Abstract
The flagellum and the injectisome enable bacterial locomotion and pathogenesis, respectively. These nanomachines assemble and function using a type III secretion system (T3SS). Exported proteins are delivered to the export apparatus by dedicated cytoplasmic chaperones for their transport through the membrane. The structural and mechanistic basis of this process is poorly understood. Here we report the structures of two ternary complexes among flagellar chaperones (FliT and FliS), protein substrates (the filament-capping FliD and flagellin FliC), and the export gate platform protein FlhA. The substrates do not interact directly with FlhA; however, they are required to induce a binding-competent conformation to the chaperone that exposes the recognition motif featuring a highly conserved sequence recognized by FlhA. The structural data reveal the recognition signal in a class of T3SS proteins and provide new insight into the assembly of key protein complexes at the export gate.
Bacterial flagella are composed of proteins secreted by a type III secretion system (T3SS), which requires the action of dedicated chaperones. Here, Xing et al. report the structures of two ternary complexes among flagellar chaperones, flagellar protein substrates, and the export gate platform protein.
Introduction
Type III secretion systems (T3SSs) are membrane-embedded nanomachines that export dedicated proteins from the bacterial cytoplasm1–6. T3SSs share the same morphology and overall structure and can be functionally classified into two classes7,8: the flagellar T3SS, which promotes bacterial locomotion and motility enabled by the flagellum, and the pathogenic (or non-flagellar) T3SS, which uses the injectisome to transport virulence proteins into human or animal host cells9,10. Both the flagellum11–13 and the injectisome5,6 are supramolecular complexes that are assembled by several different proteins. Flagella may also act as virulence factors because motility is crucial for the action of pathogenic bacteria14,15. The proteins that serve as building blocks of these organelles and the virulence proteins are typically associated with dedicated chaperones in the cytosol16,17. The chaperones bind and protect their cognate substrates from aggregation or premature interactions in the cytoplasm, and they assist in the targeting and delivery of the substrates at the export gate at the membrane18–20.
The export apparatus is formed by six integral membrane proteins (in the flagellar system FlhA, FlhB, FliO, FliP, FliQ, and FliP) that are highly conserved in the flagellar and the pathogenic T3SSs5,6. The cytoplasmic domain of FlhA (FlhAC)21 forms the export platform onto which chaperone–substrate complexes dock to deliver the substrates for their subsequent transport to the extracellular milieu, powered primarily by the proton motive force22,23 and assisted by the FliI ATPase1 (Fig. 1). Crystal structures of FlhAC (refs. 24–26) and their pathogenic T3S homologs27,28 revealed high structural similarity. Biochemical and genetic experiments have demonstrated that FlhA is a key protein for the assembly and operation of the flagellum21,29 and deletion of flhA prevents export of any flagellar protein30. Similar experiments in pathogenic T3SSs31 have indicated that the FlhA homologs (referred to collectively as SctV5,6) are crucial for the operation of the injectisome. FlhAC assembles into a nonameric ring28,32, which is positioned ~6 nm from the membrane surface2 and forms a platform that operates as the export gate. Despite its central role, how the export gate recognizes and interacts with proteins in flagellar and the pathogenic T3SS is not known.
Fig. 1.
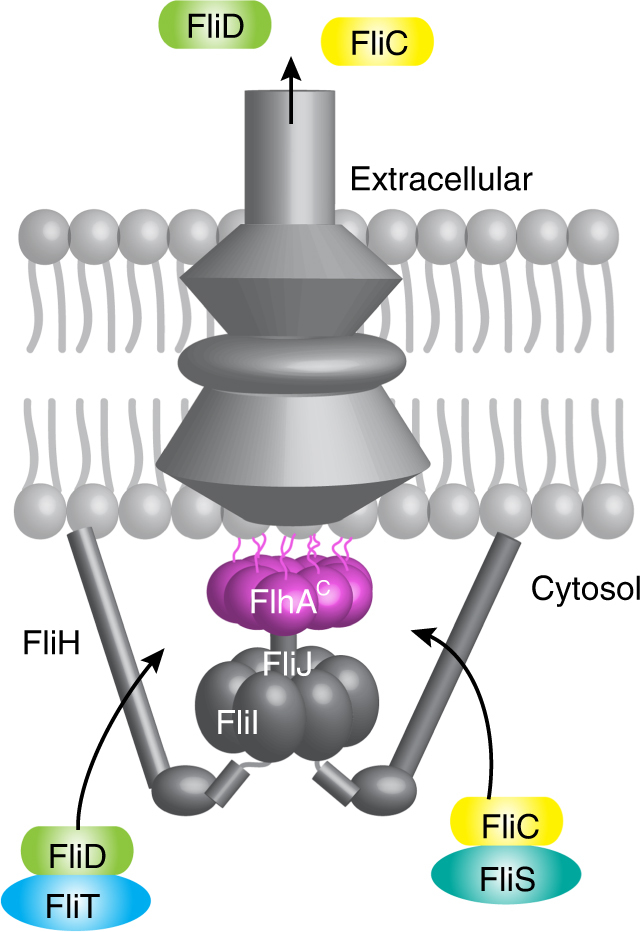
A simplified schematic of the flagellum that includes the proteins studied in this work. For detailed view, see refs. 5 13
About 25 different proteins are involved in the assembly of the flagellum, which is divided into five parts from the base to the tip: the basal body, hook, hook–filament junction, filament, and filament cap33. The assembly of the hook and the filament is strictly sequential13 and the final steps are controlled by the export apparatus through an elusive mechanism. The last steps of the flagellum assembly involve the export first of the filament-capping protein FliD, followed by the export of as many as 3000 flagellin (FliC) molecules, the main building blocks of the filament (Fig. 1). FliD and FliC are found in complex with their dedicated chaperones FliT and FliS, respectively, in the cytoplasm34,35. Biochemical and biophysical data have demonstrated that the chaperones are required for the delivery of the FliD and FliC proteins to the export gate20,24.
Here we report the structures of the ternary complexes among the FliD and FliC flagellar proteins, their cognate chaperones FliT and FliS, and the export gate protein FlhAC. The findings reveal how the export gate specifically recognizes cognate exported proteins and suggest mechanisms of operation of these protein complexes within the T3S nanomachinery.
