Abstract
Arbuscular mycorrhizal fungi (AMF) colonise roots of most plants; their extra-radical mycelium (ERM) extends into the soil and acquires nutrients for the plant. The ERM coexists with soil microbial communities and it is unresolved whether these communities stimulate or suppress the ERM activity. This work studied the prevalence of suppressed ERM activity and identified main components behind the suppression. ERM activity was determined by quantifying ERM-mediated P uptake from radioisotope-labelled unsterile soil into plants, and compared to soil physicochemical characteristics and soil microbiome composition. ERM activity varied considerably and was greatly suppressed in 4 of 21 soils. Suppression was mitigated by soil pasteurisation and had a dominating biotic component. AMF-suppressive soils had high abundances of Acidobacteria, and other bacterial taxa being putative fungal antagonists. Suppression was also associated with low soil pH, but this effect was likely indirect, as the relative abundance of, e.g., Acidobacteria decreased after liming. Suppression could not be transferred by adding small amounts of suppressive soil to conducive soil, and thus appeared to involve the common action of several taxa. The presence of AMF antagonists resembles the phenomenon of disease-suppressive soils and implies that ecosystem services of AMF will depend strongly on the specific soil microbiome.
Introduction
Soil microorganisms are fundamental for soil health and provide ecosystem services that are essential for plant production [1, 2]. Societal awareness of agricultural sustainability is continuously growing, making it increasingly important to unravel the biotic and abiotic factors in different soils that support the beneficial components of their microbiome.
Arbuscular mycorrhizal fungi (AMF) are present in most soils and form symbiotic associations with majority of crop plants [3]. The mycorrhizal symbiosis has multiple beneficial impacts on nutrient cycling and plant stress tolerance [4] and represents under-exploited potential to increase global food security [5]. Hence, a well-developed extra-radical mycelium (ERM) that proliferates in the bulk soil beyond the rhizosphere is important for plant P uptake, while the AMF depend on their host plant for carbon nutrition [6, 7]. The large surface area of the ERM provides nutrient-rich niches for colonisation and growth of other soil microorganisms, in particular bacteria [8]. Hence, several members of the Glomeraceae increase the abundance of Firmicutes, Streptomycetes and members of the family Oxalobacteraceae on hyphae, or in soil affected by hyphae [9–11]. The hyphae-associated bacteria may in return affect the AMF as exemplified by mycorrhizal helper bacteria that promote AMF hyphal growth and root colonisation [12, 13]. Yet, even bacteria with suppressive effects have been identified [14, 15]. These observations support the notion that soil harbours both stimulatory and antagonistic bacteria towards AMF. However, we know relatively little about how AMF hyphae and bacteria interact in soil outside the rhizosphere, even though those hyphae represent the major niche where interactions with soil bacteria can take place [10].
Scattered evidence suggests that natural soils can suppress AMF colonisation and plant growth responses ([16]; see also review by Nehl et al. [17]). In one study, the addition of a bacterial soil filtrate decreased the length of the ERM [18]. Furthermore, the contribution of the ERM pathway to plant P uptake was negligible in one out of five unsterile soils, which otherwise produced similar levels of root colonisation [19]. These observations led us to speculate that soils may be either suppressive or conducive to ERM activity. Whereas soils suppressive towards plant pathogenic fungi, so-called ‘disease-suppressive soils’, are well-known [20], it remains unknown whether soil-induced suppression of AMF is common. Disease suppression may be related to the total microbial community in the soil (general suppression), while in other cases, suppression is caused by the specific effects of selected soil microorganisms (specific suppression) [21]. Thus, soil pasteurisation strongly reduces both general and specific disease suppression of soils [22, 23], while transferability of small soil volumes is a key characteristic of specific suppression [21]. Importantly, suppressive effects mediated by components of the soil microbiota might well be confounded by specific characteristics of the soil physicochemical environment such as low temperature [24] or low pH [25, 26].
Microbial community analysis has revealed that suppression of fungal root pathogens may be associated with groups of bacteria or fungi in the rhizosphere microbiome [22, 27, 28]; Actinomycetes and Lysobacter strains are examples of Rhizoctonia-suppressive bacteria [29]. Interestingly, microbial consortia appear to be more suppressive than individual microbial isolates [20, 30].
The aim of this work was to test the following hypotheses: (1) that field soils exert differential effects on the activity of the ERM of AMF; and (2) that the soil contains microbial populations responsible for suppression of the ERM activity. To address these aims, 21 cultivated soils were screened for their ability to suppress AMF in a model system where P uptake from radioisotope-labelled soil was used as a proxy for activity of ERM. The ERM activity was related to a range of soil physicochemical characteristics and to the composition of the soil microbiome.
Materials and methods
Soils, plant and AMF
Plants grew in soil collected from the Risø field site at the Technical University of Denmark (referred to as ‘Risø’ hereafter; Table 1). The soil was γ-irradiated (15 kGy) and mixed 1:1 with quartz sand (w/w) and with basal nutrients as in ref. [31]. This semi-sterile standard soil was used in all pots except in Expt. 1 where half of the pots contained the corresponding unsterile soil. Soils used in hyphal compartments (HC) of the plant model system (see below) were collected from 21 Scandinavian field sites and had contrasting organic matter content, texture, pH and plant-available P concentrations (Table S1). The soils were air-dried, sieved and stored dry; one soil (‘Risø stored’) had been collected in 1982 and was subsequently stored dry.
Two different AMF were used as inoculum: experiment 1 (see below) used Funneliformis mosseae (Nicolson & Gerd.) C. Walker & A. Schüßler (BEG85), formerly known as Glomus mosseae, and experiments 2 and 3 used Rhizophagus irregularis (Blaszk., Wubet, Renker & Buscot) C. Walker & A. Schüßler (BEG87), f.k.a. Glomus intraradices. Both inocula consisted of dry soil, spores and root fragments from Trifolium subterraneum pot cultures. Mycorrhizal associations were established with Medicago truncatula cv. Jemalong (A17). Plants received supplemental N during the course of growing to suppress nodulation by Rhizobium.
Model system
The model system is modified from ref. [32] (Figure S1). M. truncatula plants colonised by AMF served as donors for the production of ERM. The system enables studies of the ability of ERM to colonise, and take up P from, a mesh-enclosed soil patch (the ‘HC’ that was buried in pots with the standard Risø soil supporting plant growth). The HC soil was sieved (<2 mm), mixed 1:1 with quartz sand and labelled with a radioisotope of P. The plant uptake of the radioisotope at harvest was subsequently quantified and used as a proxy for the P uptake activity of the ERM. The HCs were 50 mL plastic cylinders capped at both ends with 25 μm nylon mesh, which allowed for ingrowth of ERM, but not roots.
Experiment 1: do field soils suppress ERM production and activity?
A dual radioisotope labelling experiment was conducted to test whether ERM activity differs between semi-sterile and unsterile soil. Three pre-germinated M. truncatula seeds were sown into each pot, which contained 1.5 kg standard soil mixed with 20 mg P per kg (as KH2PO4). Half the pots contained semi-sterile soil, while the other half contained unsterile soil. The soil in half of each of the semi-sterile and unsterile pots was mixed thoroughly with 75 g of F. mosseae inoculum. Two HCs were placed in each pot; both contained 55 g of standard soil, either semi-sterile or unsterile that had been uniformly mixed with either 5 kBq g−1 soil of carrier-free H333PO4 (for semi-sterile soil) or 5 kBq g−1 soil of carrier-free H332PO4 (for unsterile soil). Each treatment had five replicates and plants were harvested after 40 days.
Experiment 2: how common is the AMF suppression?
Prevalence of, and variation in, the AMF-suppressive effects of field soil was studied using 21 Scandinavian field soils. All soils were labelled with 3 kBq g−1 of carrier-free H333PO4 that was uniformly mixed into the soil before packing 55 g soil into a HC. HCs of each soil, replicated four times, were buried into pots containing semi-sterile standard soil amended with 10 mg P per kg and mixed with 75 g of R. irregularis inoculum (total weight: 1.0 kg). Two M. truncatula plants were grown in each pot, and harvested after 32 days.
Experiment 3: what are the AMF-suppressive components of field soils?
