Abstract
Microorganisms adapt to frequent environmental changes through population diversification. Previous studies demonstrated phenotypic diversity in a clonal population and its important effects on microbial ecology. However, the dynamic changes of phenotypic composition have rarely been characterized. Also, cellular variations and environmental factors responsible for phenotypic diversity remain poorly understood. Here, we studied phenotypic diversity driven by metabolic heterogeneity. We characterized metabolic activities and growth kinetics of starved Escherichia coli cells subject to nutrient upshift at single-cell resolution. We observed three subpopulations with distinct metabolic activities and growth phenotypes. One subpopulation was metabolically active and immediately grew upon nutrient upshift. One subpopulation was metabolically inactive and non-viable. The other subpopulation was metabolically partially active, and did not grow upon nutrient upshift. The ratio of these subpopulations changed dynamically during starvation. A long-term observation of cells with partial metabolic activities indicated that their metabolism was later spontaneously restored, leading to growth recovery. Further investigations showed that oxidative stress can induce the emergence of a subpopulation with partial metabolic activities. Our findings reveal the emergence of metabolic heterogeneity and associated dynamic changes in phenotypic composition. In addition, the results shed new light on microbial dormancy, which has important implications in microbial ecology and biomedicine.
Introduction
Microorganisms occupy virtually every niche on earth, most of which are scarce in nutrients. The lifestyle of microorganisms can be well characterized by long periods of nutrient deprivation intercepted by short periods of nutrient excess [1]. Population diversification is an important mechanism for populations to adapt to fluctuating environments [2, 3]; with diversity, there will likely be some individuals that are well suited for a given environment. Previous studies characterized how genetic composition in a population changes slowly through mutations and becomes diverse in environments where nutrients are limited and fluctuate [4–11]. In recent years, it became clear that a genetically identical population can also diversify phenotypically [12–23]. Phenotypic diversity can have significant effects on ecological dynamics of populations and species [24]; for example, it has a critical role in population survival through catastrophic environmental changes [25], promoting sustenance of microbial species [26, 27]. Because phenotypic diversity does not involve genetic mutation, it is expected to arise on short timescales, leading to dynamic changes in phenotypic composition in a population. However, these temporal dynamics have rarely been quantified. Furthermore, cellular variations responsible for phenotypic diversity and environmental factors triggering such cellular variations have not been well characterized.
Metabolism is a central process by which cells derive components essential for basic cellular functions. Cell-to-cell variation in metabolism, if it exists, could result in phenotypic diversity. Recent studies of stochastic gene expression are supportive of the intriguing possibility of metabolic heterogeneity. For example, studies found that genetically identical cells in the same environment may produce different amounts of metabolically relevant proteins [28–31]. Recent computational work suggested that such different protein expression could give rise to metabolic heterogeneity in E. coli cells [32]. A network model based on stochastic expression of lac enzymes in E. coli cells showed how stochastic gene expression could affect carbon metabolism [33]. Similarly, the direct measurements of metabolites in carbon metabolism revealed the coupling between metabolite pools and gene expression [34, 35]. Furthermore, a recent experimental study showed isogenic Klebsiella oxytoca cells might exhibit different N2 fixation rates, meaning different metabolic activities [36].
In this study, by examining starved E. coli cells subject to nutrient upshift, we characterized the emergence of metabolic heterogeneity and its effect on phenotypic composition in a population. Metabolism can be largely divided into three processes: (i) bringing extracellular substrates into the cytoplasm (substrate uptake), (ii) breaking down the substrates into smaller units (catabolism), and (iii) constructing macromolecules from the small units (anabolism). By visualizing accumulation/depletion of fluorescently labeled substrates and production of fluorescent proteins in individual cells, we characterized these three metabolic processes at single-cell resolution. The results revealed that there exists significant cell-to-cell heterogeneity in these processes, and that this heterogeneity leads to diverse growth phenotypes, including dormancy. Also, we found that oxidative stress can induce metabolic heterogeneity and diverse growth phenotypes.
Results
Cell-to-cell heterogeneity in metabolic activities and growth phenotypes
In nature, microorganisms are often starved of carbon [1]. Numerous studies have reported that when environmental microbial samples were plated on agar plates containing rich nutrients (e.g., LB), many cells did not form colonies [37, 38]. Known as “the great plate count anomaly”, this observation is a long-standing enigma in microbial ecology, largely because cellular states (e.g., metabolic states) of those cells that failed to form colonies are unclear and under intense debate [39–41]. When we performed a similar plating assay using carbon-starved cultures under well-controlled laboratory conditions, we made the same observation. We grew E. coli cells in minimal medium with glucose and ammonium as the sole carbon and nitrogen sources, and suspended them in medium without glucose (starvation medium) at the OD600 of ~0.4; upon suspension, cell growth stopped immediately (Supplementary Fig. 1). At different times during carbon starvation, we took a 100 μl aliquot of the culture, diluted it (by 103 – 6-fold), and plated it on LB agar plates; this exposure to LB represents nutrient upshift. As quantified in Fig. 1a, the number of colony-forming units decreased over time. This decrease indicates that with longer starvation, fewer cells resumed growth upon nutrient upshift. Please see Methods section for details and Supplementary Fig. 2 for a graphic illustration of our experimental procedure.
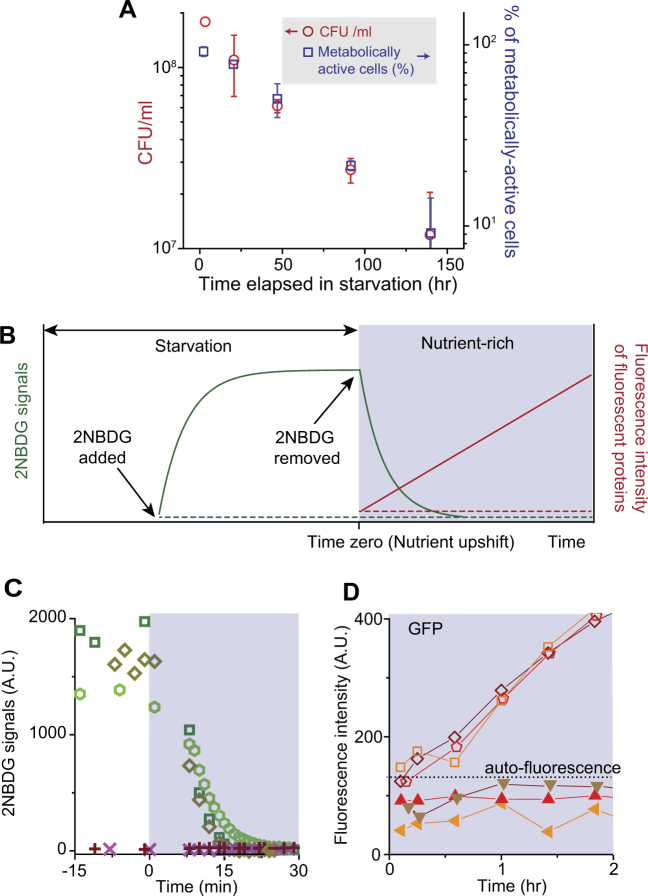
Fig. 1.
