation with the KEGG, CAZy, and the eggNOG databases at broad categories (Supplementary Figure S12). The annotation results of individual genes from the three databases at higher level categories were used to search for abundance differences of gene function between different groups with principal component analysis (PCA). PCA showed that only CAZy was useful for differentiation of group S1 and S3/4 (Supplementary Figure S13) and carbohydrate metabolism related genes in CAZy showed samples from group S1 clustering closer with only few outliers and same as group S3/4, suggesting that an important function of microbiota is to adapt to the diet transition during the development of giant panda from birth to juvenile. This was consistent with [14] who suggested that profiles of CAZy families were strong predictors of diet in mammals.
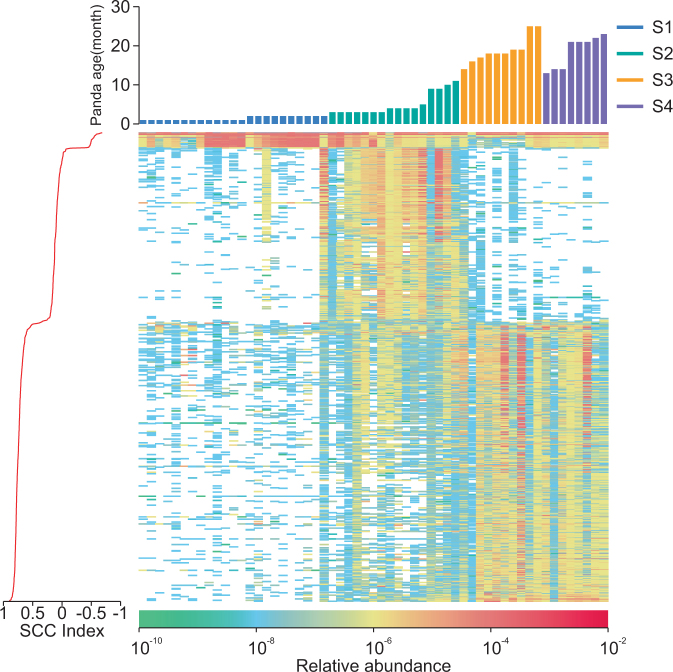
The Random Forest analysis of all 681,167 unique genes from the 57 samples showed that 1000 genes were sufficient to explain gene composition and functions among different age stages (Fig. 3; Supplementary Figure S14; Supplementary Table S8 and S9). The annotation of the 1000 genes with the CAZy database showed that 467 of the genes were assigned into carbohydrate esterases and glycoside hydrolase genes families involved in polysaccharide degradation [28]. Most (277) of the 467 genes were enriched in groups S3/4 and may be involved in the degradation of polysaccharide of bamboos and/or bamboo shoots, which is consistent with the results of PCA, suggesting that an important function of microbiota is to adapt to the diet transition (Supplementary Figure S13; Supplementary Table S8). Among the 13 GHs including cellulase genes (EC 3.2.1.4 or 3.2.1.91), only two genes were enriched in group S3/4 and belonged to GH12; though with low amino acid identity and low average abundance (Supplementary Table S8; Fig. 3). However, the KEGG annotation of the 1000 genes showed that no cellulase genes enriched in group S3/4 (Supplementary Figure S15; Supplementary Table S8).
Fig. 3.