Two soils, Møystad E2 and Risø stored, had contrasting AMF-suppressive effects in Expt. 2 and were thus selected for further investigation into the AMF-suppressive components of field soils. To determine whether soil pH affects soil suppressive activity, the strongly AMF-suppressive Møystad E2 soil, which had a baseline pH of 4.4, was supplemented with CaCO3 at the rates of 0.25, 1.0 and 4.0 g of CaCO3 per kg soil to achieve new soil pH levels of 4.7, 5.5 and 7.1, respectively. To address whether AMF suppression has a biological background, the two soils were adjusted to 10 % water and subjected to a pasteurisation in a water bath (85 °C, 90 min). Corresponding non-pasteurised treatments were included as controls. To distinguish between general or specific AMF suppression, a soil transfer experiment was set-up as follows. Two soils deemed ‘conducive’ from Expt. 2 (Møystad E7 and Risø stored) were mixed in the following ratios with the respective ‘suppressive’ partner soil (Møystad E2 and Risø): 10:0; 9:1; 1:1; 1:9; and 0:10. Following these modifications, the soils were mixed in a 1:1 ratio with sand, labelled with radioisotope at 4 kBq g−1 soil carrier-free H333PO4 and packed into HCs. Pots and seeds were prepared as for Expt. 2, with three replicates per treatment. Plants were harvested after 29 days.
Harvesting and sample analyses
Plants were harvested and analysed as follows: roots were separated from shoots; roots were washed free of soil; blotted dry; and a weighed subsample was stored in 50% EtOH for determination of mycorrhizal root colonisation by microscopy after clearing in 10% KOH and staining with Trypan blue (modified from ref. [33]). The remaining root tissue and the total shoot tissue was dried at 70 °C (>48 h) and dry weights recorded.
Shoots were ground before acid digestion, and analysis of the shoot 33P and 32P contents was conducted using a Packard liquid scintillation counter (PerkinElmer, Waltham MA, USA). Subsamples of soil from each HC were taken for quantification of soil P concentration, soil pH, AMF hyphal length density (HLD; Expt. 1 only), phospholipid-derived fatty acid (PLFA) analysis (Expt. 3) and 16S rRNA gene sequencing (Expt. 2 and 3). HLD was determined in duplicates of 2 g dry soil by a grid intersect method [34]. Soil P status was determined from P concentrations in extracts with water [35] or NaHCO3 [36], and soil pH was determined after shaking with CaCl2 [37].
PLFAs were extracted from 2 g frozen soil according to ref. [38] using nonadecanoic acid as the internal standard. Samples were analysed by gas chromatography mass spectromety using aVarian CP482fitted with a 50 m CPSIL8CB column (Ø 0.25 mm, film thickness 0.25 µm), after injection of 3 µL using a temperature programme of 50 °C for 1.89 min, 20 °C min−1 to 160 °C, hold for 5 min, 5 °C min−1 to 270 °C, hold for 0 min, 50 °C min−1 to 325 °C, hold for 3 min. The mass spectometer was operated in a scan mode, m/z 50–300.
DNA was isolated from frozen HC soil by bead-beating and phenol-chloroform extraction as described in ref. [39]. Each DNA sample contained pooled DNA isolated from two subsamples of 0.5 g soil from each HC. Subsequently, 16S rRNA gene copies were quantified by quantitative PCR (qPCR) using primers bac341F and bac907R [40, 41]. Reactions were carried out with 1× Brilliant III SYBR Green QPCR Master Mix (Agilent Technologies, USA), 0.4 µM of each primer and 1 mg ml−1 bovine serum albumin under thermal cycling conditions: 95 °C for 3 min followed by 35 cycles of 95 °C for 15 s and 57 °C for 20 s.
Illumina MiSeq 300 bp paired-end sequencing of the hypervariable V3-V4 regions of the 16S rRNA gene, amplified with primers Bakt_341F and Bakt_805R (from ref. [42]) was performed on the extracted DNA by Macrogen Inc. (Seoul, Rep. of Korea). Sequencing of soil DNA from Expt. 2 and Expt. 3 was done in two separate sequencing runs and data were analysed separately. Sequencing data were analysed using the CLC Genomics Workbench with the Microbial Genomics Module (Qiagen) using the software’s default settings. Partial 16S rRNA gene sequences were clustered and assigned to operational taxonomic units (OTUs) with 97% similarity using the SILVA 16S rRNA reference database release 119 without de novo OTU clustering.
Statistics
A two-way analysis of variance (ANOVA) was conducted on Expt. 1 data for each HC (semi-sterile or unsterile) separately, with the factors (1) soil in rooting compartment (semi-sterile or unsterile) and (2) AMF inoculation (inoculated with F. mosseae or not). Shoot 33P content, 32P content and HLD were analysed by Student’s t-test to compare between the two HCs within each treatment.
One-way ANOVA compared the shoot 33P activity in plants grown across 21 unsterile soils (Expt. 2), in Møystad E2 soil of increasing pH (Expt. 3) and in the soil dilution experiment for the pairs of Møystad and Risø soils, respectively (Expt. 3), with Tukey’s post hoc tests conducted where appropriate (JMP Pro v. 12.0.1). For the pasteurisation experiment, Student’s t-tests were used to compare shoot 33P content from soils with and without pasteurisation treatment.
Regression analyses were performed between shoot radioactive P content and HLD (Expt. 1), and between shoot 33P content and a number of HC soil physicochemical characteristics (Expt. 2). Data that were non-normal, including shoot 33P content (as per Shapiro–Wilk test), were subjected to a log-transformation prior to statistical analyses. Statistical analyses were performed using JMP Pro v. 12.0.1, except PLFA data that were normalised and analysed by principal components analysis using Minitab v. 17.
For the purpose of the 16S rRNA gene amplicon data analyses, soils were considered AMF-suppressive when plant 33P uptake <1 kBq per pot. Principal coordinates analyses (PCoAs) were performed on Bray–Curtis dissimilarity matrices originating from 97% nucleotide similarity OTU tables using the software PAST v.2.17. Statistical significance of the overall difference in microbial community composition was tested by PERMANOVA analysis on Bray–Curtis dissimilarities with 9999 permutations using the Adonis function of the vegan package implemented in R. The α-diversity, represented by Shannon H and Chao-1 estimations, was calculated in PAST and compared by Student’s t-test. The online tool Venny (http://bioinfogp.cnb.csic.es/tools/venny/index.html) was used for Venn diagram construction. Similarity percentage analysis (SIMPER) on Bray–Curtis dissimilarities of genus-level OTU tables was performed in PAST. Differences in taxa abundance between the soil groups were tested by one-way ANOVA, with Tukey’s post hoc test using STAMP v. 2.1.3. Filtered genus-level OTU tables and the environmental variables pH, SOM, P, clay, silt and sand (Table S1) were included in a canonical correspondence analysis (CCA) performed in PAST, and significant correlations between the variables were tested by Spearman correlation analysis on the full genus-level OTU table in PAST. 16S rRNA gene amplicon sequences are deposited in NCBI’s Sequence Read Archive under BioProject PRJNA408226.
Results
Do field soils suppress production and activity of ERM?
Roots growing in soil with AMF propagules in Expt. 1 were abundantly colonised by the fungi (66% colonised root length in inoculated semi-sterile soil; 65% in unsterile soil; and 68% in inoculated unsterile soil). HLD was significantly (P < 0.05) lower in the HC with unsterile than in the HC with semi-sterile soil (Fig. 1a), and radioisotope uptake into shoots was significantly lower from unsterile soil (32P) than from semi-sterile soil (33P) (Fig. 1b). Plants grown in the absence of AMF propagules in semi-sterile soil remained non-colonised, and the corresponding hyphal lengths and uptake of radioisotopes were at background levels. Regression analysis between shoot radioactive P content and HLD in the relevant HC revealed a significant (P < 0.0001) and strong, positive (R2 = 0.91) relationship. This experiment demonstrated that natural soil in the HC can inhibit growth of, and P uptake by, ERM. The high correlation between hyphal length and mycorrhizal radioisotope P uptake in shoots allowed for the latter measure to be used as a proxy for ERM activity in subsequent experiments.
Fig. 1.
