Abstract
Many Proteobacteria use quorum sensing to regulate production of public goods, such as antimicrobials and proteases, that are shared among members of a community. Public goods are vulnerable to exploitation by cheaters, such as quorum sensing-defective mutants. Quorum sensing- regulated private goods, goods that benefit only producing cells, can prevent the emergence of cheaters under certain growth conditions. Previously, we developed a laboratory co-culture model to investigate the importance of quorum-regulated antimicrobials during interspecies competition. In our model, Burkholderia thailandensis and Chromobacterium violaceum each use quorum sensing-controlled antimicrobials to inhibit the other species’ growth. Here, we show that C. violaceum uses quorum sensing to increase resistance to bactobolin, a B. thailandensis antibiotic, by increasing transcription of a putative antibiotic efflux pump. We demonstrate conditions where C. violaceum quorum-defective cheaters emerge and show that in these conditions, bactobolin restrains cheaters. We also demonstrate that bactobolin restrains quorum-defective mutants in our co-culture model, and the increase in antimicrobial-producing cooperators drives the C. violaceum population to become more competitive. Our results describe a mechanism of cheater restraint involving quorum control of efflux pumps and demonstrate that interspecies competition can reinforce cooperative behaviors by placing constraints on quorum sensing-defective mutants.
Introduction
Many Proteobacteria use acyl-homoserine lactone (AHL) quorum-sensing signals to regulate gene expression in a cell density-dependent manner [1–3]. AHLs are produced by LuxI-family synthases and when they reach a sufficient concentration, the AHLs interact with cytoplasmic LuxR-family signal receptors to affect changes in gene transcription. Many quorum-controlled genes encode factors, such as extracellular proteases, that can be shared among members of the population and constitute public goods [4]. Such public goods are cheatable by individuals that benefit without paying any costs of production [5]. In some conditions, the cheaters can ultimately cause a population crash [6], presenting a significant threat to cooperation. Yet quorum-sensing mutants are relatively rare in some natural microbial populations [7, 8], suggesting there might be mechanisms to restrain cheating in certain contexts. One such mechanism relies on the quorum sensing-dependent linkage of private goods, such as cell-localized enzymes, with public goods. These private goods benefit only producing members of the population and have been shown to restrain cheating in some AHL-based quorum-sensing systems [6].
Many bacteria use quorum sensing to regulate the production of antimicrobials, a type of public good. For example, quorum sensing regulates the production of phenazine in Pseudomonas chlororaphis [9] and bactobolin antibiotic in Burkholderia thailandensis [10, 11]. Quorum-controlled antimicrobials are thought to be important for interspecies competition [12–14]. Previously, we developed a laboratory dual-species model with the soil saprophytes Burkholderia thailandensis and Chromobacterium violaceum to investigate the relationship between quorum sensing and interspecies competition [14]. The B. thailandensis bactobolin antibiotic targets the 50 S ribosome and has activity against a range of bacterial species including C. violaceum [11, 15, 16]. Bactobolin production is regulated by one of the three B. thailandensis LuxR-LuxI-type quorum-sensing pairs, BtaR2–BtaI2, that senses and produces the signals 3-hydroxy-octanoyl-homoserine lactone and 3-hydroxy-decanoyl-homoserine lactone [10]. In our dual-species model, B. thailandensis competitiveness relies upon the production of the bactobolin antibiotic and also BtaR2 [14]. Likewise, C. violaceum uses quorum sensing to control production of as-yet unknown secreted antimicrobials that are important to compete with B. thailandensis. In C. violaceum, there is a single LuxR-LuxI pair, the CviR-CviI quorum-sensing system. The CviR-CviI system produces and responds to hexanoyl-homoserine lactone (C6-HSL) [17]. This system controls production of a purple pigment, violacein [17]. Although violacein has broad-spectrum antimicrobial activity [18], the violacein biosynthetic genes were previously shown to be dispensable for competition in our dual-species model [14].
We are interested in understanding how quorum sensing benefits bacteria in natural microbial communities, and how interspecies competition might shape the evolution of quorum-sensing systems. Here, we show that C. violaceum quorum sensing promotes resistance to bactobolin and several other antimicrobials through a CviR-dependent putative multidrug efflux pump. Because multidrug efflux pumps are cell-localized they might be considered a private good [19]. Here, we show antibiotics restrain quorum sensing-defective mutants during serial passaging experiments and in direct competition with B. thailandensis. Such antibiotic-dependent selection might contribute to the maintenance of quorum sensing-dependent behaviors important for competition, such as antimicrobial production. Our results provide a possible mechanism controlling quorum-defective cheater variants in natural microbial populations where interspecies interactions are common.
Materials and methods
Growth media, reagents and bacterial strains
Bacteria were grown in Luria-Bertani (LB) broth containing morpholinepropanesulfonic acid (50 mΜ; pH 7)(MOPS-LB), or in M9 minimal medium with 1% (wt/vol) of sodium caseinate (casein broth), or on LB with 1.5% (wt/vol) agar or skim milk agar (1/4-strength LB broth with 4% (wt/vol) skim milk and 1.5% (wt/vol) agar). All broth cultures were grown at 30°C with shaking at 230 r.p.m. except Escherichia coli, which was at 37°C. Growth on solid medium was at 30°C for C. violaceum or 37°C for B. thailandensis and for C. violaceum–E. coli matings. Growth in liquid cultures was with shaking at 30°C in 18-mm test tubes. Synthetic C6-HSL was purchased from Cayman Chemical (Ann Arbor, MI, USA) and dissolved in acidified ethyl acetate with glacial acetic acid (0.1 ml l−1). Dissolved C6-HSL was added to an empty sterile conical tube and dried by evaporation prior to the addition of LB agar that had been melted and cooled to 55°C. As a source of bactobolin, either filter-sterilized B. thailandensis culture fluid or purified bactobolin A was used (see Supplementary Information for additional information). Bactobolin A was suspended in sterile water prior to use in experiments. For casein evolution experiments, we used tetracycline at 1 μg ml−1, bactobolin at 2 μg ml−1, and gentamicin at 12 μg ml−1. For strain constructions, we used gentamicin at 50 μg ml−1 (C. violaceum) or 15 μg ml−1 (E. coli). For selection from dual-species cultures we used gentamicin at 100 μg ml−1 (B. thailandensis) and trimethoprim at 100 μg ml−1 (C. violaceum).
Bacterial strains are listed in Table S1. B. thailandensis strains were E264 (wild type) or BD20, a bactobolin-deficient derivative of E264 [20]. C. violaceum strains are derivatives of strain ATCC31532 [21]. C. violaceum CV017 (referred to as wild type) has a transposon insertion in gene CV_RS05185 causing overexpression of violacein [22, 23], CV026 is a derivative of CV017 with a second transposon insertion in the cviI gene [17], and CV026R is a derivative of CV026 with an in-frame deletion of the cviR gene [14]. The C. violaceum CdeAB-OprM− and CdeR H127Y mutants were constructed using homologous recombination as previously described [14] with synthetic gene fragments (IDT) cloned into the pEX18Gm-derived delivery plasmid [24]. For fragment cloning, we used SphI–XbaI (for ΔcdeAB-oprM) or HindIII–XbaI (for CdeR H127Y). The ΔcdeAB-oprM construct was made with ~500-bp DNA flanking cdeA and oprM, resulting in a deletion of all but the first 8 codons of cdeA and the last 13 codons of oprM. The CdeR H127Y construct was made using a ~1000-bp DNA fragment encoding cdeR with a missense C379T mutation and ~100-bp DNA flanking cdeR. All candidate mutants were screened by PCR and susceptibility testing to gentamicin, the antibiotic resistance marker on the pEX18Gm delivery plasmid. All strains and PCR-generated plasmids were verified by PCR amplification and sequencing.