Heatmap of the relative abundance of the 1000 genes obtained from Random Forest (Figure S14). SCC index between the relative gene abundance and age of giant pandas are shown for these genes (More details are shown in Supplementary Table S8)
Function enrichment analysis of 681,167 unique genes identified 22 ECs (Enzyme Commission) and 81 KOs (KEGG orthologous groups) whose proportional representation in fecal microbiomes differed significantly with age following KEGG database (Supplementary Table S10). The genes enriched in group S3/4 included some genes involved in phenylalanine metabolism, fatty acid biosynthesis, purine metabolism, glutathione metabolism, antibiotic resistance (e.g., transporters), and streptomycin biosynthesis. However, these did not include putative cellulase genes (EC 3.2.1.4 or 3.2.1.91) (Supplementary Table S10). Putative cyanate lyases (EC 4.2.1.104) were also enriched in S3/4 (Supplementary Table S10). Cyanate is toxic to all animals and widely distributed in plants, including bamboo shoots. The high abundance of cyanate lyases in group S3/4, together with the recently reported host rhodanese activity in giant pandas [29], might help giant pandas to prevent cyanide intoxication from bamboo. To our knowledge, this is the first report about cyanate lyases enriched in gut microbiota. In addition, no significant relative abundance difference of genes existed between group S3 and S4. The group S1 samples had an enrichment of carbohydrate-metabolizing genes involved in simple sugar (such as, lactose/galactose, and glucose) uptake and utilization, pyruvate and glycerolipid metabolism. These data likely reflected high quantities of breast milk in diets of giant pandas in group S1/2, which was consistent with the CAZy annotation, which differentiated S1 from the other categories (Supplementary Figure S13). Except these, some genes involved in secretion systems, antibiotic resistance (e.g., transporters), transcription factors, peptidases, and the biosynthesis of phenylalanine, tyrosine and tryptophan were also enriched in group S1 (Supplementary Table S10). These results were similar with the annotation of the 1000 genes of the Random Forest analysis (Supplementary Table S8).
The survey of gene families found three CAZyme classifications according to CAZy and 21 non-supervised orthologous groups (NOGs) according to eggNOG with significant abundance difference following age (Supplementary Table S10). CBM26, a gene family with starch-binding domains, was overrepresented in group S3/4, suggesting that giant pandas have higher capabilities to take-up and utilize starch following the development from birth to juvenile. By contrast, GH108 (EC 3.2.1.17) with putative antimicrobial activity and GT48 (EC 2.4.1.34) involving the cell wall glucan biosynthesis had higher abundance in 2-month-old giant pandas (group S1) than in 1-year-old pandas (group S3/4). Thirteen NOGs, which represented—among others—purine biosynthesis, deacetylase, transport systems, DNA ligase, transposase, heptaprenyl diphosphate synthase, were detected in significantly higher proportions in giant pandas > 1-year old compared with 2 months old pandas. In contrast, eight NOG protein families, involved in outer membrane porins, heat shock proteins, and transcriptional regulators, were depleted in group S3/4.
We also analyzed all the genes involved in starch and sucrose metabolism pathways including the genes degrading cellulose in giant pandas with the KEGG database. A total of 56 KOs were involved in the starch and sucrose metabolism pathway, 35 of the 56 KOs were detected in giant pandas. A total of 29 ECs had at least one KO with significant difference between one group and another group (P < 0.05, T-test) (Supplementary Table S11; Supplementary Figure S16). Three ECs (3.2.1.21; 2.7.1.2; 2.4.1.21) had a positive age-related change (SSC > 0.49) and were enriched in group S3/4. Nine KOs had higher abundance in group S1 than group S2 or S3 and belonged to the putative cellulase gene (EC 3.2.1.4). A putative exoglucanase (EC 3.2.1.91) was annotated only in few samples with very low relative abundance (Supplementary Figure S16). KEGG annotation of all genes in this study showed that no genes related to phenol oxidases (laccases) and peroxidases (lignin peroxidases and manganese peroxidases), the key enzymes in lignin degradation pathways [30], were present in the metagenome.
The carbohydrate-active enzymes genes among mammals
The samples of groups S3/4 were merged with metagenome data from other mammals [11, 14] to compare their putative CAZymes profiles. A total of 24,685 putative GH-encoding genes representing 108 different GH families were annotated. Among the 108 families, 36 were significantly more abundant in giant panda metagenomes than other mammals and 22 were less abundant (Supplementary Table S12; P-value < 0.05, T-test). These enriched families in giant panda metagenomes included α-amylase families (including GH13 and CBM48) that are able to bind and degrade starch [31], gene families with hemicellulose degrading activity including four families (GH1, 4, 8, and 31), and one gene family involved in both cellulose and hemicellulose degradation pathways (GH8, [32]. Other GHs involved in cellulose degradation pathways were significantly more abundant in herbivores than in carnivore and giant panda metagenomes (Supplementary Table S12; P-value < 0.05, Wilcoxon rank-sum test). Considering the total abundance of GH families related to cellulase genes suggested that carnivores had the lowest and herbivores the highest abundance and giant pandas had a slightly higher abundance than carnivores (Fig. 4). In addition, GH90 was only found in the giant panda (Supplementary Table S12), which is involved in endorhamnosidase enzymatic activity to hydrolyze the O-antigen [33].