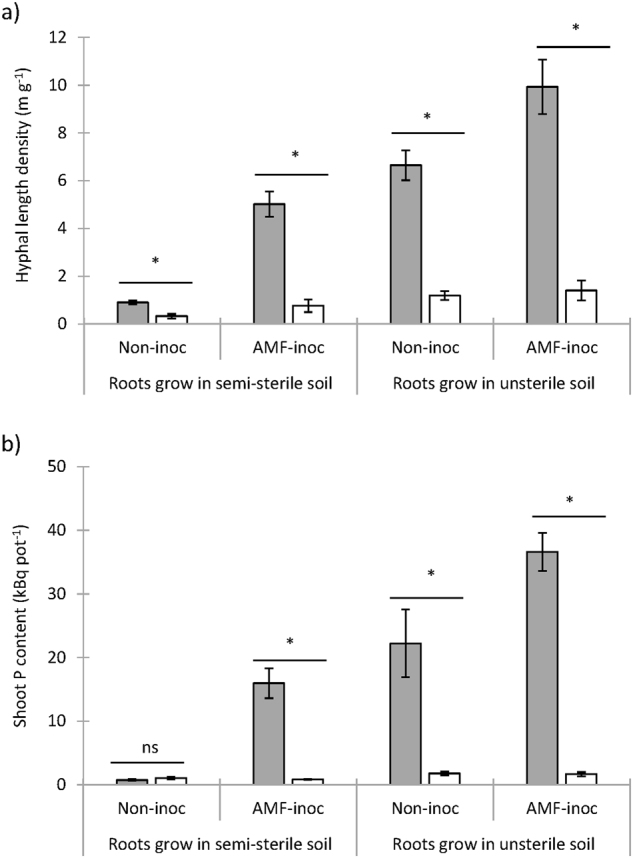
AMF hyphal length density (a) in the hyphal compartments (HCs), and shoot 33P and 32P contents (b) taken up from the HCs containing either semi-sterile (33P; grey bars) or unsterile (32P; white bars) Risø soil. The AMF mycelium was derived from the inoculant fungus (F. mosseae) in semi-sterile soil, or from the native AMF community/inoculant F. mosseae in unsterile Risø soil (Expt. 1)
How common is the AMF suppression?
The P-transfer activity of AMF was subsequently determined in an experiment where the HC contained 1 of 21 unsterile soils with contrasting properties (Table S1). Root colonisation as a result of AMF inoculation was high with little variation across treatments (86 ± 0.9%), and all HCs thus had the same potential for ERM ingrowth. However, hyphal transport of 33P to shoots varied greatly across the 21 unsterile soils (P < 0.0001; see Table S2 for ANOVA outcomes). Four soils, Roverud, Møystad E2, Rødekro and Årnes, were AMF-suppressive, while Risø stored and Trelleborg were the most AMF-conducive soils (Fig. 2). Furthermore, the Risø stored soil conferred significantly more 33P to the plant than any other soil tested. A significant (P < 0.0001), positive correlation (R2 = 0.493) was found between pH of the HC soil and shoot 33P content (Figure S2a). No other significant correlations were found between ERM activity and physicochemical properties of the soils; this includes a lack of correlation with water-extractable P (Figure S2b). Overall, these results demonstrated that the ERM activity was highly soil-dependent, and highly suppressed in four soils.
Fig. 2.
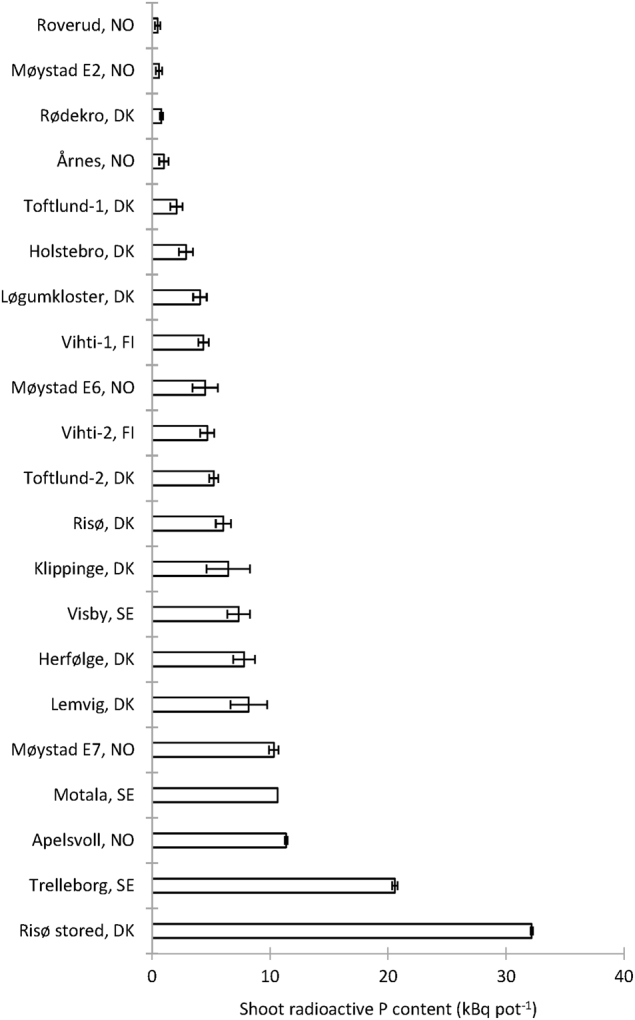
Shoot uptake of 33P by AMF from HCs containing 1 of 21 different unsterile soils (two plants per pot). Root external mycelium grew from M. truncatula roots colonised by R. irregularis (Expt. 2)
What causes AMF suppression in field soils?
Hyphal transport of 33P from the AMF-suppressive Møystad E2 soil increased when soil pH was increased from 4.4 to 7.1 (P < 0.001; Fig. 3a, Table S2). In addition, pasteurisation of this soil led to significantly higher values of shoot 33P content than were obtained from the non-pasteurised soil (Fig. 4). Additionally, the suppression of ERM activity was mitigated by pasteurisation in two other soils (data not presented), and by irradiation of Risø soil (Fig. 1). In contrast, pasteurisation of the AMF-conducive soil (Risø stored) did not lead to increased shoot 33P content (Fig. 4). These experiments showed that besides a pH effect, suppression had a biological component. Soil transplantation experiments prepared with two pairs of conducive vs. suppressive soils showed that significant suppression required at least a 1:1 mixing of suppressive with conducive soil (Fig. 5).
Fig. 3.
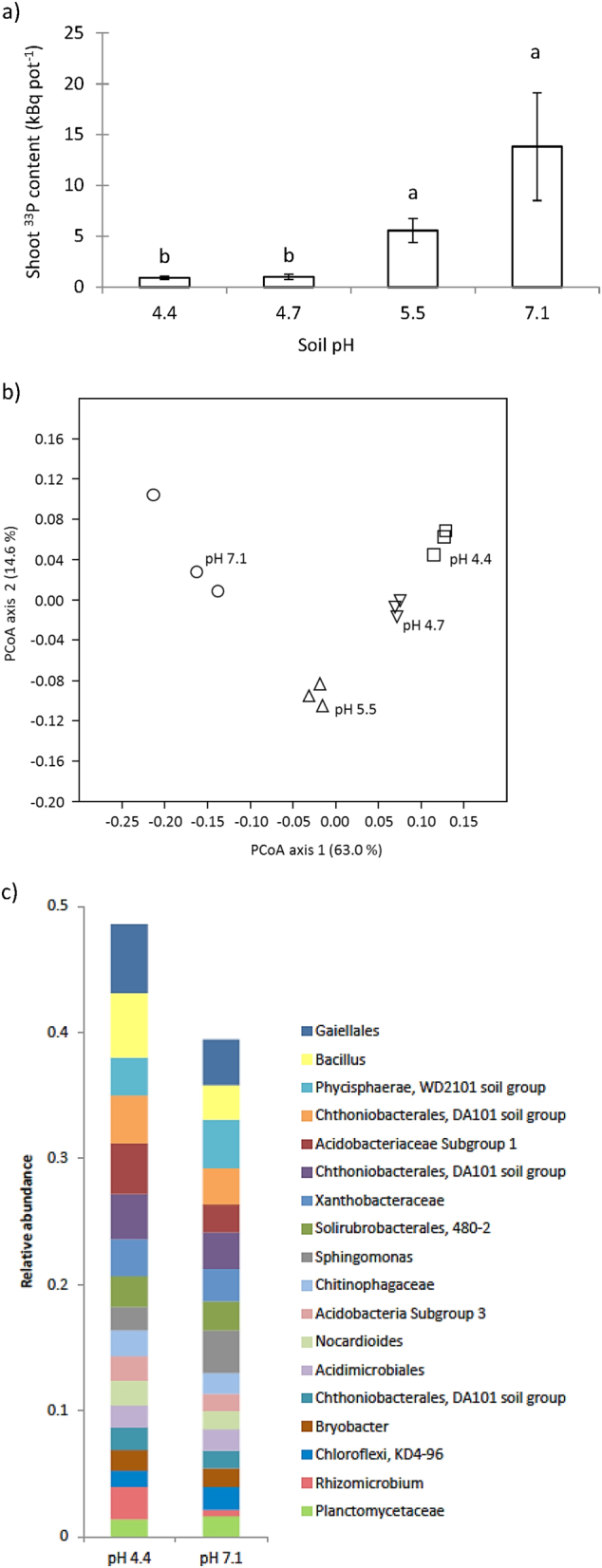
Shoot 33P uptake (a) and principal coordinates analysis (PCoA) of bacterial communities (b) from the HC containing unsterile Møystad E2 soil, amended with increasing amounts of CaCO3 to increase soil pH (Expt. 3). The PCoA was made on Bray–Curtis dissimilarities of OTUs with 97% similarity. The relative abundance of genus- or higher-level taxa (c) in Møystad E2 soils with pH 4.4 or 7.1 (Expt. 3). Only taxa constituting >1.5% of total abundance in the two soils are included in the bar plot
Fig. 4.
