Abstract
Here we used flow cytometry (FCM) and filtration paired with amplicon sequencing to determine the abundance and composition of small low nucleic acid (LNA)-content bacteria in a variety of freshwater ecosystems. We found that FCM clusters associated with LNA-content bacteria were ubiquitous across several ecosystems, varying from 50 to 90% of aquatic bacteria. Using filter-size separation, we separated small LNA-content bacteria (passing 0.4 µm filter) from large bacteria (captured on 0.4 µm filter) and characterized communities with 16S amplicon sequencing. Small and large bacteria each represented different sub-communities within the ecosystems’ community. Moreover, we were able to identify individual operational taxonomical units (OTUs) that appeared exclusively with small bacteria (434 OTUs) or exclusively with large bacteria (441 OTUs). Surprisingly, these exclusive OTUs clustered at the phylum level, with many OTUs appearing exclusively with small bacteria identified as candidate phyla (i.e. lacking cultured representatives) and symbionts. We propose that LNA-content bacteria observed with FCM encompass several previously characterized categories of bacteria (ultramicrobacteria, ultra-small bacteria, candidate phyla radiation) that share many traits including small size and metabolic dependencies on other microorganisms.
Introduction
Bacteria constitute the smallest forms of independent life, and considerable effort has been made to theoretically calculate, locate, and characterize the smallest bacterial representatives [1, 2]. Different terminologies, potentially overlapping and not always clearly defined, are used in this field including ultramicrobacteria (UMB), ultra-small bacteria (USB), ultramicrocells, nanoarcahea, and nanoplankton. Focusing within bacteria, UMB are defined as bacteria less than 0.1 μm3 in size [1]. This upper limit is 1 order of magnitude smaller than a typical Escherichia coli cell (1 μm3), and 9 orders of magnitude smaller than the largest known bacterium, Thiomargarita namibiensis, (2.2 × 108 μm3 [3]). Similarly, USB have been studied as having small genomes and have been isolated following 0.2 μm filtration [2]. The terms ultramicrocells and nanoplankton (marine ecology) are similarly defined by filterability [1]. The exact upper boundary of these groups of “small bacteria”, while defined [1], remains somewhat arbitrary. Filtration with small pore size (i.e., 0.2 μm filterable) is often used for the isolation or enrichment of these groups [2, 4–6], but many bacteria may well exist on the fringe of this border [1].
Since particle-size and nucleic acid-content can be quickly assessed using flow cytometry (FCM), it may be a useful tool for identifiying small cells. FCM has been used extensively in natural aquatic environments, including wastewater [7, 8], drinking water [9], process water [10], seawater [11], and lake water [12]. Using this method, a bimodal distribution of cells is frequently observed, with two dominant cell clusters separated by fluorescence intensity and/or light scatter signals after staining of nucleic acids. These two groups are commonly referred to as high (HNA) and low (LNA) nucleic acid-content bacteria [13–16], based on an inferred correlation between observed fluorescence intensity and cellular DNA/RNA content, the target for the fluorescent dyes. Initial studies suggested that LNA-content bacteria represented the dead or inactive fraction of microbial communities [14], but subsequent research contradicted this by showing their growth [16] and substrate uptake [15]. Moreover, using cell sorting, Bouvier et al. [15] identified LNA-content bacteria that specifically had small genome sizes, and Vila-Costa et al. [17] characterized distinct phylogenetic communities of LNA-content bacteria in marine waters. In a finding particularly relevant to the present study, Wang et al. [16] demonstrated that 0.45 µm membrane filtration essentially separated HNA- and LNA-content bacteria, thus establishing a link between LNA-content bacteria and the size and filterability of bacterial cells.
Filtration for size-separation is therefore useful for studying LNA-content bacteria as well as UMB and USB. Such techniques were previously used for the isolation of bacteria in the candidate phyla (i.e., candidate phyla radiation (CPR) lacking culturable representatives but likely representing a distinct clade) [18], and have recovered proposed symbiont bacteria and oligotrophic bacteria [19, 20]. All of these groups are proposed to resist traditional culturing for various reasons, including obligate oligotrophy and dependencies on substrates supplied by other species in nature and not typically supplied in culture media (i.e., auxotrophy). To overcome difficulty with culturing these groups of organisms, some approaches use filtration to remove large competitors and isolate small bacteria [2, 21]. Caution should be taken using filtration to isolate small bacteria since large bacteria with one small dimension (e.g. long with small diameter) can also pass through filters [22].
In this study, we approach this concept of small bacteria using FCM and filtration paired with amplicon sequencing. We hypothesize that the cluster of LNA-content bacteria observed with FCM are physically small and thus easily separated with 0.4 µm filtration, are ubiquitous across and even dominate some aquatic environments, and that these LNA-content bacteria are phylogenetically distinct from large HNA-content bacteria. We test these hypotheses both within and across several freshwater ecosystems, including lake water, river water, wastewater effluent, groundwater and non-chlorinated tap water. Comparing traits of our small bacteria to other bacteria isolated with filtration, including the broadly defined groups of UMB, USB, CPR, and symbiont bacteria, we propose that LNA-content bacteria can encompass all of these categories that in fact share many traits (including small size and metabolic dependencies on other microorganisms).
Materials and Methods
Sampling
A total of 47 samples were taken from 22 different sampling sites in Switzerland in five categories of aquatic ecosystems, i.e. groundwater, river water, lake water, (non-chlorinated) tap water, and wastewater (secondary effluent) (Fig. 1a, Table S1). One river and one lake sampling site were sampled a total of 12–15 times each to assess temporal dynamics. Samples were taken in muffled (560 °C for 3 h) glass bottles. Volumes per sampling site ranged from 500 to 25’000 ml depending on the expected concentration of bacteria in the respective ecosystem (Table S2). Samples were transported and stored at 4 °C and processed within 24 h.
Fig. 1.
Sample collection, treatment, and statistical analysis. a A total of 47 samples were collected from 22 sampling sites classified in 5 ecosystems. b Each sample was processed in duplicate, and for each duplicate, 3 different groups were collected: “All bacteria”, which was filtered directly onto a 0.2 µm filter, “Large bacteria” (red), which was filtered directly onto a 0.4 µm filter, and “Small bacteria” (blue), which was the filtrate from the 0.4 µm filter captured on a 0.2 µm filter. c Each OTU from the community sequencing data were classified into 5 categories based on its appearance in the large and small bacteria group of a filter pair or sample. For all categories, it was permissible that an OTU appeared in both the large and small bacteria groups of a filter pair. Unclassifiable was a catch-all for OTUs not meeting the criteria of the other categories, and eliminated OTUs did not meet abundance cutoffs
Filtration
Filtration volumes were adjusted between 100 and 5'000 ml per filter based on FCM total cell concentration (TCC) measurements to approximately equalize number of cells captured (Table S2). Three types of filters were captured in duplicate for each sample (Fig. 1b). The first filter captured the entire community with direct filtration onto 0.2 µm membrane filters (“all bacteria”) (NucleporeTM track-etched polycarbonate membranes, 47 mm, Whatman, UK) using sterilized filtration units (NalgeneTM, Thermo Fisher Scientific, USA) mounted on sterilized glass bottles. Separately, a two-step filtration was performed to obtain size-based groups. Another water sample was first filtered onto 0.4 µm membrane filters (large bacteria) (NucleporeTM track-etched polycarbonate membranes, 47 mm, Whatman, UK), and the resulting filtrate was subsequently filtered again on 0.2 µm filters (small bacteria). Filters from the paired filtration step (Large, small) and direct filtration (All) were then stored at −80 °C until DNA extraction.