Metabolic activities and growth of cells upon nutrient upshift. a We performed a conventional plate assay. E. coli cells were deprived of carbon (glucose) at time zero. At various time points during starvation, a fixed volume of the starved culture was taken, diluted appropriately, and spread on LB agar plates; see Supplementary Methods for details. The number of colonies was counted after 24 h of incubation, and colony-forming unit (CFU)/ml was then determined. CFU/ml decreased over time (red circles, left axis). We also plotted the percentage of metabolically active cells in a population (blue squares, right axis); a detailed description of the metabolically active cells is provided below, in the caption to c, d. Note that the scales of the left and right axes have the same fold change. CFU/ml and the percentage of metabolically active cells decreased similarly over time. The error bars indicate one standard deviation, which was obtained from at least two biological replicates. b We determined (1) the substrate uptake activity by intracellular accumulation of fluorescently labeled substrates, 2NBDG, (2) the catabolic activity by intracellular depletion of 2NBDG upon inhibition of its transport, and (3) the anabolic activity by de novo synthesis of green fluorescent proteins (GFP) or red fluorescent proteins (mCherry). When 2NBDG is available externally, metabolically active cells would transport them, exhibiting strong intracellular 2NBDG signals (solid green curve in the white region). Active catabolism rapidly decomposes 2NBDG molecules into non-fluorescent metabolites [44, 47]. Therefore, intracellular 2NBDG concentration is determined by both transport and catabolic activities, as described by Eq. (S1)–(S3) in Supplementary Note 1. When 2NBDG is removed from a culture, because 2NBDG uptake into cells would stop, intracellular 2NBDG signals are expected to decrease in metabolically active cells (solid green curve in the shaded region); see Eq. (S4) in Supplementary Note 1. Also, these metabolically active cells are expected to express fluorescent proteins, GFP or mCherry, when the expression is induced (solid red line). Conversely, metabolically inactive cells are expected to exhibit neither 2NBDG uptake (dashed green line) nor production of fluorescent proteins (dashed red line). c, d We experimentally tested these expected patterns. We starved E. coli cells of carbon (glucose) for 2.3 h and subjected them to nutrient upshift; see text or Methods for details. We then monitored ~2500 cells in four biological repeats. We observed subpopulations with different metabolic activities. 91.7% (±5.9%, one standard deviation from biological replicates) of cells exhibited the pattern of active metabolism. These cells exhibited strong 2NBDG signals before nutrient upshift, but the signals decreased rapidly upon shift (Supplementary Movie). Typical changes in 2NBDG signals were plotted in c (open symbols). In all of these cells, GFP intensity increased, indicating de novo protein synthesis; see d (open symbols) for typical changes in GFP intensity. GFP intensity typically reached its saturation levels 3–4 h after nutrient upshift. We repeated this analysis by starving a culture for different periods of time and monitoring ~1500 cells in two independent experiments (i.e., biological repeats). We counted metabolically active cells and plotted their percentage in a (blue squares). We also observed cells that exhibited the patterns of inactive metabolism: neither 2NBDG uptake (crosses in c) nor production of fluorescent proteins (solid symbols in d). The details of these metabolically inactive cells were described in Supplementary Fig. 6. See Supplementary Table 1 for the number of cells with different metabolic activities
We sought to examine how metabolic activities of cells that grew were different from those that failed to grow. To measure metabolic activities of individual cells, we adopted the following approach, which was based on recent developments in single-cell-level fluorescence imaging [42, 43]. First, we determined which cells have positive substrate uptake activity by monitoring the accumulation of fluorescently labeled substrates in the cytoplasm. Glucose is the preferred carbon source for many microbes, including E. coli. When glucose molecules become available extracellularly, starved cells with positive substrate uptake activity will transport and accumulate them in the cytoplasm. This accumulation can be visualized using the fluorescently labeled glucose molecules, 2-N-7-nitrobenz-2-oxa-1,3-diazol-4-yl amino-2-deoxyglucose (2NBDG), which would result in strong fluorescence signals inside cells [44–48]; see the solid green curve in the white region in Fig. 1b (also, see Eq. (S2) for the expected functional form). When we incubated starved cells with 10 µM of 2NBDG for various durations, we were able to detect intracellular 2NBDG signals already with ~5 min incubation (Supplementary Fig. 3A); therefore, in our experiments below, to ensure the reliable detection of signals, we pre-incubated cells with 2NBDG for ~25 min (see Supplementary Fig. 3 caption for details). Conversely, cells with no transport activity are expected to exhibit no 2NBDG signals (Fig. 1b, dashed green line in the white region).
Active catabolism leads to the intracellular breakdown of transported substrates into smaller units. Hence, we determined which cells are catabolically active by monitoring decomposition of 2NBDG molecules. Previous molecular-level studies have shown that after 2NBDG molecules are transported into bacterial cells, active catabolism rapidly decomposes these 2NBDG molecules into non-fluorescent metabolites [47], while inactivation of catabolism abolishes decomposition [44]. When 2NBDG molecules are present in the environment, the intracellular 2NBDG concentration will depend on both transport and catabolic activities; this dependence is described by Eqs. (S1)–(S3) in Supplementary Note 1 and further discussed in Supplementary Fig. 3. If 2NBDG molecules are abruptly removed from the environment, due to the cessation of transport, intracellular 2NBDG concentration will decrease, as described by Eq. (S4) in Supplementary Note 1 and discussed in Supplementary Fig. 3. Here, we are particularly interested in knowing whether starved cells could readily catabolize nutrients when these nutrients became available. Thus, if we incubate starved cells with 2NBDG for 25 min (which would result in strong intracellular 2NBDG signals as observed in Supplementary Fig. 3) and suspend them in 2NBDG-free nutrient-rich medium, active catabolism would lead to a rapid decrease in intracellular 2NBDG signals (Fig. 1b, solid green curve in the shaded region).
Next, we determined protein anabolism by characterizing de novo synthesis of fluorescent proteins. We employed a strain in which the expression of green fluorescent protein (GFP) or red fluorescent proteins (mCherry) was driven by a synthetic promoter P tet (the repressor of P tet, TetR, is constitutively expressed in this strain) [49]. An inducer of the P tet promoter, aTc, diffuses into cells rapidly [50], activating protein expression. Cells with active anabolism would synthesize fluorescent proteins, leading to an increase in fluorescence intensity (Fig. 1b, solid red line in the shaded region). With inactive anabolism, fluorescence intensity would not increase (Fig. 1b, dashed red line). We note that there exists a study that characterized protein anabolism during starvation using a similar approach [51]. Conversely, we are interested in anabolism in cells after nutrients become available. Thus, our study focuses on protein anabolism after nutrient upshift (post-starvation).