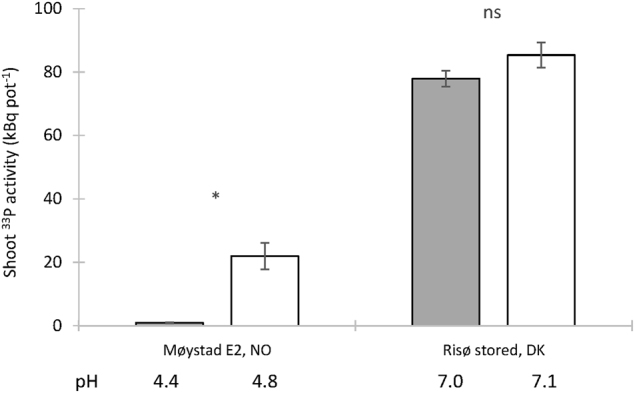
Shoot 33P uptake from HCs containing one of two unsterile soils (one AMF-suppressive and one AMF-conducive), either pasteurised by heating at 80 °C (white bars) or not heated (grey bars) (Expt. 3)
Fig. 5.
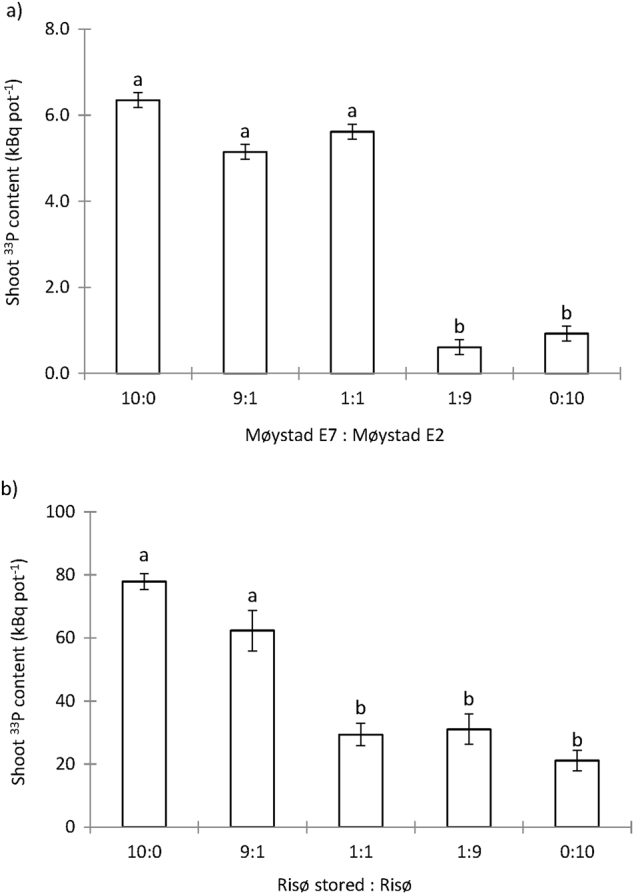
Shoot 33P uptake from HCs containing (a) either Møystad E2 or Møystad E7 soil, or three different mixes of the two soils and (b) either Risø stored or Risø soil, or three different mixtures of the two soils. Root external mycelium grew from M. truncatula roots colonised by R. irregularis (Expt. 3)
Principal component analysis of microbial PLFAs from Møystad E2 soil with different pH showed clear pH-induced changes in the bacterial community (Figure S3). Notably, five non-saturated iso- and antheiso PLFAs (r = −0.59 to −0.73; 0.04 > P > 0.01), which are markers for Gram-positive bacteria, correlated negatively with pH. Similarly, PCoA of 16S rRNA gene sequences at 97% similarity OTU level of the same soils showed a shift in bacterial community composition along the PCoA axis 1 (explaining 63.0% of the variation) in response to increasing soil pH (Fig. 3b). Species richness, estimated by Chao-1, was significantly (P < 0.05) higher in soil limed to pH 7.1 than in the untreated soil at pH 4.4, but no significant change was observed in the Shannon H-index (Figure S4a). The taxa that contributed to most of the variation between soils at pH 4.4 and 7.1 were identified by SIMPER. The largest decreases in relative abundance of the taxa being significantly influenced by liming (P < 0.05) were exhibited by Acidobacteriaceae subgroup 1 (Gp1), Bacillus, Rhizomicrobium and Gaiellales (Fig. 3c).
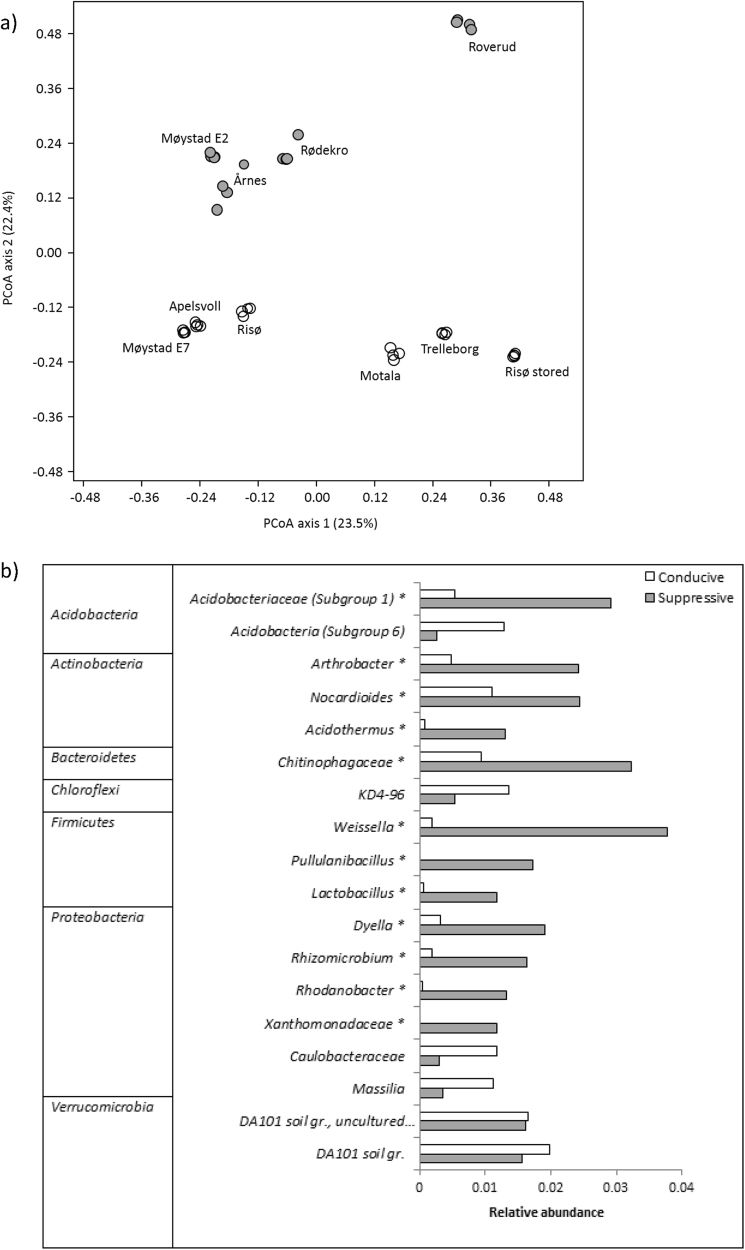
Subsequently, 16S rRNA gene-targeted analyses were used to investigate whether specific bacterial groups were associated with AMF suppression in a broader range of soils (Fig. 2); four highly AMF-suppressive (plant 33P uptake < 1 kBq per pot) soils, and six soils that conferred intermediate to high plant 33P uptake. Interestingly, PCoA at the 97% similarity OTU level revealed clear clustering of AMF-suppressive vs. AMF-conducive soils along the PCoA axis 2, explaining 22.4% of the variation (Fig. 6a). Furthermore, three of the AMF-suppressive soils clearly clustered together, while the Roverud soil diverged from the other suppressive soils on both PCoA axes. The six AMF-conducive soils clustered into two groups, mostly along PCoA axis 1. Separation of the AMF-suppressive and AMF-conducive soils was tested by PERMANOVA analysis with Bonferroni correction, which showed a significant separation (P < 0.05) up to phylum level (Table S3). The separation of AMF-suppressive and AMF-conducive soils was connected neither to differences in 16S rRNA gene copy number (data not shown) nor to differences in α-diversity as determined by species richness (Chao-1) and diversity (Shannon H-index; P > 0.05; Figure S4b). Nevertheless, as also indicated by rarefaction curves (Figure S5), diversity was distinctly lower in the divergent AMF-suppressive Roverud soil, when compared with the remaining suppressive soils, and distinctly higher in the cluster of AMF-conducive soils from Møystad E7, Apelsvoll and Risø, when compared with the three other conducive soils that clustered together in the PCoA plot.