Antibiotic sensitivity
To determine antibiotic sensitivity, stationary-phase broth cultures were suspended to an optical density at 600 nm (OD600) of 1.0 and this suspension was serially diluted in 10-fold increments in LB broth. A total of 10 μl aliquots of these successive dilutions were spotted onto LB agar plates containing antimicrobials at the concentration indicated or in a series of concentrations to determine the minimum inhibitory concentration (MIC). The agar plates were incubated at 30°C for 24 h and an additional 24 h at room temperature. The MIC was defined as the lowest concentration of antimicrobial in the plate that prevented visible growth of cells from the 10−3 dilution (~104 cells). B. thailandensis culture fluid was tested in 0.1% increments over a range from 0 to 7%, tetracycline was tested in 0.5 μg increments over a range from 0 to 2 μg ml−1, ethidium bromide (EtBr) was tested in 10 μg increments in a range from 20 to 150 μg per ml, and gentamicin was tested in 0.1 μg increments in a range from 1 to 10 μg per ml.
Droplet digital PCR
RNA was harvested from stationary-phase C. violaceum cells (OD600 of 4) using methods described previously [25]. Droplet digital PCR was performed on Bio-Rad’s QX200 Droplet Digital PCR System using Eva Green Supermix. Each reaction used 1 ng μl−1 of cDNA template, 0.25 μM primer, 10 μl Eva Green Supermix, and 8 μl H2O in a 20-μl volume. After generating 40 μl of oil droplets, 40 rounds of PCR were conducted using the following cycling conditions: 94°C for 20 sec, 58°C for 20 sec, and 72°C for 20 sec. Absolute transcript levels were determined using the Bio-Rad QuantaSoft Software. In all cases a no template control was run with no detectable transcripts. The gene encoding glyceraldehyde-3-phosphate dehydrogenase (gapdh) was used as a reference gene and the results are reported as the calculated transcript amount of a given gene per calculated gapdh transcript.
Casein evolution experiments
To inoculate C. violaceum casein evolution experiments, a starter culture was prepared by diluting stationary-phase cells from an LB broth culture to an OD600 of 0.05 in casein broth and growing this an additional 24 h. To start the experiment, 40 μl from this starter culture was transferred to 4 ml fresh casein broth and at 24 h intervals 40 μl was transferred to fresh casein broth in a new tube. When indicated, antibiotics were added to the culture medium every day except for gentamicin, which was added every other day for the first week and every day after that. Colony forming units (CFUs) were determined by spread-plating 10-fold dilutions of the 24 h culture onto LB agar plates. Cheater abundance was determined by the proportion of colonies identified as cheaters compared with the rest of the population. Cheaters were identified based on complete loss of purple pigmentation due to abolished production of the quorum-dependent purple pigment violacein, and their inability to form a zone of clearing on skim milk agar plates (skim milk clearing was described previously for P. aeruginosa [26, 27]. Although intermediate-phenotype variants were occasionally observed (partial pigmentation and protease production), only those variants with complete loss of each phenotype (similar to the quorum-sensing-defective mutants) were counted as cheaters. As described in the text, a subset of cheaters was also verified by sequencing the cviR gene.
Dual-species experiments
Dual-species experiments were conducted in 1 ml MOPS-LB broth in 18 mm test tubes. The inoculum was from logarithmic-stage pure cultures of B. thailandensis and C. violaceum. The initial OD600 in the dual-species culture was 0.05 (2–4 × 107 cells per ml) for B. thailandensis and 0.005 (2–4 × 106 cells per ml) for C. violaceum. When indicated, C. violaceum wild type and CviIR− mutant cells were combined at a 1:1 ratio just prior to mixing with B. thailandensis. Colony forming units of each species were determined by using differential antibiotic selection on LB agar plates. B. thailandensis was selected with gentamicin and C. violaceum was selected with trimethoprim. Wild type and the CviIR− mutant were differentiated by color as described above.
Results
C. violaceum quorum sensing is important for antimicrobial resistance
In transcriptomics studies of saprophytic Proteobacteria, quorum sensing commonly activates expression of genes predicted to be for antibiotic export [28–31]. Because some efflux pumps increase antimicrobial resistance, this finding suggests there might be a connection between quorum sensing and antimicrobial resistance. Using our B. thailandensis–C. violaceum laboratory model as a starting point, we sought to test the hypothesis that C. violaceum quorum sensing might increase resistance to the B. thailandensis bactobolin antibiotic. To test this hypothesis, we supplemented LB agar plates with B. thailandensis culture fluid, and compared the growth of C. violaceum wild type and two quorum sensing-deficient mutants, an AHL synthase mutant (CviI−) and an AHL synthase and receptor double mutant (CviIR−) by spotting 10-fold dilutions from stationary-phase cultures onto the plates (Fig. 1 and Table 1). We determined that wild-type C. violaceum was more resistant to B. thailandensis culture fluid than either of the quorum-sensing mutants, with an MIC of 1.7% compared with 0.6% for both mutants. We could rescue resistance to the AHL synthase mutant by supplementing the LB agar plate with synthetic C6-HSL, the AHL specific to CviR (Table 1). The wild type and each of the quorum-sensing mutants grew identically on LB agar with or without supplementation with 1 or 3% cell-free fluid from a B. thailandensis bactobolin-deficient mutant (Fig. 1 and Table 1). These results indicated C. violaceum quorum sensing is important for bactobolin resistance.
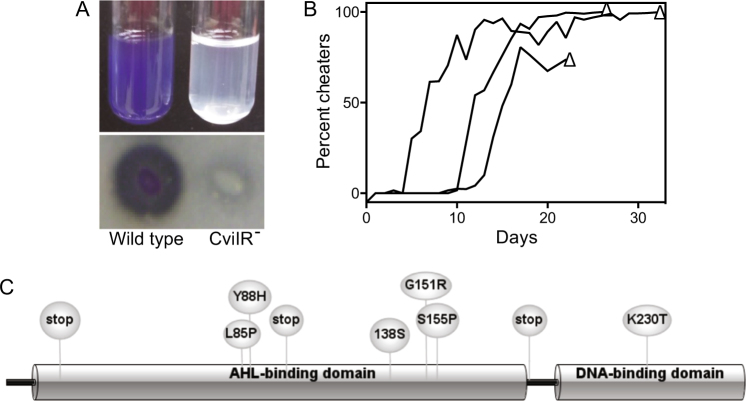
Fig. 1.