Results
Interaction of FlhA with flagellar proteins
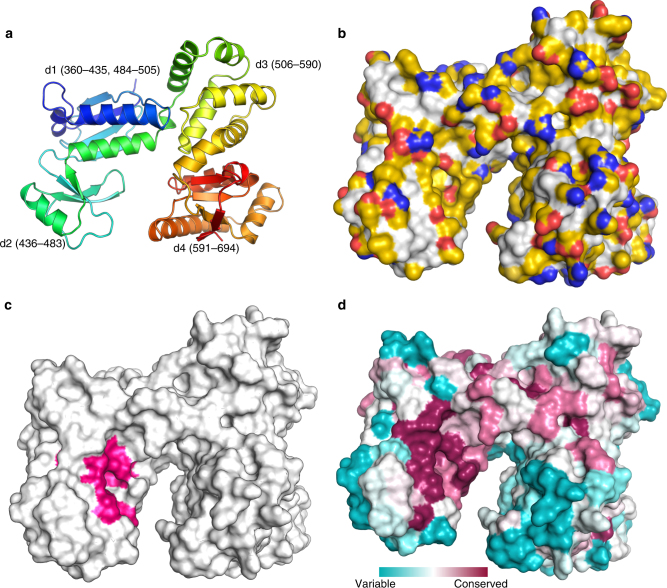
The globular cytoplasmic domain of Salmonella enterica serovar Typhimurium FlhA (FlhAC) encompasses residues 362–692 (36.4 kDa) and is tethered to the transmembrane domain via a linker (residues 328–362). We determined the crystal structure of FlhAC at 1.9 Å resolution (Fig. 2a, b), which shows high similarity to a previously reported structure solved at 2.8 Å resolution of a longer FlhAC construct that includes the linker (FlhAC−link)26 (Supplementary Fig. 1). Multiangle light scattering (MALS) data (Supplementary Fig. 2a) showed that FlhAC is monomeric in solution whereas the linker in FlhAC−link promotes a dimeric state that, based on the crystal structure24,26, is mediated almost exclusively by electrostatic contacts. Indeed, FlhAC oligomerization is very sensitive to the salt concentration in solution (Supplementary Fig. 2b). Structural comparison of FlhAC and FlhAC−link shows that the linker does not alter the three-dimensional fold of the protein. FlhAC folds in a clamp-like structure with four discrete subdomains (d1 through d4; Fig. 2a)24,26. The overall topology of FlhAC and its export gate homologs from both flagellar and pathogenic T3SSs27,28 is very similar, with the main structural variation being the opening of the clamp, as defined by the distance between the d2 and d4 domains and their relative orientation (Supplementary Fig. 1).
Fig. 2.
Structural properties of the export gate protein FlhAC. a Cartoon rendering of the crystal structure of FlhAC determined at 1.9 Å resolution. The protein is colored using a continuous-gradient color scheme from the N terminus (blue) to the C terminus (red). b Solvent-exposed surface rendering of FlhAC displayed in the YRB color scheme59 (yellow, carbons not attached to nitrogen or oxygen; red, negatively charged; blue, positively charged). c The FlhAC site determined by NMR to interact with its binding partners is highlighted in pink. d Sequence conservation of FlhAC colored according to residue identity conservation scores obtained by ConSurf60. The binding site is the most conserved surface in FlhAC
We used NMR (nuclear magnetic resonance) spectroscopy to obtain atomic insight into the interaction of FlhAC with cytosolic flagellar proteins. The 1H-15N and 1H-13C correlated spectra (Supplementary Fig. 3a, b) of FlhAC labeled in methyl-bearing amino acid residues (Ala, Ile, Met, Leu, Thr, and Val)36–39 (Supplementary Fig. 3c) are of high quality and near-complete assignment was obtained. Addition of chaperones FliT and FliS to labeled FlhAC had no effect on the NMR spectrum of FlhAC suggesting these substrate-free chaperones do not interact with FlhAC (Supplementary Fig. 3d). This observation is in agreement with previous NMR data collected using labeled FliT chaperone and unlabeled FlhAC (ref. 20). We also tested the interaction d1of free FliD and FliC with FlhAC by NMR and the data showed that neither of these substrates interacts with FlhAC in their chaperone-free form (Supplementary Fig. 3d).
Next, we tested the interaction between FlhAC and the chaperone–substrate complexes FliT−FliD and FliS−FliC. NMR analysis showed pronounced chemical shift perturbation on FlhAC caused by the addition of FliT−FliD or FliS−FliC (Supplementary Figs. 4a, 5a). NMR chemical shift analysis showed that the two chaperone–substrate complexes share the same binding site on FlhAC, which is located in a cleft at the interface between domains d1 and d2 (Fig. 2c). The location of the NMR-identified binding site is in agreement with previous biochemical data40. The cleft is hydrophobic and is lined by aliphatic and aromatic residues, as well as several Thr residues (Fig. 2b). The binding cleft is the most conserved surface in FlhAC (Fig. 2d) highlighting the importance of this region to the function of FlhA.
Analysis of the NMR data of the titration of unlabeled FlhAC to isotopically labeled FliT−FliD indicated that the FliT α4 helix constitutes the primary FlhAC-binding site (Supplementary Fig. 4b, c), confirming previous observations20. Similarly, NMR data showed that the N-terminal helix of FliS constitutes the primary FlhAC-binding site in FliS−FliC (Supplementary Fig. 5b, c). Both of these helices are part of an autoinhibitory mechanism in the free chaperones20,41, wherein they are buried and thus unavailable for binding. Substrate (FliD and FliC, respectively) dislodges these helices in FliT and FliS, thereby activating the chaperone for binding to FlhAC (Supplementary Figs. 4c, 5c).
Structure of the FlhAC−FliT−FliD ternary complex
We used NMR spectroscopy to determine the structure of the FlhAC−FliT−FliD ternary complex in solution using NMR approaches tailored for protein complexes of large molecular weight36,37,39. Each one of the three proteins (FlhAC, FliT, and FliD) was specifically labeled (1H and 13C) in methyl-bearing (Ala, Ile, Leu, Met, Thr, and Val) and aromatic (Phe and Tyr) residues. In order to mitigate the severe resonance overlap, differentially labeled samples of the ternary complexes were prepared wherein typically one of the proteins was isotopically (1H,13C, and 15N) labeled and the other two uniformly deuterated. NMR spectra of the FlhAC−FliT−FliD ternary complex showed significant line broadening, especially for residues located at the binding interface. Several constructs were tested and the highest quality NMR spectra suitable for solution structure determination were provided by a ternary complex formed between FlhAC and a fused FliT−FliDC construct, wherein FliDC consists of the last 40 C-terminal residues of FliD. Other than the superior quality of the NMR spectra of the fused construct, due to the suppression of the unfavorable exchange processes giving rise to line broadening, the chemical shifts are essentially identical to the non-fused construct. Further NMR analysis demonstrated that FliD does not directly participate in FlhAC binding and thus a shorter FliD construct only encompassing the FliT-binding site could be used to simplify biophysical studies. The NMR observations are in line with isothermal titration calorimetry (ITC) data showing that FliD fragments longer than the FliDC have no effect on the affinity (Supplementary Fig. 4d).
The high quality spectra of the ~60-kDa FlhAC−FliT−FliDC ternary complex enabled its solution structure determination by NMR. Several intermolecular NOEs were observed at the binding interface between FlhAC and FliT (Supplementary Fig. 6a) and a large number of long-distance restrains were collected using paramagnetic relaxation enhancement (PRE) experiments (see Methods) (Supplementary Fig. 6b). The structure and NMR statistics are summarized in Supplementary Table 1. The optimized protein constructs used for the NMR structure determination of the ternary complex were also used for crystallization. The ternary complex was readily crystallized and the X-ray crystallographic structure of the FlhAC−FliT−FliDC ternary complex was determined at 2.75 Å resolution (Supplementary Table 2). The solution and crystal structures are essentially identical (Supplementary Fig. 6c, d).