Flow cytometry (FCM)
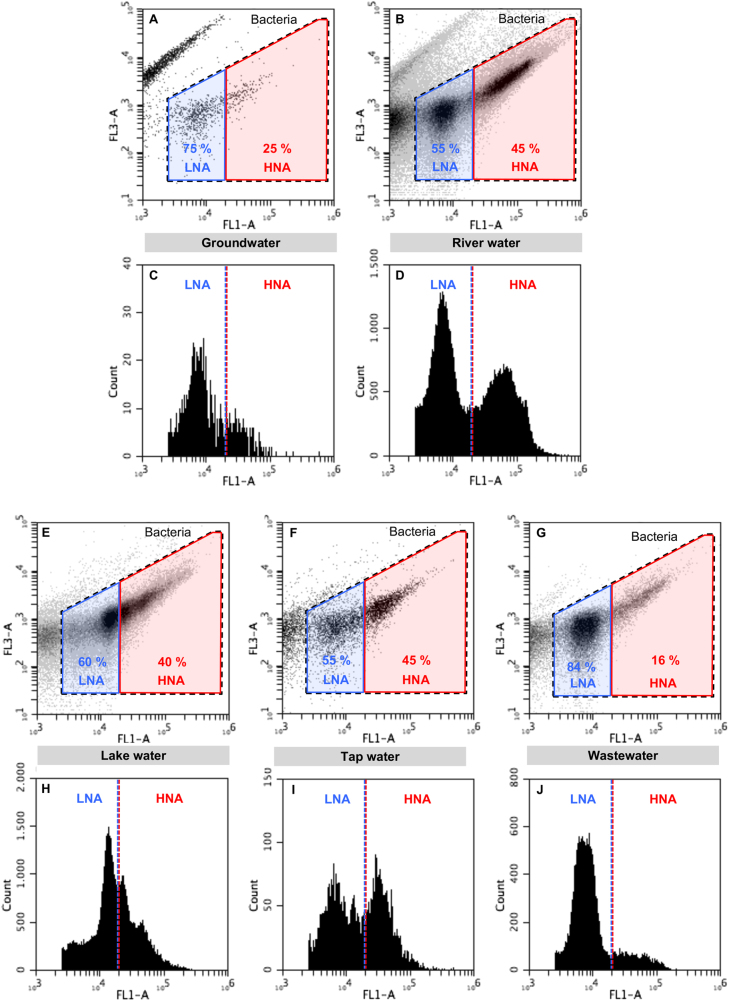
The TCC of all water samples was determined with FCM before and after 0.4 µm filtration in triplicate. FCM sample preparation and measurements were based on the standard method 333.1 accredited in Switzerland [23]. In short, 200 µl of the water samples were pre-warmed (3 min, 37 °C) and then stained with 2 µl of fluorescent stain (SYBR Green I, Life Technologies, Eugene OR, USA; final concentration 1:10’000). After 10 min of incubation at 37 °C in the dark, 50 µl were measured on a BD Accuri C6 flow cytometer (BD Accuri, San Jose CA, USA) at a flow rate of 66 µl min−1 with a lower threshold on the green fluorescence (FL1-H) channel at 1000. Fixed standard gates were applied to separate bacteria from background signals and LNA from HNA bacteria [24] (Fig. 2). A 10-fold dilution with 0.1 µm filtered Evian water was performed before measurement for samples expected to have high cell numbers.
Fig. 2.
Typical flow cytometric density and histogram plots from the five investigated natural and engineered freshwater ecosystems (groundwater a, c, river water b. d, lake water e, h, tap water f, i, wastewater g, j) stained with SYBR Green I. Dotted black lines indicate electronic gates separating bacteria from background. Blue and red gates/dotted lines indicate electronic gates separating LNA and HNA content bacteria. FL1-A indicates green fluorescence intensity, FL3-A indicates red fluorescence intensity
DNA extraction
Microbial DNA was extracted from preserved filters by enzymatic digestion and cetyltrimethyl ammoniumbromide (CTAB) extraction following a published protocol with minor adaptations [25]. In short, enzymatic cell lysis was performed on filters by subsequent incubations with Lysozyme, Proteinase K, and RNase A (Proteinase K volume was increased to 10 µl, and RNase to 5 µl). Cells were lysed with a CTAB buffer, and unwanted materials were extracted with chloroform isoamyl alcohol (we used 49:1 instead of 24:1 v:v ratio). DNA was precipitated with ethanol and DNA redissolved in TE buffer. Sample replicates were extracted separately. DNA concentrations in the extracts ranged from 0.8 to 50 ng µl−1.
Amplicon sequencing with Illumina MiSeq
16S rRNA amplicon sequencing was performed as described previously [26]. Bacterial primers 341F and 785R [27] were used, adapted with a tail incorporating frame-shifts that were used to separate replicates during PCR amplification. Products were purified with the Agencort AMPure beads XP system (Beckman Coulter, Inc., Bera, CA) and Nextera index primers were added with Index PCR. Index PCR product was purified, quality controlled and quantified by qPCR. Details for all steps are in Table S3. Equal amounts (4 nM) of PCR product were then pooled for sequencing on the Illumina MiSeq platform following standard protocols for the MiSeq Reagent Kit v3 600 cycles (MS-102-3003).
Multiple algorithms were used for sequence quality control and merging, trimming, and filtering reads, as well as OTU clustering (Table S4), using preferred elements of established pipelines. FastQC v0.11.2 [28] was used for quality control. FLASH v1.2.9 was used for merging reads with a minimum overlap of 14, maximum overlap of 250, and max mismatch density of 0.25. Cutadapt v1.5 [29] was used with an error rate of 0 at full-length to trim adaptor sequences and sort frame shifts. Quality filtering was done with PRINSEQ-lite v0.20.4 [30] with a size range of 390–440 bp, mean quality score of 25, maximum of 1 ambiguous nucleotide, GC range of 30–70, and low complexity filter dust/25. Finally, OTU clustering was done with usearch v7.0.1090 [31] with identity cutoff of 97%, abundance sorting with minimum size of 2, and chimera filtering applied.
Sequences were classified taxonomically according to greengenes v.13.5 [32] using usearch v10.0.240 linux 64 and sintax (classifier). In R, phyloseq [33] was used for processing. Sequences identified as Archaea and Chloroplasts were removed from the data set. All samples considered had more than 4'000 reads in each of the six related samples (3 groups and 2 replicates). Raw sequence data are available under accession number PRJEB23669.
Exclusivity analysis
For exclusivity analysis (Fig. 1c), rare OTUs that did not reach at least 20 reads in at least two samples were excluded from the data set (i.e. 43'616 OTUs reduced to 5'029 OTUs for consideration). Only large and small bacteria were considered for this analysis (not the “all bacteria” group). Relative abundances of OTUs per filter were calculated from the total number of reads, and OTUs with abundances <0.25% per filter were ignored on that filter.
Each OTU was categorized into one of five categories: “exclusively small”, “exclusively large”, “non-exclusive”, “unclassifiable”, or “eliminated” (Fig. 1c). For these definitions, we considered occurrence of OTUs on the following: (1) corresponding filter pairs, i.e. a pair of a 0.2 µm filter and 0.4 µm filters used to process the same sample replicate and (2) filter replicates, i.e. two filters of the same pore size (either 0.2 µm or 0.4 µm) that both received the same sample and thus should in theory contain the same number and composition of bacteria (technical replicates, Fig. 1b,c). Exclusivity was determined by whether an OTU was present in only one group (large or small bacteria) on both technical duplicate filters from at least one sample. If this criteria was met, it was tolerated that in other samples the OTU (1) was only present in that same group on only one of two technical replicates (e.g., low abundance preventing reproducibility), (2) was present in both groups of a filter pair (e.g., matrix effects trapping small bacteria on 0.4 µm filter, dimensions near border of filterability), or (3) was not present at all. OTUs that were present only in the small bacteria group of a filter pair in one sample and only in the large bacteria group of a filter pair in another sample were classified as non-exclusive. These non-exclusive OTUs could be further divided as to whether this (1) occurred in two separate samples with duplicate filters matching, or (2) occurred without dupilicate filters matching. Eliminated OTUs were all those not meeting any of the abundance criteria (all filters having low abundance (less than 0.25%)). Unclassifiable OTUs were either (1) in too low abundance (nearly all filters having less than 0.25%) or (2) too often co-occurring to be considered exclusive. Exclusivity analysis was performed in excel using exported OTU tables (for sample calculations and excel formulas used to assess exclusivity, see Supplementary Information—Exclusivity Analysis).
A phylogenetic tree constructed with greengenes v13.9 was plotted to reflect these classifications using the plot_tree function in ggplot2 [34]. The phylogenetic tree was constructed using OTU sequence alignment created with PyNAST, and with gaps removed. Phylogenetic trees were then constructed using FastTree using gamma 20 likelihood for boot-strap values. For ease of interpretation, many OTUs not meeting certain exclusivity benchmarks were removed from trees.
Community analysis
For non-metric multidimensional scaling (NMDS) community analyses, a different OTU filtration was used. Reads from duplicate filters were merged and all OTUs that did not reach three or more reads in three or more samples were removed (i.e., 43'616 OTUs reduced to 16'254 OTUs). The data set were then rarefied to the minimum number of reads in the merged data set (9’781 reads, representing 16'049 OTUs). In phyloseq, NMDS was performed for visualization of community similarities using Bray-Curtis dissimilarity. In R, from the vegan package [35], adonis analysis was performed to quantify relative importance of each factor for community composition.
Scanning electron microscopy (SEM)
Water samples from a small artificial experimental pond system (ecosystem not included in any other analyses) that had a naturally high proportion of LNA bacteria (90%), were filtered directly onto a 0.2 µm filter. Samples were fixed with 2.5% gluteraldehyde solution. Final preparation and imaging was done by the Center for Microscopy and Image Analysis (University of Zurich).
Results and Discussion
In this study, we demonstrate that so-called LNA-content bacteria are ubiquitous across several freshwater ecosystems (3.1) and that they are small in size and thus separatable by filtration with 0.4 µm filters (3.2). Using amplicon sequencing, we demonstrate that size-filtration accounts for some deviation in community composition (3.3), and that this could be attributed to some exclusive OTUs (3.4), which had a particular phylogenetic make-up (3.5).