Next, we show experimental data confirming these predicted patterns for metabolically active or inactive cells. Following the procedure described above, we starved cells of glucose for 2.3 h (at OD600 of 0.4), spread ~5 µl aliquot on LB agarose plates (i.e., nutrient upshift), and monitored ~2500 cells using time-lapse microscopy in four independent experiments. 91.7 (±5.9)% of cells showed the predicted pattern of active metabolism. First, when 2NBDG molecules became available extracellularly during starvation, these cells accumulated them intracellularly, exhibiting strong 2NBDG signals; compare the solid green curve in Fig. 1b and Supplementary Fig. 3A. When we subjected the starved cells to nutrient upshift (i.e., LB medium containing no 2NBDG), their 2NBDG signals decreased rapidly and became undetectable within 20 min (Supplementary Movie). In Fig. 1c, we quantified 2NBDG signals for a few cells (open symbols), which again showed strong signals before and an immediate decrease after nutrient upshift: ~100-fold decrease in 30 min. This rapid decrease cannot be accounted for by dilution due to cell growth; even for cells that are actively growing in LB medium (whose doubling time is ~20 min), the dilution would lead to ~3-fold decrease over 30 min. Rather, the rapid decrease in 2NBDG signals indicates active 2NBDG degradation. This decrease in 2NBDG signals upon nutrient upshift agreed with the predicted pattern; compare Fig. 1c (open symbols) and Fig. 1b (solid green curve in the shaded region), indicating active substrate uptake and catabolism. The re-plot of 2NBDG signals in a semi-log scale indicates that this decrease is exponential (Supplementary Fig. 4), as predicted by Eq. (S4). Furthermore, the activation of protein production (using aTc as described above) led to an increase in fluorescence intensity (open symbols in Fig. 1d), agreeing with the predicted pattern from active anabolism (solid red line in the shaded region in Fig. 1b). All of these metabolically active cells recovered their growth within 100 min after nutrient upshift (see Supplementary Fig. 5 for the distribution of growth recovery time), indicating that active metabolism is associated with active cell growth.
We also observed cells showing the predicted patterns of metabolic inactivity. These cells exhibited neither 2NBDG uptake (compare dashed green line in Fig. 1b and crosses in Fig. 1c) nor production of fluorescent proteins (compare dashed red line in Fig. 1b and solid symbols in Fig. 1d). None of these cells resumed growth after nutrient upshift. These cells either had diffuse cell boundaries (i.e., loss of refractivity) or were stained by propidium iodide (PI), which are signs of the loss of viability. These cells were described in greater details in Supplementary Fig. 6.
Our observation above was made using cells starved for 2.3 h. When we starved cells for different durations and counted the number of metabolically active cells, we found that their percentage decreased monotonically (blue squares in Fig. 1a). Interestingly, this decrease was very similar to the decrease in the number of colony-forming units obtained from conventional plate assays above (compare red circles and blue squares in Fig. 1a), further supporting that active metabolism is a good indicator for a growth phenotype. The percentage of metabolically inactive non-viable cells was plotted in Supplementary Fig. 6B, which showed a monotonic increase with longer starvation periods.
Emergence of dormant cells with partial metabolic activities
In cultures starved longer, we observed the emergence of a subpopulation with a unique metabolic state. Cells in this subpopulation showed high 2NBDG intensity before nutrient upshift and a rapid decrease in intensity after upshift (open triangles in Fig. 2a), exhibiting the predicted pattern of active substrate uptake and catabolism. However, they did not produce fluorescent proteins. Thus, metabolically, they were partially active. Above, we described that metabolically active cells resumed growth within 100 min after nutrient upshift. In contrast, these partially active cells did not resume growth in this time window; see Supplementary Fig. 7 for an exemplary image sequence. They exhibited clear cell boundaries and were not stained by PI, suggesting that they were viable. Their percentage was initially very low and peaked after ~2 days of starvation (Fig. 2b).
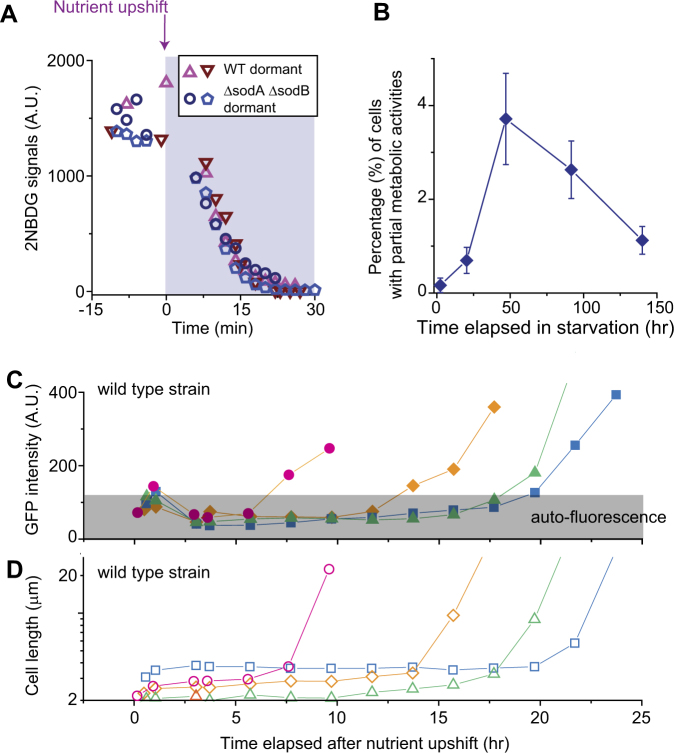
Fig. 2.
Characterization of metabolically partially active, dormant cells. a, b We starved cells of glucose for different periods of time and monitored ~1500 cells in two biological replicates. We observed dormant cells exhibiting partial metabolic activities; see text for detail. The number of these cells varied depending on the duration of starvation and was always lower than 5% of a population (b; the error bars indicate one standard deviation from two biological replicates). In all of these cells, 2NBDG signals decreased immediately upon nutrient upshift. In a, we plotted exemplary changes in 2NBDG signals for wild-type (WT) cells starved for 3 days (triangles) and ΔsodAΔsodB cells starved for 8 h (other symbols); the data showed a typical, rapid decrease in 2NBDG signals upon nutrient upshift. c, d We subjected a 3-day-starved culture to nutrient upshift. Dormant cells did not produce fluorescent proteins for several hours or a day; see the initial plateau in the fluorescence intensity c. Fluorescence intensity increased suddenly at later times c, indicating the spontaneous recovery of protein anabolism. This recovery was associated with growth resumption d. Note that we used ampicillin to avoid unrestricted growth of metabolically active cells (see text). With ampicillin, cells elongate without division until they eventually lyse [54], which is why cells appear unusually long in the plot. Out of ~4500 cells monitored, we observed 24 cells exhibiting such late recoveries. We observed similar fractions in two biological repeats
We wondered whether these cells were dormant. Dormancy refers to a reversible non-growth state [52], meaning that dormant cells can later revert back to a growth state. Thus, we wished to monitor these cells for longer periods of time. However, the metabolically active cells that initiated growth early multiplied exponentially and overwhelmed the microscope field of view within a few hours (Supplementary Fig. 8). Thus, a long-term observation of non-growing cells with partial metabolic activities was challenging. To overcome this issue, we used an antibiotic, ampicillin, which selectively kills growing cells [53]. With ampicillin, growing cells elongate without dividing and eventually lyse [54]. Hence, the unrestricted exponential growth described above can be avoided. Our control experiments with and without ampicillin indicated no significant effects of ampicillin on our findings of metabolic activities (Supplementary Note 2). A long-term observation of the non-growing cells with partial metabolic activities showed no production of fluorescent proteins in them for several hours or a day after nutrient upshift; see the initial plateau in Fig. 2c. However, at later times, protein productions suddenly began (Fig. 2c), indicating spontaneous recovery of anabolic activities. This recovery was accompanied by growth resumption (Fig. 2d). Therefore, these metabolically partially active cells were dormant cells, and they were capable of reverting to a growth state through metabolic recovery. These data show a unique metabolic state associated with dormancy. Also, they reveal metabolic heterogeneity underlying heterogeneous growth phenotypes.