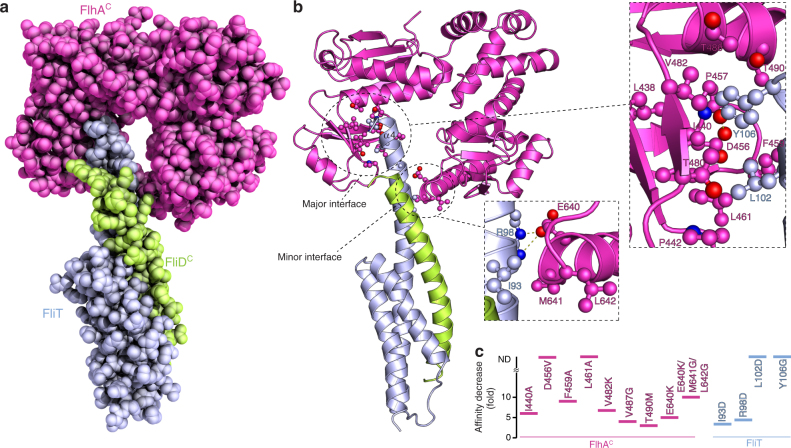
The structure of the FlhAC−FliT−FliDC complex is shown in Fig. 3a. Two distinct interfaces mediate the formation of the ternary complex. In the major interface, which buries a total surface of 350 Å2, the FliT helix α4 juxtaposes with a hydrophobic cleft of FlhAC located between domains d1 and d2 (Fig. 3b). This mode of interaction is in agreement with the NMR chemical shift perturbation data and previous biochemical findings40. Two bulky non-polar residues, Leu102 and Tyr106, emanating from FliT helix α4 bury their side chains into the FlhAC hydrophobic dimple. In addition to a network of intimate non-polar contacts, FliT Tyr106 forms an optimal hydrogen bond with the side chain of FlhAC Asp456 (O–H⋯O distance 2.3 Å). Amino acid substitutions that perturbed the binding interface decreased the ternary complex stability, with FliT residues Leu102 and Tyr106 and FlhA residues Asp456 and Leu461 being essential for complex formation (Fig. 3c). All of these residues are highly conserved (Fig. 2d and Supplementary Fig. 7).
Fig. 3.
Structure of the FlhAC−FliT−FliDC ternary complex. a Crystal structure of the ternary complex shown as a space-filling model. b Cartoon rendering of the structure and expanded views of the two interfaces, major and minor, between FlhAC and FliT. Residues participating in intermolecular contacts are shown as ball-and-stick. Hydrogen bonds and salt bridges are shown as broken lines. c Effect of the indicated amino acid substitutions on the affinity of FlhAC for FliT−FliDC. The effect is given as a fold decrease relative to the affinity of the wild-type proteins (Kd ~20 μM; Supplementary Fig. 4d). Non-detected binding is indicated as ND
The minor binding interface features a salt bridge between FliT Arg98 and FlhA Glu640 (Fig. 3b). Disruption of this electrostatic contact decreases the affinity 5-fold (Fig. 3c). Non-polar contacts mediated by FliT Ile93 with FlhA Met641 and Leu642 also contribute significantly to the complex stability (Fig. 3b, c). Of note, the structural data revealed that there is no direct interaction between FliD and FlhA. To maximize the binding interface with FlhAC, FliT undergoes a dramatic conformational change with helices α3 and α4 merging to one continuous, long α helix (Supplementary Fig. 4e).
Structure of the FlhAC−FliS−FliC ternary complex
We used a similar approach to determine the structure of the FlhAC−FliS−FliC ternary complex. FliS recognizes and binds the last 40 C-terminal residues of FliC41,42 and FliS−FliC forms a stable ternary complex with FlhAC (Kd ~8 μM). FliS−FliC and FliS−FliCC (where FliCC is a construct consisting of residues 454–495) have the same affinity for FlhA indicating that the first 156 residues of FliC do not contribute to FlhA binding. Similarly to FlhAC−FliT−FliD, NMR analysis showed that the highest quality spectra were yielded by a ternary complex formed between FlhAC and a fused construct wherein FliCC was covalently linked to FliS. The structure of the FlhAC−FliS−FliCC ternary complex was determined by X-ray crystallography at a resolution of 2.6 Å (Fig. 4a, b).
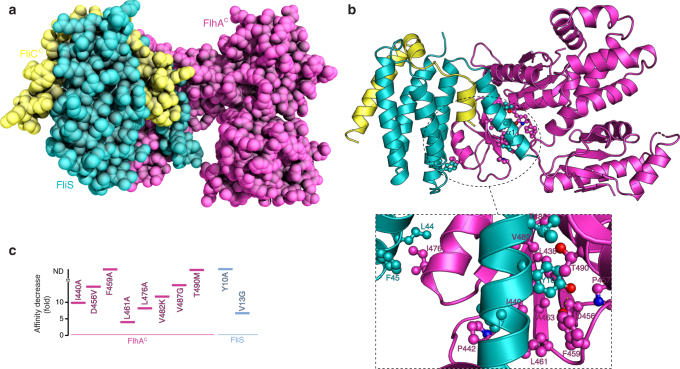
Fig. 4.
Structure of the FlhAC−FliS−FliCC ternary complex. a Crystal structure of the ternary complex shown as a space-filling model. b Cartoon rendering of the structure and expanded view of the interacting regions between FlhAC and FliT. Residues participating in intermolecular contacts are shown as ball-and-stick. Hydrogen bond is shown as broken lines. c Effect of the indicated amino acid substitutions on the affinity of FlhAC for FliS−FliCC. The effect is given as a fold decrease relative to the affinity of the wild-type proteins (Kd ~10 μM; Supplementary Fig. 5d). Non-detected binding is indicated as ND
All contacts within the ternary complex are formed between FliS and FlhAC, with FliC making no direct interaction with FlhAC. Two distinct binding interfaces mediate complex formation. The most significant appears to be the juxtaposition of the FliS N-terminal helix (α1) with the FlhAC hydrophobic cleft located between the d1 and d2 domains (Fig. 4b). Thus, FliS and FliT share a common binding site on FlhA in agreement with the NMR data (Fig. 2c). Three FliS residues (Ile7, Tyr10, and Val13) emanating from helix α1 form intimate hydrophobic contacts with multiple non-polar residues within the FlhAC cleft. The binding interface buries a total surface of 680 Å2. In addition, a hydrogen bond is formed between the hydroxyl group of FliS Tyr10 and FlhAC Asp456 (Fig. 4b). A minor binding interface is formed between FlhA Ile476 and FliS Leu44 and Phe45, with the three residues participating in favorable hydrophobic contacts. The structure of FlhAC in the two ternary complexes is essentially identical with a root-mean-square deviation of backbone atoms of 0.67 Å.
Amino acid substitutions that perturbed the binding interface decreased the ternary complex stability, with FliS residue Tyr10 and several FlhA residues in the binding cleft being essential for complex formation (Fig. 4c).