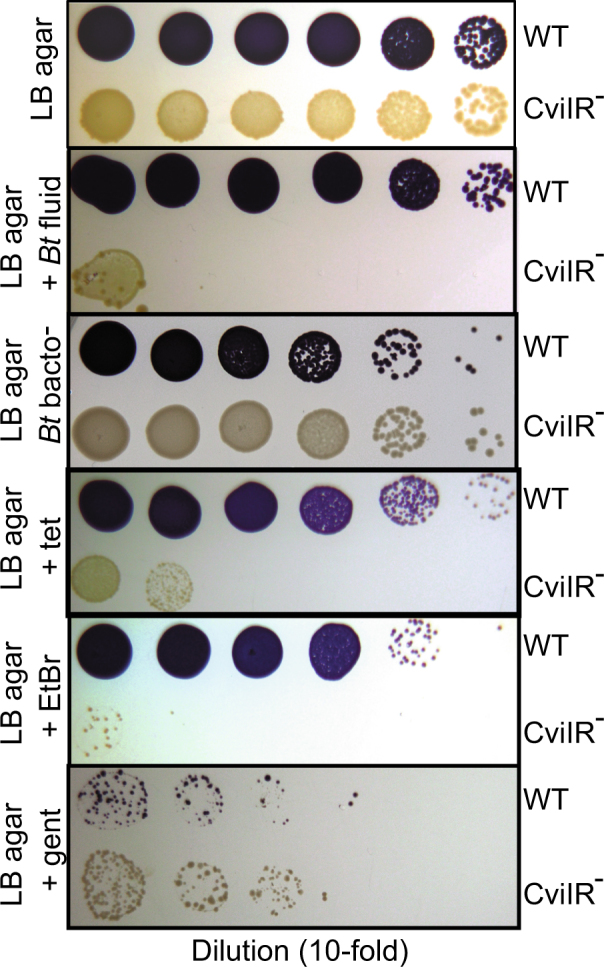
Quorum sensing increases C. violaceum resistance to antimicrobials. Growth of C. violaceum wild type (WT) or the AHL synthase, receptor double mutant (CviIR−). The wild type is purple due to production of the quorum sensing-dependent purple pigment violacein. Stationary-phase cultures were prepared in 10-fold serial dilutions in Luria-Bertani broth, and 10 μl of each successive dilution was spotted from left to right onto LB agar alone (top panel), LB agar containing 0.5% B. thailandensis culture fluid (as a source of bactobolin, see Materials and Methods), 1% culture fluid from the bactobolin-deficient B. thailandensis strain (BD20), 0.4 μg per ml tetracycline (tet), 60 μg per ml ethidium bromide (EtBr), or 8 μg per ml gentamicin (gent). Photos were taken after 24 h growth at 30°C and an additional 24 h growth at room temperature
Table 1.
Antimicrobial susceptibility of C. violaceum strains
| Cv strain | MICa | |||
|---|---|---|---|---|
| Bt fluidb (%) | Tet (μg ml−1) | EtBr (μg ml−1) | Gent (μg ml−1) | |
| Wild type | 1.6 (0.1) | 1.2 | >150 | 9.3 (1.2) |
| CviIR− | 0.6* | 0.4 (0.1)* | 60 | 9.3 (1.2) |
| CviI− | 0.5 (0.1)* | 0.4 (0.1)* | 60 | 7.7 (2.1) |
| CviI− + AHLs | 1.7 | 1.0* | 90 | NDc |
| CviIR− BRd | 3.3 (0.2)* | 2.5* | >150 | 10 |
| CviI− CdeR H127Y | 3.4 (0.1)* | 2.5* | >150 | 10 |
| CdeAB-OprM− | 0.3* | 0.1* | 30 | 8.7 (1.2) |
| CviIR− CdeAB-OprM− | 0.3* | 0.3* | ND | 8.7 (1.2) |
aThe minimum inhibitory concentration (MIC) of B. thailandensis culture fluid (Bt fluid), tetracycline (Tet), ethidium bromide (EtBr), or gentamicin (Gent) was determined by spotting ~104° C. violaceum stationary-phase cells of the indicated strain onto LB agar containing a range of antimicrobial concentrations as described in Materials and Methods. Results are the average of three independent experiments and the standard deviation is in parentheses when it was not zero. Statistical analysis by t-test compared with wild type under each treatment condition: *, p ≤ 0.0001. For ethidium bromide, no p-value was given because there was no variation (standard error is 0). For gentamicin, none of the averages significantly differed from wild type
bResults are from a single preparation of B. thailandensis fluid. Results with other preparations were similar. There were no observed growth defects in identical experiments with 3% culture fluid from a B. thailandensis bactobolin-deficient mutant
cND, not determined
dCviIR− BR refers to spontaneous bactobolin-resistant isolates. Two isolates were tested and results were identical
We also evaluated the importance of quorum sensing for resistance to other antimicrobials. We tested growth of our C. violaceum strains on plates supplemented with the antimicrobials tetracycline, gentamicin, or ethidium bromide (EtBr). Our results showed that quorum sensing increases C. violaceum resistance to tetracycline and EtBr but had no effect on gentamicin susceptibility (Fig. 1 and Table 1). Altogether our results support the conclusion that C. violaceum quorum sensing regulates resistance to several antimicrobials, including bactobolin. Our results are consistent with the idea that the quorum sensing-controlled resistance factor is an efflux pump. Antibiotic efflux pumps commonly confer resistance to a subset of antibiotics with unrelated mechanisms of action, and they are frequently associated with EtBr resistance [32].
Quorum sensing controls antimicrobial resistance through a putative efflux pump
To identify the genetic factors responsible for quorum sensing-dependent antibiotic resistance, we isolated spontaneous bactobolin-resistant mutants and defined the mutations leading to resistance using a whole-genome sequencing approach. We chose this approach because antibiotic resistance mechanisms are not well understood in C. violaceum [23], and because genetic mutations that increase resistance often occur through efflux pumps, providing a relatively simple approach to identify efflux pumps with specificity for bactobolin. We used a previously described approach [16], detailed in the Supplementary Information, to isolate two bactobolin-resistant variants in the otherwise sensitive CviIR− mutant. We assessed the antimicrobial susceptibility profiles of each variant relative to the parent strain. Results were identical for each variant, and are described in Table 1 (strain CviIR− BR). Both variants were more resistant than the parent to bactobolin, tetracycline, and EtBr, but there was no difference in resistance to gentamicin, similar to the quorum sensing-dependent antimicrobial resistance profile described above. To define the mutations involved in bactobolin resistance, we sequenced the genomes of our bactobolin-resistant variants using an Illumina platform (see Supplementary Information). The variants both had 11 identical single-nucleotide polymorphisms (SNPs) when compared to the sequenced strain CV017 [23]. Two of these were in putative antibiotic resistance genes. When we PCR-amplified and sequenced the two SNP sites, only one was mutated in both variants relative to CV026R. This was a C379T mutation in a predicted TetR-family transcriptional regulator CV017_15440, coding for the mutation H127Y. CV017_15440 is upstream and divergent to a 3-gene cluster with homology to resistance nodulation division (RND) efflux pumps in other Proteobacteria, such as mexAB-oprM from P. aeruginosa [33]. We named the TetR-family gene cdeR and the downstream genes cdeAB-oprM (Chromobacterium drug exporter).