Fig. 4.
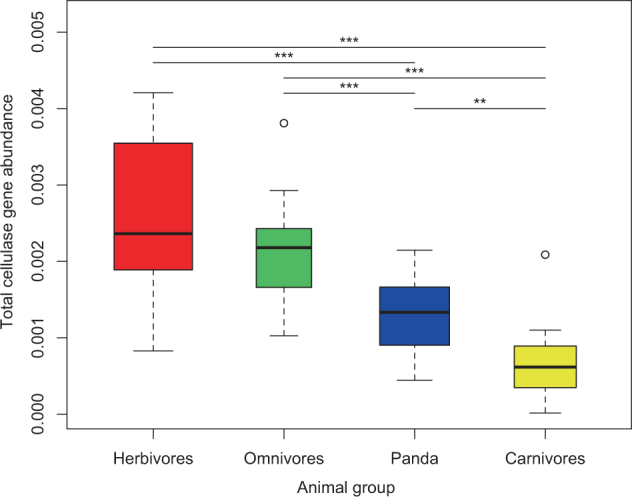
Comparisons of the total abundance of putative cellulase gene families of gut microbiomes of giant pandas and other mammals by Wilcoxon signed-rank test. The lines and squares inside boxes represent the median and mean, respectively. ** P < 0.01, *** P < 0.001
UPGMA clustering of CAZymes abundance profiles (Supplementary Figure S17) showed that the giant panda microbiota grouped separately from herbivore and carnivore microbiota, but clustered most closely with the Ursidae family (Supplementary Figure S17–18), which reflected that the giant panda and Ursidae microbiomes might have similar polysaccharide utilization potential.
Amylase gene copy number
The amylase sequences showed that some heterozygosity sites exist in giant pandas (Supplementary Figure S19 and Supplementary Table S13), which showed that at least two amylase gene copies are present in giant pandas. This was verified by qPCR that proved the median number of pancreatic amylase gene copies was two (mean = 1.89) in Ursidae (including giant pandas, black bears, and brown bears) and one (mean = 1.12) in Felidae (including tigers, lions, and leopards) (Supplementary Table S14). The complete genome analysis also showed one copy of amylase in strict carnivores (Such as, cheetah, leopard, seal, and others) and more than one amylase copy in Ursidae or omnivores (Such as polar bear, giant panda, and dog) (Supplementary Table S15).
Discussion
Our analysis showed that, although few genes in giant pandas microbiota are involved in cellulose degradation—with low abundance in juveniles and cubs (Supplementary Figure S15–16, Supplementary Table S11)—the abundance of putative cellulase genes is significantly lower in giant pandas than in omnivores or herbivores (Fig. 4), and no genes were annotated by KEGG among lignin degradation pathways, which suggests that giant pandas do not depend on cellulose or lignin to obtain energy. Then, which carbohydrates might provide the giant panda with energy?
Amylase is the digestive enzyme for starch and its copy number is positively correlated with the amylase concentration and enzymatic activity level [16]. Our results showed two gene copies of pancreatic amylase in Ursidae and one in Felidae, which proved that giant pandas have a higher capability to digest starch than strict carnivores. Following the development of giant pandas from birth to juvenile stages, more α-amylase family genes were found in the metagenome (Fig. 3, Supplementary Table S8), suggesting increasing abilities to digest starch following the dietary transition from breast milk to bamboo. Compared with other mammals, giant pandas had significantly more α-amylase families in their gut microbiota (Supplementary Table S12), suggesting that the giant panda is better able to digest starch than other mammals. In line with this, high levels of amylase activity in the giant panda intestine and strong abilities to digest starch-rich food were described recently [34].