LNA-content bacteria are ubiquitous
Forty-seven samples from 22 sampling sites in five different natural and engineered aquatic ecosystems (Fig. 1a) contained distinct clusters in FCM data, which were identified as LNA-content bacteria (Fig. 2, Supplementary Figure S1 for quantification, Supplementary Figure S2 for additional examples). We defined LNA-content bacteria as the bacterial cluster(s) with green fluorescence intensity below a defined instrument-specific threshold in the FCM density plots, and HNA-content bacteria as the cluster(s) above that threshold (Fig. 2). To ensure comparability, all samples were analyzed with the exact same protocol and the same FCM gate was used for all samples to select for HNA- and LNA-content bacteria [24]. The FCM detection of LNA-content bacteria was robust even with different staining protocols, variables, instrumentation, and operators (Supplementary Figure S3, Supplementary Figure S4). Additionally, similar LNA-content bacteria clusters have been observed when only considering intact cell counts [36], indicating that these ubiquitous cells are likely alive.
River water and tap water samples showed distinct LNA- and HNA-content bacteria clusters with similar relative abundance (around 50%, Fig. 2), although river water samples had approximately ten times higher absolute abundance (Supplementary Figure S1). Groundwater and wastewater effluent samples were both dominated by LNA-content bacteria (up to 90%, Fig. 2), although wastewater effluent samples had nearly 100 times higher absolute abundance than groundwater (Supplementary Figure S1). The data concurs with the previous studies describing LNA-content bacteria in diverse ecosystems, including river water [16], and seawater [14, 15]. Despite relative consistency within ecosystems, the underlying factors contributing to differences in LNA-content bacteria relative abundance in different ecosystems remain elusive. For example, it goes without argument that groundwater and wastewater effluent samples have vastly different environmental conditions, yet both ecosystems show similar dominance of LNA-content bacteria.
Interestingly, lake water samples did not show clear separation between LNA- and HNA-content bacterial clusters (Fig. 2e, Supplementary Figure S2 for additional examples). In general, the lake water samples showed more FCM clusters than the other ecosystem samples, and the cluster within the LNA gate had particularly high median fluorescence relative to the other samples (Fig. 2h). The lake water data challenges the perspective of a simplistic separation between only two major groups (i.e. LNA- and HNA-content bacteria). In fact, several previous studies have observed multiple FCM clusters in complex microbial communities (e.g., Supplementary Figure S4 with DAPI stained samples and more sophisticated optical instrumentation) [7, 37–39]. Still, most freshwater environments had a nearly bimodal distribution of fluorescence intensity.
Filtration separates small LNA- and large HNA-content bacteria
Filtration selects for bacteria with a sufficiently small diameter to pass through the filter pores. Filtration (0.4 µm) retained the majority of HNA-content bacteria from samples but allowed passage of LNA-content bacteria, substantially increasing the relative abundance of LNA-content bacteria in the filtrate (Supplementary Figure S5). Subsequent filtration of the 0.4 µm filtrate on 0.2 µm filters thus enabled the separate collection of communities dominated by HNA-content bacteria (0.4 µm filter) and LNA-content bacteria (0.2 µm filter) respectively (Fig. 1b). For simplicity we will from here on refer to these as large bacteria and small bacteria. This filtration approach was used previously for the enrichment of particularly small LNA-content bacteria [16], and similar sequential size-separating filtration techniques have been used to study differences between attached and free-living biomass [40, 41] and to enrich for small bactria of interest [42].
Supplementary Figure 3 shows SEM images of a pond sample rich in small LNA-content bacteria on a 0.2 µm filter (i.e., all cells colored in Fig. 3 can be considered “small”). These bacteria had an average diameter of 0.18 µm, an average length of 0.57 µm, and an average volume of 0.016 µm3 (n = 12); they would thus easily pass a 0.4 µm filter. This follows the small cell sizes for LNA-content bacteria (0.05 µm3) and 0.2 µm filterable bacteria (0.009 µm3) previously shown [2, 16] and fits within the theoretical limits of minimum cell sizes for bacteria [2]. In addition to this microscopy evidence, there was also a strong correlation between qPCR and FCM cell counts throughout ecosystems (including both archaea and bacterial primers, Spearman’s ρ = 0.72, p < 0.001, Supplementary Figure S6). Archaea contributed an average of 16% to the directly filtered (all) 16S rRNA gene copies, indicating that archaea (i.e., nanoarchaea [43]) may be of interest in future studies. These results indicate that LNA-content cells as measured by FCM are in fact bacteria and archaea, rather than an FCM artifact or non-bacterial particles such as viruses, free DNA, or auto-fluorescent particles. The qPCR data further suggest that small bacteria may have slightly fewer 16S rRNA gene copies per cell than large bacteria (Supplementary Figure S6). Low rRNA operon copy number has been linked with oligotrophic bacteria like S. alaskensis [44]. S. alaskensis was also identified as passing 0.2-µm filtration in ocean water [45], and is often studied as a UMB.
Fig. 3.
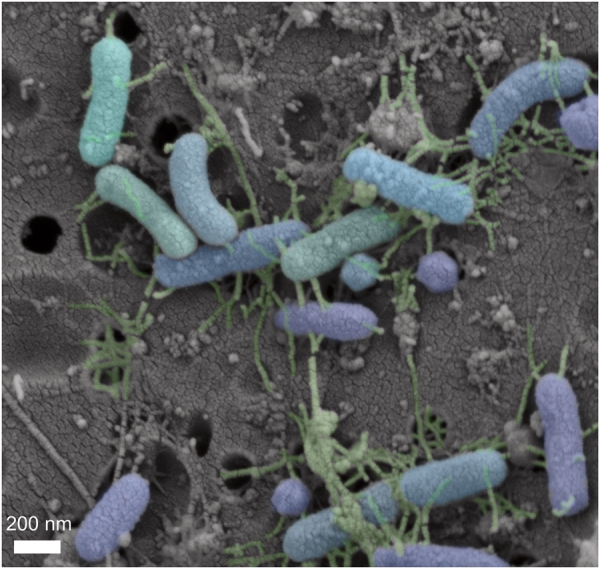
Scanning electron microscopy (SEM) image of bacteria from a stagnant pond sample rich in LNA content bacteria (>90%), filtered onto a 0.2 µm pore-size filter. Filter pores are visible as black holes, bacteria are highlighted in blue/purple shades and extracellular filaments are highlighted in green. Colors were added articifially, and the original image can be found in Supplementary Figure S13
Thus, our data links low fluorescence after nucleic acid staining (Fig. 2), low FCM scatter (Supplementary Figure S4), filterability (Supplementary Figure S5), small cell size (Fig. 3), and low DNA content (Supplementary Figure S6) to LNA-content cells. These links are supported by literature, where low fluorescence is linked to low DNA content [46], and low scatter is linked to small cell size [13, 14, 16, 47]. Filterability further confirmed the small cell size (at least diameter) [2, 16, 48], while small cell sizes have also been linked to small genome sizes [1] and low DNA content. Since many of the physical similarities were initially proposed to be linked to temporary physiological state (e.g., starvation [49]), further characterization was required to determine if these distinct physical characterisics linked to a phylogenetically distinct community.
Bacterial community differences are driven by environmental conditions and filtration
Bacterial communities captured on all filter sizes (Fig. 1b) were characterized with 16S amplicon sequencing to determine the differences in community composition attributable to size and other factors. Ecosystem (e.g., lake water vs. river water) was the most important factor for community composition (Fig. 4), accounting for 46% of all community variations (Adonis, p < 0.001). The five freshwater ecosystems were chosen to be diverse, so this outcome was expected. While not quantified in this study, multiple factors including nutrient conditions, hydraulics, and temperature vary dramatically different between these five ecosystems. Notably, two similar and linked ecosystems, tap water and its primary source in this study, groundwater, clustered close to each other.
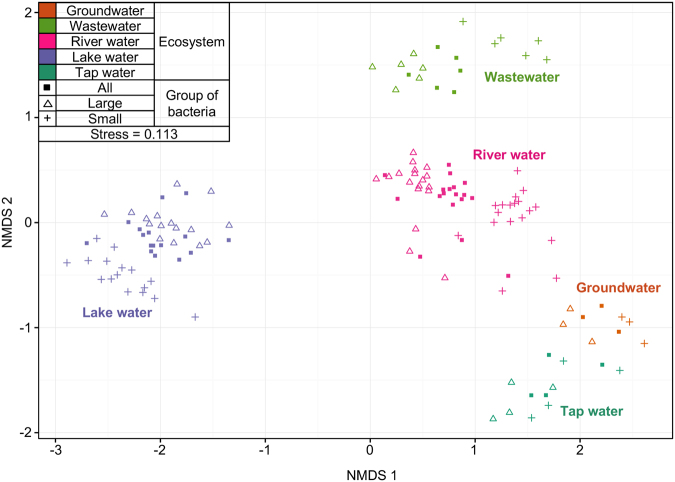
Fig. 4.