Recognition mechanisms of flagellar proteins by the export gate
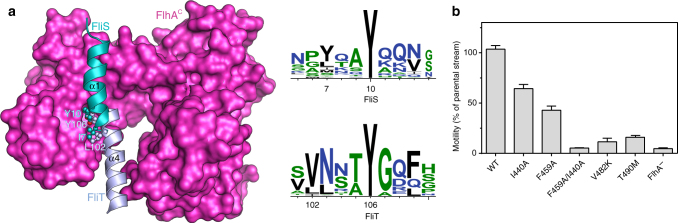
Comparison of the structures of the two ternary complexes reported here illuminates the most salient features that underlie recognition of a T3S chaperone–substrate complex by the export gate (Fig. 5a). Although the FlhA-binding helix in FliT (α4) is positioned very differently along the FlhAC hydrophobic cleft than the corresponding helix in FliS (α1), their two key residues that form crucial contacts with FlhAC have the same topology. Specifically, FliS Tyr10 and FliT Tyr106 interact very similarly with the FlhAC cleft and the Tyr residue is absolutely conserved in both FliT and FliS (Fig. 5a). A Tyr residue in this position is favored because it can form intimate hydrophobic contacts with all the non-polar residues lining the cleft, and at the same time hydrogen bond with the only charged residue in the cleft (Asp456) to neutralize its charge. The second key position is that occupied by FliS Ile7 and FliT Leu102, which form extensive, favorable hydrophobic contacts with the FlhAC cleft (Fig. 5a). The hydrophobic nature of the residue in this position is also highly conserved in both FliT and FliS (Fig. 5a).
Fig. 5.
Recognition mechanisms and functional importance of the FlhA ternary complexes. a Superposition of the two ternary complexes (FlhAC−FliT−FliDC and FlhAC−FliS−FliCC) on FlhAC. For clarity only the recognition helices are shown (FliT helix α4 and FliS helix α1). The two key residues recognized by FlhA is a Tyr (Tyr106 in FliT and Tyr10 in FliS) and a hydrophobic one (Leu102 in FliT and Ile7 in FliS). Sequence conservation of the recognition helices in FliT and FliS is also shown. b Effect of amino acid substitutions at the binding interface on the kinetics of bacterial motility. Bar graphs represent the mean value of the colony diameter and error bars represent standard deviation (n = 6)
In addition to FliD and FliC, the hook–filament junction associated proteins FlgK and FlgL are required for the assembly of the extracellular part of the flagellum. FlgK and FlgL are delivered to the export gate by their cognate chaperone FlgN. NMR analysis (Supplementary Fig. 8a–c) revealed that FlgN, with or without substrate, binds to the same cleft in FlhAC where FliT−FliD and FliS−FliC bind. Thus, FlhA uses a single binding site to engage all of the flagellar chaperones (Fig. 2c). Our NMR data indicated that the C-terminal region of FlgN is responsible for binding to FlhA. By analogy to the mode of binding of FliT and FliS, we hypothesized that Tyr122 in the C-terminal region of FlgN is the key residue for mediating complex formation with FlhA. Indeed, substitution of FlgN Tyr12243 abolished its interaction with FlhA (Supplementary Fig. 8d), confirming the crucial role of the Tyr residue in mediating recognition between all flagellar chaperones and FlhA.
Substrate binding to the export gate is required for motility
To test the functional importance of the interactions we observed in the ternary complexes reported herein, we sought to determine their effect on bacterial motility. Several amino acid residues identified as crucial, based on the structural and ITC data, for complex formation were mutated and their effect on bacterial motility was assessed by complementing a FlhA knockout (FlhA−) strain with either wild type or mutant forms of FlhA32. The effect was quantified by measuring the diameter of the bacteria colony in soft agar after incubation at 37 ℃. FlhA mutants I440A and F459A that decrease significantly the stability of the ternary complexes (Figs. 3c and 4c) have a strong effect on motility (Fig. 5b). Overexpression of the mutated FlhA proteins restored motility, suggesting that the phenotype is due to the low affinity of the chaperone–substrate complexes for FlhA (Supplementary Fig. 9). Previous reports43 showed that substitution of FlhA Asp456, FliS Tyr10, and FliT Tyr106 all have pronounced defect on the motility of the bacterium, in agreement with the present structural data highlighting the key role of these residues. Therefore, proper formation of the ternary complexes mediated by the binding interfaces reported here are important to the function of the flagellum.
Discussion
Delivery of the filament-forming proteins at the export gate is an indispensable step for their export and subsequent assembly of the flagellum. Secretion of the late injectisome components and effector proteins in pathogenic bacteria employing T3SSs requires a similar targeting mechanism. While it has been known that delivery of these proteins to the export gate is assisted by dedicated chaperones, the structural basis of the process had been hitherto unknown. The present data provide atomic view of the structural features underlying the recognition mechanisms of flagellar proteins by the export gate platform. The key residue appears to be a Tyr amino acid in one of the terminal helices in chaperones, which can form extensive hydrophobic contacts with the non-polar residues lining the major binding cleft and at the same time hydrogen bond to the only charged residue that is buried in the cleft. The chaperone–substrate binding cleft is highly conserved in all flagellar systems suggesting that the recognition and targeting mechanisms are evolutionary conserved (Fig. 2d and Supplementary Fig. 10a).
The chaperone–substrate binding cleft in FlhA, which is located at the interface between the d1 and d2 domains, is not conserved in the FlhA analog (SctV) in pathogenic T3SS (Supplementary Fig. 10b). Two surfaces appear to be highly conserved in SctV: the first mediates the formation of the nonameric ring and the second one is located at the interface between the d3 and d4 domains (Supplementary Fig. 10b). The latter can possibly serve as the chaperone–substrate binding site as mutations in this region decreased secretion28. This putative binding surface in SctV, in contrast to the hydrophobic nature of the binding site in FlhA, is made of highly conserved charged residues (Glu and Arg). Thus, although FlhA and SctV share very similar structures and they both bind to cognate chaperone–substrate complexes31, the recognition motif is likely distinct in pathogenic and flagellar T3SSs. The oligomerization-mediated interface is highly conserved in SctV, but not significantly conserved in FlhA.
Interestingly, the exported proteins (e.g., FliD and FliC) do not directly interact with FlhA. Nevertheless, their binding to the chaperone is required for targeting as it poises them for binding to FlhA by relieving the autoinhibitory conformation and dislodging the recognition helix. Of note, although FliT and FliS both adopt an autoinhibitory conformation in the absence of their substrate, FlgN does not and thus is capable of binding to FlhA even in the absence of its substrates (FlgK and FlgL). Whether these different binding properties of the chaperones is physiologically significant is unclear.
A hallmark of the functionality of the flagellum and the injectisome is the hierarchical transport of proteins. In the flagellar system, FlgK and FlgL are exported first, followed by FliD and finally FliC. The mechanism underlying this process is unknown. Because the affinity of the substrate-loaded chaperones for FlhA is very similar, it is unlikely that the hierarchical transport is determined by the energetics of the various ternary complexes. The export gate platform protein forms a nonamer; hence, saturation of all nine of the binding sites would require very large differences in the relative affinities of the various chaperone–substrate complexes for the export gate.
