Abstract
Cancer-testis (CT) antigens, rarely in normal tissues except testis, are expressed in many tumor types. In recent years, DDX43 has been shown to be expressed in several malignancies. However, the role of DDX43 during tumorigenesis is not well established. In the present study, we explored the function of DDX43 in chronic myeloid leukemia (CML). We found that DDX43 overexpression in CML cell lines enhanced survival and colony formation, inhibited cell apoptosis, promoted tumorigenesis, and CML progression. In contrast, silencing of DDX43 inhibited cell survival and tumorigenesis. Upregulated H19 and downregulated miR-186 were identified in DDX43-transfected cells. Furthermore, we demonstrated that miR-186 targeted DDX43, and overexpressed miR-186 increased apoptosis and decreased cell survival. We also showed that DDX43 regulated the expression of H19 through demethylation and silencing H19 inhibited cell survival. Taken together, these results indicate that DDX43 provides critical support to the progression of CML by enhancing cell survival, colony formation, and inhibiting cell apoptosis, thereby implicating DDX43 as a potential therapeutic target in CML.
Introduction
Chronic myeloid leukemia (CML), present of a distinct translocation t(9;22)(q34;q11), is a malignant myeloproliferative disorder arising from pluripotent stem cells [1, 2]. The t(9;22) rearrangement generates the BCR-ABL1 fusion gene [3, 4]. According to the natural course of disease, three stages of CML are clinically recognized: an initial chronic phase (CP), then an accelerated phase (AP), and a terminal blast crisis (BC) [1]. During this course, gene mutations and chromosomal abnormalities are involved in the progression of CML [5–7]. However, the mechanisms underlying the evolution need to be further explored. Our team has reported that low methylation of DDX43 may be related to the development of CML [8].
DDX43 (also known as HAGE) is initially found as a cancer/testis antigen overexpressed in many solid tumors but absent in normal tissues except testis [9], which indicates this gene has highly tissue-specific expression. These important characteristics make it become a hot spot in the tumor biological treatment and supposed to be an ideal target molecule for cancer therapy.
In hematologic malignancies, DDX43 is overexpressed in acute myeloid leukemia (AML), CML, multiple myeloma and a variety of malignant cell lines derived from B or T lymphocytic cells [8, 10–13]. Our group found that DDX43 was highly expressed and associated with the hypomethylation of its promoter in AML and CML [8, 10]. In this study we found that DDX43 promoted tumorigenesis and CML progression by enhancing cell adhesion, survival, and colony formation.
H19 is a long chain non-coding RNA and its length is about 2.3 kb. Abnormal H19 expression, identified in many solid tumors, such as bladder cancer, breast cancer, liver cancer, lung cancer, plays an important role in tumor growth [14–20]. Several studies have reported that miR-186 influences tumor development by inhibiting tumor cell growth and regulating cell cycle, as a tumor suppressor miRNA [21–23]. We had reported that low expression of miR-186 was associated with poor prognosis in AML [24].
In the present study, we discovered that DDX43 overexpression in CML cell lines enhanced survival and colony formation, inhibited cell apoptosis, promoted tumorigenesis, and CML progression. In contrast, silencing of DDX43 inhibited cell survival and tumorigenesis. Furthermore, we demonstrated that miR-186 targeted DDX43, and DDX43 regulated H19 expression through demethylation. All of these play a very important role in the progression of CML.
Results
DDX43 affects apoptosis and survival of leukemia cells
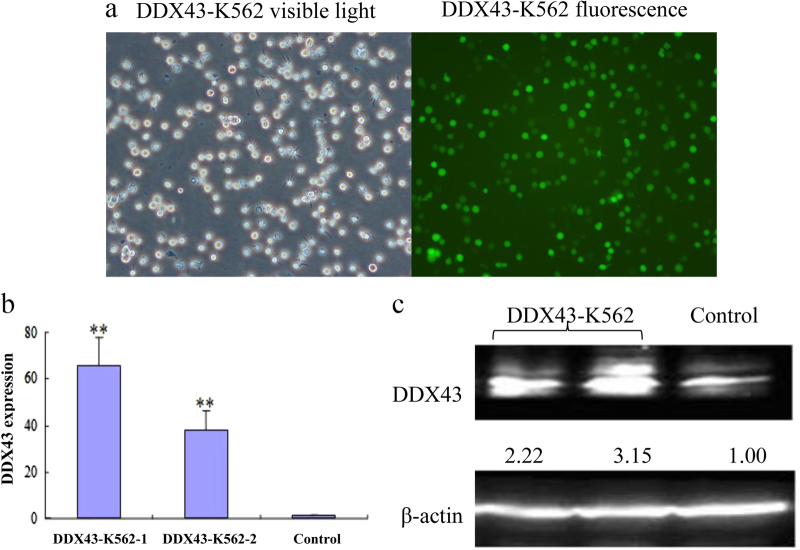
To explore the role of DDX43, a construct expressing DDX43 was generated and stably expressed in leukemia line K562 (DDX43-K562). DDX43 mRNA and protein were detected using real-time quantitative PCR (RQ-PCR) and western blot, respectively. DDX43 level significantly increased in DDX43-transfected cell lines compared with the mock transfection (Fig. 1).
Fig. 1.