Fig. 6.
Principal coordinate analysis (a) of bacterial communities in soils suppressive (filled circles) for AMF and in non-suppressive (open circles) soils (Expt. 2). Suppressive soils are defined as the soils where plant 33P uptake was <1 kBq per pot. The analysis was made on Bray–Curtis dissimilarities between OTUs with 97% similarity. Relative abundances (b) of genus- or higher-level taxa that contribute to most of the variation between the bacterial communities in AMF-suppressive and non-suppressive soils identified by SIMPER analysis and are present in significantly different abundances in suppressive and conducive soils (Expt. 2). Taxa marked with an asterisk (*) were observed in significantly (ANOVA; P < 0.05) higher abundance in AMF-suppressive soils than in conducive soils
Only a small proportion (3%) of the total number of bacterial genera-level OTUs was unique to the AMF-suppressive soils, while the majority (82%) of the genera was shared between the suppressive and conducive soils (see Venn diagram, Figure S6). The most abundant of the genera unique to the suppressive soils had a relative abundance of 0.02 % (Table S4), and the remaining unique genera were even less abundant. This suggests that AMF suppression is coupled with a higher relative abundance of one or more taxa. To identify such taxa, a SIMPER analysis was performed. The taxa that contributed most to the variation (cumulative 40%) between suppressive and conducive soils, and that were present in significantly different relative abundances in the two groups of soils, are presented in Fig. 6b. Of these, taxa belonging to Acidobacteriaceae Gp1, Acidothermus, Arthrobacter, Norcardioides, Chitinophagaeceae, Lactobacillus, Pullanibacillus, Weissella, Dyella, Rhizomicrobium, Rhodanobacter and Xanthomonadaceae were significantly (ANOVA; P < 0.05%) more abundant in AMF-suppressive than in AMF-conducive soils. The largest differences in relative abundances were found for Weissella and Acidobacteriaceae Gp1. Furthermore, CCA (Fig. 7) and regression analysis revealed that the relative abundance of Acidobacteriaceae Gp1 was significantly (P < 0.01), negatively, correlated to soil pH. No other taxa identified by SIMPER analysis correlated significantly with any of the measured soil physicochemical properties.
Fig. 7.
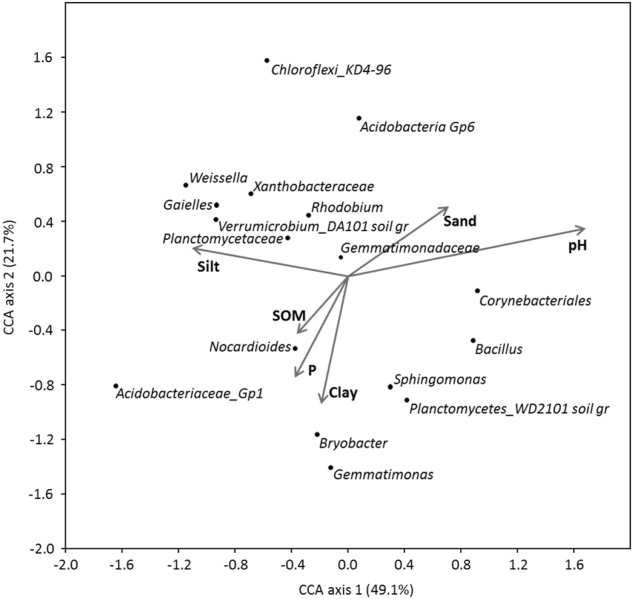
Canonical correspondence analysis (CCA) between genus- or higher-level relative abundance and environmental variables (Expt. 2). Only taxa contributing to >1 % relative abundance of all taxa are included in the plot
Discussion
This is the first systematic study to show that the activity of AMF mycelium (ERM) is inhibited in natural soil, and that the degree of suppression varies between soils. This suppression resembles previous observations made of disease-suppressive soils that minimise plant diseases caused by pathogenic fungi. While disease suppression in soil is often associated with specific groups or consortia of microorganisms [29, 30], our study suggests that suppression of AMF involves both abiotic and biotic components. Importantly, suppression was observed with ERM growing from roots colonised by each of two individual AMF isolates: F. mosseae and R. irregularis, and also with a native AMF field community. The results hence suggest that soil-associated suppression of ERM activity occurs across different AMF taxa.
The suppression of ERM activity is a bulk soil phenomenon as the observed effects were exerted on hyphae >0.5–1 mm from the root surface (being the approx. extension of root hairs penetrating the HC meshes). Yet, AMF colonisation of roots was not suppressed by a soil that was otherwise suppressive to ERM activity.
While plant P sufficiency is known to suppress AMF colonisation of roots [43] and soil [44], and thereby the symbiotic C costs, plants grown at sub-optimal P conditions will depend on AMF for maintaining P sufficiency. In the current experiments, soil suppressiveness did not correlate to water-extractable soil P. Likewise, the specific activity of 33P in aqueous extracts of HC soil was rather constant across soils that were suppressive and conducive and decreased only little with increasing soil P (data not shown). Thus, reduced uptake of 33P in some soils is not likely to have resulted from competition for 33P with soil microbes. ERM suppression could interfere with a range of soil processes that are stimulated by ERM: uptake of P and other nutrients [45–48]; distribution of plant C to bulk soil bacteria [49]; soil aggregate formation [50]; SOM mineralisation [51]; and formation of mycorrhizal networks [52].
The reduced suppression of ERM activity in semi-sterile soils shows that suppression involves a biotic component. For comparison, reduction of suppression by steaming, pasteurisation or fumigation is a key property used to define disease-suppressive soils [23]. Further, the observation that suppression against AMF could not be transferred by adding small amounts of suppressive soil to conducive soil indicates that the suppression of AMF is so-called general suppression rather than a specific suppression, for which transferability is a hallmark [23].
Soil pasteurisation mitigated AMF suppression much more than liming treatments, and the biotic component therefore dominated over an abiotic pH effect. The observed weak correlation between pH and ERM activity among the 21 field soils agrees with reports that low pH is suppressive to the Glomeracea, possibly via a direct effect on growth [25, 53]. However, the composition of the bacterial microbiome is strongly pH-dependent [54]; here 16S rRNA gene sequencing and PLFA analysis indeed showed a clear effect of liming on the bacterial community composition in a selected soil. Hence, the observed pH effect on ERM activity may well work via effects on ERM-suppressive bacteria. We found significantly higher abundance of Acidobacteria, Gp1, Bacillus, Rhizomicrobium and Gaiellales in un-limed soil at pH 4.4 than in limed soil at pH 7.1. For the Acidobacteria Gp1, a broader analysis of 10 different soils confirmed their higher abundance at low pH, in agreement with previous observations [54, 55]. Although the correlation of Acidobacteria Gp1 abundance with pH may confound a direct correlation with suppression, OTUs belonging to this group are frequently associated with disease-suppressive soil. For instance, they are observed in higher frequencies in the rhizosphere outside patches of Rhizoctonia-diseased plants, and in soil suppressive to Fusarium wilt or take-all disease caused by Gaeumannomyces [1, 56, 57].
