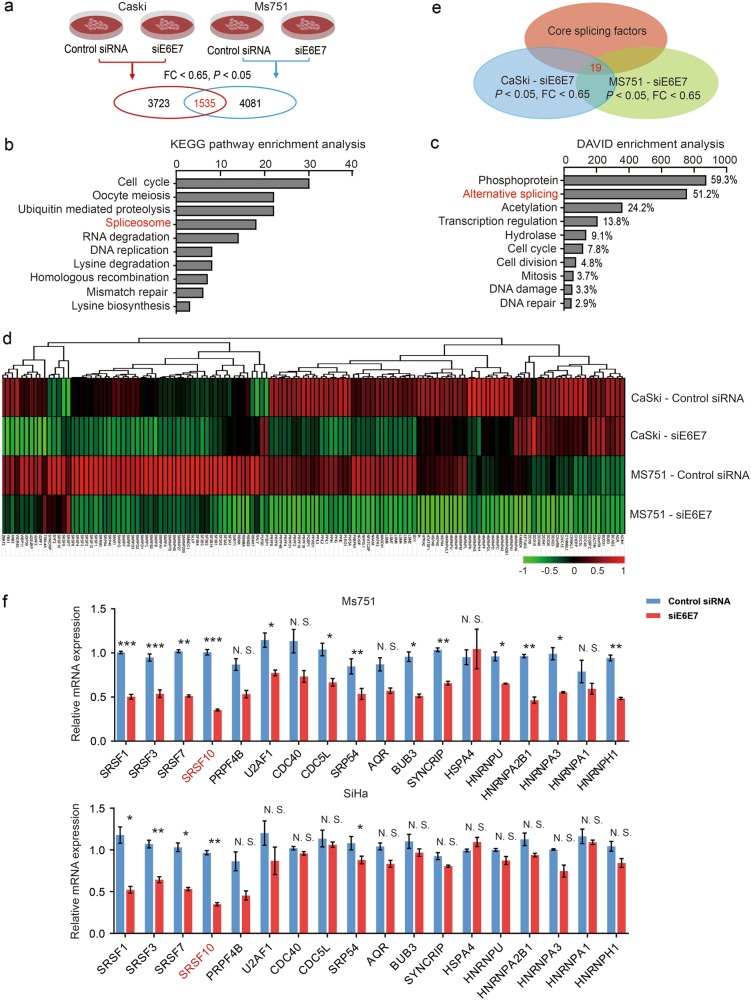
Fig. 1.
E6 and E7 regulate the expression of splicing factors in CC cells. a Summary of microarray analyses. In all, microarray analyses were performed in Caski and Ms751 cells transiently transfected with either siE6E7 or control siRNA to screen the differentially expressed genes. A total of 1535 genes were identified after overlap in Caski and Ms751 cells. b Top significant pathways for overlapped differentially expressed genes in E6E7-knockdown cells analyzed by KEGG pathway enrichment analysis. Spliceosome ranked fourth. c Top significant pathways for overlapped differentially expressed genes in E6E7-knockdown cells analyzed by DAVID enrichment analysis. Alternative splicing is the second pathway affected by E6E7 knockdown. d The heatmap of differentially expressed genes in AS pathway in Caski and Ms751 groups. e The schematic diagram of differentially expressed core splicing factors. In all, 19 genes were identified by overlapping differentially expressed genes related to AS pathway in microarray and core splicing factors in Ref. [16]. f Validation of the microarray results by RT-PCR of the 19 core splicing factors in Ms751 and SiHa cells. The expression of HNRPDL is undetectable. The experiments were repeated three times, and the results are shown as mean ± s.d. *P < 0.05; **P < 0.01; ***P < 0.001 (Student’s t-test)