Expression of DDX43 in CML cells. a The expression of DDX43 from DDX43-K562 cells under a fluorescent microscope (×100). b Results from RQ-PCR of control cells and two DDX43-K562 clones. c Western blot of DDX43-K562 and control cells. **P < 0.01. Error bars indicate the standard deviation (SD) (n = 3)
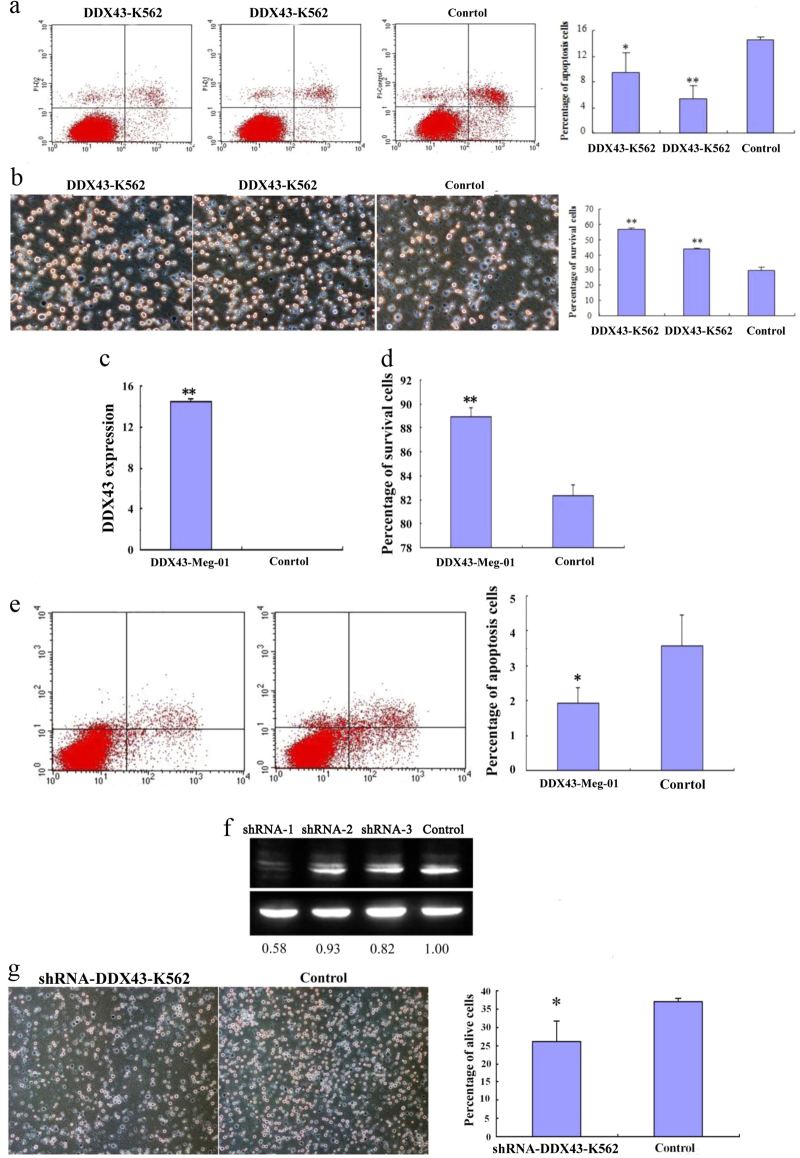
Apoptosis was analyzed for DDX43-K562 cells and a control vector transfected K562 cells in serum-free culture conditions for 48 h. Cells were subjected to staining, followed by flow cytometry analysis. DDX43-K562 showed less apoptosis than the control cells (Fig. 2a). Survival was also analyzed for these cells cultured in serum-free conditions for 3 days. DDX43-K562 cells showed significantly increased survival as compared with control (Fig. 2b). Cell apoptosis and survival were also assayed for the leukemia cell line Meg-01 (Fig. 2c–e), and similar results were observed, confirming that DDX43 increased survival and reduced apoptosis of these cells. To further confirm the functions of DDX43, we used three shRNAs to silence DDX43 expression in DDX43-K562 cells, and significantly downregulated expression of DDX43 was observed in shRNA-1-transfected DDX43-K562 (Fig. 2f). The effects of shRNA on DDX43-K562 survival were also tested. Transfection of the shRNA-DDX43 decreased survival as compared with control (Fig. 2g).
Fig. 2.
K562 cell apoptosis and survival affected by DDX43. a DDX43-K562 or control cells were cultured for 48 h. Expression of DDX43 inhibited cell apoptosis. b DDX43-K562 or control cells were cultured in serum-free conditions. DDX43 promoted the survival of transfected cells. c Expression of DDX43 in DDX43-Meg-01 or control cells were detected by RQ-PCR. d, e Cell survival and apoptosis were assayed in Meg-01. f Expression of DDX43 in DDX43 shRNA or control vector transfected DDX43-K562 cells was detected by western blot. Lanes 1–3: DDX43-K562 transfected with three shRNAs; Lane 4: DDX43-K562 transfected with a negative control. ShRNA-1 significantly decreased DDX43 expression. g Cell survival was assayed in DDX43 shRNA-1-transfected K562. Error bars indicate SD (n = 3). *P < 0.05; **P < 0.01
DDX43 facilitates colony formation and tumorigenesis
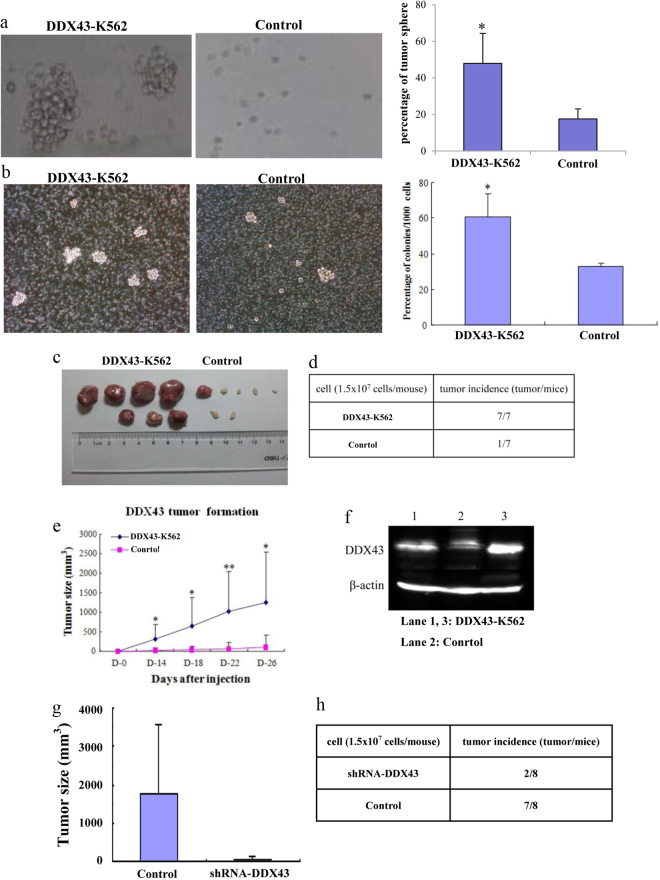
To further identify the role of DDX43 in tumorigenesis, K562 cells were inoculated into soft agarose. Colonies were numbered and photographed after 4 weeks. More and larger colonies were found in DDX43 group, compared with control (Fig. 3a). Clone formation in methylcellulose was also tested. DDX43-K562 and vector-K562 cells were plated in 1% methylcellulose with 10% fetal bovine serum (FBS). The colonies formed by the cells transfected with DDX43 were more and larger than control cells (Fig. 3b). These results suggested that DDX43 could promote colony formation.
Fig. 3.