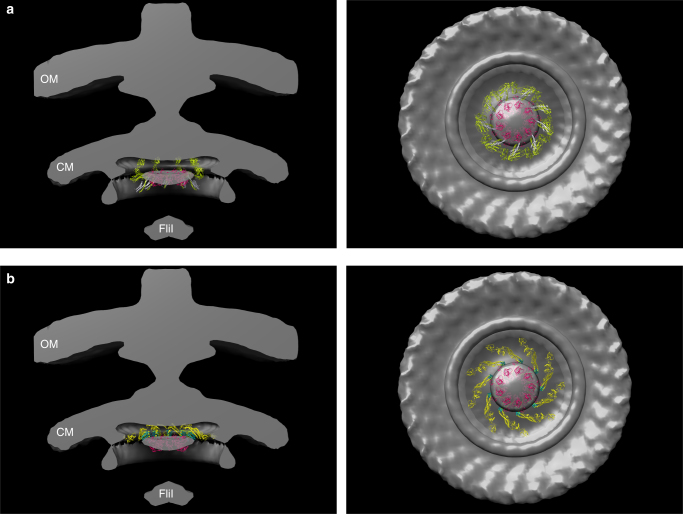
We have fitted the structures of the ternary complexes into the cryoEM density map of the flagellum basal body44 (Fig. 6). As noted before44, the nonameric ring of FlhAC fits nicely into the density map below the membrane basal base where the export gate platform is located. The bound FliT−FliD and FliS−FliC extend into the void space and these interactions bring the substrate very close to the membrane opening. How exactly the substrate dissociates from the chaperone for its export is unclear. The FliI ATPase, which is located far away from the export gate is unlikely to be the one directly disrupting the complex. The present structural data demonstrate that neither FliD nor FliC bind to FlhA. Hence, another proteinaceous factor in the export apparatus should be responsible for interacting with these substrates and assist with positioning them for export.
Fig. 6.
Fitting of the FlhAC ternary complexes into the cryoEM density map of the flagellum base body44. The nonameric oligomeric structure of FlhAC was generated using the structure of the homologous MxiA protein28. The ternary structures obtained in this work were then used to model the complete oligomeric nonameric ring of FlhAC bound to nine molecules of a FliT−FliD and b FliS−FliC. Non-cooperative binding of the chaperone–substrate molecules on FlhAC is assumed. The full-length FliD (5H5V) and FliC (PDB ID 3A5X) structures were used. FliI is the hexameric flagellar ATPase. CM cytoplasmic membrane; OM outer membrane. Side views of half-cut sections and bottom views are shown. The colors of the FlhAC-chaperone–substrate complexes are as in Figs. 2 and 3
Methods
Expression and preparation of proteins
All constructs of Salmonella enterica serovar Typhimurium ST313 FliT, FliD, FlgN, and FlhAC were cloned into the pET16b vector (Novagen) with His10-GB1 or His10-MBP and tobacco etch virus (TEV) protease site in between (Supplementary Table 3). The fusion constructs were prepared by fusing FliDC to the N terminus of FliT and FliCC to the C terminus of FliS. Fusion constructs with varied linker lengths were prepared to ensure that the fusion process does not bias the conformation of the complex in any way. A 20-residue-long linker (VLFQGPSAGLVPRGSGGIEG) was selected from the pCold vector (Takara Bio). All mutants were constructed by site-directed mutagenesis using PfuTurbo high-fidelity DNA polymerase (Agilent). Unlabeled proteins were expressed in BL21(DE3) cells grown in Luria-Bertani (LB) medium in the presence of ampicillin (100 μg ml−1) at 37 ℃, and protein expression was induced at 18 ℃ with 0.4 mM isopropyl-β-D-1-thiogalactopyranoside (IPTG) at OD600 ≈ 0.5 for ~48 h. Cells were harvested at OD600 ≈ 1.5 and were suspended in lysis buffer containing 50 mM Tris-HCl (pH 7.5), 100 mM NaCl, 1% EDTA-free protease inhibitor cocktail (Sigma-Aldrich), and 20 mM imidazole. Cells were disrupted by a high-pressure homogenizer and centrifuged at 20,000 r.p.m. for 1 h. Proteins were purified using Ni Sepharose 6 Fast Flow resin (GE Healthcare), followed by tag removal by TEV protease at 4 ℃ for 12–20 h and gel filtration using Superdex 75 16/60. Proteins were buffer-exchanged and concentrated in Amicon filters (Millipore). Protein concentration was determined spectrophotometrically at 280 nm using the corresponding extinction coefficient.
MALS experiments
MALS was measured by using DAWN HELEOS-II (Wyatt Technology Corporation) downstream of a Shimadzu liquid chromatography system connected to a Superdex 200 10/300 GL (GE Healthcare) gel-filtration column. The running buffer was 50 mM NaPi (pH 6.8), 0.1 M NaCl, 0.05% NaN3. Protein samples at a concentration of 0.05–0.5 mM were used. The flow rate was set to 0.5 ml min−1 with an injection volume of 200 μl and the light scattering signal was collected at room temperature. The data were analyzed with ASTRA version 6.0.5 (Wyatt Technology Corporation).
ITC experiments
All ITC experiments were carried out on an iTC200 microcalorimeter (GE Healthcare) at 25 ℃. Protein samples prepared were extensively dialyzed against the ITC buffer containing 50 mM NaPi (pH 6.8), 300 mM NaCl, 0.05% NaN3. All solutions were prepared by filtering with membrane filters (pore size, 0.45 μm) and thoroughly degassing for 20 min. The sample cell (200 μL) was filled with 0.1–0.2 mM protein (FlhAC and variants), and the 60-μL injection syringe was filled with 1.0–2.0 mM protein (FliT-FiD, FliS-FliC, and variants). The titration was initiated with a preliminary 0.2-μL injection, followed by 15–25 injections of 1.9-3.9 μL, separated by a time interval of 150 s. The solution was stirred at 1000 r.p.m. Data for the first injection were discarded as it is affected by diffusion of the solution from and into the injection syringe during the initial equilibrium period. Binding isotherms profiles were generated by plotting heats of reaction normalized by the modes of injectant vs. the ratio of total injectant to total protein per injection. The data were fitted with Origin 7.0 (OriginLab Corporation) using one-site binding mode.
Protein isotope labeling for NMR studies
Isotopically enriched protein samples were expressed in BL21 (DE3) cells grown in minimal (M9) medium supplied with 99.9%-2H2O. Cells were induced at 18 ℃ with 0.4 mM IPTG at OD600 ≈ 0.4 and typically harvested at OD600 ≈ 1.2. U-[2H, 15N, 13C]-labeled samples were prepared by supplementing the growth medium with 15NH4Cl (1 g L−1) and 2H7-13C-glucose (2 g L−1) and 200 μL L−1 IsoGrow (Isotec). Methyl-protonated samples were prepared as described previously36–39,45 using 50 mg L−1 alpha-ketobutyric acid, 85 mg L−1 alpha-ketoisovaleric acid, 50 mg L−1 of 13CH3-Met, 50 mg L−1 2H2, 13CH3-Ala, and 50 mg L−1 U-2H, Thr-γ2[13CH3]. For selective labeling of Phe and Tyr residues, 50 mg L−1 of U-[13C,15N]-Phe and U-[13C,15N]-Tyr were added to the cell culture. Labeled media and compounds were purchased by Cambridge Isotope Laboratories and Isotec.