Bacillus was also significantly more prevalent in the Møystad E2 soil at pH 4.4 than at pH 7.1, but its abundance did not correlate to pH in the broader analysis of 10 soils. Bacillus is well-known for its antifungal properties and high abundance in soils suppressive for fungal diseases [22, 58–60]. As Bacillus produces heat-resistant endospores [61], the lower suppression in the Møystad E2 soil after pasteurisation suggests that AMF suppression in this soil could not be explained by the abundance of Bacillus alone. Rhizomicrobium and Gaiellales have not, to our knowledge, been associated with suppressive soils.
As general suppression is considered to be caused by the collective competitive and antagonistic effects of the soil microbial community [21], we compared the microbiomes of 10 selected AMF-suppressive or AMF-conducive soils to identify additional groups potentially contributing to the suppression. We found significant differences in community composition between the soils defined as suppressive or conducive; suppressive soils had increased abundance of not only Acidobacteria Gp1, but also several indicator taxa, primarily affiliated with the Actinobacteria, Bacteriodetes, Firmicutes and Proteobacteria. Members of these groups have repeatedly been associated with the suppression of plant diseases [59, 62, 63]. Although some studies found few specific groups dominating in disease-suppressive soils [60, 64], the relatively high number of indicator taxa in the current study supports the hypothesis that AMF suppression results from the interactive effects of several microbial groups, as found for some disease-suppressive soils [59, 65, 66]. Indeed, putative fungal antagonists were most abundant in the AMF-suppressive soils; in addition to Acidobacteria Gp1, Firmicutes OTUs were particularly more abundant. Among these, Weissella can be abundant in Fusarium-suppressive soil [67], and both Weissella and Lactobacillus are antagonistic to several fungi in vitro [68–70].
Production of antimicrobial metabolites, even organic acids, may be involved in a putative antagonism. The Actinobacteria OTUs with increased abundance in the AMF-suppressive soils included OTUs from the genera Acidothermus, Nocardioides and Arthobacter. Actinobacteria are recognised for their ability to produce an array of antibiotics and enzymes [71] that may play a role in antagonistic interactions with other soil microorganisms. Members of the family Chitinophagaceae produce antifungal metabolites when co-cultured with other bacteria [72], and may associate with the decomposing mycelia of ectomycorrhizal fungi [73]. From the Proteobacteria, OTUs affiliated with two genera of the Xanthomonaceae, Rhodanobacter and Dyella, were far more abundant in AMF-suppressive soils. Rhodanobacter has previously been associated with the suppression of compost-amended potting mix, and isolates show antagonism against a root-rot pathogen [74]. Dyella was more abundant in soil patches that had recovered from Rhizoctonia bare patch disease [57], but without strong correlation to suppression.
Conclusions and wider perspectives
Soils that suppress ERM P uptake activity appear to be abundant. Suppression involves both an abiotic (pH) component, and a biotic component related to so-called general suppression. Hence, the mechanisms behind suppression of ERM activity are most likely due to the combined action of several members of the soil microbial communities, and our study points to several taxa that could play a role in AMF suppression. Further studies need to isolate the candidate suppressive bacteria and perform manipulation experiments (with various bacterial isolates, alone or in combination) to test their suppressive effects on foraging activities by ERM. Future studies should also consider other soil microbiome members with potential suppressive effects such as fungal taxa, and aim to elicit the specific antagonistic or competitive mechanisms involved. This study emphasises that extrapolations to field conditions from AMF model systems using semi-sterile soil could be misleading, and that the ecosystem services of the AMF community will depend strongly on the specific soil microbiome.
Electronic supplementary material
Acknowledgements
The authors are thankful to Mette Flodgaard, Lena A Byrgesen, Lene Vigh and Morten L Stephensen for skilful technical assistance and to Simon Mundus for providing access to a range of Scandinavian agricultural soils. Funding for this work was provided by the Novo Nordisk Foundation, grant NNF16OC0021576.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
The online version of this article (10.1038/s41396-018-0059-3) contains supplementary material, which is available to authorized users.
References
- 1.Shen Z, Ruan Y, Xue C, Zhong S, Li R, Shen Q. Soils naturally suppressive to banana Fusarium wilt disease harbor unique bacterial communities. Plant Soil. 2015;393:21–33. doi: 10.1007/s11104-015-2474-9. [DOI] [Google Scholar]
- 2.van der Voort M, Kempenaar M, van Driel M, Raaijmakers JM, Mendes R. Impact of soil heat on reassembly of bacterial communities in the rhizosphere microbiome and plant disease suppression. Ecol Lett. 2016;19:375–82. doi: 10.1111/ele.12567. [DOI] [PubMed] [Google Scholar]
- 3.Smith SE, Read DJ. Mycorrhizal symbiosis. 3rd edn. New York, NY: Academic Press; 2008. [Google Scholar]
- 4.van der Heijden MGA, Martin FM, Selosse MA, Sanders IR. Mycorrhizal ecology and evolution: the past, the present, and the future. New Phytol. 2015;205:1406–23. doi: 10.1111/nph.13288. [DOI] [PubMed] [Google Scholar]
- 5.Rodriguez A, Sanders IR. The role of community and population ecology in applying mycorrhizal fungi for improved food security. ISME J. 2015;9:1053–61. doi: 10.1038/ismej.2014.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Munkvold L, Kjøller R, Vestberg M, Rosendahl S, Jakobsen I. High functional diversity within species of arbuscular mycorrhizal fungi. New Phytol. 2004;164:357–64. doi: 10.1111/j.1469-8137.2004.01169.x. [DOI] [PubMed] [Google Scholar]
- 7.Roth R, Paszkowski U. Plant carbon nourishment of arbuscular mycorrhizal fungi. Curr Opin Plant Biol. 2017;39:50–6. doi: 10.1016/j.pbi.2017.05.008. [DOI] [PubMed] [Google Scholar]
- 8.Qin H, Brookes PC, Xu J. Arbuscular mycorrhizal fungal hyphae alter soil bacterial community and enhance polychlorinated biphenyls dissipation. Front Microbiol. 2016;7:939. doi: 10.3389/fmicb.2016.00939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Andrade G, Mihara KL, Linderman RG, Bethlenfalvay GJ. Bacteria from rhizosphere and hyphosphere soils of different arbuscular-mycorrhizal fungi. Plant Soil. 1997;192:71–79. doi: 10.1023/A:1004249629643. [DOI] [Google Scholar]
- 10.Nuccio EE, Hodge A, Pett-Ridge J, Herman DJ, Weber PK, Firestone MK. An arbuscular mycorrhizal fungus significantly modifies the soil bacterial community and nitrogen cycling during litter decomposition. Environ Microbiol. 2013;15:1870–81. doi: 10.1111/1462-2920.12081. [DOI] [PubMed] [Google Scholar]
- 11.Scheublin TR, Sanders IR, Keel C, van der Meer JR. Characterisation of microbial communities colonising the hyphal surfaces of arbuscular mycorrhizal fungi. ISME J. 2010;4:752–63. doi: 10.1038/ismej.2010.5. [DOI] [PubMed] [Google Scholar]
- 12.Frey-Klett P, Garbaye J, Tarkka M. The mycorrhiza helper bacteria revisited. New Phytol. 2007;176:22–36. doi: 10.1111/j.1469-8137.2007.02191.x. [DOI] [PubMed] [Google Scholar]