Microbial fermentative processes are essential for cellulose degradation but are relatively slow, so animals that rely on those processes typically possess enlarged GI compartment(s) to maintain a specific microbiota and to decrease digesta flow [35]. But the giant panda has a comparatively short small intestine and a very rapid passage time of digesta ranging from only 5 to 11 h [2], resulting in a weak ability to degrade cellulose with only ~8% degradation coefficients [36]. Our study showed that the gut microbiota of giant pandas included a large and diverse set of genes for hemicellulose-hydrolysis enriched in juveniles, but only low abundance of genes for cellulose degradation (Supplementary Tables S8, S10-S12; Figs. 3–4; Supplementary Figures S16–S17). These results are clearly consistent with the observation that giant pandas have much higher digestibility for hemicellulose than cellulose [36], and that the giant panda prefers high hemicellulose bamboo shoots, tender leaves and first-year bamboo [37], and adult giant pandas living on bamboo shoots as diet gained weight steadily (0.23 ± 0.24 kg per day, n = 15) and the same individuals lived on only bamboo stems diet lost weight (0.07 ± 0.40 kg per day, n = 15; [38]. Moreover, Senshu et al. [39] reported that no bamboo cell wall digestion occurred by microbes from the digestive tract of the panda following incubating the panda feces with bamboo as a substrate in vitro. Thus, sufficient bamboo shoots are one key factor for giant pandas’ breeding for energy need and more attention should be payed to protect bamboo shoots during mating and parturition season of giant pandas in the wild. Lignin and cellulose in bamboo form a covalently cross-linked matrix with hemicellulose, which hampers the degradation of hemicellulose. Thus, we suggest that the low abundant enzymes degrading lignin and cellulose in the giant panda gut microbiome help releasing the more easily digestible hemicellulose and starch from bamboo cell walls, instead of being responsible for complete cellulose degradation.
In conclusion, our data strongly suggest that the giant pandas’ gut microbiota is not well adapted to the degradation of cellulose and lignin in their highly fibrous bamboo diet, but has evolved to utilize more readily digestible carbohydrates to maximize nutritional and energetic intake from bamboo. This indicates that the digestive physiology of the giant panda overrides the importance of diet in the assembly of the gut microbiota.
Electronic supplementary material
Acknowledgements
This study was supported by the National Key Technology R&D Program (2012BAC01B06), Chengdu Giant Panda Breeding Research Foundation (CPF08002, CPF2015-01, and CPF2015-09), Science and Technology Department of Sichuan Province (2015JY0236), and Chengdu research base of giant panda breeding. We thank Lena Li, Hong Liu, Bo Zhao and other staff at the Chengdu Research Base of Giant Panda Breeding for assistance with sampling, Benhua Zeng, Wujiao Li, Ting Huang, Xuhao Song, and Chengsheng Xue for discussion.
Conflict of interest
The authors declare that they have no conflict of interest.
Contributor Information
Wenping Zhang, Email: zhang_zoology@163.com.
Zhihe Zhang, Email: zzh@panda.org.cn.
Electronic supplementary material
The online version of this article (10.1038/s41396-018-0051-y) contains supplementary material, which is available to authorized users.
References
- 1.Zhang Y, Ryder O. Molecular phylogeny of the superfamily Arctoidea. J Genet Genom. 1997;24:15–22. [Google Scholar]
- 2.Wildt DE, Zhang A, Janssen DL, Ellis S (2006). Giant pandas: biology, veterinary medicine and management. In: Wildt DE, Zhang A, Janssen DL, Ellis S (eds). Cambridge University Press: Cambridge.