NMR spectroscopy
All NMR experiments were performed on Bruker AVANCE III 700, 850, and 900 MHz instruments equipped with cryogenic probes at 25 ℃. Typically, 0.3 mM isotopically labeled protein samples were prepared in 50 mM NaPi (pH 6.8), 300 mM NaCl, 0.05% NaN3, and 10% 2H2O. All recorded spectra were processed with NMRPipe46 and analyzed with Sparky47. Backbone assignment was accomplished using transverse relaxation optimized spectroscopy (TROSY)-based triple resonance experiment. 13Cα, 13Cβ, 13C′, and backbone 1H and 15N chemical shifts were used to compute dihedral angle (ψ, φ) restraints using TALOSN48. Assignment of selectively [1H-methyl-13C] labeled methyl groups was initiated with HMCM(CG)CBCA49, and completed using a combination of 3D (1H)-13C-HMQC-NOESY-1H-15N-HMQC, 3D 15N-edited NOESY-TROSY, and (1H)-13C-HMQC-NOESY-1H-13C-HMQC experiments50.
PRE experiments
PRE experiments were designed to confirm the solution structure of the ternary complex. Based on the NOE data and the binding interface we obtained, a Cys residue was introduced to three positions in FlhAC (S389C, Q473C, S637C), one position in FliT (Q84C), and one position in FliD (T441C). Protein samples with single-point cysteine substitution were expressed in M9-minimal medium with 1 g L−1 U-15NH4Cl, induced at OD600 ≈ 0.5 with 0.4 mM IPTG at 18 ℃ for 2 days, and purified as detailed above in the presence of 5 mM reducing agent beta-mercaptoethanol (βME) in the buffer. The single cysteine mutant proteins were desalted into the reaction buffer containing 50 mM Tris (pH 6.8), 100 mM NaCl, 0.5 mM EDTA, and fivefold molar excess of N-[S-(2-pyridylthio)cysteaminyl]ethylene-diamineN,N,N',N'-tetraacetic acid (Toronto Research Chemicals), and tenfold molar excess of divalent cation (paramagnetic: Mn2+, diamagnetic: Ca2+), incubating for about 24 h at 4 ℃51. Proteins conjugated and chelated with probes were further purified with a Mono-Q column and extensively buffer-exchanged and concentrated with Amicon filters (Millipore). Intermolecular PRE data were collected using 2D 1H-13C HMQC spectra at 25 ℃ on a Bruker Avance III 850 MHz spectrometer equipped with a cryogenic probe. Resonances experiencing significant NMR signal intensity reduction (>50% intensity loss) were identified as sites being within 20 Å of the paramagnetic center, whereas residues experiencing more than 90% intensity loss were identified as sites being within 14 Å of the paramagnetic center.
Structure determination
The structure calculation of FlhAC in complex with FliT–FliDC was performed with CYANA 3.97 (ref. 52), using dihedral restraints extracted as described above, NOE derived distance restraints and H-bond derived distance restraints from SO-FAST 3D (1H)-13C-HMQC-NOESY-1H-15N-HMQC, SO-FAST 3D 15N-edited NOESY-TROSY, SO-FAST 1H-(13C)-HMQC-NOESY-1H-13C-HMQC, and SO-FAST (1H)-13C-HMQC-NOESY-1H-13C-HMQC experiments50 and PRE derived distance restraints from SO-FAST 2D-1H-13C-HMQC. To obtain pure intermolecular NOEs, SO-FAST (1H)-13Carom-HMQC-NOESY-1H-13C-HMQC50 was recorded by using U-[13C,15N]-Tyr-specific labeled FliT–FliDC and U-2H, Ala-13CH3, Met-13CH3, Ile-δ1-13CH3, Leu/Val-13CH3/12C2H3, and Thr-13CH3-labeled FlhAC. Hydrogen bond restraints were obtained from analysis of NOE data and chemical shift information. Twenty structures with lowest target function were subjected to restrained molecular dynamics energy water refinement using CNS53. The percentage of residues falling in favored and disallowed regions of the Ramachandran plot from Procheck is: 99.7% and 0.3%, respectively. The ensemble of the 15 lowest-energy conformers are shown in Supplementary Fig. 11.
Crystallization and data collection
Proteins were purified and concentrated to 10 g L−1. Crystallization was setup using the CrystalTrak system (Rigaku). A total of seven screens were setup on Intelli-Plate 96 trays (Art Robbins Instruments). Crystals were transferred into cryo-protectant containing the corresponding well solution and 20% v/v glycerol using the CryoLoop from Hampton Research, and flash frozen in liquid nitrogen. The crystals were screened at the Advanced Photon Source Northeastern Collaborative Access Team beamlines (24-ID-C). FlhAC and FlhAC–FliT–FliDC crystallized in space group P1, whereas FlhAC–FliS–FliCC crystallized in space group P212121. X-ray diffraction data were subsequently collected and data images were processed with XDS54 . Matthews coefficient55 calculation indicated that there are three complexes in the asymmetric unit. Using the published FlhAC structure (PDB ID 3A5I) as a searching model, PHASER56 located three copies of FlhAC monomers in the asymmetric unit by molecular replacement. Subsequent iterative refinement with the PHENIX suite57, followed by model inspection/building using COOT58 and molecular replacement using the structure of FliT−FliDC (PDB ID 5KRW)20 generated three complete copies of the FlhAC−FliT−FliDC ternary complex, resulting in Rwork/Rfree 22.13%/26.86%. Ramachandran analysis shows that 97.0%, 3.0%, and 0% of the protein residues are in the most favored, allowed, and disallowed region, respectively. Similar procedures were used to determine the structure of the FlhAC−FliS−FliCC ternary complex. The summary of data collection and refinement statistics is shown in Supplementary Table 2. The 2Fo–Fc electron density maps are shown in Supplementary Fig. 11.
Secretion and motility assays
Motility assays were performed as described previously32. In brief, Salmonella enterica serovar Typhimurium ΔflhA− strain was freshly transformed with pKG116 plasmids containing wild-type flhA or its mutants. Bacteria were stabbed in 0.2% LB-agar plates (1 μl from overnight pre-culture) and incubated for 6 h at 37 ℃. When indicated, plates were supplemented with 5 mM sodium salicylate to induce gene expression. Plates were scanned using Las 4000 (GE healthcare). Images were processed with Adobe Photoshop to adjust the background color and colony diameter was measured using Image Quant (GE Healthcare). Strains and plasmids were a kind gift from Marc Erhardt.
Data availability
Atomic coordinates for the structures have been deposited in the Protein Data Bank under accession numbers 6CH1 (FlhAC), 6CH2 (FlhAC−FliT−FliDC ternary complex), and 6CH3 (FlhAC−FliS−FliCC ternary complex). Other data are available in this article and its Supplementary Information files, or from the corresponding author upon request.
Electronic supplementary material
Acknowledgements
We thank Marc Erhardt for providing the ΔflhA-strain and the pKG116 flhA plasmid, and for help in setting up the motility assays. This work was supported by NIH grant AI094623 (to C.G.K.) and T3RecS (Vlaanderen Onderzoeksprojecten; #G002516N; FWO) and DIP-BiD (#AKUL/15/40—G0H2116N; Hercules/FWO) (to A.E.). This work is based upon research conducted at the Northeastern Collaborative Access Team beamlines, which are funded by the US National Institutes of Health (NIGMS P41-GM103403). The Pilatus 6 M detector on 24-ID-C beamline is funded by a NIH-ORIP HEI grant (S10 RR029205). This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357.