DDX43 overexpression affected clone formation and tumor formation. a Firstly, 2% agarose was plated into six-well plates, and DDX43-K562 or control cells were mixed with 0.3% low melting agarose, respectively. More and larger colonies were observed in the DDX43-K562 group than in the control group after 4 weeks. b K562 cells stably transfected with DDX43 or the control vector were plated in 1% methylcellulose with 10% FBS. c, d DDX43- or mock-transfected K562 cells were inoculated into CD1 strain nude mice subcutaneously. Mice were photographed and killed after 4 weeks of injection. e We measured tumor sizes and the tumor growth curves were drawn. Data showed the mean ± SD from three experiments. f DDX43 protein detected by western blot in tumors from mice inoculated with DDX43-K562 or control cells. g K562 cells transfected with DDX43 shRNA-1 or a control vector were inoculated into CD1 strain nude mice subcutaneously. Mice were photographed and killed after 4 weeks of injection. Tumor sizes were measured. h Tumor incidence from K562 cells transfected with DDX43 shRNA-1 or a control vector. *P < 0.05; **P < 0.01
To further confirm the role of DDX43 on tumorigenesis in vivo, DDX43-K562 or vector-K562 cells were injected subcutaneously into CD1 nude mice. The growth of tumor and the size of tumor were monitored every 4 days. The mice inoculated with DDX43-K562 cells developed more and larger tumors than those with control cells (Fig. 3c–e). DDX43 expression in tumor was detected by western blot, which showed that DDX43 was highly expressed in the DDX43-K562 group (Fig. 3f). DDX43-K562 cells transfected with the control vector or shRNA-1 were inoculated into nude mice subcutaneously. Mice were killed after 4 weeks and tumor sizes were measured. We found less and smaller tumors in the DDX43 shRNA-1-transfected group, compared with the control vector-transfected group (Fig. 3g, h). All mentioned above confirmed the role of DDX43 in tumorigenesis.
DDX43 promotes CML progression
In a previous study, the expression of DDX43 transcript was assayed in bone marrow samples from CML patients [25]. DDX43 overexpression was identified more frequently in patients at AP/BC stages (4/4, 100%) than those at CP stage (5/22, 23%). These results suggested that DDX43 expression was relevant to the progression of CML [25].
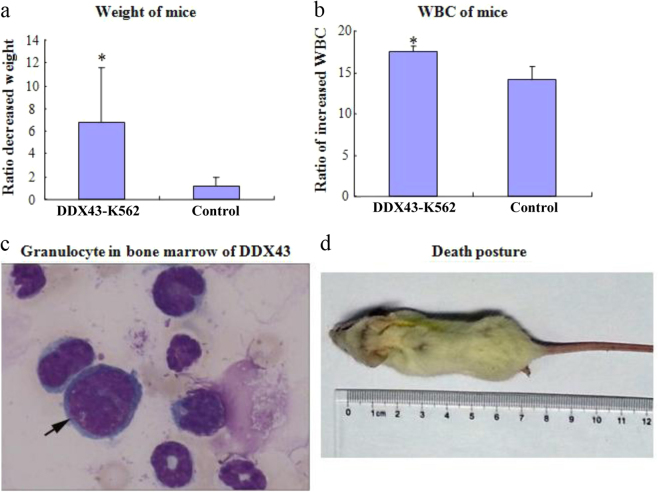
To further test the role of DDX43 in CML progression, DDX43-K562 or vector-K562 cells were injected via tail vein into SCID mice. We found that the ratio of weight loss was higher in SCID mice injected with DDX43-transfected cells than control (Fig. 4a). The ratio of white blood cell (WBC) increase was higher in mice injected with DDX43-transfected cells than control (Fig. 4b). Mice were photographed (Fig. 4d) and killed after 4 weeks of injection. Bone marrow was separated and stained with Wright−Giemsa; myeloblasts were easily found in the group injected with DDX43-transfected cells, but not in the control group (Fig. 4c). These results suggested the role of DDX43 in CML progression.
Fig. 4.
CML progression affected by DDX43 overexpression. DDX43-K562 or control cells were injected via tail vein into SCID mice. a The ratio of decreased weight of SCID mice injected with K562 transfected cells. Data shown were mean ± SD from three experiments. b The ratio of increased WBC of SCID mice injected with K562 transfected cells. c Mice were killed after 4 weeks of injection and blasts in bone marrow of SCID mice injected with DDX43-K562 transfected cells were detected with Wright−Giemsa stain. d Mice were photographed and killed after 4 weeks of injection. *P < 0.05. Error bars indicate SD (n = 5)
Release of H19 by demethylation mediates DDX43 function
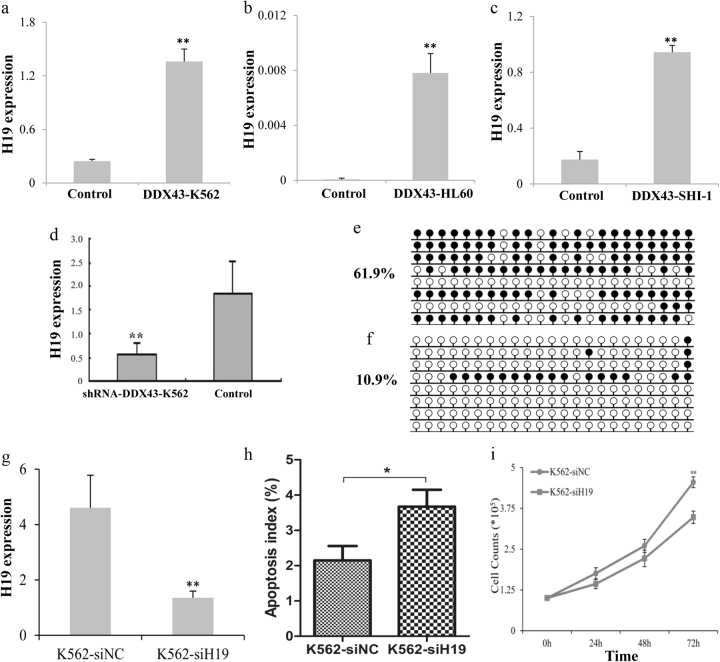
To explore the molecular mechanism of the DDX43 effects, we performed deep sequencing assay to identify related genes for DDX43. A number of genes and microRNAs were up- or down- regulated, including H19 (Table 1). Our study found that the H19 expression increased in DDX43-transfected K562, HL60, and SHI-1 cells (Fig. 5a–c), and decreased after DDX43 silencing (Fig. 5d), suggesting DDX43 could increase H19 expression. Furthermore, the methylation level of H19 promoter was identified to be decreased in DDX43-transfected cells (Fig. 5e, f). Moreover, clinical sample analysis found H19 overexpression was associated with promoter hypomethylation and was upregulated during CML progression [26]. These results indicated that DDX43 may regulate the expression of H19 through demethylation.
Table. 1.