- 3.Figueirido B, Serrano-Alarcón FJ, Palmqvist P. Geometric morphometrics shows differences and similarities in skull shape between the red and giant pandas. J Zool. 2012;286:293–302. doi: 10.1111/j.1469-7998.2011.00879.x. [DOI] [Google Scholar]
- 4.Hirayama K, Kawamura S, Mitsuoka T, Tashiro K. The faecal flora of the giant panda (Ailuropoda melanoleuca) J Appl Bacteriol. 1989;67:411–5. doi: 10.1111/j.1365-2672.1989.tb02511.x. [DOI] [PubMed] [Google Scholar]
- 5.Li R, Fan W, Tian G, Zhu H, He L, Cai J, et al. The sequence and de novo assembly of the giant panda genome. Nature. 2010;463:311–7. doi: 10.1038/nature08696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou Z, Zhou X, Li J, Zhong Z, Li W, Liu X, et al. Transcriptional regulation and adaptation to a high-fiber environment in Bacillus subtilis HH2 isolated from feces of the giant panda. Plos ONE. 2015;10:e0116935. doi: 10.1371/journal.pone.0116935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu L, Wu Q, Dai J, Zhang S, Wei F. Evidence of cellulose metabolism by the giant panda gut microbiome. Proc Natl Acad Sci USA. 2011;108:17714–9. doi: 10.1073/pnas.1017956108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wei G, Lu H, Zhou Z, Xie H, Wang A, Nelson K, et al. The microbial community in the feces of the giant panda (Ailuropoda melanoleuca) as determined by PCR-TGGE profiling and clone library analysis. Microb Ecol. 2007;54:194–202. doi: 10.1007/s00248-007-9225-2. [DOI] [PubMed] [Google Scholar]
- 9.Xue Z, Zhang W, Wang L, Hou R, Zhang M, Fei L, et al. The bamboo-eating giant panda harbors a carnivore-like gut microbiota, with excessive seasonal variations. MBio. 2015;6:e00022–00015. doi: 10.1128/mBio.00022-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nie Y, Speakman J, Wu Q, Zhang C, Hu Y, Xia M, et al. Exceptionally low daily energy expenditure in the bamboo-eating giant panda. Science. 2015;349:171–4. doi: 10.1126/science.aab2413. [DOI] [PubMed] [Google Scholar]
- 11.Muegge BD, Kuczynski J, Knights D, Clemente JC, Gonzalez A, Fontana L, et al. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science. 2011;332:970–4. doi: 10.1126/science.1198719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med. 2009;1:6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–8. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sanders JG, Beichman AC, Roman J, Scott JJ, Emerson D, McCarthy JJ, et al. Baleen whales host a unique gut microbiome with similarities to both carnivores and herbivores. Nat Commun. 2015;6:8285. doi: 10.1038/ncomms9285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilke A, Bischof J, Gerlach W, Glass E, Harrison T, Keegan KP, et al. The MG-RAST metagenomics database and portal in 2015. Nucleic Acids Res. 2016;44:D590–D594. doi: 10.1093/nar/gkv1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perry GH, Dominy NJ, Claw KG, Lee AS, Fiegler H, Redon R, et al. Diet and the evolution of human amylase gene copy number variation. Nat Genet. 2007;39:1256–60. doi: 10.1038/ng2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Knights D, Costello EK, Knight R. Supervised classification of human microbiota. FEMS Microbiol Rev. 2011;35:343–59. doi: 10.1111/j.1574-6976.2010.00251.x. [DOI] [PubMed] [Google Scholar]
- 18.Liaw A, Weiner M (2002). Classification and regression by randomForest. R News. 2002:2/3.
- 19.Feng Q, Liang S, Jia H, Stadlmayr A, Tang L, Lan Z, et al. Gut microbiome development along the colorectal adenoma-carcinoma sequence. Nat Commun. 2015;6:6528. doi: 10.1038/ncomms7528. [DOI] [PubMed] [Google Scholar]
- 20.Bahl MI, Bergstrom A, Licht TR. Freezing fecal samples prior to DNA extraction affects the Firmicutes to Bacteroidetes ratio determined by downstream quantitative PCR analysis. FEMS Microbiol Lett. 2012;329:193–7. doi: 10.1111/j.1574-6968.2012.02523.x. [DOI] [PubMed] [Google Scholar]
- 21.Fouhy F, Deane J, Rea MC, O’Sullivan O, Ross RP, O’Callaghan G et al. (2015). The effects of freezing on faecal microbiota as determined using MiSeq sequencing and culture-based investigations. Plos ONE. 2015;10:e0119355. [DOI] [PMC free article] [PubMed]