Author contributions
C.G.K. conceived the study. Q.X., K.S., A.P., P.R., A.E., and C.G.K. designed the experiments and interpreted the data. Q.X. and C.G.K. wrote the paper. All authors reviewed and approved the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary Information accompanies this paper at 10.1038/s41467-018-04137-4.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Galan JE, Lara-Tejero M, Marlovits TC, Wagner S. Bacterial type III secretion systems: specialized nanomachines for protein delivery into target cells. Annu. Rev. Microbiol. 2014;68:415–438. doi: 10.1146/annurev-micro-092412-155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Radics J, Konigsmaier L, Marlovits TC. Structure of a pathogenic type 3 secretion system in action. Nat. Struct. Mol. Biol. 2014;21:82–87. doi: 10.1038/nsmb.2722. [DOI] [PubMed] [Google Scholar]
- 3.Diepold, A. & Armitage, J. P. Type III secretion systems: the bacterial flagellum and the injectisome. Philos. Trans. R. Soc. Lond. B Biol. Sci. 370, 20150020 (2015). [DOI] [PMC free article] [PubMed]
- 4.Lee PC, Rietsch A. Fueling type III secretion. Trends Microbiol. 2015;23:296–300. doi: 10.1016/j.tim.2015.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Portaliou AG, Tsolis KC, Loos MS, Zorzini V, Economou A. Type III secretion: building and operating a remarkable nanomachine. Trends Biochem. Sci. 2016;41:175–189. doi: 10.1016/j.tibs.2015.09.005. [DOI] [PubMed] [Google Scholar]
- 6.Deng W, et al. Assembly, structure, function and regulation of type III secretion systems. Nat. Rev. Microbiol. 2017;15:323–337. doi: 10.1038/nrmicro.2017.20. [DOI] [PubMed] [Google Scholar]
- 7.Pallen MJ, Bailey CM, Beatson SA. Evolutionary links between FliH/YscL-like proteins from bacterial type III secretion systems and second-stalk components of the FoF1 and vacuolar ATPases. Protein Sci. 2006;15:935–941. doi: 10.1110/ps.051958806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abby SS, Rocha EP. The non-flagellar type III secretion system evolved from the bacterial flagellum and diversified into host-cell adapted systems. PLoS Genet. 2012;8:e1002983. doi: 10.1371/journal.pgen.1002983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blocker A, Komoriya K, Aizawa S. Type III secretion systems and bacterial flagella: insights into their function from structural similarities. Proc. Natl Acad. Sci. USA. 2003;100:3027–3030. doi: 10.1073/pnas.0535335100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saier MH., Jr. Evolution of bacterial type III protein secretion systems. Trends Microbiol. 2004;12:113–115. doi: 10.1016/j.tim.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 11.Evans LD, Hughes C, Fraser GM. Building a flagellum outside the bacterial cell. Trends Microbiol. 2014;22:566–572. doi: 10.1016/j.tim.2014.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Minamino T. Protein export through the bacterial flagellar type III export pathway. Biochim. Biophys. Acta. 2014;1843:1642–1648. doi: 10.1016/j.bbamcr.2013.09.005. [DOI] [PubMed] [Google Scholar]
- 13.Altegoer F, Bange G. Undiscovered regions on the molecular landscape of flagellar assembly. Curr. Opin. Microbiol. 2015;28:98–105. doi: 10.1016/j.mib.2015.08.011. [DOI] [PubMed] [Google Scholar]
- 14.Duan Q, Zhou M, Zhu L, Zhu G. Flagella and bacterial pathogenicity. J. Basic Microbiol. 2013;53:1–8. doi: 10.1002/jobm.201100335. [DOI] [PubMed] [Google Scholar]
- 15.Chaban B, Hughes HV, Beeby M. The flagellum in bacterial pathogens: for motility and a whole lot more. Semin. Cell Dev. Biol. 2015;46:91–103. doi: 10.1016/j.semcdb.2015.10.032. [DOI] [PubMed] [Google Scholar]
- 16.Parsot C, Hamiaux C, Page AL. The various and varying roles of specific chaperones in type III secretion systems. Curr. Opin. Microbiol. 2003;6:7–14. doi: 10.1016/S1369-5274(02)00002-4. [DOI] [PubMed] [Google Scholar]
- 17.Evans LD, Stafford GP, Ahmed S, Fraser GM, Hughes C. An escort mechanism for cycling of export chaperones during flagellum assembly. Proc. Natl Acad. Sci. USA. 2006;103:17474–17479. doi: 10.1073/pnas.0605197103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen L, et al. Structural instability tuning as a regulatory mechanism in protein-protein interactions. Mol. Cell. 2011;44:734–744. doi: 10.1016/j.molcel.2011.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen L, et al. Substrate-activated conformational switch on chaperones encodes a targeting signal in type III secretion. Cell Rep. 2013;3:709–715. doi: 10.1016/j.celrep.2013.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khanra N, Rossi P, Economou A, Kalodimos CG. Recognition and targeting mechanisms by chaperones in flagellum assembly and operation. Proc. Natl Acad. Sci. USA. 2016;113:9798–9803. doi: 10.1073/pnas.1607845113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saijo-Hamano Y, Minamino T, Macnab RM, Namba K. Structural and functional analysis of the C-terminal cytoplasmic domain of FlhA, an integral membrane component of the type III flagellar protein export apparatus in Salmonella. J. Mol. Biol. 2004;343:457–466. doi: 10.1016/j.jmb.2004.08.067. [DOI] [PubMed] [Google Scholar]
- 22.Minamino T, Namba K. Distinct roles of the FliI ATPase and proton motive force in bacterial flagellar protein export. Nature. 2008;451:485–488. doi: 10.1038/nature06449. [DOI] [PubMed] [Google Scholar]
- 23.Paul K, Erhardt M, Hirano T, Blair DF, Hughes KT. Energy source of flagellar type III secretion. Nature. 2008;451:489–492. doi: 10.1038/nature06497. [DOI] [PubMed] [Google Scholar]
- 24.Bange G, et al. FlhA provides the adaptor for coordinated delivery of late flagella building blocks to the type III secretion system. Proc. Natl Acad. Sci. USA. 2010;107:11295–11300. doi: 10.1073/pnas.1001383107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moore SA, Jia Y. Structure of the cytoplasmic domain of the flagellar secretion apparatus component FlhA from Helicobacter pylori. J. Biol. Chem. 2010;285:21060–21069. doi: 10.1074/jbc.M110.119412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saijo-Hamano Y, et al. Structure of the cytoplasmic domain of FlhA and implication for flagellar type III protein export. Mol. Microbiol. 2010;76:260–268. doi: 10.1111/j.1365-2958.2010.07097.x. [DOI] [PubMed] [Google Scholar]