We sought to determine whether the H127Y mutation in CdeR increased bactobolin resistance and whether cdeAB-oprM is responsible for the quorum-dependent antibiotic resistance phenotype we had observed. We introduced the allele encoding CdeR H127Y into the genome of the C. violaceum AHL synthase mutant at the native cdeR locus. The engineered CdeR H127Y mutant demonstrated the same antimicrobial susceptibility profile as the bactobolin-resistant variants (Table 1), confirming the increased antimicrobial resistance is due to the CdeR H127Y mutation. We subsequently deleted the cdeAB-oprM genes in the wild type and quorum sensing-defective CviIR− mutant. Deletion of cdeAB-oprM resulted in an approximately sixfold decrease in bactobolin resistance relative to the wild type and similar decreases in resistance for tetracycline and EtBr (Table 1). Importantly, the loss of quorum sensing did not further decrease antibiotic sensitivity of the CdeAB-OprM-deficient mutant (Table 1, compare the CdeAB-OprM− mutant to the CdeAB-OprM−, CviIR− mutant), supporting the idea that quorum sensing-dependent antibiotic resistance occurs through the CdeAB-OprM putative efflux pump. In support of this idea, both cdeA and cdeB transcripts were ~15-fold higher in wild-type cells compared with identically grown CviIR− mutant cells (Fig. 2), demonstrating that quorum sensing activates transcription of these genes. The genes were also activated in one of the spontaneous bactobolin-resistant variants, suggesting that increased resistance in this strain might have been due to increased transcription of the cdeAB-oprM genes.
Fig. 2.
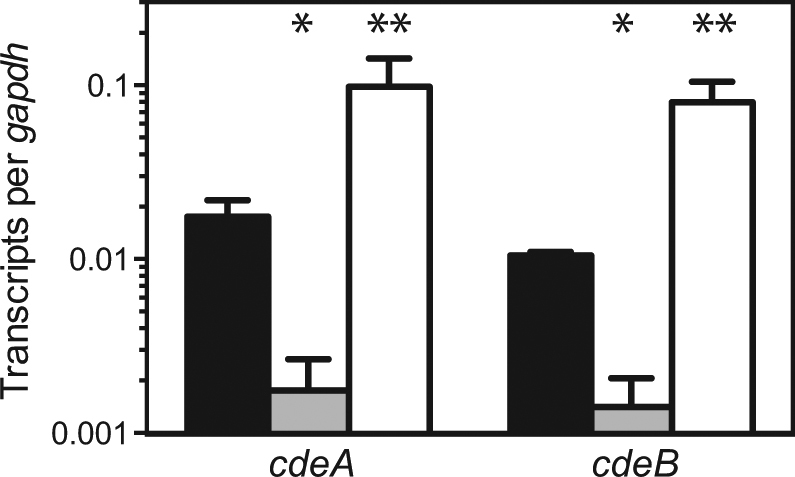
Quorum-sensing induction of cdeA and cdeB transcription. Quantitative digital PCR was used to quantify cdeA and cdeB transcripts in cells from stationary-phase cultures (optical density of 600 nm [OD600] of 4). Shown are cdeA and cdeB transcripts from wild type (WT, black bars), the AHL synthase, receptor double mutant (CviIR−, gray bars), and the bactobolin-resistant double mutant (CviIR− BR, white bars). In all cases, results were normalized to the housekeeping gene encoding glyceraldehyde-3-phosphatase dehydrogenase (gapdh). The values represent the average of three independent experiments and the error bars represent the standard deviation. Statistical analysis by t-test compared with wild type: *, p ≤ 0.005; **, p ≤ 0.05
Antibiotics suppress the emergence of C. violaceum cheaters during passage in casein broth
Because CdeAB-OprM is a predicted efflux pump, and efflux pumps are membrane-localized, we hypothesized that CdeAB-OprM might be a type of quorum-controlled private good that can restrain cheating during growth with antibiotics. Previous studies of P. aeruginosa and Vibrio cholerae demonstrated the necessity of secreted quorum sensing-dependent proteases for growth on casein as the sole carbon and energy source and, after prolonged passaging in these conditions, quorum sensing-deficient cheaters emerge [26, 27, 34]. Initially, we assessed the ability of wild type and a quorum sensing-deficient (CviIR−) C. violaceum mutant to grow in minimal casein broth and proteolyze skim milk protein (Fig. 3a). Wild type, but not the CviIR− mutant could grow on casein and form a zone of clearing on skim milk agar (Fig. 3a), consistent with the conclusion that a quorum-dependent protease is required for C. violaceum to grow on casein.
Fig. 3.
Protein degradation and cheater emergence during C. violaceum growth on protein. a Growth of C. violaceum wild type (WT) and the AHL synthase, receptor double mutant (CviIR−) after growth in minimal casein broth or on skim milk agar. Photos were taken after 24 h growth at 30°C. b Emergence of cheaters in C. violaceum populations passaged in casein broth. Cultures were transferred daily, and every 2–3 days, cheater abundance was determined just prior to subculture by dilution plating onto Luria-Bertani agar and enumerating the yellow colonies that also did not form a zone of proteolysis when patched onto skim milk agar plates. Each black line represents an independent experiment. In all three experiments, cheaters emerged after 2–10 days and their abundance rose to > 80% of the population, followed by termination of the experiment when there was no growth after transfer. c Location of nucleotide substitutions, deletions and insertions in C. violaceum cheaters mapped to the CviR amino-acid sequence. Each unique amino-acid substitution or early termination is shown in the illustration and indicated in Table S2. The AHL- and DNA-binding domains of CviR are indicated
Having established that C. violaceum requires quorum sensing to grow on casein, we assessed the emergence of quorum sensing-defective cheaters by passaging a C. violaceum population every day into fresh casein growth media and spread-plating dilutions of the culture onto LB agar every 2–3 days to enumerate the frequency of cheaters (see Materials and Methods). Cheaters were identified as yellow colonies (owing to loss of violacein production) that also did not form a zone of proteolysis when patched onto skim milk agar plates. We also sequenced the quorum-sensing receptor cviR in 10 of these putative mutants and in each case there was a mutation in cviR (Table S2 and Fig. 3c). In three independent experiments (shown as three separate black lines in Fig. 3b), cheaters emerged between 2 and 10 days. At a frequency of > 80% cheaters, which occurred between 22 and 33 days, the culture failed to grow after transfer due to a population collapse (Fig. 3b, open triangles). The emergence of quorum sensing receptor-mutated cheaters in the population and ultimate population collapse was similar to that previously reported for P. aeruginosa [6, 26, 27].