Upregulated and downregulated genes in DDX43-K562 by deep sequencing assay
| 20 upregulated genes | DDX43-K562/Control | 20 downregulated genes | DDX43-K562/Control |
|---|---|---|---|
| DDX43 | 432.5745 | EPB41L3 | 0.001285 |
| H19 | 91.57027 | MGST1 | 0.006269 |
| ITGBL1 | 83.46942 | PPFIBP2 | 0.006 |
| PPARGC1A | 53.39036 | SPARC | 0.017029 |
| SPATA17 | 34.96689 | ARHGAP15 | 0.010071 |
| LAPTM5 | 20.31337 | TUBA3C | 0.008135 |
| SPANXA2 | 19.73602 | FAM5C | 0.022559 |
| KIF1A | 17.12457 | STXBP6 | 0.020508 |
| GPRC5C | 13.10999 | PRDM9 | 0.02692 |
| PDE2A | 12.78386 | NXF2 | 0.025782 |
| LPL | 11.50522 | PASD1 | 0.02479 |
| RBPMS2 | 10.95067 | TDRG1 | 0.030693 |
| COL1A2 | 10.80316 | NXF2B | 0.029298 |
| TMEM163 | 9.399711 | C6orf145 | 0.028922 |
| SHANK1 | 8.798151 | TBX15 | 0.028922 |
| HLA-DOB | 7.444569 | C9orf125 | 0.033839 |
| ANKRD62 | 7.37238 | MAML3 | 0.039916 |
| TTYH3 | 6.834143 | ADAM19 | 0.039347 |
| XKR8 | 6.821518 | MALT1 | 0.045117 |
| RASGRP2 | 6.69587 | GNG11 | 0.044672 |
Fig. 5.
DDX43 promoted H19 expression by demethylation. a–d Expression of H19 in DDX43-transfected K562, HL60, SHI-1 and DDX43-silencing K562 cells was detected by RQ-PCR. e, f Promoter methylation levels of H19 in control vector or DDX43 transfected cells were detected by bisulfite sequencing. g Expression of H19 was detected by RQ-PCR. h K562 cells transfected with H19 siRNA or a control was used to apoptotic assays in serum-free media for 48 h. Cells were subjected to staining, followed by flow cytometry analysis. i K562 cells transfected with H19 siRNA or a control were subjected to proliferation assays in 10% FBS media and the live cells were counted at 24, 48, and 72 h. *P < 0.05; **P < 0.01. Error bars indicate SD (n = 3)
Many DDX43 related genes were involved in tumor growth, including H19. To explore the role of H19 in leukemia, the siRNA of H19 was transfected into K562 cells (K562-siH19). Expression of H19 was detected by RQ-PCR and decreased H19 was found in H19 siRNA-transfected cells, compared with control (Fig. 5g). Apoptosis was assayed for K562 cells transfected with H19 siRNA or a control in serum-free media for 48 h. K562-siH19 showed increased apoptosis rate compared with control (Fig. 5h). Moreover, K562-siH19 cells achieved reduced proliferation, compared to the control group (Fig. 5i). These results suggested that silencing H19 could promote K562 apoptosis and inhibit proliferation.
Arresting of miR-186 by DDX43 and miR-186 mediates K562 functions
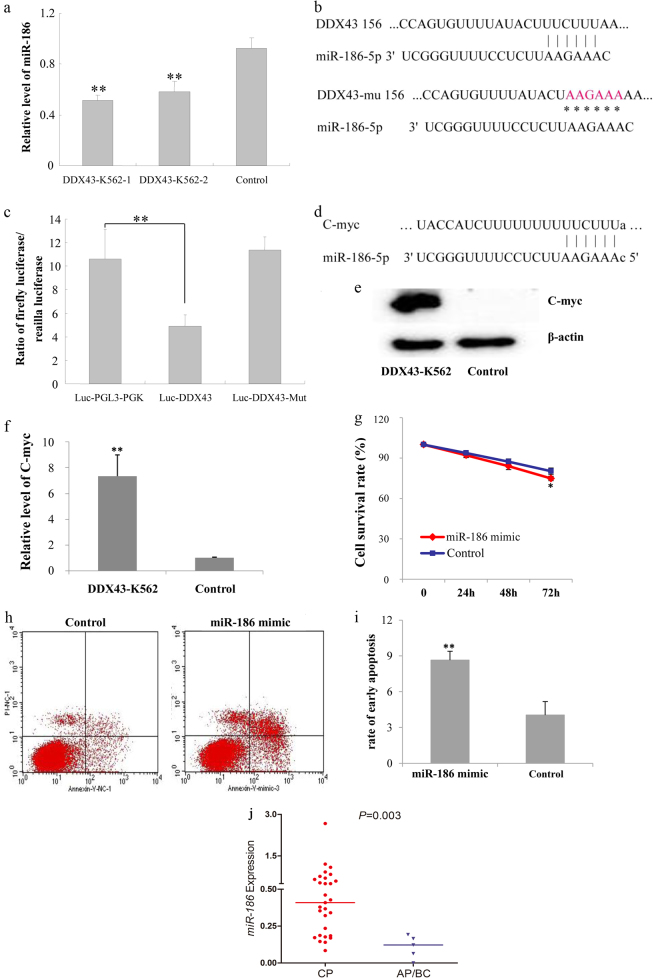
The deep sequencing also identified that miR-186 was downregulated in DDX43-transfected K562 cells (Table 2). RQ-PCR results further confirmed the decreased miR-186 expression in DDX43-transfected cells (Fig. 6a), suggesting that DDX43 may be involved in the reverse regulation to miR-186.
Table. 2.
Upregulated and downregulated microRNAs in DDX43-K562 by deep sequencing assay
| 20 upregulated microRNAs | DDX43-K562/Control | 20 downregulated microRNAs | DDX43-K562/Control |
|---|---|---|---|
| miR-218-5p | 396.223 | miR-1268a | 0.001539 |
| miR-520 | 131.4816 | miR-151a-5p | 0.002198 |
| miR-103a-3p | 107.1052 | hsa-miR-4429 | 0.012104 |
| miR-107 | 107.1052 | miR-135b-5p | 0.020591 |
| miR-9-3p | 74.74683 | miR-34c-5p | 0.044525 |
| miR-517c-3p | 65.74081 | miR-3180 | 0.060846 |
| miR-373-3p | 48.38775 | miR-4301 | 0.074321 |
| miR-520b | 47.69739 | miR-320c | 0.084726 |
| miR-526b-3p | 47.69739 | miR-320b | 0.084726 |
| miR-520e | 47.69739 | miR-22-3p | 0.085581 |
| miR-522-3p | 47.06979 | miR-3180-3p | 0.091131 |
| miR-3529-3p | 43.59387 | miR-1285-3p | 0.092263 |
| miR-3180-5p | 37.18514 | miR-4448 | 0.09414 |
| miR-1972 | 29.81087 | miR-30c-5p | 0.129583 |
| miR-1255b-5p | 26.35908 | miR-151a-3p | 0.134717 |
| miR-518d-5p | 22.5935 | miR-499a-5p | 0.150078 |
| miR-520c-5p | 22.5935 | miR-32-5p | 0.294186 |
| miR-526a | 22.5935 | miR-1254 | 0.313799 |
| miR-518f-5p | 22.5935 | miR-186-5p | 0.362075 |
| miR-1304-5p | 16.47443 | miR-10b-5p | 0.362075 |
Fig. 6.