Non-metric multidimensional scaling (NMDS) of bacterial communities (characterized with 16 S amplicon sequencing) calculated with Bray–Curtis dissimilarity between samples from five different ecosystems (marked by color: Groundwater, Wastewater, River water, Lake water, and Tap water), with three different size groups by shape. ‘All bacteria’ is the total community, directly filtered onto a 0.2 µm filter. Large bacteria is the HNA-dominated community collected on a 0.4 µm filter, and Small bacteria is the LNA-dominated community in the 0.4 µm filtrate, collected on a 0.2 µm filter. In NMDS plots, points that are closer together represent bacterial communities more similar to each other than those further away. A low stress value indicates a robust diagram
It was clear that clustering of bacterial communities by size (i.e., large/small) only occurred within individual ecosystems. The community composition of the combined community (“all bacteria”) consistently clustered in between the small and large bacterial groups that contributed to it. However, the community composition of the small and large groups separated considerably from each other. When only considering these two groups (i.e., leaving “all” out of analysis), size and its interactions with ecosystem accounted for 27% of community variations (9.3 and 18%, respectively, Adonis, p < 0.001). Looking at each individual ecosystem (i.e., Fig. 5, focusing on river water), the separation of communities by size is even more apparent. Size (small vs. large) accounts for 24% of community variation within one ecosystem, and is a significant factor (p < 0.001) in all ecosystems with more than four samples (Adonis, Table 1; Table S5 for analysis including “all bacteria” samples). However, within any ecosystem, the sampling site (e.g., River A vs. River B) was a significant factor, often accounting for more varation than size. Sampling site is important amongst rivers for similar reasons to why ecosystem is important when looking at all samples—different environmental conditions select for the total community. Thus, large and small bacteria have distinct community compositions, but they are not completely distinct subsets of the total community.
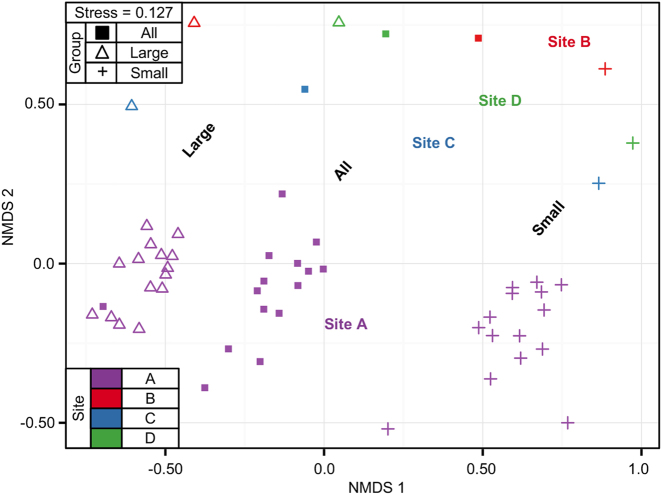
Fig. 5.
Non-metric multidimensional scaling (NMDS) of bacterial communities (characterized with 16 S amplicon sequencing) calculated with Bray–Curtis dissimilarity between samples from four different rivers (Site A-D), with three different groups by filter pore size. Color is by sampling site, and shape is by size group. ‘All bacteria’ is the total community, directly filtered onto a 0.2 µm filter. Large bacteria is the HNA-dominated community collected on a 0.4 µm filter, and Small bacteria is the LNA-dominated community in the 0.4 µm filtrate, collected on a 0.2 µm filter. In NMDS plots, points that are closer together represent bacterial communities more similar to each other than those further away. A low stress value indicates a robust diagram
Table 1.
Relative importance of various factors in bacterial communities within each ecosystem, calculated by Adonis
| Explanatory factor for Adonis analysis | Number in analysis (filters)—samples | |||
|---|---|---|---|---|
| Portion of variation accountable to factor (p value) | ||||
| Ecosystem | Sampling Site | Size | Site × size | |
| Lake water | 0.31 (0.001) | 0.24 (0.001) | 0.08 (0.628) | (32)—16 |
| River water | 0.14 (0.001) | 0.37 (0.001) | 0.08 (0.007) | (36)—18 |
| Groundwater | 0.38 (1.00) | 0.38 (1.00) | 0.24(1.00) | (8)—4 |
| Wastewater | 0.33 (0.001) | 0.33 (0.001) | 0.18 (0.065) | (12)—6 |
| Tap water | 0.51 (1.00) | 0.31 (1.00) | 0.18 (1.00) | (8)—4 |
For each explanatory variable (sampling site, size, and interactions between these two factors), the portion of variation accounted for by each variable and the p value for statistical significance of the variable are expressed. Sampling site refers to the specific location for each sample (i.e., River A vs. River B). For this analysis, only large and small bacteria groups were included for size (“All bacteria” was excluded from analysis). Large bacteria is the HNA-dominated community collected on a 0.4 µm filter, and Small bacteria is the LNA-dominated community in the 0.4 µm filtrate, collected on a 0.2 µm filter
It has been suggested that HNA and LNA taxonomy is dependent on location and time (freshwater springs, [50]), and that percentage of LNA-content bacteria as measured by FCM varies seasonally (rivers, [51]). As a test for temporal stability within a sampling site, samples taken over 4 months (June–September) from River Site A were further analyzed (Site A, Fig. 5). Both community composition by size (as measured with 16S amplicon sequencing) (Supplementary Figure S7) and the percentage of LNA-content bacteria (as measured with FCM) (Supplementary Figure S8) remained relatively stable in this site. This may indicate that samples were representative in terms of their size groups.
While there was a clear separation between the two size groups, the small and large bacteria were not completely independent, indicating species overlap and a common dependence on environmental conditions. In some previous studies, separation through cell sorting failed to see a clear separation of LNA- and HNA-content bacteria communities [50, 52], which may be due to OTU overlap between sizes. While our filtration approach to separate small and large bacteria has imperfections (i.e., filter cross contamination (Supplementary Figure S9)), we were able to characterize a wide array of ecosystems at a great depth, identifying approximately 108 cells for each sample. Alternative methods, like cell sorting, are limited and time-intensive for capturing rare organisms in low-biomass environments (e.g., tap water, ground water). The depth of our sequencing data allowed us to further investigate the causes for community differences and overlap as well as discrepancies with previous studies. In the next section, an analysis of individual OTUs was used to determine which bacteria were truly phylogenetically distinct and exclusive by size.
Individual OTUs are exclusive to each size across five diverse ecosystems
The forthcoming analysis is based on the classification of all OTUs into five categories namely (1) exclusively small, (2) exclusively large, (3) non-exclusive, (4) eliminated, and (5) unclassifiable (Fig. 1c). Of the 5'029 OTUs that passed the first abundance cutoff (>20 reads in >2 samples), 434 OTUs were classified as exclusive to the small bacteria and 441 OTUs were classified as exclusive to the large bacteria (Fig. 6). These OTUs occurred exclusively in one size group (small or large), and appeared on both technical duplicate filters from at least one sample. The relative abundance of these two categories reflected expected trends, with exclusively small OTUs more abundant in the small bacteria community and exclusively large OTUs more abundant in the large bacteria community (Fig. 7), and these size-exclusive OTUs contributed to a substantial portion of the community on each filter.
Fig. 6.
Phylogenetic tree colored by OTU occurrences in each size constructed with 1'224 of >40'000 OTUs found in water samples. Circle area represents the number of samples (both technical duplicates) in which an OTU was consistently exclusively appearing with either large bacteria (0.4 µm filter, red) or small bacteria (0.2 µm filtered after 0.4 µm filter, blue), with this number ranging from 1 to 16 samples. OTUs, which were at times exclusive to both sizes in a filter pair (non-exclusive) are marked in green. Extraneous OTU branches that never met these criteria (unclassifiable, eliminated) were removed from the figure. Several phyla and a class of interest are labeled. For more detailed phylogenetic identification, see Supplementary Figure S10a
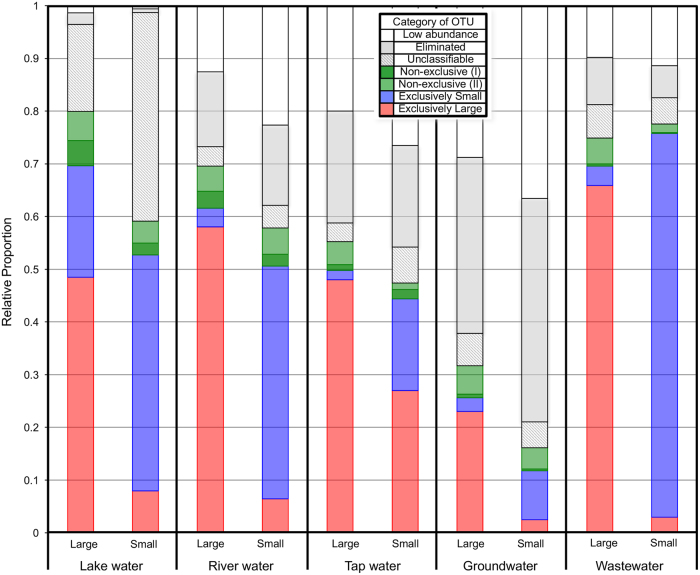
Fig. 7.