In P. aeruginosa, quorum sensing co-regulates the expression of a secreted protease and a cellular enzyme, nuh (nucleoside hydrolase), that promotes adenosine utilization, and growth on adenosine restrains the emergence of cheaters [6]. To test our hypothesis that antibiotics might similarly restrain cheater emergence in C. violaceum, we added tetracycline or purified bactobolin A to our casein growth experiments (Fig. 4). Wild-type cultures could be passaged serially with a maximum antibiotic concentration of either 2 μg ml−1 bactobolin A or 0.25 μg ml−1 tetracycline, which caused a maximum of a 1-log reduction of the final population yield relative to cultures grown with no antibiotic (Fig. S1). In the presence of either antibiotic, cheaters remained below the limit of detection (Fig. 4, dashed gray line) and the population yield remained consistent throughout the experiment (Fig. S1). However, it appeared that only antibiotic substrates of CdeAB-OprM were able to restrain cheaters, because cheaters emerged in populations grown with growth-inhibitive concentrations of gentamicin, which is not targeted by quorum sensing or CdeAB-OprM (Table 1). Interestingly, in the course of our experiments, populations grown with gentamicin never crashed as we observed in populations grown in the absence of any antibiotics (Fig. 4). These results might suggest a potentially unmeasurable quorum sensing-dependent effect on gentamicin resistance or an indirect effect of gentamicin, for example, through increased selection of mutations that increase cheater tolerance [35].
Fig. 4.
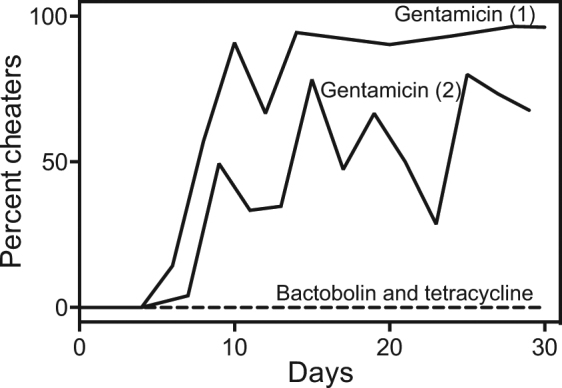
Antibiotics suppress C. violaceum cheaters in casein broth. Cultures were grown as described in Fig. 3 except the culture media was supplemented with antibiotics. The black lines show two independent experiments of populations passaged with 12 μg ml−1 gentamicin. The dashed gray line shows populations passaged with 2 μg ml−1 bactobolin A or 0.25 μg ml−1 tetracycline (two independent experiments were conducted with each antibiotic and results were identical in all cases). All antibiotic-treated populations were passaged in parallel with populations grown without antibiotics (partially represented in Fig. 3). In all cases, in populations grown without antibiotics cheaters emerged within 2–10 days and reached >80% of the population by the end of the experiment
Bactobolin restricts growth of quorum sensing-deficient C. violaceum mutants in dual-species cultures
The sensitivity of C. violaceum quorum sensing-defective mutants to bactobolin suggests these mutants may be growth-inhibited during direct competition with B. thailandensis. To test this hypothesis, we used our genetically engineered C. violaceum AHL synthase, receptor double mutant (CviIR−). Our approach was a modification of the methods used previously to grow C. violaceum-B. thailandensis cultures [14]. In our experiments, C. violaceum wild type and CviIR− mutant were mixed at a 1:1 ratio prior to combining with B. thailandensis (see Materials and Methods and [14]). As controls, we also grew the C. violaceum mixture without B. thailandensis or with a B. thailandensis bactobolin-deficient mutant. At the start and end of each experiment we determined the proportion of each C. violaceum strain by first spread-plating the entire C. violaceum population onto agar containing trimethoprim to select for C. violaceum and then enumerating the yellow (CviIR− mutant) colonies as a proportion of the total C. violaceum population (Fig. 5). During growth with B. thailandensis, the CviIR− mutant decreased from 50 to 17% of the total C. violaceum population. In contrast, when grown with no B. thailandensis or with the bactobolin-defective B. thailandensis, the CviIR− mutant population increased from 50% to more than 90% of the total C. violaceum population. These results show B. thailandensis-produced bactobolin selects against CviIR− mutants, supporting the idea that bactobolin stabilizes C. violaceum quorum sensing during competition.
Fig. 5.
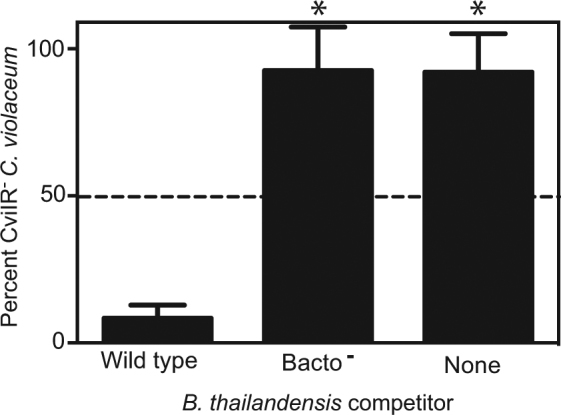
Competition increases the proportion of wild-type C. violaceum relative to quorum sensing-defective mutants. C. violaceum wild type or a quorum sensing-deficient mutant (CviIR−) were mixed at a 1:1 ratio then either combined with wild-type B. thailandensis or a bactobolin-deficient B. thailandensis mutant (bacto−, strain BD20), or grown with no B. thailandensis as described in the text. The percent CviIR− mutants within the C. violaceum population was determined by counting the yellow colonies as a percent of the total C. violaceum population isolated on selective agar. The dashed line indicates the initial percent CviIR−. Shown is the mean of four independent experiments and the error represents the standard deviation. Statistical analysis by t-test compared with wild type: *, p ≤ 0.0001
Because bactobolin increases the frequency of C. violaceum quorum-sensing cells (Fig. 5), and C. violaceum quorum sensing controls production of antimicrobials [14], we hypothesized that changes in the frequency of C. violaceum quorum-sensing cells would influence the outcome of competition with B. thailandensis. To test this hypothesis, we varied the starting ratio of wild type and CviIR− C. violaceum cells in our competition experiments with B. thailandensis. We used a C. violaceum starting ratio of 9:1 and 1:9 wild type to CviIR−, similar to the ratios observed at the end of the competition experiments described in Fig. 5. Our results showed that the higher frequency of wild-type C. violaceum correlated with lower B. thailandensis growth (Fig. 6). Thus altering the frequency of wild-type C. violaceum can influence the competitive ability of the C. violaceum population. Together, our results support the idea that antibiotic stabilization of quorum sensing can increase competitiveness.
Fig. 6.