miR-186-mediated DDX43 functions. a Expression of miR-186 in DDX43 or control vector transfected K562 cells was detected by RQ-PCR. b Binding of miR-186 to DDX43 3′ UTR was detected by luciferase assay. c The wild-type (Luc-DDX43) and mutant (Luc-DDX43-Mut) 3′ UTR sequences of DDX43 for luciferase activity assays. d Bioinformatics analysis showed the binding of miR-186 to c-Myc 3′ UTR. e, f Expression of c-Myc in DDX43- or control vector-transfected cells was detected by western blot and RQ-PCR. g K562 cells transfected with miR-186 mimic or control vector were cultured in serum-free media. Cells were harvested and surviving cells were counted. h, i K562 cells transfected with miR-186 mimic or a control were harvested for apoptotic assays in serum-free media after 48 h. Cells were subjected to staining, followed by flow cytometry analysis. j Relative expression level of miR-186 in CP and AP/BC stage of CML patients was presented with scatter plots. The median level of miR-186 in each group was shown with horizontal line. *P < 0.05; **P < 0.01. Error bars indicate SD (n = 3)
Bioinformatics analysis showed that miR-186 could bind to DDX43 3′ UTR. In order to confirm this result, the 3′ UTR of DDX43 was cloned to the downstream of luciferase reporter gene. Luciferase activity assay showed that the luciferase relative expression of the group co-transfected with DDX43-3′ UTR and miR-186 was lower than the control groups (PGL3-PGK + miR-186, negative control; DDX43-3′ UTR MUT + miR-186, negative control) (Fig. 6b, c). The results supported the direct binding of miR-186 to DDX43 3′ UTR region, and DDX43 as a potential target of miR-186. Combining with the effect of DDX43 on miR-186 expression, we speculate that DDX43 may be involved in the reverse regulation of miR-186. Further study is needed to determine the detailed dual-regulatory mechanisms. Moreover, bioinformatics analysis showed that c-Myc was another target of miR-186 (Fig. 6d). Our results identified that c-Myc increased in DDX43-transfected cells (Fig. 6e, f).
We transfected miR-186 mimic into K562 cells, and analyzed the role of miR-186 on K562 cells. The survival rate of miR-186-K562 cell was decreased (Fig. 6g) and the apoptosis rate was increased (Fig. 6h, i), compared to the control group, suggesting that miR-186 could inhibit K562 survival and promote apoptosis. In addition, the level of miR-186 in CML patients at AP/BC stages (median 0.12, range 0–0.19) was lower than that at the CP stage (median 0.41, range 0.08–2.66) (P = 0.003) (Fig. 6j).
Discussion
Our previous study confirmed that DDX43 overexpression, regulated by the hypomethylation of its promoter, was associated with the progression of CML. In this study, we found that DDX43 enhanced survival and colony formation, inhibited cell apoptosis, promoted tumorigenesis, and CML progression in vitro and in vivo.
To elucidate the molecular mechanisms of DDX43 in leukemia, the deep sequencing assay was performed to screen related genes for DDX43. Various mRNAs related to DDX43 were up- or down- regulated. H19, RASGRP2, ITGBL1, and PPARGC1A were upregulated, the roles of which are identified in tumorigenesis, cell growth, cell adhesion, and nucleotide binding; MGST1, SPARC, and FAM5C were downregulated, whose functions are involved in the response to drug, cell proliferation, cell cycle and so on. At the same time, some microRNAs related with DDX43 were up- or down- regulated. Some tumor-promoting microRNAs, such as miR-218-5p, miR-107, and miR-9-3p, were upregulated; some tumor-suppressing microRNAs, such as miR-186, miR-22-3p, and miR-1285-3p, were downregulated. We confirmed that H19 expression was increased and miR-186 expression was decreased in DDX43-transfected cells using RQ-PCR. Other genes and microRNAs changed in DDX43-transfected cells might be studied in a future study.
We further showed that DDX43 induced H19 promoter demethylation, and then increased its expression. H19 has been revealed to be upregulated in various carcinomas and to play important roles in the occurrence, development, metastasis, and prognosis of tumors [27–30]. Our previous study indicated that H19 expression, associated with its promoter methylation, was significantly upregulated in CML patients, and its expression increased as disease progressed [26]. H19 acts mainly through two ways: first, miR-675, encoded by the first exon of H19, regulates the relevant tumor suppressor genes [28, 31–33]; second, the total length of H19 combines with microRNAs or related proteins to regulate their functions [28, 34, 35]. We also detected miR-675 expression in DDX43- transfected K562, HL60, and SHI-1 cells. However, miR-675 was downregulated in DDX43-transfected K562 cells, upregulated in DDX43-transfected SHI-1 cells and showed no difference in DDX43-transfected HL60 cells. It is suggested that miR-675 regulation might not be the main factor in leukemic cell function.
In addition, we also focused on miR-186 which was decreased in DDX43-transfected cells. Luciferase activity test showed that miR-186 might bind to the DDX43 3′ UTR region, and DDX43 may be a potential target gene of miR-186. We also detected the effect of miR-186 on K562 cells. Our results suggested that miR-186 could inhibit K562 survival and promote apoptosis. Recently, miR-186 has been considered as a tumor suppressive microRNA in many solid cancers, such as prostate cancer and non-small cell lung cancer (NSCLC) [21, 36, 37]. However, it was also reported that miR-186 acted as an oncogenic microRNA by modulating PTTG1 expression and accelerating migration and invasion in human NSCLC cells [38]. As to hematologic malignancies, Ferrer G identified the downregulation of miR-186 expression in chronic lymphocytic leukemia [39]. Our group previously identified that miR-186 transcript frequently decreased in AML. Moreover, in this study, miR-186 decreased in AP/BC stage compared with the CP stage in CML, suggesting that miR-186 downregulation may promote the progression of CML. Downregulated expression of miR-186 also increased the expression of c-Myc. We speculate that it may involve complex regulatory mechanisms that need further study.
Now, we will propose the following hypothesis: miR-186 expression is downregulated in the progression of CML, leading to the upregulation of target gene—DDX43 in CML; DDX43 further upregulates the expression of H19 through demethylation. In summary, the signal of miR-186/DDX43/H19 may be involved in regulating the function of leukemia cells.
In conclusion, our results indicate that DDX43 enhances survival and colony formation, inhibits cell apoptosis, and promotes tumorigenesis and CML progression. The potential mechanism involves the arresting of miR-186 and releasing of H19 by DDX43.