Taken together, our observations above indicate that a viable population can be divided into a metabolically active subpopulation and a partially active subpopulation. We determined the relative size of the latter subpopulation using the percentage of each subpopulation reported in Figs. 1a and 2b. The fraction of cells with partial metabolic activities was initially negligible, but increased with longer periods of starvation (Fig. 3a), revealing a dynamic change in phenotypic composition with starvation.
Fig. 3.
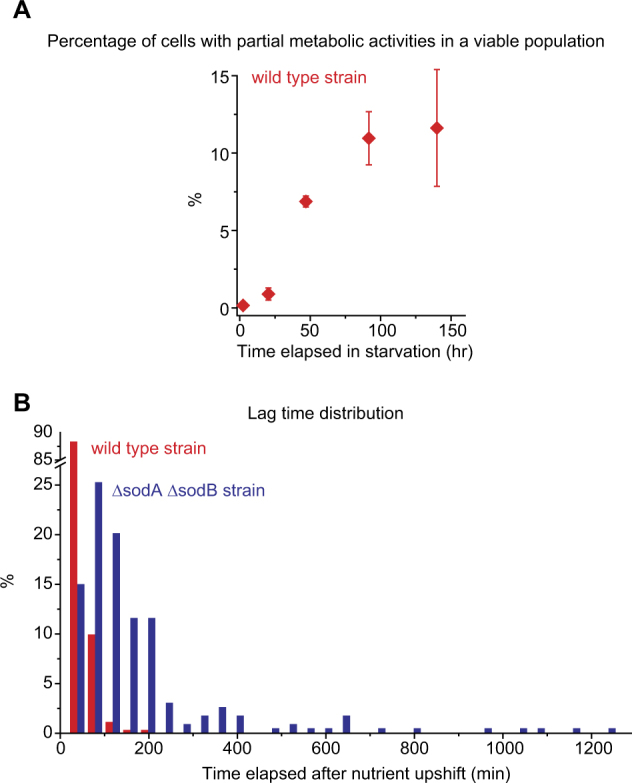
Effects of oxidative stress on the emergence of metabolically partially active, dormant cells. a Our observations indicated that a viable population could be divided into a metabolically active subpopulation and a partially active subpopulation. When we calculated the relative size of the partially active subpopulation using the percentage of each subpopulation reported in Figs. 1a and 2b, we observed that its size was initially negligible, but increased with longer periods of starvation. b We examined the metabolic activities and growth of oxidative-stress-prone cells (ΔsodAΔsodB); see text. We starved these cells and WT cells (as a control) for 8 h, subjected them to nutrient upshift, determined the time points at which cells resumed growth (lag time), and plotted the lag-time distribution. ~350 WT cells and ~250 stress-prone cells were analyzed. We performed two biological repeats and these distributions were reproducible. We re-plotted these distributions as scatter plots in a semi-log scale in Supplementary Fig. 13
Oxidative stress induces the emergence of dormant cells with partial metabolic activities
Starvation induces complex physiological responses in cells [1], and thus, the mechanistic determination of how starvation results in the emergence of a subpopulation with partial metabolic activities is a challenging task. However, the observation that the duration of starvation has significant effects on the size of this subpopulation may guide us to identify innate factors involved in the emergence of this subpopulation. Importantly, the results described above showed that these factors must lead to the loss of anabolic activity, but not substrate uptake and catabolic activities. Furthermore, this loss must be transient and its recovery lead to growth resumption. We next sought to find a potential factor that satisfies these specific requirements.
Oxidative stress is a universal stress that most aerobic organisms experience to varying degrees. Previous studies have shown that prolonged starvation inflicts oxidative stress to bacterial cells, and the stress leads to failure of cells to form visible colonies on nutrient-rich agar plates after overnight incubation [55–58]. These studies assumed that the failure to form visible colonies is due to viability loss. However, our observation above showed that prolonged starvation leads to the emergence of dormant cells, which can spontaneously resume growth at later times; thus they are unlikely to form visible colonies after overnight incubation. On the basis of these results, we wondered whether oxidative stress could cause the emergence of dormant cells through transient inactivation of anabolism, meaning it could be a potential factor that satisfies the specific requirements put forth above. Below, we present the data supporting this argument.
To test the effect of oxidative stress, we generated a strain prone to oxidative stress, and characterized its metabolic activities and growth resumption kinetics. The sodA and sodB genes in E. coli encode superoxide dismutases, enzymes that detoxify superoxide [59–62]. Deletion of these genes disables key defense mechanisms, and renders cells prone to oxidative stress, thereby amplifying the effect of oxidative stress; we refer to this double deletion mutant as the oxidative-stress-prone strain. As discussed above, starvation inflicts oxidative stress to cells [55–58]. In the case of wild-type (WT) cells, as previously described, we had to starve cells for ~2 days to see a noticeable increase in the number of cells with partial metabolic activities (Figs. 2b and 3a). However, in the oxidative-stress-prone strain, a shorter period of starvation was expected to be sufficient. Thus, we starved a stress-prone population of carbon for 8 h and then spread them on LB agarose plates. We indeed observed many dormant cells with partial metabolic activities. These cells exhibited strong 2NBDG signals and a rapid decrease in signals upon nutrient upshift (Fig. 2a), but did not produce fluorescent proteins (Supplementary Fig. 9A). Thus, these cells are metabolically partially active. Importantly, these cells began protein production spontaneously at later times (several hours or a day after nutrient upshift), and this production was accompanied by growth recovery (Supplementary Fig. 9B). Therefore, these cells with partial metabolic activities were dormant cells and could revert to a growth state.
To demonstrate the abundance of such dormant cells in the stress-prone population, we plotted the distribution of time points at which cells resumed growth, i.e., lag-time distribution (blue in Fig. 3b); as a control, we plotted the lag-time distribution for the WT population subjected to the same condition (red in Fig. 3b). The stress-prone cells exhibited a much wider distribution than WT cells; its tail stretched significantly farther, showing an increase in the fraction of dormant cells (see Supplementary Fig. 13 for further discussion). We quantified this tail by determining what fraction of cells in a population have the lag time >100 min. The fraction was more than 10-fold higher in the stress-prone population (63.6 ± 16.2%) compared to the WT population (4.6 ± 4.6 %); the error represents one standard deviation from two biological replicates. Next, we performed another experiment in which we exogenously induced oxidative stress in WT cells by using H2O2. As shown in Supplementary Fig. 10, the fraction of cells with the lag time >100 min increased markedly with H2O2 treatment, consistent with our finding above for the stress-prone strain.