- 27.Worrall LJ, Vuckovic M, Strynadka NC. Crystal structure of the C-terminal domain of the Salmonella type III secretion system export apparatus protein InvA. Protein Sci. 2010;19:1091–1096. doi: 10.1002/pro.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abrusci P, et al. Architecture of the major component of the type III secretion system export apparatus. Nat. Struct. Mol. Biol. 2013;20:99–104. doi: 10.1038/nsmb.2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McMurry JL, Van Arnam JS, Kihara M, Macnab RM. Analysis of the cytoplasmic domains of Salmonella FlhA and interactions with components of the flagellar export machinery. J. Bacteriol. 2004;186:7586–7592. doi: 10.1128/JB.186.22.7586-7592.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Minamino T, Macnab RM. Components of the Salmonella flagellar export apparatus and classification of export substrates. J. Bacteriol. 1999;181:1388–1394. doi: 10.1128/jb.181.5.1388-1394.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Portaliou AG, et al. Hierarchical protein targeting and secretion is controlled by an affinity switch in the type III secretion system of enteropathogenic Escherichia coli. EMBO J. 2017;36:3517–3531. doi: 10.15252/embj.201797515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Erhardt M, et al. Mechanism of type-III protein secretion: regulation of FlhA conformation by a functionally critical charged-residue cluster. Mol. Microbiol. 2017;104:234–249. doi: 10.1111/mmi.13623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chevance FF, Hughes KT. Coordinating assembly of a bacterial macromolecular machine. Nat. Rev. Microbiol. 2008;6:455–465. doi: 10.1038/nrmicro1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Auvray F, Thomas J, Fraser GM, Hughes C. Flagellin polymerisation control by a cytosolic export chaperone. J. Mol. Biol. 2001;308:221–229. doi: 10.1006/jmbi.2001.4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aldridge P, Karlinsey J, Hughes KT. The type III secretion chaperone FlgN regulates flagellar assembly via a negative feedback loop containing its chaperone substrates FlgK and FlgL. Mol. Microbiol. 2003;49:1333–1345. doi: 10.1046/j.1365-2958.2003.03637.x. [DOI] [PubMed] [Google Scholar]
- 36.Saio T, Guan X, Rossi P, Economou A, Kalodimos CG. Structural basis for protein antiaggregation activity of the trigger factor chaperone. Science. 2014;344:1250494. doi: 10.1126/science.1250494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang C, Rossi P, Saio T, Kalodimos CG. Structural basis for the antifolding activity of a molecular chaperone. Nature. 2016;537:202–206. doi: 10.1038/nature18965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Monneau YR, et al. Exploiting E. coli auxotrophs for leucine, valine, and threonine specific methyl labeling of large proteins for NMR applications. J. Biomol. NMR. 2016;65:99–108. doi: 10.1007/s10858-016-0041-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang C, Kalodimos CG. Structures of large protein complexes determined by nuclear magnetic resonance spectroscopy. Annu Rev. Biophys. 2017;46:317–336. doi: 10.1146/annurev-biophys-070816-033701. [DOI] [PubMed] [Google Scholar]
- 40.Kinoshita M, Hara N, Imada K, Namba K, Minamino T. Interactions of bacterial flagellar chaperone-substrate complexes with FlhA contribute to co-ordinating assembly of the flagellar filament. Mol. Microbiol. 2013;90:1249–1261. doi: 10.1111/mmi.12430. [DOI] [PubMed] [Google Scholar]
- 41.Evdokimov AG, et al. Similar modes of polypeptide recognition by export chaperones in flagellar biosynthesis and type III secretion. Nat. Struct. Biol. 2003;10:789–793. doi: 10.1038/nsb982. [DOI] [PubMed] [Google Scholar]
- 42.Ozin AJ, Claret L, Auvray F, Hughes C. The FliS chaperone selectively binds the disordered flagellin C-terminal D0 domain central to polymerisation. FEMS Microbiol. Lett. 2003;219:219–224. doi: 10.1016/S0378-1097(02)01208-9. [DOI] [PubMed] [Google Scholar]
- 43.Minamino T, et al. Interaction of a bacterial flagellar chaperone FlgN with FlhA is required for efficient export of its cognate substrates. Mol. Microbiol. 2012;83:775–788. doi: 10.1111/j.1365-2958.2011.07964.x. [DOI] [PubMed] [Google Scholar]
- 44.Kawamoto A, et al. Common and distinct structural features of Salmonella injectisome and flagellar basal body. Sci. Rep. 2013;3:3369. doi: 10.1038/srep03369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tzeng SR, Pai MT, Kalodimos CG. NMR studies of large protein systems. Methods Mol. Biol. 2012;831:133–140. doi: 10.1007/978-1-61779-480-3_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Delaglio F, et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 47.Lee W, Tonelli M, Markley JL. NMRFAM-SPARKY: enhanced software for biomolecular NMR spectroscopy. Bioinformatics. 2015;31:1325–1327. doi: 10.1093/bioinformatics/btu830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shen Y, Bax A. Protein backbone and sidechain torsion angles predicted from NMR chemical shifts using artificial neural networks. J. Biomol. NMR. 2013;56:227–241. doi: 10.1007/s10858-013-9741-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tugarinov V, Kay LE. Ile, Leu, and Val methyl assignments of the 723-residue malate synthase G using a new labeling strategy and novel NMR methods. J. Am. Chem. Soc. 2003;125:13868–13878. doi: 10.1021/ja030345s. [DOI] [PubMed] [Google Scholar]
- 50.Rossi P, Xia Y, Khanra N, Veglia G, Kalodimos CG. 15N and 13C- SOFAST-HMQC editing enhances 3D-NOESY sensitivity in highly deuterated, selectively [1H,13C]-labeled proteins. J. Biomol. NMR. 2016;66:259–271. doi: 10.1007/s10858-016-0074-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tang C, Iwahara J, Clore GM. Visualization of transient encounter complexes in protein-protein association. Nature. 2006;444:383–386. doi: 10.1038/nature05201. [DOI] [PubMed] [Google Scholar]
- 52.Guntert P. Automated NMR structure calculation with CYANA. Methods Mol. Biol. 2004;278:353–378. doi: 10.1385/1-59259-809-9:353. [DOI] [PubMed] [Google Scholar]
- 53.Brunger AT. Version 1.2 of the crystallography and NMR system. Nat. Protoc. 2007;2:2728–2733. doi: 10.1038/nprot.2007.406. [DOI] [PubMed] [Google Scholar]
- 54.Kabsch W. Xds. Acta Crystallogr. D Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Matthews BW. Solvent content of protein crystals. J. Mol. Biol. 1968;33:491–497. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- 56.McCoy AJ, et al. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed]
- 58.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hagemans D, van Belzen IA, Moran Luengo T, Rudiger SG. A script to highlight hydrophobicity and charge on protein surfaces. Front. Mol. Biosci. 2015;2:56. doi: 10.3389/fmolb.2015.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ashkenazy H, et al. ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 2016;44:W344–W350. doi: 10.1093/nar/gkw408. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates for the structures have been deposited in the Protein Data Bank under accession numbers 6CH1 (FlhAC), 6CH2 (FlhAC−FliT−FliDC ternary complex), and 6CH3 (FlhAC−FliS−FliCC ternary complex). Other data are available in this article and its Supplementary Information files, or from the corresponding author upon request.