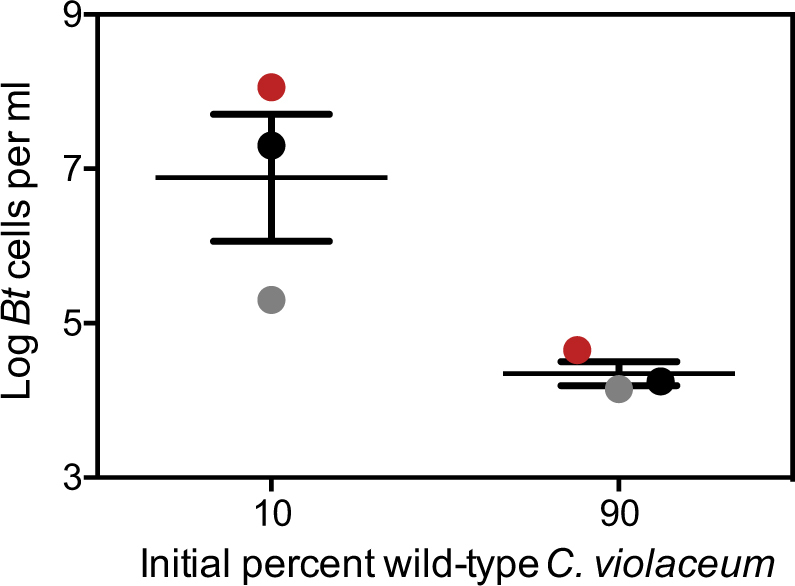
C. violaceum competitiveness correlates with the frequency of wild type relative to quorum sensing-deficient mutants. C. violaceum wild type or a quorum sensing-deficient mutant (CviIR−) cells were mixed together at a 1:9 or 9:1 ratio and then combined with wild-type B. thailandensis and grown for 24 h. The final B. thailandensis density was determined by selective plating and colony counts. The solid lines represent the means for each group. The vertical bars represent the standard deviation of each group. Statistical analysis by paired t-test compared with wild type: *, p = 0.0468. Paired groups are indicated by color and represent experiments performed in parallel on different days.
Discussion
In many Proteobacteria, quorum sensing activates dozens to hundreds of genes, including those coding for secreted or excreted public goods that are important for cooperation. Cooperating populations are threatened by quorum sensing-deficient cheaters, individuals utilizing the public goods without paying any cost for their production. Cheaters typically have a growth advantage over cooperators and their proliferation may ultimately cause the cooperative behavior to be lost. However, the maintenance of cooperation in natural populations suggests there are diverse mechanisms of cheater control across different bacterial groups. Indeed, several such mechanisms have been described, including kin selection through spatial structure [36, 37], mechanisms that allow cooperators to specifically recognize and interact with other cooperators (e.g., greenbeard traits) [38, 39], and pleiotropy, or co-regulation of genes coding for self-benefiting private activities and genes coding for freely shared public goods [6, 40]. Results of this study demonstrate a type of pleiotropy dependent on antibiotics produced by other species. Cheater control by other species has also been shown to occur through nutrient competition [41], and through selective grazing of cheaters by protists [42, 43]. Our study expands the list of known mechanisms of cheater control and provides additional support of the idea that cheaters can be controlled through interspecies or interkingdom interactions.
The results of this study demonstrate cheater control by antibiotics occurs through a quorum-controlled antibiotic efflux pump. What factors might contribute to maintenance of quorum-controlled resistance? The ecological or physiological factors that favor quorum-control of efflux pumps are as-yet unknown, however, it is unlikely that cheater control is sufficient for such selection [44]. Efflux pumps are known to have some fitness cost [45] thus using quorum sensing to control efflux pump production might mitigate fitness costs at low density as a means to optimize growth. Quorum sensing also commonly controls secreted toxin production, and quorum-sensing regulation of efflux pumps might serve to prevent self-toxicity. C. violaceum itself produces several antimicrobials, including the quorum sensing-regulated antibiotic violacein [17, 46]. However, in our experiments, CdeAB-OprM does not appear to prevent self-toxicity, as the quorum-defective variants are not growth-inhibited by the wild-type parent when the two strains are grown in co-culture, in fact, the mutants are more fit than the parent (Figs. 3 and 5).
Quorum sensing-dependent antibiotic resistance might also be important for competing with other strains or species in multispecies communities. Quorum sensing might be one of several systems involved in ‘competition sensing’ [47], providing an indirect measurement of the potential for competitors in the surrounding environment. As the population density increases, nutrients and space become limited and there is an increased potential for conflicts with neighbors. Thus quorum-sensing systems might coordinate production of several factors, including antibiotic resistance determinants, which promote survival when competition is likely. Quorum sensing might also activate the production of efflux pumps when they directly detect competitors by sensing and responding to their signals through eavesdropping [14]. Broad signal-specificity receptors in some saprophytic species such as C. violaceum and P. aeruginosa might serve in this capacity [17, 48].
There is some evidence suggesting quorum-sensing control of antimicrobial resistance is not limited to C. violaceum. Quorum sensing has also been reported to control resistance to tobramycin in biofilm-grown P. aeruginosa [49]. In Escherichia coli, a quorum-sensing receptor increases resistance to quinolones and several other antibiotics [50, 51]. Quorum sensing also activates expression of genes predicted to encode multidrug efflux pumps in other Proteobacteria [28–31], which may have important implications in treating infections if these antibiotics contribute to the selection or maintenance of quorum sensing or ‘rewiring’ of quorum-sensing systems during infections [52].
Our results indicate quorum sensing in C. violaceum controls antibiotic resistance through the CdeAB-OprM putative efflux pump. The genes encoding CdeAB-OprM share sequence and structural similarity to other RND efflux pump genes. RND efflux pumps have three proteins, an inner membrane transporter (efflux) protein (CdeB), a periplasmic accessory protein (CdeA), and an outer membrane channel (OprM) (for a review, see ref. [32]. RND efflux pump genes are frequently linked to a TetR-family regulator gene, and in the case of CdeAB-OprM, this appears to be CdeR. TetR-family members typically act as repressors by binding to a site in the promoter of the first efflux pump gene through a N-terminal DNA-binding domain. De-repression occurs when the C-terminal ligand-binding domain interacts with ligand (e.g., tetracycline). In many bacteria with RND efflux pumps, antibiotic resistance mutations map to the efflux pump regulator, typically the N-terminal DNA-binding domain, and cause de-repression (reviewed in [32]). The H127Y mutation in CdeR is in the N-terminal domain and might similarly affect CdeR, causing de-repression of cdeAB-oprM, although the role of CdeR as a repressor has not been experimentally determined. The hypothesis that CviR directly regulates cdeR, cdeA, or both can be tested experimentally, and such studies might provide insight into the mechanism of efflux pump regulation by quorum sensing in C. violaceum and other bacteria.
Electronic supplementary material
Acknowledgements
We thank Nikolas Skye Robins, Amy Schaefer, Ben Kerr, Chris Waters, Ajai Dandekar. and John Henry Kimbrough for helpful discussions and Stuart Macdonald for technical expertize. This work was supported by startup funds from the University of Kansas to JRC and a NIH COBRE Center for Molecular Analysis of Disease Pathways Research Project Award to JRC (P20GM103638). KCE was supported by the NIH Chemical Biology Training program (T32 GM08545). LC was supported by the NIH Post-Baccalaureate Research Education program (R25GM078441). EBN was supported by a KU Undergraduate Research Award. BN was supported by the NIH KU Legacy Chemical Methodologies and Library Development program (R24GM111385) and the COBRE CMADP Chemical Biology Core (P20GM103638 and P20GM113117). XW was supported by the NIH K-INBRE program (P20GM103418). We thank the COBRE CMADP Genome Sequencing Core (P20GM103638) for library preparation and the K-INBRE Bioinformatics Core (P20GM103418) for providing help with mutant analysis. The content in this manuscript is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
The online version of this article (10.1038/s41396-018-0047-7) contains supplementary material, which is available to authorized users.