- 13.Battini F, Grønlund M, Agnolucci M, Giovannetti M, Jakobsen I. Facilitation of phosphorus uptake in maize plants by mycorrhizosphere bacteria. Sci Rep. 2017;7:4686. doi: 10.1038/s41598-017-04959-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Larsen J, Cornejo P, Barea JM. Interactions between the arbuscular mycorrhizal fungus Glomus intraradices and the plant growth promoting rhizobacteria Paenibacillus polymyxa and P. macerans in the mycorrhizosphere of Cucumis sativus. Soil Biol Biochem. 2009;41:286–92. doi: 10.1016/j.soilbio.2008.10.029. [DOI] [Google Scholar]
- 15.Li B, Ravnskov S, Xie G, Larsen J. Differential effects of Paenibacillus spp. on cucumber mycorrhizas. Mycol Prog. 2008;7:277–84. doi: 10.1007/s11557-008-0570-4. [DOI] [Google Scholar]
- 16.Wilson GWT, Hetrick BAD, Kitt DG. Suppression of mycorrhizal growth response of big bluestem by non-sterile soil. Mycologia. 1988;80:338–43. doi: 10.1080/00275514.1988.12025547. [DOI] [Google Scholar]
- 17.Nehl DB, Allen SJ, Brown JF. Mycorrhizal colonisation, root browning and soil properties associated with a growth disorder of cotton in Australia. Plant Soil. 1996;179:171–82. doi: 10.1007/BF00009326. [DOI] [Google Scholar]
- 18.Leigh J, Fitter AH, Hodge A. Growth and symbiotic effectiveness of an arbuscular mycorrhizal fungus in organic matter in competition with soil bacteria. FEMS Microbiol Ecol. 2011;76:428–38. doi: 10.1111/j.1574-6941.2011.01066.x. [DOI] [PubMed] [Google Scholar]
- 19.Jakobsen I, Gazey C, Abbott LK. Phosphate transport by communities of arbuscular mycorrhizal fungi in intact soil cores. New Phytol. 2001;149:95–103. doi: 10.1046/j.1469-8137.2001.00006.x. [DOI] [PubMed] [Google Scholar]
- 20.Henry AW. The natural microflora of the soil in relation to the foot-rot problem of wheat. Can J Res. 1931;4:69–77. doi: 10.1139/cjr31-006. [DOI] [Google Scholar]
- 21.Weller DM, Raaijmakers JM, Gardener BBM, Thomashow LS. Microbial populations responsible for specific soil suppressiveness to plant pathogens. Annu Rev Phytopathol. 2002;40:309–48. doi: 10.1146/annurev.phyto.40.030402.110010. [DOI] [PubMed] [Google Scholar]
- 22.Mendes R, Kruijt M, de Bruijn I, Dekkers E, van der Voort M, Schneider JHM, et al. Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science. 2011;332:1097–100. doi: 10.1126/science.1203980. [DOI] [PubMed] [Google Scholar]
- 23.Schlatter D, Kinkel LL, Thomashow LS, Weller DM, Paulitz TC. Disease suppressive soils: new insights from the soil microbiome. Phytopathology. 2017;107:1284–97. doi: 10.1094/PHYTO-03-17-0111-RVW. [DOI] [PubMed] [Google Scholar]
- 24.Gavito ME, Schweiger P, Jakobsen I. P uptake by arbuscular mycorrhizal hyphae: effect of soil temperature and atmospheric CO2 enrichment. Glob Change Biol. 2003;9:106–16. doi: 10.1046/j.1365-2486.2003.00560.x. [DOI] [Google Scholar]
- 25.Abbott L, Robson A. The effect of soil pH on the formation of VA mycorrhizas by two species of Glomus. Soil Res. 1985;23:253–61. doi: 10.1071/SR9850253. [DOI] [Google Scholar]
- 26.van Aarle IM, Olsson PA. Fungal lipid accumulation and development of mycelial structures by two arbuscular mycorrhizal fungi. Appl Environ Microbiol. 2003;69:6762–7. doi: 10.1128/AEM.69.11.6762-6767.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Penton CR, Gupta VVSR, Tiedje JM, Neate SM, Ophel-Keller K, Gillings M, et al. Fungal community structure in disease suppressive soils assessed by 28S LSU gene sequencing. PLoS ONE. 2014;9:e93893. doi: 10.1371/journal.pone.0093893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chapelle E, Mendes R, Bakker PAHM, Raaijmakers JM. Fungal invasion of the rhizosphere microbiome. ISME J. 2016;10:265–8. doi: 10.1038/ismej.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cordovez V, Carrion VJ, Etalo DW, Mumm R, Zhu H, van Wezel GP, et al. Diversity and functions of volatile organic compounds produced by Streptomyces from a disease-suppressive soil. Front Microbiol. 2015;6:1081. doi: 10.3389/fmicb.2015.01081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raaijmakers JM, Mazzola M. Soil immune responses. Science. 2016;352:1392–3. doi: 10.1126/science.aaf3252. [DOI] [PubMed] [Google Scholar]
- 31.Merrild MP, Ambus P, Rosendahl S, Jakobsen I. Common arbuscular mycorrhizal networks amplify competition for phosphorus between seedlings and established plants. New Phytol. 2013;200:229–40. doi: 10.1111/nph.12351. [DOI] [PubMed] [Google Scholar]
- 32.Smith SE, Smith FA, Jakobsen I. Mycorrhizal fungi can dominate phosphate supply to plants irrespective of growth responses. Plant Physiol. 2003;133:16–20. doi: 10.1104/pp.103.024380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Phillips JM, Hayman DS. Improved procedures for clearing roots and staining parasitic and vesicular-arbuscular mycorrhizal fungi for rapid assessment of infection. T Br Mycol Soc. 1970;55:158–61. doi: 10.1016/S0007-1536(70)80110-3. [DOI] [Google Scholar]
- 34.Jakobsen I, Abbott LK, Robson AD. External hyphae of vesicular-arbuscular mycorrhizal fungi associated with Trifolium subterraneum L 2. Hyphal transport of 32P over defined distances. New Phytol. 1992;120:509–16. doi: 10.1111/j.1469-8137.1992.tb01800.x. [DOI] [Google Scholar]
- 35.Olsen S, Sommers L, Page A. Methods of soil analysis. Part 2. Chemical and microbiological properties of phosphorus. Agron. Mongr. 9 (2nd ed.), ASA and SSSA, Madison. WI (1982), pp 403–430.
- 36.Olsen S, Cole C, Watanabe F, Dean L. Estimation of available phosphorus in soils by extraction with sodium bicarbonate. Washington, DC: USDA Circular 939; 1954. [Google Scholar]
- 37.Schofield RK, Taylor AW. The measurement of soil pH. Soil Sci Soc Am J. 1955;19:164–7. doi: 10.2136/sssaj1955.03615995001900020013x. [DOI] [Google Scholar]
- 38.Frostegård Aring, Tunlid A, Bååth E. Microbial biomass measured as total lipid phosphate in soils of different organic content. J Microbiol Methods. 1991;14:151–63. doi: 10.1016/0167-7012(91)90018-L. [DOI] [Google Scholar]
- 39.Nicolaisen MH, Bælum J, Jacobsen CS, Sørensen J. Transcription dynamics of the functional tfdA gene during MCPA herbicide degradation by Cupriavidus necator AEO106 (pRO101) in agricultural soil. Environ Microbiol. 2008;10:571–9. doi: 10.1111/j.1462-2920.2007.01476.x. [DOI] [PubMed] [Google Scholar]
- 40.Muyzer G, de Waal EC, Uitterlinden AG. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Muyzer G, Teske A, Wirsen CO, Jannasch HW. Phylogenetic relationships of Thiomicrospira species and their identification in deep-sea hydrothermal vent samples by denaturing gradient gel electrophoresis of 16S rDNA fragments. Arch Microbiol. 1995;164:165–72. doi: 10.1007/BF02529967. [DOI] [PubMed] [Google Scholar]
- 42.Herlemann DPR, Labrenz M, Jürgens K, Bertilsson S, Waniek JJ, Andersson AF. Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J. 2011;5:1571–9. doi: 10.1038/ismej.2011.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Breuillin F, Schramm J, Hajirezaei M, Ahkami A, Favre P, Druege U, et al. Phosphate systemically inhibits development of arbuscular mycorrhiza in Petunia hybrida and represses genes involved in mycorrhizal functioning. Plant J. 2010;64:1002–17. doi: 10.1111/j.1365-313X.2010.04385.x. [DOI] [PubMed] [Google Scholar]
- 44.Abbott LK, Robson AD, De Boer G. The effect of phosphorus on the formation of hyphae in soil by the vesicular-arbuscular mycorrhizal fungus Glomus fasciculatum. New Phytol. 1984;97:437–46. doi: 10.1111/j.1469-8137.1984.tb03609.x. [DOI] [Google Scholar]
- 45.Garcia K, Doidy J, Zimmermann SD, Wipf D, Courty PE. Take a trip through the plant and fungal transportome of mycorrhiza. Trends Plant Sci. 2016;21:937–50. doi: 10.1016/j.tplants.2016.07.010. [DOI] [PubMed] [Google Scholar]
- 46.Sawers RJH, Svane SF, Quan C, Grønlund M, Wozniak B, Gebreselassie MN, et al. Phosphorus acquisition efficiency in arbuscular mycorrhizal maize is correlated with the abundance of root-external hyphae and the accumulation of transcripts encoding PHT1 phosphate transporters. New Phytol. 2017;214:632–43. doi: 10.1111/nph.14403. [DOI] [PubMed] [Google Scholar]