We further probed the effects of oxidative stress using a conventional colony formation assay. We starved oxidative-stress-prone cells and WT cells for 8 h, plated them, and recorded the time at which individual colonies became visible to the naked eye. Then, we compared their time distributions of the colony formation. The distribution for the WT cells was narrow (Fig. 4, red columns), showing that all WT cells formed visible colonies within ~12 h after plating. The distribution for the oxidative-stress-prone cells was much wider, with its tail stretching to ~ 50 h (Fig. 4, blue columns); this means that some cells formed visible colonies ~50 h after plating. This long tail in the time distribution agrees with our single-cell-level data above. Collectively, these data reveal that oxidative stress leads to the emergence of dormant cells through transient inactivation of anabolism.
Fig. 4.
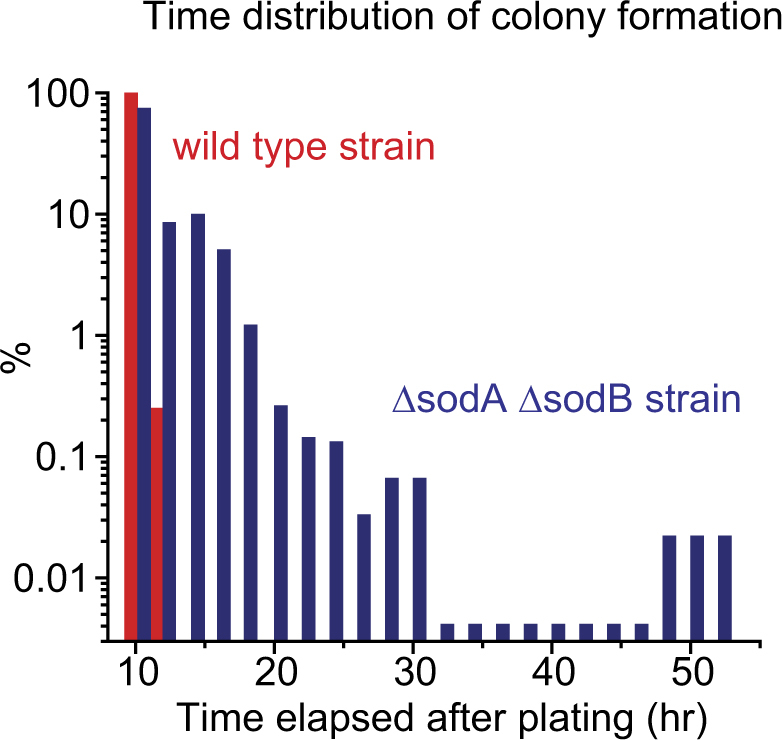
Wide distribution of colony formation time for the oxidative-stress-prone cells. The wild-type strain and ΔsodAΔsodB strain were starved for 8 h and plated on LB agar plates. Subsequently, we recorded the time at which visible colonies were newly formed (~500 colonies were examined for each strain). Compared to the wild-type strain, the ΔsodAΔsodB strain exhibited a much wider distribution, with its tail stretching to ~50 h. We observed similar distributions in two biological replicates
Discussion
Population diversification is a critical adaptation mechanism to changing environments [2, 3]. Previous studies of genetic diversity extensively characterized how genetic composition in a population changes slowly. In this study, we characterized the metabolic heterogeneity and its effect on phenotypic diversity in a clonal population. The results revealed the dynamic changes in phenotypic composition in a clonal population, cellular variations driving such changes (metabolic heterogeneity), and a factor triggering such cellular variations (oxidative stress).
Importantly, our findings provide a fresh metabolic perspective of dormancy. Dormant bacteria are common in nature, having significant ecological consequences [52]. However, our understanding of dormancy remains limited. For example, although it has been generally assumed that dormant cells are metabolically inactive [63], their metabolism has not been characterized. Our study reveals a metabolic state of dormant cells; dormant cells are metabolically partially active, exhibiting active substrate uptake and catabolism but inactive anabolism. This understanding sheds new light on their maintenance of viability. Dormant cells are ecologically important because they can remain viable for long. For example, due to this property, they can contribute to recovery of a microbial population after disturbance [25, 27]. However, bacteria, just to stay viable, must take up and catabolize nutrients. This “maintenance requirement” arises due to the fact that it takes substrates and energy to repair chemical wear and tear of cellular materials, maintain the membrane potential, and fulfill other non-growth-related functions [64, 65]. Previously, with the assumption of inactive metabolism, it was not clear how dormant cells could meet this maintenance requirement. Yet, our findings of active substrate uptake and catabolism in dormant cells suggest how this maintenance requirement could be satisfied. Also, our results showed that oxidative stress induces dormancy. Oxidative stress is a common stress that most aerobic microbes experience in nature, and thus could contribute to the common occurrence of dormancy.
Oxidative stress induces complex cellular responses in cells, and studies have extensively characterized how oxidative stress leads to changes in gene expression [60, 66, 67]. For example, one study showed that oxidative stress increases the expression of the efflux pump AcrAB, and the increased expression leads to extrusion of toxic compounds, resulting in better survival of cells [68]. Another study showed that oxidative stress activates OxyR and phage-shock responses, enhancing bacterial survival in stress conditions [69]. Our findings reveal that oxidative stress also has significant effects on metabolic activities and thereby on phenotypic states of cells.
Our findings raise a concern about the colony formation assay, one of the most frequently performed techniques in microbial research. In this assay, microbial samples are spread on a nutrient-rich agar plate, and then the number of viable colonies formed after overnight incubation is counted to determine the number of viable cells in the original samples. This assay is routinely used to determine microbial soil contamination or the presence of pathogens in drinking water. Studies of microbial dynamics critically rely on this assay as well. Our finding showed how cells with partial metabolic activities are viable but fail to grow in nutrient-rich conditions. In fact, when environmental microbial samples are plated on agar plates containing rich nutrients (e.g., LB), many cells fail to grow to form colonies, a long-standing problem known as “great plate count anomaly” [37, 38]. Our findings provide a fresh metabolic perspective on this problem and also prompt a cautious interpretation of assay results.
Lastly, we want to emphasize that phenotypic diversity can have significant effects on evolutionary dynamics and vice versa. For example, recent studies of experimental evolution showed that phenotypic diversity may evolve under fluctuating selection [70] and further accelerates evolutionary adaptation to various other environmental challenges [71, 72]. Therefore, our findings would be useful for understanding the evolution of microbes in nutrient-limiting conditions, e.g., emergence of mutants expressing the growth advantage in stationary phase (GASP) during long-term starvation conditions [5, 11].
Methods
Strain and culture
The strains used in this study are derived from E. coli K12 strain NCM3722 [73–75]; see the Supplementary Table 2 for the strains used. We cultured cells in N-C-minimal media [76], supplemented with 20 mM glucose (the sole carbon source) and 20 mM ammonium chloride (the sole nitrogen source). We starved these cells of carbon by suspending them in the same medium but without glucose (starvation medium). At the times indicated, starved cells were collected and plated on Luria–Bertani (LB) solid medium for CFU assays (Supplementary Methods) or for microscope experiments (see below); this exposure to LB represents nutrient upshift. We further described the details of strains and cultures in Supplementary Methods.