References
- 1.Fuqua WC, Winans SC, Greenberg EP. Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J Bacteriol. 1994;176:269–75. doi: 10.1128/jb.176.2.269-275.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Papenfort K, Bassler BL. Quorum sensing signal-response systems in Gram-negative bacteria. Nat Rev Microbiol. 2016;14:576–88. doi: 10.1038/nrmicro.2016.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schuster M, Sexton DJ, Diggle SP, Greenberg EP. Acyl-homoserine lactone quorum sensing: from evolution to application. Annu Rev Microbiol. 2013;67:43–63. doi: 10.1146/annurev-micro-092412-155635. [DOI] [PubMed] [Google Scholar]
- 4.West SA, Griffin AS, Gardner A, Diggle SP. Social evolution theory for microorganisms. Nat Rev Microbiol. 2006;4:597–607. doi: 10.1038/nrmicro1461. [DOI] [PubMed] [Google Scholar]
- 5.Ghoul M, Griffin AS, West SA. Toward an evolutionary definition of cheating. Evolution. 2014;68:318–31. doi: 10.1111/evo.12266. [DOI] [PubMed] [Google Scholar]
- 6.Dandekar AA, Chugani S, Greenberg EP. Bacterial quorum sensing and metabolic incentives to cooperate. Science. 2012;338:264–6. doi: 10.1126/science.1227289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cabrol S, Olliver A, Pier GB, Andremont A, Ruimy R. Transcription of quorum-sensing system genes in clinical and environmental isolates of Pseudomonas aeruginosa. J Bacteriol. 2003;185:7222–30. doi: 10.1128/JB.185.24.7222-7230.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campbell ME, Farmer SW, Speert DP. New selective medium for Pseudomonas aeruginosa with phenanthroline and 9-chloro-9-[4-(diethylamino)phenyl]-9,10-dihydro-10- phenylacridine hydrochloride (C-390) J Clin Microbiol. 1988;26:1910–2. doi: 10.1128/jcm.26.9.1910-1912.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pierson LS, 3rd, Keppenne VD, Wood DW. Phenazine antibiotic biosynthesis in Pseudomonas aureofaciens 30-84 is regulated by PhzR in response to cell density. J Bacteriol. 1994;176:3966–74. doi: 10.1128/jb.176.13.3966-3974.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duerkop BA, Varga J, Chandler JR, Peterson SB, Herman JP, Churchill ME, et al. Quorum-sensing control of antibiotic synthesis in Burkholderia thailandensis. J Bacteriol. 2009;191:3909–18. doi: 10.1128/JB.00200-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seyedsayamdost MR, Chandler JR, Blodgett JA, Lima PS, Duerkop BA, Oinuma K, et al. Quorum-sensing-regulated bactobolin production by Burkholderia thailandensis E264. Org Lett. 2010;12:716–9. doi: 10.1021/ol902751x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.An D, Danhorn T, Fuqua C, Parsek MR. Quorum sensing and motility mediate interactions between Pseudomonas aeruginosa and Agrobacterium tumefaciens in biofilm cocultures. Proc Natl Acad Sci USA. 2006;103:3828–33. doi: 10.1073/pnas.0511323103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smalley NE, An D, Parsek MR, Chandler JR, Dandekar AA. Quorum sensing protects Pseudomonas aeruginosa against cheating by other species in a laboratory coculture model. J Bacteriol. 2015;197:3154–9. doi: 10.1128/JB.00482-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chandler JR, Heilmann S, Mittler JE, Greenberg EP. Acyl-homoserine lactone-dependent eavesdropping promotes competition in a laboratory co-culture model. ISME J. 2012;6:2219–28. doi: 10.1038/ismej.2012.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Amunts A, Fiedorczuk K, Truong TT, Chandler J, Peter Greenberg E, Ramakrishnan V. Bactobolin A binds to a site on the 70S ribosome distinct from previously seen antibiotics. J Mol Biol. 2015;427:753–5. doi: 10.1016/j.jmb.2014.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chandler JR, Truong TT, Silva PM, Seyedsayamdost MR, Carr G, Radey M et al. Bactobolin resistance is conferred by mutations in the L2 ribosomal protein. mBio. 2012;3. [DOI] [PMC free article] [PubMed]
- 17.McClean KH, Winson MK, Fish L, Taylor A, Chhabra SR, Camara M, et al. Quorum sensing and Chromobacterium violaceum: exploitation of violacein production and inhibition for the detection of N-acylhomoserine lactones. Microbiology. 1997;143:3703–11. doi: 10.1099/00221287-143-12-3703. [DOI] [PubMed] [Google Scholar]
- 18.Lichstein HC, Van De Sand VF. Violcein, an antibiotic pigment produced by Chromobacterium violaceum. J Infect Dis. 1945;76:47–51. doi: 10.1093/infdis/76.1.47. [DOI] [Google Scholar]
- 19.Conlin PL, Chandler JR, Kerr B. Games of life and death: antibiotic resistance and production through the lens of evolutionary game theory. Curr Opin Microbiol. 2014;21:35–44. doi: 10.1016/j.mib.2014.09.004. [DOI] [PubMed] [Google Scholar]
- 20.Brett PJ, DeShazer D, Woods DE. Burkholderia thailandensis sp. nov., a Burkholderia pseudomallei-like species. Int J Syst Bacteriol. 1998;48:317–20. doi: 10.1099/00207713-48-1-317. [DOI] [PubMed] [Google Scholar]
- 21.Wells JS, Trejo WH, Principe PA, Bush K, Georgopapadakou N, Bonner DP, et al. SQ 26,180, a novel monobactam. I Taxonomy, fermentation and biological properties. J Antibiot (Tokyo) 1982;35:184–8. doi: 10.7164/antibiotics.35.184. [DOI] [PubMed] [Google Scholar]
- 22.Chernin LS, Winson MK, Thompson JM, Haran S, Bycroft BW, Chet I, et al. Chitinolytic activity in Chromobacterium violaceum: substrate analysis and regulation by quorum sensing. J Bacteriol. 1998;180:4435–41. doi: 10.1128/jb.180.17.4435-4441.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang X, Hinshaw KC, Macdonald SJ, Chandler JR (2016). Draft genome sequence of Chromobacterium violaceum strain CV017. Genome Announc. 2016; 4. [DOI] [PMC free article] [PubMed]
- 24.Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene. 1998;212:77–86. doi: 10.1016/S0378-1119(98)00130-9. [DOI] [PubMed] [Google Scholar]
- 25.Chugani S, Greenberg EP. LuxR homolog-independent gene regulation by acyl-homoserine lactones in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2010;107:10673–8. doi: 10.1073/pnas.1005909107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Diggle SP, Griffin AS, Campbell GS, West SA. Cooperation and conflict in quorum-sensing bacterial populations. Nature. 2007;450:411–4. doi: 10.1038/nature06279. [DOI] [PubMed] [Google Scholar]