Relative abundances of OTUs classified with the described exclusivity criteria (exclusively small [blue], exclusively large [red], non-exclusive [green], and unclassifiable. eliminated and rare OTUs[white/gray]) in each size group of each ecosystem. For each ecosystem (Groundwater, Wastewater, River water, Lake water, and Tap water), the total relative abundance for all filters in a particular size (small, large bacteria) is shown. Large bacteria is the HNA-dominated community collected on a 0.4 µm filter, and Small bacteria is the LNA-dominated community in the 0.4 µm filtrate, collected on a 0.2 µm filter. Non-exclusive OTUs are further divided into whether they occur in duplicate (I) or not (II). OTUs not meeting initial cutoffs are marked as “rare”, and not meeting secondary cutoffs are marked as “eliminated”. Overlap (e.g., exclusively large OTUs in the small bacteria community) is due to leniency that OTUs may occur on both filters (0.2 and 0.4 µm filters) of a filter-pair, so long as it does not appear exlcusviely on the opposite fitler anywhere (e.g., non-exclusive OTU)
The chosen exclusivity levels were lenient enough to allow presence on both sized filters in some samples, and thus sometimes exclusively small OTUs appeared with the large bacteria community and vice versa. Applying a higher level of exclusivity, wherein co-occurrence in both size filters was never allowed (i.e., not allowing for the cross-contamination described above) or including rare abundances (<0.25%) resulted in far fewer OTUs for analysis. However, we accepted the contamination risk and potential bias, given the likelihood of cross-contamination on filters (Supplementary Figure S9) and the specificity desired when comparing across ecosystems.
Another 38 OTUs, which we called non-exclusive, were classified as small in some filter pairs and large in other filter pairs. For 12 of these 38 OTUs, this occurred in duplicate for both small and large fractions (i.e., dark green Fig. 7). This could be due to differences between ecosystem or sampling site conditions (i.e., the same OTU has different characteristics dependent on environmental conditions). It could also indicate small species that are exclusively intracellular symbonts in some samples (appearing large), while exclusively free-living in other samples (appearing as small). These non-exclusive OTUs could be quite abundant, especially in lake and river samples (Fig. 7).
The remainder of OTUs were either eliminated due to low relative abundance (eliminated—3'805 OTUs), or could not be classified in any of the above categories (unclassifiable—264 OTUs). Most of these unclassifiable OTUs had consistent, but not complete low abundance (i.e., 262 OTUs had <0.25% on 87% or more of filters considered). Another 2 OTUs were often in high abundance, but were always co-occurring (i.e., appeared in >0.25% in both large and small filters consistently). These 2 OTUs were taxonomically identified as Pelagibacterales and ACK-M1 of Actinomycetes. These remaining groups, together with the OTUs failing to meet the first abundance cutoff (38,587 OTUs) represented a large portion of the community (Fig. 7), and may represent a bias in our analysis methods. For example, the large percentage of non analyzable OTUs in groundwater may owe to the high diversity (making relative abundances for each OTU lower) and low number of samples (only 3 distinct samples).
This data aligns with previous arguments that LNA-content bacteria are viable unique microorganisms [15, 16] and refutes the notion that small LNA-content bacteria are simply dead/inactive cells [14, 53]. It seems unlikely for an entire species (OTU) to be consistently dead/inactive across many samples and ecosystems with vastly different nutrient conditions. However, since some OTUs were not clearly or consistently classified exclusively as small or large OTUs, this may follow the theory of Bouvier et al. [15] that while some bacteria are “intrinsic to each fraction” (small or large OTUs), others can “exchange between fractions” (non-exclusive OTUs).
Small and large OTUs cluster on phylum level
When looking at the phylogenetic classification of exclusively small and large OTUs, a remarkably clear pattern emerged (Fig. 6, Supplementary Figure S10, Supplementary Figure S11, Supplementary Figure S12). The OTUs classified as exclusive to each size were frequently grouped at a high taxonomic level (i.e., phylum). This provides further evidence that a bacterium’s size (filterability), and thus its classification as a LNA- or HNA-content bacterium, is part of a fundamental and evolutionarily well-preserved trait, rather than linked to its temporary physiological state. Moreover, this separation of the two bacteria classes occurred even when considering five diverse ecosystems. Correlation between some phenotypic traits and phylogenetic relationships has been suggested in bacteria previously [54], and thus this strong relationship between phylogeny and log-scale differences in size is not entirely suprising.
It should be noted that the high level clustering between sizes, while remarkable, was not entirely consistent (e.g., phyla distributions not exclsuvie between large and small fractions (Supplementary Figure S11)). Small OTUs could be found within phyla dominated by large OTUs and vice versa (e.g., Bacteroidetes, Deltaproteobacteria, Supplementary Figure S11). Especially when considering the phyla-level relative abundance of OTUs falling into different size-exclusive categories (Supplementary Figure S12), it is clear that (1) much of several phyla could not be easily divided into the two size-based groups (i.e., low abundance, non-exclusive, eliminated, and unclassifiable OTUs), (2) that some phyla have considerable variability in sizes (e.g., Proteobacteria, Verrumicrobia), and (3) that phyla were not exclusively found in either one fraction or another. Previous studies linking cell size to phylogeny have also noted variability in size within a phylum [55]. As many OTUs were discarded from our analysis (i.e., low abundance), even more size varation is also possible within each phylum. Nonetheless, more information about the phyla that were dominated by one size or another provides some deeper insight into why this size-based phylogenetic clustering occurs.
Phyla associated with small OTUs
Many of the 434 OTU associated with small bacteria were attributed to the so-called “candidate phyla radiation” (CPR), which do not yet have cultivated representatives (i.e., Parcubacteria (OD1), Gracilibacteria (GN02), Saccharibacteria (TM7), Dependentiae (TM6), and Omnitrophica (OP3)). Many CPR bacteria have been associated with small genomes and have similarities with symbiotic bacteria [18, 56]. Reduced genomes would be consistent with observations of UMB [1] and with low fluorescence after staining of nucleic acids. Furthermore, several taxa were associated with symbiotic or predatory relationships with other microorganisms. Symbionts are associated with genome reduction which may reduce their ability to live independently [57]. Although shown to be growing with an innovative metagenomic approach [58], growth rates among CPR are slow, which may further contribute to difficulties with isolating and culturing these small bacteria. Like these groups, LNA-content bacteria are also difficult to culture [16]. Altogether, this may suggest that many observed exclusively small OTUs (i.e., bacteria passing a 0.4 µm filter, LNA-content bacteria) lack sufficient genomes to produce all necessary cellular building blocks, and rather depend on metabolites from other cells. Rapid FCM observation of LNA-content bacteria may offer an easy method to quantify these otherwise difficult to study bacteria.
Many exclusively small OTUs fell into the proposed Patescibacteria superpylum. Parcubacteria (OD1) has previously been associated with a small size (ultra-small bacteria passing 0.2 µm filter) [2, 59], a reduced genome (<1 Mb) with reduced functionality compared to cells with large genomes (e.g., lacking ATP synthase [60]), and ectosymbiosis or parasitism towards other organisms [20, 61]. Gracilibacteria (GN02) have also been reported to possess small genomes [62]. Saccharibacteria (TM7) recently achieved one cultivated representative bacteria from a human host. It had a small coccus shape, small genome (with reduced capacity), and was an epibiont of Actinomyces odontolyticus with parasitic tendencies [63]. Metagenomic reconstructions of Saccharibacteria genomes from activated sludge and other sources confirm small genomes (<1 Mb), and indicate a fermented microaerophilic lifestyle and small cell size (<0.7 µm) [56, 64]. Dependentiae (TM6), has been suggested as an LNA-content bacterial taxa previously [52]. This phylum is thought to contain widespread parasitism and endosymbiosis, as it has been associated with small genomes (0.5–1.5 Mb), a lack of complete essential synthetic pathways, and endosymbiosis with amoebae [65–67]. Many of these CPR bacteria have high and variable abundance reported across freshwater ecosystems (e.g., tap water dominated by Parcubacteria, with more diversity in CPR in groundwater) [43, 68, 69]. The consistency in size and features within this superphylum may indicate that cellular size, on a log-scale, is a complex and deeply conserved phylogenetic trait.
Deltaproteobacteria deviated from the rest of the Proteobacteria phylum, with many OTUs identified as small. Some belonged to the Spirobacillales order, so named because they are associated with a spiral shape [70]. This could indicate a bias in our results, as only the smallest dimension determines filterability, and these cells may otherwise be considered large. However, many others belonged to Bdellovibrionales, including the predatory genus Bdellovibrio, which is known to be small (e.g., 0.2 × 0.5 µm) [71]. Other orders, including Myxococcalles, did not follow the trend of the class and were identified as HNA. Interestingly, it has been speculated that Deltaproteobacteria have a close evolutionary relationship with Omnitrophica (OP3), a candidate phylum associated with small bacteria in this study, due to similar metabolic capabilities and genes [72].
Although only 20 OTUs in the Actinobacteria phylum could be identified as small, this represented a large proportion of the community (Supplementary Figure S12), especially in lakes. Actinobacteria and Microbacteriaceae were previously associated with LNA-content bacteria [4, 73]. While the AC1 lineage of Actinobacteria was not specifically found in high abundance, this association may be interesting, as the AC1 lineage of Actinobacteria has many similarities to the CPR [42]. For the AC1 lineage, dependencies on metabolites from other organisms (auxotrophies) are proposed to develop through genome-streamlining [42], and thus the small cell-size may be linked to a more recently evolved and less conserved trait than for the CPR.