Microscope experiments
Our typical procedure for a microscope experiment is as follows (also see Supplementary Fig. 2 for the graphic illustration of the experimental procedure). At the times indicated, 100 µl of a culture was collected and transferred to a 1.5 ml Eppendorf tube. 2NBDG (Thermo-Fisher) was added into this aliquot. We prepared a 2NBDG stock solution by dissolving it in N-C-medium (stock concentration: 0.5 mM) and added it to the aliquot at a final concentration of 10 µM; this concentration of 2NBDG does not support cell growth (Supplementary Fig. 11). In some cases (Supplementary Fig. 6), to visualize dead cells, PI (Thermo-Fisher) was added to the aliquot at a final concentration of 10 µM (its stock solution was prepared by dissolving it in N-C-medium at a concentration of 2 mM). After incubation for 25 min in the dark at 37°C, a 5–8 µl aliquot from this sample was spread onto a pre-warmed 35 mm glass-bottom Petri dish (InVitro Scientific). Then, the dish was moved into a pre-warmed (at 37°C) inverted microscope (Olympus IX83). A pre-warmed (~1.6 cm2×1 mm) LB agarose pad was gently placed on top (i.e., nutrient upshift). The microscope has an automated mechanical XY stage and auto-focus, and is controlled by MetaMorph software (Molecular Devices). Furthermore, it is housed in a microscope incubator (InVivo Scientific) which maintains the temperature of samples at 37°C during experiments. An oil immersion 60× objective was used to obtain phase-contrast and fluorescence images of cells. FITC and TRITC filter sets (Olympus) were used to collect fluorescence signals. Images were captured using a Neo 5.5 sCMOS camera (Andor). In some experiments, there were minor variations in the procedure. These variations were described in the Supplementary Methods.
Image analysis
Image analysis was performed using MicrobeJ [77], a freely available plug-in for the ImageJ software [78]. This program can automatically segment cell boundaries from phase-contrast microscope images (“segmentation”) and measure the cell length (“shape descriptors”). Also, it can measure intracellular fluorescence intensities (“intensity”). To obtain a lag-time distribution, we ran this program for time-lapse images, obtained cell sizes at different times, and determined the time points at which cell sizes changed. We confirmed these time points by re-examining them manually. In some cases, we simply had to determine how many cells resumed growth within a certain time window (e.g., Supplementary Figs. 10 and 12). Then, we placed two images, one at time zero (the first image taken after nutrient upshift) and the other at the end of the time window, side by side. We visually compared the sizes of cells in these two images and determined which cells became larger.
Electronic supplementary material
Acknowledgements
We are grateful to Ilya Nemenman and Hyun Youk for helpful discussion. This work was supported by Emory Start-up Funds.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
The online version of this article (10.1038/s41396-017-0036-2) contains supplementary material, which is available to authorized users.
References
- 1.Morita, RY. Bacteria in oligotrophic environments: starvation-survival lifestyle. New York, NY: Chapman & Hall; 1997.
- 2.Cordero OX, Polz MF. Explaining microbial genomic diversity in light of evolutionary ecology. Nat Rev Microbiol. 2014;12:263–73. doi: 10.1038/nrmicro3218. [DOI] [PubMed] [Google Scholar]
- 3.Wolf DM, Vazirani VV, Arkin AP. Diversity in times of adversity: probabilistic strategies in microbial survival games. J Theor Biol. 2005;234:227–53. doi: 10.1016/j.jtbi.2004.11.020. [DOI] [PubMed] [Google Scholar]
- 4.Acinas SG, Haverkamp THA, Huisman J, Stal LJ. Phenotypic and genetic diversification of Pseudanabaena spp. (cyanobacteria) ISME J. 2008;3:31–46. doi: 10.1038/ismej.2008.78. [DOI] [PubMed] [Google Scholar]
- 5.Finkel SE. Long-term survival during stationary phase: evolution and the GASP phenotype. Nat Rev Microbiol. 2006;4:113–20. doi: 10.1038/nrmicro1340. [DOI] [PubMed] [Google Scholar]
- 6.Krismer J, Tamminen M, Fontana S, Zenobi R, Narwani A. Single-cell mass spectrometry reveals the importance of genetic diversity and plasticity for phenotypic variation in nitrogen-limited Chlamydomonas. ISME J. 2017;11:988–98. doi: 10.1038/ismej.2016.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lenski RE. Experimental evolution and the dynamics of adaptation and genome evolution in microbial populations. ISME J. 2017;11:2181–94. doi: 10.1038/ismej.2017.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maharjan R, Seeto S, Notley-Mcrobb L, Ferenci T. Clonal adaptive radiation in a constant environment. Science. 2006;313:514–7. doi: 10.1126/science.1129865. [DOI] [PubMed] [Google Scholar]
- 9.Mitri S, Clarke E, Foster KR. Resource limitation drives spatial organization in microbial groups. ISME J. 2016;10:1471–82. doi: 10.1038/ismej.2015.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rendueles O, Velicer GJ. Evolution by flight and fight: diverse mechanisms of adaptation by actively motile microbes. ISME J. 2017;11:555–68. doi: 10.1038/ismej.2016.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zambrano MM, Siegele DA, Almiron M, Tormo A, Kolter R. Microbial competition: Escherichia coli mutants that take over stationary phase cultures. Science. 1993;259:1757–60. doi: 10.1126/science.7681219. [DOI] [PubMed] [Google Scholar]
- 12.Choi PJ, Cai L, Frieda K, Xie XS. A stochastic single-molecule event triggers phenotype switching of a bacterial cell. Science. 2008;322:442–6. doi: 10.1126/science.1161427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frankel NW, Pontius W, Dufour YS, Long J, Hernandez-Nunez L, Emonet T. Adaptability of non-genetic diversity in bacterial chemotaxis. eLife. 2014;3:e03526. doi: 10.7554/eLife.03526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guantes R, Benedetti I, Silva-Rocha R, De Lorenzo V. Transcription factor levels enable metabolic diversification of single cells of environmental bacteria. ISME J. 2016;10:1122–33. doi: 10.1038/ismej.2015.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ackermann M. A functional perspective on phenotypic heterogeneity in microorganisms. Nat Rev Microbiol. 2015;13:497–508. doi: 10.1038/nrmicro3491. [DOI] [PubMed] [Google Scholar]
- 16.Jõers A, Tenson T. Growth resumption from stationary phase reveals memory in Escherichia coli cultures. Sci Rep. 2016;6:24055. doi: 10.1038/srep24055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaern M, Elston TC, Blake WJ, Collins JJ. Stochasticity in gene expression: from theories to phenotypes. Nat Rev Genet. 2005;6:451–64. doi: 10.1038/nrg1615. [DOI] [PubMed] [Google Scholar]