- 47.Smith SE, Jakobsen I, Grønlund M, Smith FA. Roles of arbuscular mycorrhizas in plant phosphorus nutrition: interactions between pathways of phosphorus uptake in arbuscular mycorrhizal roots have important implications for understanding and manipulating plant phosphorus acquisition. Plant Physiol. 2011;156:1050–7. doi: 10.1104/pp.111.174581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Watts-Williams SJ, Jakobsen I, Cavagnaro TR, Grønlund M. Local and distal effects of arbuscular mycorrhizal colonization on direct pathway Pi uptake and root growth in Medicago truncatula. J Exp Bot. 2015;66:4061–73. doi: 10.1093/jxb/erv202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kaiser C, Kilburn MR, Clode PL, Fuchslueger L, Koranda M, Cliff JB, et al. Exploring the transfer of recent plant photosynthates to soil microbes: mycorrhizal pathway vs direct root exudation. New Phytol. 2015;205:1537–51. doi: 10.1111/nph.13138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rillig MC, Aguilar-Trigueros CA, Bergmann J, Verbruggen E, Veresoglou SD, Lehmann A. Plant root and mycorrhizal fungal traits for understanding soil aggregation. New Phytol. 2015;205:1385–8. doi: 10.1111/nph.13045. [DOI] [PubMed] [Google Scholar]
- 51.Paterson E, Sim A, Davidson J, Daniell TJ. Arbuscular mycorrhizal hyphae promote priming of native soil organic matter mineralisation. Plant Soil. 2016;408:243–54. doi: 10.1007/s11104-016-2928-8. [DOI] [Google Scholar]
- 52.Montesinos-Navarro A, Segarra-Moragues JG, Valiente-Banuet A, Verdú M. The network structure of plant–arbuscular mycorrhizal fungi. New Phytol. 2012;194:536–47. doi: 10.1111/j.1469-8137.2011.04045.x. [DOI] [PubMed] [Google Scholar]
- 53.van Aarle IM, Olsson PA, Söderström B. Arbuscular mycorrhizal fungi respond to the substrate pH of their extraradical mycelium by altered growth and root colonization. New Phytol. 2002;155:173–82. doi: 10.1046/j.1469-8137.2002.00439.x. [DOI] [PubMed] [Google Scholar]
- 54.Rousk J, Baath E, Brookes PC, Lauber CL, Lozupone C, Caporaso JG, et al. Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J. 2010;4:1340–51. doi: 10.1038/ismej.2010.58. [DOI] [PubMed] [Google Scholar]
- 55.Lauber CL, Strickland MS, Bradford MA, Fierer N. The influence of soil properties on the structure of bacterial and fungal communities across land-use types. Soil Biol Biochem. 2008;40:2407–15. doi: 10.1016/j.soilbio.2008.05.021. [DOI] [Google Scholar]
- 56.Sanguin H, Sarniguet A, Gazengel K, Moënne-Loccoz Y, Grundmann GL. Rhizosphere bacterial communities associated with disease suppressiveness stages of take-all decline in wheat monoculture. New Phytol. 2009;184:694–707. doi: 10.1111/j.1469-8137.2009.03010.x. [DOI] [PubMed] [Google Scholar]
- 57.Yin C, Hulbert SH, Schroeder KL, Mavrodi O, Mavrodi D, Dhingra A, et al. Role of bacterial communities in the natural suppression of Rhizoctonia solani bare patch disease of wheat (Triticum aestivum l.) Appl Environ Microbiol. 2013;79:7428–38. doi: 10.1128/AEM.01610-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim DS, Cook RJ, Weller DM. Bacillus sp. L324-92 for biological control of three root diseases of wheat grown with reduced tillage. Phytopathology. 1997;87:551–8. doi: 10.1094/PHYTO.1997.87.5.551. [DOI] [PubMed] [Google Scholar]
- 59.Klein E, Ofek M, Katan J, Minz D, Gamliel A. Soil suppressiveness to Fusarium disease: shifts in root microbiome associated with reduction of pathogen root colonization. Phytopathology. 2012;103:23–33. doi: 10.1094/PHYTO-12-11-0349. [DOI] [PubMed] [Google Scholar]
- 60.Xue C, Ryan Penton C, Shen Z, Zhang R, Huang Q, Li R, et al. Manipulating the banana rhizosphere microbiome for biological control of Panama disease. Sci Rep. 2015;5:11124. doi: 10.1038/srep11124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nicholson WL, Munakata N, Horneck G, Melosh HJ, Setlow P. Resistance of Bacillus endospores to extreme terrestrial and extraterrestrial environments. Microbiol Mol Biol Rev. 2000;64:548–72. doi: 10.1128/MMBR.64.3.548-572.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu X, Zhang S, Jiang Q, Bai Y, Shen G, Li S, et al. Using community analysis to explore bacterial indicators for disease suppression of tobacco bacterial wilt. Sci Rep. 2016;6:36773. doi: 10.1038/srep36773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rosenzweig N, Tiedje JM, Quensen JF, Meng Q, Hao JJ. Microbial communities associated with potato common scab-suppressive soil determined by pyrosequencing analyses. Plant Dis. 2011;96:718–25. doi: 10.1094/PDIS-07-11-0571. [DOI] [PubMed] [Google Scholar]
- 64.Stutz EW, Défago G, Kern H. Naturally occurring fluorescent pseudomonads involved in suppression of black root rot of tobacco. Phytopathology. 1986;76:181–5. doi: 10.1094/Phyto-76-181. [DOI] [Google Scholar]
- 65.Kyselkova M, Kopecky J, Frapolli M, Defago G, Sagova-Mareckova M, Grundmann GL, et al. Comparison of rhizobacterial community composition in soil suppressive or conducive to tobacco black root rot disease. ISME J. 2009;3:1127–38. doi: 10.1038/ismej.2009.61. [DOI] [PubMed] [Google Scholar]
- 66.Wu K, Yuan S, Wang L, Shi J, Zhao J, Shen B, et al. Effects of bio-organic fertilizer plus soil amendment on the control of tobacco bacterial wilt and composition of soil bacterial communities. Biol Fertil Soils. 2014;50:961–71. doi: 10.1007/s00374-014-0916-9. [DOI] [Google Scholar]
- 67.Huang X, Wen T, Zhang J, Meng L, Zhu T, Cai Z. Toxic organic acids produced in biological soil disinfestation mainly caused the suppression of Fusarium oxysporum f. sp. cubense. BioControl. 2015;60:113–24. doi: 10.1007/s10526-014-9623-6. [DOI] [Google Scholar]
- 68.Fhoula I, Najjari A, Turki Y, Jaballah S, Boudabous A, Ouzari H. Diversity and antimicrobial properties of lactic acid bacteria isolated from rhizosphere of olive trees and desert truffles of Tunisia. BioMed Res Int. 2013;2013:14. doi: 10.1155/2013/405708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yang EJ, Chang HC. Purification of a new antifungal compound produced by Lactobacillus plantarum AF1 isolated from kimchi. Int J Food Microbiol. 2010;139:56–63. doi: 10.1016/j.ijfoodmicro.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 70.Cortés-Zavaleta O, López-Malo A, Hernández-Mendoza A, García HS. Antifungal activity of lactobacilli and its relationship with 3-phenyllactic acid production. Int J Food Microbiol. 2014;173:30–5. doi: 10.1016/j.ijfoodmicro.2013.12.016. [DOI] [PubMed] [Google Scholar]
- 71.Shivlata L, Satyanarayana T. Thermophilic and alkaliphilic Actinobacteria: biology and potential applications. Front Microbiol. 2015;6:1014. doi: 10.3389/fmicb.2015.01014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Loudon AH, Holland JA, Umile TP, Burzynski EA, Minbiole KPC, Harris RN. Interactions between amphibians’ symbiotic bacteria cause the production of emergent anti-fungal metabolites. Front Microbiol. 2014;5:441. doi: 10.3389/fmicb.2014.00441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Brabcová V, Nováková M, Davidová A, Baldrian P. Dead fungal mycelium in forest soil represents a decomposition hotspot and a habitat for a specific microbial community. New Phytol. 2016;210:1369–81. doi: 10.1111/nph.13849. [DOI] [PubMed] [Google Scholar]
- 74.De Clercq D, Van Trappen S, Cleenwerck I, Ceustermans A, Swings J, Coosemans J, et al. Rhodanobacter spathiphylli sp. nov., a gammaproteobacterium isolated from the roots of Spathiphyllum plants grown in a compost-amended potting mix. Int J Syst Evol Microbiol. 2006;56:1755–9. doi: 10.1099/ijs.0.64131-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.