Materials and methods
Construct generation
A DDX43-expressing construct was designed by our laboratory. Briefly, a cDNA sequence containing DDX43 was inserted into a lenti-virus vector containing green fluorescent protein in the restriction enzyme site BamHI.
RNA isolation, reverse transcription, and RQ-PCR
Total RNA was isolated (mirVana miRNA Isolation Kit, Ambion, USA) and then reverse transcribed into cDNA. RQ-PCR was performed as described previously [40]. DDX43 was amplified with primers of 5′-CCTTTCAATGTTATCCTGAG-3′ (forward) and 5′-TATTCTTCAGATTGACGAAG-3′ (reverse). The specific forward primer of miR-186 was 5′-CAAAGAATTCTCCTTTTGGGCT-3′, while H19 was amplified with primers of 5′-GGGTCAGACAGGGACATGG-3′ (forward) and 5′-GAGCGGTGAGGGCATACA-3′ (reverse). MiR-186 and H19 expressions were determined as reported previously [21, 41].
DNA isolation, modification, and methylation detection
DNA extraction and modification were performed as described previously [8]. The level and density of H19 promoter methylation were detected using real-time quantitative MSP and bisulfite sequencing PCR as previously reported [41].
Cell survival and apoptosis assays
To analyze cell survival, 1×106 K562 or Meg-01cells (ATCC, Manassas, USA) were cultured in 25 cm2 tissue culture flasks for 72 h in Iscove’s Modified Dulbecco’s Medium (IMDM, Wisent) without FBS (ExCell Bio). The viable cells were numbered using trypan blue exclusion. To study cell apoptosis, 1×106 cells were cultured in 25 cm2 tissue culture flasks for 48 h in IMDM without FBS. The cultured cells were also stained for apoptosis assay using the apoptosis detection Kit (Annexin V-FITC, BD, 556547; or Annexin V-PE/7AAD, BD, 559763), followed by flow cytometry analysis.
Colony formation
Soft agarose assay: 2% agarose was plated on six-well plates, and then 1×103 cells were inoculated into 0.3% agarose containing 10% FBS. Colonies were numbered and photographed after 4 weeks. Methylcellulose assay: 1×103 cells in IMDM were mixed in 1% methylcellulose supplemented with 10% FBS and plated on six-well tissue culture plates. Colonies were numbered and photographed 2 weeks after cell inoculation.
Tumorigenesis assays in nude mice
The animal protocol was approved by the Animal Care Committee of the Affiliated People’s Hospital of Jiangsu University. The DDX43- or mock-transfected K562 cells were injected randomly into 5-week-old CD1 strain nude mice subcutaneously (1.5×107 cells per mouse, seven mice per group). Tumor growth was assessed every 4 days. The mice were killed and the tumors were removed at day 30 post inoculation as described previously [42]. Tumors were stored in −80 °C for PQ-PCR and western blot to detect DDX43 expression. DDX43 shRNA- or a control vector-transfected DDX43-K562 cells were injected randomly into CD1 strain nude mice subcutaneously. Eight mice were included in each group. Mice were photographed and killed after 4 weeks of injection. Tumor sizes were measured.
CML progression in SCID mice
In the CML progression assay, the DDX43- or mock-transfected K562 cells were injected into 5-week-old SCID mice randomly via tail vein (1.5×107 cells per mouse, five mice per group). Mouse weight was monitored weekly. The peripheral WBC was counted weekly. When the mice suffered from CML symptoms (4 weeks after injection), they were killed and bone marrows were obtained to detect blasts with Wright−Giemsa stain.
Transfection of tumor cells with shRNA or siRNA
The siRNAs and shRNAs were designed and synthesized (GenePharma, Shanghai, China). Three shRNAs targeting DDX43 expression and one negative control were used in this experiment: shRNA-1 (DDX43-homo-909 GCAGATTTACCACCAATTAAG); shRNA-2 (DDX43-homo-1066 GCACATTTGATGACGCCTTTC); shRNA-3 (DDX43-homo-1175 GCAAGGAATAGATCTTATAGG); and negative control (TTCTCCGAACGTGTCACGT). SiH19-1977 (CCCGUCCCUUCUGAAUUUATT) and H19 siRNA-negative control (UUCUCCGAACGUGUCACGUTT) were used for H19-silencing experiments. ShRNA or siRNA transfection was performed using Lipofectamine 2000 (Invitrogen). Western blot and RQ-PCR were carried out 72 h after transfection.
Western blot analysis
Cells were lysed with 100 µl of lysis buffer containing protease inhibitors (Beyotime, Shanghai, China). The lysates were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to a polyvinylidene fluoride (PVDF) membrane, and then incubated with an anti-DDX43 primary antibody (H00055510-M07, Abnova) overnight at 4 °C. Horseradish peroxidase (HRP)-conjugated goat-anti-mouse secondary antibody (sc-2005, Santa Cruz Biotechnology) was used. The protein bands were detected by enhanced chemiluminescence detection. Anti-β-actin antibody (AA128, Beyotime, Shanghai, China) was used as a loading control.
Luciferase activity assay
Luciferase activity assays were carried out (Promega Luciferase Assay System, USA). The vector was purchased from HuiJun (Guangzhou, China). The wild-type (Luc-DDX43) or mutant (Luc-DDX43-Mut) sequences of DDX43 3′ UTR was inserted into a dual luciferase reporter vector (Promega, USA). A total of 5×104 HepG2 cells were cultured in Dulbecco's Modified Eagle Medium (DMEM) containing 10% FBS in 24-well plates. The cells were co-transfected with 0.8 μg of Luc-PGL3-PGK or Luc-DDX43 or Luc-DDX43-Mut and 24 pmol of miR-186 mimic or scramble miR mimic, and the pRL-TK plasmid for internal normalization using Lipofectamine 2000. We measured Firefly and Renilla luciferase activities immediately using dual-luciferase assays (Promega, USA) 24 h after transfection.
Patients
This study was approved by Institutional Ethics Committee of the Affiliated People’s Hospital of Jiangsu University. Bone marrow samples from 34 CML patients (29 cases at CP stage, 3 at AP stage, and 2 at BC stage) were obtained with written informed consent. Bone marrow samples were enriched for mononuclear cells using density gradient centrifugation.
Statistical analysis
All assays were performed at least in triplicate. Statistical analyses were performed as previously described [25, 43]. In brief, results expressed as mean values ± SD of all experiments were used for statistical analysis. Student’s t test or one-way analysis of variance was carried out to compare the difference among two or multiple groups. The level of significance was set at P < 0.05.
Funding
This study was supported by National Natural Science Foundation of China (81172592, 81270630), Medical Innovative Team of “Ke Jiao Qiang Wei” Project of Jiangsu Province (CXTDB2017002), six talent peaks project in Jiangsu Province (2015-WSN-115), China Postdoctoral Science Foundation (2016M601747), and Jiangsu Provincial “Innovative & entrepreneurial talent team” Program.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no competing interests.