- 18.Kuchina A, Espinar L, Çağatay T, Balbin AO, Zhang F, Alvarado A, et al. Temporal competition between differentiation programs determines cell fate choice. Mol Syst Biol. 2011;7:557. [DOI] [PMC free article] [PubMed]
- 19.Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. Bacterial persistence as a phenotypic switch. Science. 2004;305:1622–5. doi: 10.1126/science.1099390. [DOI] [PubMed] [Google Scholar]
- 20.Sheik AR, Muller EEL, Audinot JN, Lebrun LA, Grysan P, Guignard C, et al. In situ phenotypic heterogeneity among single cells of the filamentous bacterium Candidatus Microthrix parvicella. ISME J. 2016;10:1274–9. doi: 10.1038/ismej.2015.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bartumeus F, Campos D, Ryu WS, Lloret-Cabot R, Méndez V, Catalan J. Foraging success under uncertainty: search tradeoffs and optimal space use. Ecol Lett. 2016;19:1299–313. doi: 10.1111/ele.12660. [DOI] [PubMed] [Google Scholar]
- 22.Venturelli OS, Zuleta I, Murray RM, El-Samad H. Population diversification in a yeast metabolic program promotes anticipation of environmental shifts. PLoS Biol. 2015;13:e1002042. doi: 10.1371/journal.pbio.1002042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Belete MK, Balázsi G. Optimality and adaptation of phenotypically switching cells in fluctuating environments. Phys Rev E. 2015;92:062716. doi: 10.1103/PhysRevE.92.062716. [DOI] [PubMed] [Google Scholar]
- 24.Ackermann M. Microbial individuality in the natural environment. ISME J. 2013;7:465–7. doi: 10.1038/ismej.2012.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kalamees R, Zobel M. The role of the seed bank in gap regeneration in a calcareous grassland community. Ecology. 2002;83:1017–25. doi: 10.1890/0012-9658(2002)083[1017:TROTSB]2.0.CO;2. [DOI] [Google Scholar]
- 26.Chesson PL, Warner RR. Environmental variability promotes coexistence in lottery competitive systems. Am Nat. 1981;117:923–43. doi: 10.1086/283778. [DOI] [Google Scholar]
- 27.Jones SE, Lennon JT. Dormancy contributes to the maintenance of microbial diversity. Proc Natl Acad Sci USA. 2010;107:5881–6. doi: 10.1073/pnas.0912765107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.De Martino M, Ershov D, Van Den Berg PJ, Tans SJ, Meyer, AS. Single-cell analysis of the Dps response to oxidative stress. J Bacteriol. 2016;198:1662–74. [DOI] [PMC free article] [PubMed]
- 29.Nikolic N, Barner T, Ackermann M. Analysis of fluorescent reporters indicates heterogeneity in glucose uptake and utilization in clonal bacterial populations. BMC Microbiol. 2013;13:258. doi: 10.1186/1471-2180-13-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Silander OK, Nikolic N, Zaslaver A, Bren A, Kikoin I, Alon U, et al. A genome-wide analysis of promoter-mediated phenotypic noise in Escherichia coli. PLoS Genet. 2012;8:e1002443. doi: 10.1371/journal.pgen.1002443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taniguchi Y, Choi PJ, Li GW, Chen H, Babu M, Hearn J, et al. Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science. 2010;329:533–8. doi: 10.1126/science.1188308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Labhsetwar P, Cole JA, Roberts E, Price ND, Luthey-Schulten ZA. Heterogeneity in protein expression induces metabolic variability in a modeled Escherichia coli population. Proc Natl Acad Sci. 2013;110:14006–11. doi: 10.1073/pnas.1222569110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kiviet DJ, Nghe P, Walker N, Boulineau S, Sunderlikova V, Tans SJ. Stochasticity of metabolism and growth at the single-cell level. Nature. 2014;514:376–9. doi: 10.1038/nature13582. [DOI] [PubMed] [Google Scholar]
- 34.Kotte O, Volkmer B, Radzikowski JL, Heinemann, M. Phenotypic bistability in Escherichia coli’s central carbon metabolism. Mol Syst Biol. 2014;10:736. [DOI] [PMC free article] [PubMed]
- 35.Radzikowski JL, Vedelaar S, Siegel D, Ortega AacuteD, Schmidt A, Heinemann M. Bacterial persistence is an active σS stress response to metabolic flux limitation. Mol Syst Biol. 2016;12:882. doi: 10.15252/msb.20166998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schreiber F, Littmann S, Lavik G, Escrig S, Meibom A, Kuypers MMM, et al. Phenotypic heterogeneity driven by nutrient limitation promotes growth in fluctuating environments. Nat Microbiol. 2016;1:16055. doi: 10.1038/nmicrobiol.2016.55. [DOI] [PubMed] [Google Scholar]
- 37.Epstein SS. Microbial awakenings. Nature. 2009;457:1083. doi: 10.1038/4571083a. [DOI] [PubMed] [Google Scholar]
- 38.JT Staley A, Konopka A. Measurement of in situ activities of nonphotosynthetic microorganisms in aquatic and terrestrial habitats. Annu Rev Microbiol. 1985;39:321–46. doi: 10.1146/annurev.mi.39.100185.001541. [DOI] [PubMed] [Google Scholar]
- 39.Bogosian G, Bourneuf EV. A matter of bacterial life and death. EMBO Rep. 2001;2:770–4. doi: 10.1093/embo-reports/kve182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li L, Mendis N, Trigui H, Oliver JD, Faucher SP. The importance of the viable but non-culturable state in human bacterial pathogens. Front Microbiol. 2014;5:258. doi: 10.3389/fmicb.2014.00258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nyström T. Nonculturable bacteria: programmed survival forms or cells at death’s door? BioEssays. 2003;25:204–11. doi: 10.1002/bies.10233. [DOI] [PubMed] [Google Scholar]
- 42.Huang KC. Applications of imaging for bacterial systems biology. Curr Opin Microbiol. 2015;27:114–20. doi: 10.1016/j.mib.2015.08.003. [DOI] [PubMed] [Google Scholar]
- 43.Taheri-Araghi S, Brown SD, Sauls JT, Mcintosh DB, Jun S. Single-cell physiology. Annu Rev Biophys. 2015;44:123–42. doi: 10.1146/annurev-biophys-060414-034236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Natarajan A, Srienc F. Dynamics of glucose uptake by single Escherichia coli cells. Metab Eng. 1999;1:320–33. doi: 10.1006/mben.1999.0125. [DOI] [PubMed] [Google Scholar]
- 45.Tao J, Diaz RK, Teixeira CRV, Hackmann TJ. Transport of a fluorescent analogue of glucose (2-NBDG) versus radiolabeled sugars by rumen bacteria and Escherichia coli. Biochemistry. 2016;55:2578–89. doi: 10.1021/acs.biochem.5b01286. [DOI] [PubMed] [Google Scholar]
- 46.Wang P, Robert L, Pelletier J, Dang WL, Taddei F, Wright A, et al. Robust growth of Escherichia coli. Curr Biol. 2010;20:1099–103. doi: 10.1016/j.cub.2010.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yoshioka K, Saito M, Oh KB, Nemoto Y, Matsuoka H, Natsume M, et al. Intracellular fate of 2-NBDG, a fluorescent probe for glucose uptake activity, in Escherichia coli cells. Biosci Biotechnol Biochem. 1996;60:1899–901. doi: 10.1271/bbb.60.1899. [DOI] [PubMed] [Google Scholar]
- 48.Yoshioka K, Takahashi H, Homma T, Saito M, Oh KB, Nemoto Y, et al. A novel fluorescent derivative of glucose applicable to the assessment of glucose uptake activity of Escherichia coli. Biochim Et Biophys Acta (BBA)-Gen Subj. 1996;1289:5–9. doi: 10.1016/0304-4165(95)00153-0. [DOI] [PubMed] [Google Scholar]