Other taxa that were identified as predominantly small include SR1, Mollicutes, Endomicrobia, and Fibrobacteria. A previously suggested LNA-content bacterium, Polynucleobacter [16], was confirmed as small in this study, even though it was classified as an HNA-content bacterium in a cell-sorting study [50]. Other suggested LNA taxa, including AC1, Alphaproteobacteria—LD12 [12], SAR11 [74, 75], SAR86 [76], Katanobacteria (WWE3), and Microgenomates (OP11) [2] were not abundant enough for analysis in this study. Some of these taxa are not expected in this freshwater data (e.g., SAR 11 is predominantly marine), and others may have had specific primer bias against their identification (e.g., LD12 only had low coverage with the selected primers). It can not be excluded that that other particular phylotypes were biased against with the primers.
Phyla associated with large OTUs
Phyla associated with large size had diverse descriptions, perhaps consistent with the much larger size range associated with HNA-content bacteria. Predominantly large phyla included Bacteroidetes, Proteobacteria (with the exception of Deltaproteobacteria), Planctomycetes, Firmicutes, Chlorobi, Verrucomicrobia, and Fusobacteria.
These taxa often overlapped with taxa suggested to be HNA-content bacteria in literature (e.g., Bacteroidetes [17, 52, 73, 77] and Gammaproteobacteria [15]. Several Proteobacteria previously identified as LNA were identified as large here, including Methylobacteriaceae, Pseudomonas, and Alteromonodaceae [52].
Taxa associated with non-exclusive OTUs
Only 38 OTUs were identified as non-exclusive, meaning they were sometimes categorized as large and sometimes small. These belonged to Bacteroidetes (8), Actinobacteria (6), Nitrospirae (2), TM6 (3), Verrucomicrobia (3), Chlamydiae (1), and Proteobacteria (Betaproteobacteria (11), Gammaproteobacteria (3), and Alphaproteobacteria (1)). Interestingly, Bacteroidetes has previously also been identified as recovering from a starved form that can pass 0.2 µm filter [78], which may indicate its ability to change sizes across a wide range. Our results suggest that environment-dependent variations in cell size are not common, but appear to be present in certain bacteria.
Implications
In this study we showed that FCM clusters identified as LNA-content bacteria are found across diverse natural and engineered aquatic ecosystems at varying relative and absolute abundances. Moreover, we link the concepts of LNA-content bacteria [15], USB [2], small genome size, and UMB [45] to 0.4 µm filterability, small cell size, and low green fluoresence. Individual OTUs could be classified as exclusively small or large based on filterability, even across five diverse ecosystems. These data strongly support the previous suggestions that LNA-content bacteria are viable microorganisms relevant to our understanding of microbial communities in natural and engineered ecosystems. The fact that individual OTUs exclusive to large and small sizes classified distinctly on phylum level, suggests that bacteria’s size and classification as LNA- or HNA-content bacteria is part of a fundamental and evolutionarily well-preserved trait. Additionally, since many OTUs exclusively filterable through the 0.4 µm filter were members of clades with non-culturable or parasitic bacteria, this may point to a limited capacity for independent life for some of these species. Finally, observing LNA with FCM, for example by using FCM fingerprinting to track spatio-temporal dynamics in enigneered [24, 79] or natural freshwater [80] systems, offers an easy way to quantify these abundant small bacteria that are otherwise rather difficult to culture and study.
Electronic supplementary material
Acknowledgements
We acknowledge financial support from MERMAID, a Marie Sklodowska-Curie Initial Training Network under grant no. 607492, from Canton Basel-Landschaft, Switzerland in the framework of the project “Regionale Wasserversorgung Basel-Landschaft 21”, and from internal Eawag Discretionary Funding. Comparative FCM data were generated by Susann Müller and Thomas Hübschmann at the Helmholtz Centre for Environmental Research—UFZ, Leipzig, Germany. Sequencing data were generated in collaboration with the Genetic Diversity Centre (GDC), ETH Zurich, with support from Aria Minder and Silvia Kobel. SEM data were generated at the Center for Microscopy and Image Analysis, University of Zurich. We acknowledge Judith Riedo and David Cayon for literature and laboratory support and Teresa Colangelo for assistance with SEM imaging.
Author Contributions:
CP: data analysis; manuscript writing; MB: Experimental design, field and laboratory work; data analysis; manuscript writing; TL: data analysis; JC: bioinformatics support; KB: Field and laboratory work; data analysis; HB: experimental design, field and laboratory work; data analysis; manuscript writing; FH: experimental design; data analysis; manuscript writing.
Compliance with Ethical Standards
Conflict of interest:
The authors declare that they have no conflict of interest.
Footnotes
Joint last authors: Helmut Bürgmann, Frederik Hammes.
Contributor Information
Helmut Bürgmann, Email: helmut.buergmann@eawag.ch.
Frederik Hammes, Email: frederik.hammes@eawag.ch.
Electronic supplementary material
The online version of this article (10.1038/s41396-018-0070-8) contains supplementary material, which is available to authorized users.
References
- 1.Duda VI, Suzina NE, Polivtseva VN, Boronin AM. Ultramicrobacteria: Formation of the concept and contribution of ultramicrobacteria to biology. MICROBIOLOGY. 2012;81:379–90. doi: 10.1134/S0026261712040054. [DOI] [PubMed] [Google Scholar]
- 2.Luef B, Frischkorn KR, Wrighton KC, Holman HYN, Birarda G, Thomas BC, et al. Diverse uncultivated ultra-small bacterial cells in groundwater. Nat Commun. 2015;6:6372. doi: 10.1038/ncomms7372. [DOI] [PubMed] [Google Scholar]
- 3.Schulz HN, Brinkhoff T, Ferdelman TG, Mariné MH, Teske A, Jørgensen BB. Dense populations of a giant sulfur bacterium in namibian shelf sediments. Science (80-). 1999;284.. [DOI] [PubMed]
- 4.Hahn MW, Lunsdorf H, Wu QL, Schauer M, Hofle MG, Boenigk J, et al. Isolation of novel ultramicrobacteria classified as Actinobacteria from five freshwater habitats in Europe and Asia. Appl Environ Microbiol. 2003;69:1442–51. doi: 10.1128/AEM.69.3.1442-1451.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.MACDonell MT, Hood MA. Isolation and characterization of ultramicrobacteria from a Gulf Coast Estuary. Appl Envir Microbiol. 1982;43:566–71. doi: 10.1128/aem.43.3.566-571.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miteva VI, Brenchley JE. Detection and isolation of ultrasmall microorganisms from a 120,000-year-old Greenland glacier ice core. Appl Environ Microbiol. 2005;71:7806–18. doi: 10.1128/AEM.71.12.7806-7818.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Günther S, Faust K, Schumann J, Harms H, Raes J, Müller S. Species-sorting and mass-transfer paradigms control managed natural metacommunities. Environ Microbiol. 2016;18:4862–77. doi: 10.1111/1462-2920.13402. [DOI] [PubMed] [Google Scholar]
- 8.Harry ISK, Ameh E, Coulon F, Nocker A. Impact of treated sewage effluent on the microbiology of a small brook using flow cytometry as a diagnostic tool. Water Air Soil Pollut. 2016;227:57. doi: 10.1007/s11270-015-2723-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van Nevel S, Koetzsch S, Proctor CR, Besmer MD, Prest EI, Vrouwenvelder JS, et al. Flow cytometric bacterial cell counts challenge conventional heterotrophic plate counts for routine microbiological drinking water monitoring. Water Res. 2017. 10.1016/j.watres.2017.01.065. [DOI] [PubMed]
- 10.Props R, Monsieurs P, Mysara M, Clement L, Boon N. Measuring the biodiversity of microbial communities by flow cytometry Hodgson D (ed) Methods Ecol Evol. 2016;7:1376–85. doi: 10.1111/2041-210X.12607. [DOI] [Google Scholar]
- 11.Marie D, Rigaut-Jalabert F, Vaulot D. An improved protocol for flow cytometry analysis of phytoplankton cultures and natural samples. Cytom Part A. 2014;85:962–8. doi: 10.1002/cyto.a.22517. [DOI] [PubMed] [Google Scholar]
- 12.Neuenschwander SM, Salcher MM, Pernthaler J. Fluorescence in situ hybridization and sequential catalyzed reporter deposition (2C-FISH) for the flow cytometric sorting of freshwater ultramicrobacteria. Front Microbiol. 2015;6. 10.3389/fmicb.2015.00247. [DOI] [PMC free article] [PubMed]