Contributor Information
J. Qian, Phone: +86 511 88915303, Email: qianjun0007@hotmail.com
Z.-Q. Deng, Phone: +86 511 88915586, Email: zqdeng2002@163.com
References
- 1.Hehlmann R, Hochhaus A, Baccarani M, European L. Chronic myeloid leukaemia. Lancet. 2007;370:342–50. doi: 10.1016/S0140-6736(07)61165-9. [DOI] [PubMed] [Google Scholar]
- 2.Giustacchini A, Thongjuea S, Barkas N, Woll PS, Povinelli BJ, Booth CAG, et al. Single-cell transcriptomics uncovers distinct molecular signatures of stem cells in chronic myeloid leukemia. Nat Med. 2017;23:692–702. doi: 10.1038/nm.4336. [DOI] [PubMed] [Google Scholar]
- 3.Ren R. Mechanisms of BCR-ABL in the pathogenesis of chronic myelogenous leukaemia. Nat Rev Cancer. 2005;5:172–83. doi: 10.1038/nrc1567. [DOI] [PubMed] [Google Scholar]
- 4.Savona M, Talpaz M. Getting to the stem of chronic myeloid leukaemia. Nat Rev Cancer. 2008;8:341–50. doi: 10.1038/nrc2368. [DOI] [PubMed] [Google Scholar]
- 5.Calabretta B, Perrotti D. The biology of CML blast crisis. Blood. 2004;103:4010–22. doi: 10.1182/blood-2003-12-4111. [DOI] [PubMed] [Google Scholar]
- 6.Wang W, Cortes JE, Lin P, Beaty MW, Ai D, Amin HM, et al. Clinical and prognostic significance of 3q26.2 and other chromosome 3 abnormalities in CML in the era of tyrosine kinase inhibitors. Blood. 2015;126:1699–706. doi: 10.1182/blood-2015-05-646489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sidaway P. Haematological cancer: ponatinib in CML—keeping PACE with multiple mutations. Nat Rev Clin Oncol. 2016;13:135. doi: 10.1038/nrclinonc.2016.13. [DOI] [PubMed] [Google Scholar]
- 8.Chen Q, Lin J, Qian J, Deng ZQ, Qian W, Yang J, et al. The methylation status of the DDX43 promoter in Chinese patients with chronic myeloid leukemia. Genet Test Mol Biomark. 2013;17:508–11. doi: 10.1089/gtmb.2012.0530. [DOI] [PubMed] [Google Scholar]
- 9.Martelange V, De Smet C, De Plaen E, Lurquin C, Boon T. Identification on a human sarcoma of two new genes with tumor-specific expression. Cancer Res. 2000;60:3848–55. [PubMed] [Google Scholar]
- 10.Lin J, Chen Q, Yang J, Qian J, Deng ZQ, Qian W, et al. DDX43 promoter is frequently hypomethylated and may predict a favorable outcome in acute myeloid leukemia. Leuk Res. 2014;38:601–7. doi: 10.1016/j.leukres.2014.02.012. [DOI] [PubMed] [Google Scholar]
- 11.Chen Q, Lin J, Yao DM, Qian J, Qian W, Yang J, et al. Aberrant hypomethylation of DDX43 promoter in myelodysplastic syndrome. Br J Haematol. 2012;158:293–6. doi: 10.1111/j.1365-2141.2012.09138.x. [DOI] [PubMed] [Google Scholar]
- 12.Adams SP, Sahota SS, Mijovic A, Czepulkowski B, Padua RA, Mufti GJ, et al. Frequent expression of HAGE in presentation chronic myeloid leukaemias. Leukemia. 2002;16:2238–42. doi: 10.1038/sj.leu.2402732. [DOI] [PubMed] [Google Scholar]
- 13.Liggins AP, Lim SH, Soilleux EJ, Pulford K, Banham AH. A panel of cancer-testis genes exhibiting broad-spectrum expression in haematological malignancies. Cancer Immun. 2010;10:8. [PMC free article] [PubMed] [Google Scholar]
- 14.Luo M, Li Z, Wang W, Zeng Y, Liu Z, Qiu J. Long non-coding RNA H19 increases bladder cancer metastasis by associating with EZH2 and inhibiting E-cadherin expression. Cancer Lett. 2013;333:213–21. doi: 10.1016/j.canlet.2013.01.033. [DOI] [PubMed] [Google Scholar]
- 15.Conigliaro A, Costa V, Lo Dico A, Saieva L, Buccheri S, Dieli F, et al. CD90 + liver cancer cells modulate endothelial cell phenotype through the release of exosomes containing H19 lncRNA. Mol Cancer. 2015;14:155. doi: 10.1186/s12943-015-0426-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou W, Ye XL, Xu J, Cao MG, Fang ZY, Li LY, et al. The lncRNA H19 mediates breast cancer cell plasticity during EMT and MET plasticity by differentially sponging miR-200b/c and let-7b. Sci Signal. 2017;10:eaak9557. doi: 10.1126/scisignal.aak9557. [DOI] [PubMed] [Google Scholar]
- 17.Fanale D, Amodeo V, Corsini LR, Rizzo S, Bazan V, Russo A. Breast cancer genome-wide association studies: there is strength in numbers. Oncogene. 2012;31:2121–8. doi: 10.1038/onc.2011.408. [DOI] [PubMed] [Google Scholar]
- 18.Wang Q, Cheng N, Li X, Pan H, Li C, Ren S, et al. Correlation of long non-coding RNA H19 expression with cisplatin-resistance and clinical outcome in lung adenocarcinoma. Oncotarget. 2017;8:2558–67. doi: 10.18632/oncotarget.13708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee DF, Su J, Kim HS, Chang B, Papatsenko D, Zhao R, et al. Modeling familial cancer with induced pluripotent stem cells. Cell. 2015;161:240–54. doi: 10.1016/j.cell.2015.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kallen AN, Zhou XB, Xu J, Qiao C, Ma J, Yan L, et al. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol Cell. 2013;52:101–12. doi: 10.1016/j.molcel.2013.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cai J, Wu J, Zhang H, Fang L, Huang Y, Yang Y, et al. miR-186 downregulation correlates with poor survival in lung adenocarcinoma, where it interferes with cell-cycle regulation. Cancer Res. 2013;73:756–66. doi: 10.1158/0008-5472.CAN-12-2651. [DOI] [PubMed] [Google Scholar]
- 22.Zhu X, Shen H, Yin X, Long L, Xie C, Liu Y, et al. miR-186 regulation of Twist1 and ovarian cancer sensitivity to cisplatin. Oncogene. 2016;35:323–32. doi: 10.1038/onc.2015.84. [DOI] [PubMed] [Google Scholar]