- 49.Lutz R, Bujard H. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 1997;25:1203–10. doi: 10.1093/nar/25.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Iyer S, Park BR, Kim M. Absolute quantitative measurement of transcriptional kinetic parameters in vivo. Nucleic Acids Res. 2016;44:e142. doi: 10.1093/nar/gkw596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gefen O, Fridman O, Ronin I, Balaban NQ. Direct observation of single stationary-phase bacteria reveals a surprisingly long period of constant protein production activity. Proc Natl Acad Sci USA. 2014;111:556–61. doi: 10.1073/pnas.1314114111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lennon JT, Jones SE. Microbial seed banks: the ecological and evolutionary implications of dormancy. Nat Rev Microbiol. 2011;9:119–30. doi: 10.1038/nrmicro2504. [DOI] [PubMed] [Google Scholar]
- 53.Tuomanen E, Cozens R, Tosch W, Zak O, Tomasz A. The rate of killing of Escherichia coli by beta-lactam antibiotics is strictly proportional to the rate of bacterial growth. J Gen Microbiol. 1986;132:1297–304. doi: 10.1099/00221287-132-5-1297. [DOI] [PubMed] [Google Scholar]
- 54.Yao Z, Kahne D, Kishony R. Distinct single-cell morphological dynamics under beta-lactam antibiotics. Mol Cell. 2012;48:705–12. doi: 10.1016/j.molcel.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dukan S, Nystrom T. Bacterial senescence: stasis results in increased and differential oxidation of cytoplasmic proteins leading to developmental induction of the heat shock regulon. Genes Dev. 1998;12:3431–41. doi: 10.1101/gad.12.21.3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gonidakis S, Finkel SE, Longo VD. Genome-wide screen identifies Escherichia coli TCA-cycle-related mutants with extended chronological lifespan dependent on acetate metabolism and the hypoxia-inducible transcription factor ArcA. Aging Cell. 2010;9:868–81. doi: 10.1111/j.1474-9726.2010.00618.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kram KE, Finkel SE. Culture volume and vessel affect long-term survival, mutation frequency, and oxidative stress of Escherichia coli. Appl Environ Microbiol. 2014;80:1732–8. doi: 10.1128/AEM.03150-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ballesteros M, Fredriksson A, Henriksson J, Nystrom T. Bacterial senescence: protein oxidation in non-proliferating cells is dictated by the accuracy of the ribosomes. EMBO J. 2001;20:5280–9. doi: 10.1093/emboj/20.18.5280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hassan HM, Fridovich I. Enzymatic defenses against the toxicity of oxygen and of streptonigrin in Escherichia coli. J Bacteriol. 1977;129:1574–83. doi: 10.1128/jb.129.3.1574-1583.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Imlay JA. The molecular mechanisms and physiological consequences of oxidative stress: lessons from a model bacterium. Nat Rev Micro. 2013;11:443–54. doi: 10.1038/nrmicro3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Keele BB, Mccord JM, Fridovich I. Superoxide dismutase from Escherichia coli B: a new manganese-containing enzyme. J Biol Chem. 1970;245:6176–81. [PubMed] [Google Scholar]
- 62.Yost FJ, Fridovich I. An iron-containing superoxide dismutase from Escherichia coli. J Biol Chem. 1973;248:4905–8. [PubMed] [Google Scholar]
- 63.Ayrapetyan M, Williams TC, Oliver JD. Bridging the gap between viable but non-culturable and antibiotic persistent bacteria. Trends Microbiol. 2015;23:7–13. doi: 10.1016/j.tim.2014.09.004. [DOI] [PubMed] [Google Scholar]
- 64.Tempest DW, Neijssel OM. The status of YATP and maintenance energy as biologically interpretable phenomena. Annu Rev Microbiol. 1984;38:459–513. doi: 10.1146/annurev.mi.38.100184.002331. [DOI] [PubMed] [Google Scholar]
- 65.van Bodegom P. Microbial maintenance: a critical review on its quantification. Microb Ecol. 2007;53:513–23. doi: 10.1007/s00248-006-9049-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chiang SM, Schellhorn HE. Regulators of oxidative stress response genes in Escherichia coli and their functional conservation in bacteria. Arch Biochem Biophys. 2012;525:161–9. doi: 10.1016/j.abb.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 67.Demple B. Regulation of bacterial oxidative stress genes. Annu Rev Genet. 1991;25:315–37. doi: 10.1146/annurev.ge.25.120191.001531. [DOI] [PubMed] [Google Scholar]
- 68.Wu Y, Vulić M, Keren I, Lewis K. Role of oxidative stress in persister tolerance. Antimicrob Agents Chemother. 2012;56:4922–6. doi: 10.1128/AAC.00921-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vega NM, Allison KR, Khalil AS, Collins JJ. Signaling-mediated bacterial persister formation. Nat Chem Biol. 2012;8:431–3. doi: 10.1038/nchembio.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Beaumont HJE, Gallie J, Kost C, Ferguson GC, Rainey PB. Experimental evolution of bet hedging. Nature. 2009;462:90. doi: 10.1038/nature08504. [DOI] [PubMed] [Google Scholar]
- 71.Bódi Z, Farkas Z, Nevozhay D, Kalapis D, Lázár V, Csörgő B, et al. Phenotypic heterogeneity promotes adaptive evolution. PLoS Biol. 2017;15:e2000644. doi: 10.1371/journal.pbio.2000644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.González C, Ray JCJ, Manhart M, Adams RM, Nevozhay D, Morozov AV, et al. Stress‐response balance drives the evolution of a network module and its host genome. Mol Syst Biol. 2015;11:827. [DOI] [PMC free article] [PubMed]
- 73.Brown SD, Jun S. Complete genome sequence of Escherichia coli NCM3722. Genome Announc. 2015;3:e00879-15. [DOI] [PMC free article] [PubMed]
- 74.Lyons E, Freeling M, Kustu S, Inwood W. Using genomic sequencing for classical genetics in E. coli K12. PLoS ONE. 2011;6:e16717. doi: 10.1371/journal.pone.0016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Soupene E, Van Heeswijk WC, Plumbridge J, Stewart V, Bertenthal D, Lee H, et al. Physiological studies of Escherichia coli strain MG1655: Growth defects and apparent cross-regulation of gene expression. J Bacteriol. 2003;185:5611–26. doi: 10.1128/JB.185.18.5611-5626.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Csonka LN, Ikeda TP, Fletcher SA, Kustu S. The accumulation of glutamate is necessary for optimal growth of Salmonella typhimurium in media of high osmolality but not induction of the proU operon. J Bacteriol. 1994;176:6324–33. doi: 10.1128/jb.176.20.6324-6333.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ducret A, Quardokus EM, Brun YV. MicrobeJ, a tool for high throughput bacterial cell detection and quantitative analysis. Nat Microbiol. 2016;1:16077. doi: 10.1038/nmicrobiol.2016.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Meth. 2012;9:671–5. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.