- 13.Gasol JM, Zweifel UL, Peters F, Fuhrman JA, Hagström A. Significance of size and nucleic acid content heterogeneity as measured by flow cytometry in natural planktonic bacteria. Appl Environ Microbiol. 1999;65:4475–83. doi: 10.1128/aem.65.10.4475-4483.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lebaron P, Servais P, Baudoux AC, Bourrain M, Courties C, Parthuisot N. Variations of bacterial-specific activity with cell size and nucleic acid content assessed by flow cytometry. Aquat Microb Ecol. 2002;28:131–40. doi: 10.3354/ame028131. [DOI] [Google Scholar]
- 15.Bouvier T, Del Giorgio PA, Gasol JM. A comparative study of the cytometric characteristics of High and Low nucleic-acid bacterioplankton cells from different aquatic ecosystems. Environ Microbiol. 2007;9:2050–66. doi: 10.1111/j.1462-2920.2007.01321.x. [DOI] [PubMed] [Google Scholar]
- 16.Wang Y, Hammes F, Boon N, Chami M, Egli T. Isolation and characterization of low nucleic acid (LNA)-content bacteria. ISME J. 2009;3:889–902. doi: 10.1038/ismej.2009.46. [DOI] [PubMed] [Google Scholar]
- 17.Vila-Costa M, Gasol JM, Sharma S, Moran MA. Community analysis of high- and low-nucleic acid-containing bacteria in NW Mediterranean coastal waters using 16S rDNA pyrosequencing. Environ Microbiol. 2012;14:1390–402. doi: 10.1111/j.1462-2920.2012.02720.x. [DOI] [PubMed] [Google Scholar]
- 18.Brown CT, Hug LA, Thomas BC, Sharon I, Castelle CJ, Singh A, et al. Unusual biology across a group comprising more than 15% of domain Bacteria. Nature. 2015;523:208–11. doi: 10.1038/nature14486. [DOI] [PubMed] [Google Scholar]
- 19.Hood MA, MacDonell MT. Distribution of ultramicrobacteria in a gulf coast estuary and induction of ultramicrobacteria. Microb Ecol. 1987;14:113–27. doi: 10.1007/BF02013017. [DOI] [PubMed] [Google Scholar]
- 20.Nelson WC, Stegen JC. The reduced genomes of Parcubacteria (OD1) contain signatures of a symbiotic lifestyle. Front Microbiol. 2015;6. 10.3389/fmicb.2015.00713. [DOI] [PMC free article] [PubMed]
- 21.Hahn MW, Stadler P, Wu QL, Pockl M. The filtration-acclimatization method for isolation of an important fraction of the not readily cultivable bacteria. J Microbiol Methods. 2004;57:379–90. doi: 10.1016/j.mimet.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 22.Wang Y, Hammes F, Düggelin M, Egli T. Influence of size, shape, and flexibility on bacterial passage through micropore membrane filters. Environ Sci Technol. 2008;42:6749–54. doi: 10.1021/es800720n. [DOI] [PubMed] [Google Scholar]
- 23.SLMB. Determining the total cell count and ratios of high and low nucleic acid content cells in freshwater using flow cytometry. Analysis Method 333.1, Swiss Food Book. Federal Office of Public Health, Swiss Confederation (2012)
- 24.Prest EI, Hammes F, Kötzsch S, van Loosdrecht MCM, Vrouwenvelder JS. Monitoring microbiological changes in drinking water systems using a fast and reproducible flow cytometric method. Water Res. 2013;47:7131–42. doi: 10.1016/j.watres.2013.07.051. [DOI] [PubMed] [Google Scholar]
- 25.Llirós M, Casamayor EO, Borrego C, EF D, ES L, D N, et al. High archaeal richness in the water column of a freshwater sulfurous karstic lake along an interannual study. FEMS Microbiol Ecol. 2008;66:331–42. doi: 10.1111/j.1574-6941.2008.00583.x. [DOI] [PubMed] [Google Scholar]
- 26.Proctor CR, Gächter M, Kötzsch S, Rölli F, Sigrist R, Walser JC, et al. Biofilms in shower hoses—choice of pipe material influences bacterial growth and communities. Environ Sci Water Res Technol. 2016;2:670–82. doi: 10.1039/C6EW00016A. [DOI] [Google Scholar]
- 27.Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M, et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013;41:e1. doi: 10.1093/nar/gks808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Magoc T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–63. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17:10. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 30.Schmieder R, Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics. 2011;27:863–4. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–1. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 32.DeSantis TZ, Dubosarskiy I, Murray SR, Andersen GL. Comprehensive aligned sequence construction for automated design of effective probes (CASCADE-P) using 16S rDNA. Bioinformatics. 2003;19:1461–8. doi: 10.1093/bioinformatics/btg200. [DOI] [PubMed] [Google Scholar]
- 33.McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wickham H. Elegant graphics for danaanalysis. In: ggplot2. New York, NY: Springer New York; 2009. p. 1–7.
- 35.Oksanen J, Blanchet FG, Kindt R, PP Legendre, Minchin PR, O’Hara RB, et al. (2013). vegan: community ecology package. R Package. http//cranr-projectorg/web/packages/vega/indexhtml. Accessed on 2015.03.24.
- 36.Ramseier MK, von Gunten U, Freihofer P, Hammes F. Kinetics of membrane damage to high (HNA) and low (LNA) nucleic acid bacterial clusters in drinking water by ozone, chlorine, chlorine dioxide, monochloramine, ferrate(VI), and permanganate. Water Res. 2011;45:1490–500. doi: 10.1016/j.watres.2010.11.016. [DOI] [PubMed] [Google Scholar]
- 37.Günther S, Koch C, Hübschmann T, Röske I, Müller RA, Bley T, et al. Correlation of community dynamics and process parameters as a tool for the prediction of the stability of wastewater treatment. Environ Sci Technol. 2012;46:84–92. doi: 10.1021/es2010682. [DOI] [PubMed] [Google Scholar]
- 38.Koch C, Günther S, Desta AF, Hübschmann T, Müller S. Cytometric fingerprinting for analyzing microbial intracommunity structure variation and identifying subcommunity function. Nat Protoc. 2013;8:190–202. doi: 10.1038/nprot.2012.149. [DOI] [PubMed] [Google Scholar]
- 39.Nishimura Y, Kim C, Nagata T. Vertical and seasonal variations of bacterioplankton subgroups with different nucleic Acid contents: possible regulation by phosphorus. Appl Environ Microbiol. 2005;71:5828–36. doi: 10.1128/AEM.71.10.5828-5836.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ganesh S, Parris DJ, DeLong EF, Stewart FJ. Metagenomic analysis of size-fractionated picoplankton in a marine oxygen minimum zone. ISME J. 2014;8:187–211. doi: 10.1038/ismej.2013.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mohit V, Archambault P, Toupoint N, Lovejoy C. Phylogenetic differences in attached and free-living bacterial communities in a temperate coastal lagoon during summer, revealed via high-throughput 16S rRNA gene sequencing. Appl Environ Microbiol. 2014;80:2071–83. doi: 10.1128/AEM.02916-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Neuenschwander SM, Ghai R, Pernthaler J, Salcher MM. Microdiversification in genome-streamlined ubiquitous freshwater Actinobacteria. ISME J. 2017. 10.1038/ismej.2017.156. [DOI] [PMC free article] [PubMed]
- 43.Ludington WB, Seher TD, Applegate O, Li X, Kliegman JI, Langelier C, et al. Assessing biosynthetic potential of agricultural groundwater through metagenomic sequencing: A diverse anammox community dominates nitrate-rich groundwater Yang S (ed) PLoS ONE. 2017;12:e0174930. doi: 10.1371/journal.pone.0174930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roller BRK, Stoddard SF, Schmidt TM, McNeill K, Ackermann M. Exploiting rRNA operon copy number to investigate bacterial reproductive strategies. Nat Microbiol. 2016;1:16160. doi: 10.1038/nmicrobiol.2016.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nakai R, Abe T, Takeyama H, Naganuma T. Metagenomic Analysis of 0.2-mu m-Passable Microorganisms in Deep-Sea Hydrothermal Fluid. Mar Biotechnol. 2011;13:900–8. doi: 10.1007/s10126-010-9351-6. [DOI] [PubMed] [Google Scholar]
- 46.Veldhuis MJW, Cucci TL, Sieracki ME. Cellular DNA content of marine phytoplankton using two new fluorochromes: taxonomic and ecologocial implications. J Phycol. 1997;33:527–41. doi: 10.1111/j.0022-3646.1997.00527.x. [DOI] [Google Scholar]
- 47.Shapiro HM. Practical Flow Cytometry. Hoboken, NJ: John Wiley & Sons, Inc.; 2003. [Google Scholar]
- 48.Gasol JM, Moran XA. Effects of filtration on bacterial activity and picoplankton community structure as assessed by flow cytometry. Aquat Microb Ecol. 1999;16:251–64. doi: 10.3354/ame016251. [DOI] [Google Scholar]
- 49.Morita RY. Bacteria in oligotrophic environments: starvation-survival lifestyle. New York: Chapman & Hall; 1997.