- 22.Cai S, Dong X. Cellulosilyticum ruminicola gen. nov., sp. nov., isolated from the rumen of yak, and reclassification of Clostridium lentocellum as Cellulosilyticum lentocellum comb. nov. Int J Syst Evol Microbiol. 2010;60:845–9. doi: 10.1099/ijs.0.014712-0. [DOI] [PubMed] [Google Scholar]
- 23.He YL, Ding YF, Long YQ. Two cellulolytic Clostridium species: Clostridium cellulosi sp. nov. and Clostridium cellulofermentans sp. nov. Int J Syst Bacteriol. 1991;41:306–9. doi: 10.1099/00207713-41-2-306. [DOI] [PubMed] [Google Scholar]
- 24.Sirohi SK, Singh N, Dagar SS, Puniya AK. Molecular tools for deciphering the microbial community structure and diversity in rumen ecosystem. Appl Microbiol Biotechnol. 2012;95:1135–54. doi: 10.1007/s00253-012-4262-2. [DOI] [PubMed] [Google Scholar]
- 25.Sanchez C. Lignocellulosic residues: biodegradation and bioconversion by fungi. Biotechnol Adv. 2009;27:185–94. doi: 10.1016/j.biotechadv.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 26.Zhou XW, Cong WR, Su KQ, Zhang YM. Ligninolytic enzymes from Ganoderma spp: current status and potential applications. Crit Rev Microbiol. 2013;39:416–26. doi: 10.3109/1040841X.2012.722606. [DOI] [PubMed] [Google Scholar]
- 27.Tun HM, Mauroo NF, Yuen CS, Ho JCW, Wong MT, Leung FCC. Microbial diversity and evidence of novel homoacetogens in the gut of both geriatric and adult Giant Pandas (Ailuropoda melanoleuca) Plos ONE. 2014;9:e79902. doi: 10.1371/journal.pone.0079902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Biely P. Microbial carbohydrate esterases deacetylating plant polysaccharides. Biotechnol Adv. 2012;30:1575–88. doi: 10.1016/j.biotechadv.2012.04.010. [DOI] [PubMed] [Google Scholar]
- 29.Huang H, Yie S, Liu Y, Wang C, Cai Z, Zhang W, et al. Dietary resources shape the adaptive changes of cyanide detoxification function in giant panda (Ailuropoda melanoleuca) Sci Rep. 2016;6:34700. doi: 10.1038/srep34700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fang W, Fang Z, Zhou P, Chang F, Hong Y, Zhang X, et al. Evidence for lignin oxidation by the giant panda fecal microbiome. Plos ONE. 2012;7:e50312. doi: 10.1371/journal.pone.0050312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Machovič M, Janeček Scaron. Domain evolution in the GH13 pullulanase subfamily with focus on the carbohydrate-binding module family 48. Biologia. 2008;63:1057. doi: 10.2478/s11756-008-0162-4. [DOI] [Google Scholar]
- 32.Petersen L, Ardevol A, Rovira C, Reilly PJ. Mechanism of cellulose hydrolysis by inverting GH8 endoglucanases: a QM/MM metadynamics study. J Phys Chem B. 2009;113:7331–9. doi: 10.1021/jp811470d. [DOI] [PubMed] [Google Scholar]
- 33.Iwashita S, Kanegasaki S. Enzymic and molecular properties of base-plate parts of bacteriophage P22. Eur J Biochem. 1976;65:87–94. doi: 10.1111/j.1432-1033.1976.tb10392.x. [DOI] [PubMed] [Google Scholar]
- 34.Zheng YC, Fei LS, Li F, Niu LL, Zhang ZH. Analysis of digestive enzyme activities in the digestive tract of giant pandas. Sichuan J Zool. 2009;28:397–400. [Google Scholar]
- 35.Karasov WH, Douglas AE. Comparative digestive physiology. Compr Physiol. 2013;3:741–83. doi: 10.1002/cphy.c110054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dierenfeld ES, Hintz HF, Robertson JB, Van Soest PJ, Oftedal OT. Utilization of bamboo by the giant panda. J Nutr. 1982;112:636–41. doi: 10.1093/jn/112.4.636. [DOI] [PubMed] [Google Scholar]
- 37.Nie Y, Zhang Z, Raubenheimer D, Elser JJ, Wei W, Wei F. Obligate herbivory in an ancestrally carnivorous lineage: the giant panda and bamboo from the perspective of nutritional geometry. Funct Ecol. 2015;29:26–34. doi: 10.1111/1365-2435.12302. [DOI] [Google Scholar]
- 38.Wang HR, Zhong HJ, Hou R, Ayala J, Liu GM, Yuan SB et al. (2017). A diet diverse in bamboo parts is important for giant panda (Ailuropoda melanoleuca) metabolism and health. Sci Rep. 2017;7:3377. [DOI] [PMC free article] [PubMed]
- 39.Senshu T, Miyata K, Ohya A, Mikogai J, Morita M, Nakao T, et al. Procedure and mechanisms of bamboo cell wall digestion in the Giant Panda, Ailuropoda melanoleuca. Mamm Study. 2014;39:219–28. doi: 10.3106/041.039.0405. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.