- 27.Sandoz KM, Mitzimberg SM, Schuster M. Social cheating in Pseudomonas aeruginosa quorum sensing. Proc Natl Acad Sci USA. 2007;104:15876–81. doi: 10.1073/pnas.0705653104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Majerczyk CD, Brittnacher MJ, Jacobs MA, Armour CD, Radey MC, Bunt R, et al. Cross-species comparison of the Burkholderia pseudomallei, Burkholderia thailandensis, and Burkholderia mallei quorum-sensing regulons. J Bacteriol. 2014;196:3862–71. doi: 10.1128/JB.01974-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O’Grady EP, Viteri DF, Malott RJ, Sokol PA. Reciprocal regulation by the CepIR and CciIR quorum sensing systems in Burkholderia cenocepacia. BMC Genomics. 2009;10:441. doi: 10.1186/1471-2164-10-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ramachandran R, Burke AK, Cormier G, Jensen RV, Stevens AM. Transcriptome-based analysis of the Pantoea stewartii quorum-sensing regulon and identification of EsaR direct targets. Appl Environ Microbiol. 2014;80:5790–5800. doi: 10.1128/AEM.01489-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schuster M, Lostroh CP, Ogi T, Greenberg EP. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J Bacteriol. 2003;185:2066–79. doi: 10.1128/JB.185.7.2066-2079.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Piddock LJ. Clinically relevant chromosomally encoded multidrug resistance efflux pumps in bacteria. Clin Microbiol Rev. 2006;19:382–402. doi: 10.1128/CMR.19.2.382-402.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poole K, Tetro K, Zhao Q, Neshat S, Heinrichs DE, Bianco N. Expression of the multidrug resistance operon mexA-mexB-oprM in Pseudomonas aeruginosa: mexR encodes a regulator of operon expression. Antimicrob Agents Chemother. 1996;40:2021–8. doi: 10.1128/AAC.40.9.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Katzianer DS, Wang H, Carey RM, Zhu J. “Quorum Non-Sensing”: social cheating and deception in Vibrio cholerae. Appl Environ Microbiol. 2015;81:3856–62. doi: 10.1128/AEM.00586-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Asfahl KL, Walsh J, Gilbert K, Schuster M. Non-social adaptation defers a tragedy of the commons in Pseudomonas aeruginosa quorum sensing. ISME J. 2015;9:1734–46. doi: 10.1038/ismej.2014.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kummerli R, Griffin AS, West SA, Buckling A, Harrison F. Viscous medium promotes cooperation in the pathogenic bacterium Pseudomonas aeruginosa. Proc Biol Sci. 2009;276:3531–8. doi: 10.1098/rspb.2009.0861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mund A, Diggle SP, Harrison F (2017). The fitness of Pseudomonas aeruginosa quorum sensing signal cheats is influenced by the diffusivity of the environment. mBio. 2017;8. [DOI] [PMC free article] [PubMed]
- 38.Sinervo B, Chaine A, Clobert J, Calsbeek R, Hazard L, Lancaster L, et al. Self-recognition, color signals, and cycles of greenbeard mutualism and altruism. Proc Natl Acad Sci USA. 2006;103:7372–7. doi: 10.1073/pnas.0510260103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Veelders M, Bruckner S, Ott D, Unverzagt C, Mosch HU, Essen LO. Structural basis of flocculin-mediated social behavior in yeast. Proc Natl Acad Sci USA. 2010;107:22511–6. doi: 10.1073/pnas.1013210108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Foster KR, Shaulsky G, Strassmann JE, Queller DC, Thompson CR. Pleiotropy as a mechanism to stabilize cooperation. Nature. 2004;431:693–6. doi: 10.1038/nature02894. [DOI] [PubMed] [Google Scholar]
- 41.Celiker H, Gore J. Competition between species can stabilize public-goods cooperation within a species. Mol Syst Biol. 2012;8:621. doi: 10.1038/msb.2012.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Friman VP, Buckling A. Effects of predation on real-time host-parasite coevolutionary dynamics. Ecol Lett. 2013;16:39–46. doi: 10.1111/ele.12010. [DOI] [PubMed] [Google Scholar]
- 43.Jousset A, Rochat L, Pechy-Tarr M, Keel C, Scheu S, Bonkowski M. Predators promote defence of rhizosphere bacterial populations by selective feeding on non-toxic cheaters. ISME J. 2009;3:666–74. doi: 10.1038/ismej.2009.26. [DOI] [PubMed] [Google Scholar]
- 44.Schuster M, Sexton DJ, Hense BA. Why quorum sensing controls private goods. Front Microbiol. 2017;8:885. doi: 10.3389/fmicb.2017.00885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sanchez P, Linares JF, Ruiz-Diez B, Campanario E, Navas A, Baquero F, et al. Fitness of in vitro selected Pseudomonas aeruginosa nalB and nfxB multidrug resistant mutants. J Antimicrob Chemother. 2002;50:657–64. doi: 10.1093/jac/dkf185. [DOI] [PubMed] [Google Scholar]
- 46.Duran N, Menck CF. Chromobacterium violaceum: a review of pharmacological and industiral perspectives. Crit Rev Microbiol. 2001;27:201–22. doi: 10.1080/20014091096747. [DOI] [PubMed] [Google Scholar]
- 47.Cornforth DM, Foster KR. Competition sensing: the social side of bacterial stress responses. Nat Rev Microbiol. 2013;11:285–93. doi: 10.1038/nrmicro2977. [DOI] [PubMed] [Google Scholar]
- 48.Lee JH, Lequette Y, Greenberg EP. Activity of purified QscR, a Pseudomonas aeruginosa orphan quorum-sensing transcription factor. Mol Microbiol. 2006;59:602–9. doi: 10.1111/j.1365-2958.2005.04960.x. [DOI] [PubMed] [Google Scholar]
- 49.Bjarnsholt T, Jensen PO, Burmolle M, Hentzer M, Haagensen JA, Hougen HP, et al. Pseudomonas aeruginosa tolerance to tobramycin, hydrogen peroxide and polymorphonuclear leukocytes is quorum-sensing dependent. Microbiology. 2005;151:373–83. doi: 10.1099/mic.0.27463-0. [DOI] [PubMed] [Google Scholar]
- 50.Rahmati S, Yang S, Davidson AL, Zechiedrich EL. Control of the AcrAB multidrug efflux pump by quorum-sensing regulator SdiA. Mol Microbiol. 2002;43:677–85. doi: 10.1046/j.1365-2958.2002.02773.x. [DOI] [PubMed] [Google Scholar]
- 51.Wei Y, Lee JM, Smulski DR, LaRossa RA. Global impact of sdiA amplification revealed by comprehensive gene expression profiling of Escherichia coli. J Bacteriol. 2001;183:2265–72. doi: 10.1128/JB.183.7.2265-2272.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Feltner JB, Wolter DJ, Pope CE, Groleau MC, Smalley NE, Greenberg EP et al (2016). LasR variant cystic fibrosis isolates reveal an adaptable quorum-sensing hierarchy in Pseudomonas aeruginosa. mBio. 2016;7. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.