- 50.Martinez-Garcia M, Swan BK, Poulton NJ, Gomez ML, Masland D, Sieracki ME, et al. High-throughput single-cell sequencing identifies photoheterotrophs and chemoautotrophs in freshwater bacterioplankton. ISME J. 2012;6:113–23. doi: 10.1038/ismej.2011.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu J, Hao Z, Ma L, Ji Y, Bartlam M, Wang Y. Spatio-Temporal Variations of High and Low Nucleic Acid Content Bacteria in an Exorheic River. PLoS One. 2016;11. 10.1371/journal.pone.0153678. [DOI] [PMC free article] [PubMed]
- 52.Longnecker K, Sherr BF, Sherr EB. Activity and phylogenetic diversity of bacterial cells with high and low nucleic acid content and electron transport system activity in an upwelling ecosystem. Appl Environ Microbiol. 2005;71:7737–49. doi: 10.1128/AEM.71.12.7737-7749.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Servais P, Casamayor EO, Courties C, Catala P, Parthuisot N, Lebaron P. Activity and diversity of bacterial cells with high and low nucleic acid content. Aquat Microb Ecol. 2003;33:41–51. doi: 10.3354/ame033041. [DOI] [Google Scholar]
- 54.Martiny JBH, Jones SE, Lennon JT, Martiny AC. Microbiomes in light of traits: A phylogenetic perspective. Science. 2015;350:aac9323. doi: 10.1126/science.aac9323. [DOI] [PubMed] [Google Scholar]
- 55.Stepanauskas R, Fergusson EA, Brown J, Poulton NJ, Tupper B, Labonté JM, et al. Improved genome recovery and integrated cell-size analyses of individual uncultured microbial cells and viral particles. Nat Commun. 2017;8:84. doi: 10.1038/s41467-017-00128-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kantor RS, Wrighton KC, Handley KM, Sharon I, Hug LA, Castelle CJ, et al. Small genomes and sparse metabolisms of sediment-associated bacteria from four candidate phyla. MBio. 2013;4:e00708–13. doi: 10.1128/mBio.00708-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McCutcheon JP, Moran NA. Extreme genome reduction in symbiotic bacteria. Nat Rev Microbiol. 2011;10:13. doi: 10.1038/nrmicro2670. [DOI] [PubMed] [Google Scholar]
- 58.Brown CT, Olm MR, Thomas BC, Banfield JF. Measurement of bacterial replication rates in microbial communities. Nat Biotechnol. 2016;34:1256–63. doi: 10.1038/nbt.3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Miyoshi T, Iwatsuki T, Naganuma T. Phylogenetic characterization of 16S rRNA gene clones from deep-groundwater microorganisms that pass through 0.2-micrometer-pore-size filters. Appl Environ Microbiol. 2005;71:1084–8. doi: 10.1128/AEM.71.2.1084-1088.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Suzuki S, Ishii S, Hoshino T, Rietze A, Tenney A, Morrill PL, et al. Unusual metabolic diversity of hyperalkaliphilic microbial communities associated with subterranean serpentinization at The Cedars. ISME J. 2017;11:2584–98. doi: 10.1038/ismej.2017.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wrighton KC, Thomas BC, Sharon I, Miller CS, Castelle CJ, VerBerkmoes NC, et al. Fermentation, Hydrogen, and Sulfur Metabolism in Multiple Uncultivated Bacterial Phyla. Science (80-) 2012;337:1661–5. doi: 10.1126/science.1224041. [DOI] [PubMed] [Google Scholar]
- 62.Rinke C, Schwientek P, Sczyrba A, Ivanova NN, Anderson IJ, Cheng JF, et al. Insights into the phylogeny and coding potential of microbial dark matter. Nature. 2013;499:431–7. doi: 10.1038/nature12352. [DOI] [PubMed] [Google Scholar]
- 63.He X, McLean JS, Edlund A, Yooseph S, Hall AP, Liu SY, et al. Cultivation of a human-associated TM7 phylotype reveals a reduced genome and epibiotic parasitic lifestyle. Proc Natl Acad Sci USA. 2015;112:244–9. doi: 10.1073/pnas.1419038112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Albertsen M, Hugenholtz P, Skarshewski A, Nielsen KL, Tyson GW, Nielsen PH. Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes. Nat Biotechnol. 2013;31:533–8. doi: 10.1038/nbt.2579. [DOI] [PubMed] [Google Scholar]
- 65.McLean JS, Lombardo MJ, Badger JH, Edlund A, Novotny M, Yee-Greenbaum J, et al. Candidate phylum TM6 genome recovered from a hospital sink biofilm provides genomic insights into this uncultivated phylum. Proc Natl Acad Sci USA. 2013;110:E2390–9. doi: 10.1073/pnas.1219809110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yeoh YK, Sekiguchi Y, Parks DH, Hugenholtz P. Comparative genomics of candidate phylum tm6 suggests that parasitism is widespread and ancestral in this lineage. Mol Biol Evol. 2016;33:915–27. doi: 10.1093/molbev/msv281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Delafont V, Samba-Louaka A, Bouchon D, Moulin L, Héchard Y. Shedding light on microbial dark matter: a TM6 bacterium as natural endosymbiont of a free-living amoeba. Environ Microbiol Rep. 2015;7:970–8. doi: 10.1111/1758-2229.12343. [DOI] [PubMed] [Google Scholar]
- 68.Borrel G, Lehours AC, Bardot C, Bailly X, Fonty G. Members of candidate divisions OP11, OD1 and SR1 are widespread along the water column of the meromictic Lake Pavin (France) Arch Microbiol. 2010;192:559–67. doi: 10.1007/s00203-010-0578-4. [DOI] [PubMed] [Google Scholar]
- 69.Bruno A, Sandionigi A, Rizzi E, Bernasconi M, Vicario S, Galimberti A, et al. Exploring the under-investigated "microbial dark matter" of drinking water treatment plants. Sci Rep. 2017;7:44350. doi: 10.1038/srep44350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rodrigues JLM, Duffy MA, Tessier AJ, Ebert D, Mouton L, Schmidt TM. Phylogenetic characterization and prevalence of ‘Spirobacillus cienkowskii,’ a red-pigmented, spiral-shaped bacterial pathogen of freshwater Daphnia species. Appl Environ Microbiol. 2008;74:1575–82. doi: 10.1128/AEM.02438-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rendulic S, Jagtap P, Rosinus A, Eppinger M, Baar C, Lanz C et al. A predator unmasked: life cycle of Bdellovibrio bacteriovorus from a genomic perspective. Science. 2004;303:689-92. [DOI] [PubMed]
- 72.Glöckner J, Kube M, Shrestha PM, Weber M, Glöckner FO, Reinhardt R, et al. Phylogenetic diversity and metagenomics of candidate division OP3. Environ Microbiol. 2010;12:1218–29. doi: 10.1111/j.1462-2920.2010.02164.x. [DOI] [PubMed] [Google Scholar]
- 73.Read DS, Gweon HS, Bowes MJ, Newbold LK, Field D, Bailey MJ, et al. Catchment-scale biogeography of riverine bacterioplankton. ISME J. 2015;9:516–26. doi: 10.1038/ismej.2014.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hill PG, Zubkov MV, Purdie DA. Differential responses of Prochlorococcus and SAR11-dominated bacterioplankton groups to atmospheric dust inputs in the tropical Northeast Atlantic Ocean. FEMS Microbiol Lett. 2010;306:82–9. doi: 10.1111/j.1574-6968.2010.01940.x. [DOI] [PubMed] [Google Scholar]
- 75.Mary I, Heywood J, Fuchs B, Amann R, Tarran G, Burkill P, et al. SAR11 dominance among metabolically active low nucleic acid bacterioplankton in surface waters along an Atlantic meridional transect. Aquat Microb Ecol. 2006;45:107–13. doi: 10.3354/ame045107. [DOI] [Google Scholar]
- 76.Fuchs B, Woebken D, Zubkov M, Burkill P, Amann R. Molecular identification of picoplankton populations in contrasting waters of the Arabian Sea. Aquat Microb Ecol. 2005;39:145–57. doi: 10.3354/ame039145. [DOI] [Google Scholar]
- 77.Schattenhofer M, Wulf J, Kostadinov I, Glöckner FO, Zubkov MV, Fuchs BM. Phylogenetic characterisation of picoplanktonic populations with high and low nucleic acid content in the North Atlantic Ocean. Syst Appl Microbiol. 2011;34:470–5. doi: 10.1016/j.syapm.2011.01.008. [DOI] [PubMed] [Google Scholar]
- 78.Haller CM, Rolleke S, Vybiral D, Witte A, Velimirov B. Investigation of 0.2 mu m filterable bacteria from the Western Mediterranean Sea using a molecular approach: dominance of potential starvation forms. FEMS Microbiol Ecol. 2000;31:153–61. doi: 10.1111/j.1574-6941.2000.tb00680.x. [DOI] [PubMed] [Google Scholar]
- 79.Prest EI, El-Chakhtoura J, Hammes F, Saikaly PE, van Loosdrecht MCM, Vrouwenvelder JS. Combining flow cytometry and 16S rRNA gene pyrosequencing: a promising approach for drinking water monitoring and characterization. Water Res. 2014;63:179–89. doi: 10.1016/j.watres.2014.06.020. [DOI] [PubMed] [Google Scholar]
- 80.Besmer MD, Weissbrodt DG, Kratochvil BE, Sigrist JA, Weyland MS, Hammes F. The feasibility of automated online flow cytometry for in-situ monitoring of microbial dynamics in aquatic ecosystems. Front Microbiol. 2014;5. 10.3389/fmicb.2014.00265. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.