- 23.Zheng J, Li XD, Wang P, Liu XB, Xue YX, Hu Y, et al. CRNDE affects the malignant biological characteristics of human glioma stem cells by negatively regulating miR-186. Oncotarget. 2015;6:25339–55. doi: 10.18632/oncotarget.4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang TJ, Wang YX, Yang DQ, Yao DM, Yang L, Zhou JD, et al. Down-regulation of miR-186 correlates with poor survival in de novo acute myeloid leukemia. Clin Lab. 2016;62:113–20. doi: 10.7754/clin.lab.2015.150606. [DOI] [PubMed] [Google Scholar]
- 25.Chen Q, Lin J, Qian J, Yao DM, Qian W, Li Y, et al. Gene expression of helicase antigen in patients with acute and chronic myeloid leukemia. Zhongguo shi yan xue ye xue za zhi. 2011;19:1171–5. [PubMed] [Google Scholar]
- 26.Zhou JD, Lin J, Zhang TJ, Ma JC, Li XX, Wen XM, et al. Hypomethylation-mediated H19 overexpression increases the risk of disease evolution through the association with BCR-ABL transcript in chronic myeloid leukemia. J Cell Physiol. 2017;233:2444–50. doi: 10.1002/jcp.26119. [DOI] [PubMed] [Google Scholar]
- 27.Si X, Zang R, Zhang E, Liu Y, Shi X, Zhang E, et al. LncRNA H19 confers chemoresistance in ERalpha-positive breast cancer through epigenetic silencing of the pro-apoptotic gene BIK. Oncotarget. 2016;7:81452–62. doi: 10.18632/oncotarget.13263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raveh E, Matouk IJ, Gilon M, Hochberg A. The H19 long non-coding RNA in cancer initiation, progression and metastasis—a proposed unifying theory. Mol Cancer. 2015;14:184. doi: 10.1186/s12943-015-0458-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matouk IJ, DeGroot N, Mezan S, Ayesh S, Abu-lail R, Hochberg A, et al. The H19 non-coding RNA is essential for human tumor growth. PLoS One. 2007;2:e845. doi: 10.1371/journal.pone.0000845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rokavec M, Horst D, Hermeking H. Cellular model of colon cancer progression reveals signatures of mRNAs, miRNA, lncRNAs, and epigenetic modifications associated with metastasis. Cancer Res. 2017;77:1854–67. doi: 10.1158/0008-5472.CAN-16-3236. [DOI] [PubMed] [Google Scholar]
- 31.Dey BK, Pfeifer K, Dutta A. The H19 long noncoding RNA gives rise to microRNAs miR-675-3p and miR-675-5p to promote skeletal muscle differentiation and regeneration. Genes Dev. 2014;28:491–501. doi: 10.1101/gad.234419.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vennin C, Spruyt N, Dahmani F, Julien S, Bertucci F, Finetti P, et al. H19 non coding RNA-derived miR-675 enhances tumorigenesis and metastasis of breast cancer cells by downregulating c-Cbl and Cbl-b. Oncotarget. 2015;6:29209–23. doi: 10.18632/oncotarget.4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang Y, Zheng Y, Jia L, Li W. Long Noncoding RNA H19 Promotes Osteoblast Differentiation Via TGF-beta1/Smad3/HDAC Signaling Pathway by Deriving miR-675. Stem Cells. 2015;33:3481–92. doi: 10.1002/stem.2225. [DOI] [PubMed] [Google Scholar]
- 34.Song Y, Liu C, Liu X, Trottier J, Beaudoin M, Zhang L, et al. H19 promotes cholestatic liver fibrosis by preventing ZEB1-mediated inhibition of EpCAM. Hepatology. 2017;66:1183–96. doi: 10.1002/hep.29209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou J, Yang L, Zhong T, Mueller M, Men Y, Zhang N, et al. H19 lncRNA alters DNA methylation genome wide by regulating S-adenosylhomocysteine hydrolase. Nat Commun. 2015;6:10221. doi: 10.1038/ncomms10221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cui G, Cui M, Li Y, Liang Y, Li W, Guo H, et al. MiR-186 targets ROCK1 to suppress the growth and metastasis of NSCLC cells. Tumour Biol: J Int Soc Oncodev Biol Med. 2014;35:8933–7. doi: 10.1007/s13277-014-2168-6. [DOI] [PubMed] [Google Scholar]
- 37.Erdmann K, Kaulke K, Thomae C, Huebner D, Sergon M, Froehner M, et al. Elevated expression of prostate cancer-associated genes is linked to down-regulation of microRNAs. BMC Cancer. 2014;14:82. doi: 10.1186/1471-2407-14-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li H, Yin C, Zhang B, Sun Y, Shi L, Liu N, et al. PTTG1 promotes migration and invasion of human non-small cell lung cancer cells and is modulated by miR-186. Carcinogenesis. 2013;34:2145–55. doi: 10.1093/carcin/bgt158. [DOI] [PubMed] [Google Scholar]
- 39.Ferrer G, Navarro A, Hodgson K, Aymerich M, Pereira A, Baumann T, et al. MicroRNA expression in chronic lymphocytic leukemia developing autoimmune hemolytic anemia. Leuk Lymphoma. 2013;54:2016–22. doi: 10.3109/10428194.2012.763123. [DOI] [PubMed] [Google Scholar]
- 40.Ma J, Lin J, Qian J, Qian W, Yin J, Yang B, et al. MiR-378 promotes the migration of liver cancer cells by down-regulating Fus expression. Cell Physiol Biochem: Int J Exp Cell Physiol, Biochem, Pharmacol. 2014;34:2266–74. doi: 10.1159/000369669. [DOI] [PubMed] [Google Scholar]
- 41.Zhou JD, Lin J, Zhang TJ, Ma JC, Li XX, Wen XM, et al. Hypomethylation-mediated H19 overexpression increases the risk of disease evolution through the association with BCR-ABL transcript in chronic myeloid leukemia. J Cell Physiol. 2018;233:2444–50. doi: 10.1002/jcp.26119. [DOI] [PubMed] [Google Scholar]
- 42.Zheng PS, Wen J, Ang LC, Sheng W, Viloria-Petit A, Wang Y, et al. Versican/PG-M G3 domain promotes tumor growth and angiogenesis. FASEB J. 2004;18:754–6. doi: 10.1096/fj.03-0545fje. [DOI] [PubMed] [Google Scholar]
- 43.Bernaudo S, Salem M, Qi X, Zhou W, Zhang C, Yang W, et al. Cyclin G2 inhibits epithelial-to-mesenchymal transition by disrupting Wnt/beta-catenin signaling. Oncogene. 2016;35:4816–27. doi: 10.1038/onc.2016.15. [DOI] [PMC free article] [PubMed] [Google Scholar]