Abstract
Differential display reverse transcription-polymerase chain reaction was used to detect the induction of gene expression during adventitious root formation in loblolly pine (Pinus taeda) after treatment with the exogenous auxin indole-3-butyric acid. A BLAST search of the GenBank database using one of the clones obtained revealed very strong similarity to the α-expansin gene family in angiosperms. A near-full-length loblolly pine α-expansin sequence was obtained using 5′- and 3′-rapid amplification of cDNA end cloning, and the deduced amino acid sequence was highly conserved relative to those of angiosperm expansins. Northern analysis indicates that α-expansin mRNA expression increases 50- to 100-fold in the base of hypocotyl stem cuttings from loblolly pine seedlings in response to indole-3-butyric acid, with peak expression occurring 24 to 48 h after induction.
Conifers show a decline in the ability to form adventitious roots that is associated with maturation, an age-related developmental process that also affects reproductive competence, morphology, and growth rate (Poethig, 1990; Greenwood and Hutchison, 1993). The rate and the extent of the loss of rooting ability are species dependent. For example, in eastern larch we showed that the frequency of cuttings that root declines from 100% to 50% during the course of 20 years (Greenwood et al., 1989). In contrast, loss of rooting ability occurs abruptly and early in loblolly pine (Pinus taeda). Greenwood and Weir (1995) showed that 20-d-old hypocotyl cuttings from loblolly pine root readily within 3 weeks in the presence of exogenous auxin. However, woody cuttings from 1- to 2-year-old plants root poorly even after 2 to 3 months. In the latter case, exogenous auxin has little effect. We have extended the observations of Greenwood and Weir (1995) and shown that, whereas hypocotyl cuttings made from 20- or 50-d-old seedlings rapidly form adventitious roots, epicotyl cuttings from 50-d-old seedlings root poorly, if at all, and only after 2 to 3 months (Diaz-Sala et al., 1996). This rapid decline in rooting ability in loblolly pine provides us the opportunity to develop a woody plant model system for the study of adventitious rooting, specifically, and maturation, generally.
Rooting of loblolly pine hypocotyl cuttings is dependent on the addition of exogenous auxin and is inhibited by the auxin polar transport inhibitor NPA. However, NPA must be applied during the first 2 d of root induction. By d 3 the cuttings are fully committed to root formation and are insensitive to NPA inhibition. Despite the clear role for auxin in adventitious rooting, auxin uptake and metabolism do not account for the differences in the ability of hypocotyls and epicotyls to form adventitious roots. Uptake and polar transport of exogenous auxin was comparable in rooting-competent hypocotyls and rooting-incompetent epicotyls. Furthermore, there was no difference in auxin tissue distribution and metabolism (Diaz-Sala et al., 1996).
During the first 2 d of the rooting process the cambium layer of the hypocotyl dedifferentiates into parenchyma cells in both hypocotyls and epicotyls. Although some mitotic figures can be detected within 2 to 4 d, localized rapid cell division is not seen until d 6 and then only in hypocotyl cuttings. Cells competent to form roots are confined to the vascular parenchyma region of the hypocotyl, immediately centrifugal to the resin canals. These observations suggest that the decline in rooting ability is a result of a loss of cells capable of fully responding to the induction of adventitious root formation by auxin. It is not known whether this is due to a loss of a specific cell type, the inability of individual cells to perceive auxin signals that are specific for root meristem organization, or the suppression of gene expression needed for cells to enter the root-formation pathway.
To extend our knowledge of the induction of adventitious roots in loblolly pine, we sought genes whose expression in hypocotyl cuttings responded in the first 24 to 48 h to the application of exogenous auxin. Using differential display RT-PCR (Liang and Pardee, 1992) we have identified a gene family whose mRNA levels increase in hypocotyl cuttings in response to the application of the auxin IBA. This gene family is the loblolly pine homolog of the α-expansins, which have previously been found in monocots and dicots (McQueen-Mason et al., 1992; Shcherban et al., 1995). Recently, expansins were thoroughly reviewed (Cosgrove, 1996, 1997, 1998). They are thought to be responsible for acid-induced loosening of cellulose-hemicellulose networks (McQueen-Mason et al., 1992; Rayle and Cleland, 1992; Cosgrove and Li, 1993; McQueen-Mason, 1995). Their expression in angiosperms has been reported in rapidly growing areas of the plant (McQueen-Mason et al., 1992; Cosgrove and Li, 1993; Cho and Kende, 1997) and in ripening fruit (Rose et al., 1997). We report here that in loblolly pine the α-expansin-related sequences are induced in nongrowing regions of the stem prior to the resumption of cell division leading to adventitious roots.
MATERIALS AND METHODS
Plant Material
Growth of seedlings and induction of rooting in hypocotyl or epicotyl stem segments were previously described (Diaz-Sala et al., 1996). Hypocotyl cuttings were made from 20-d-old seedlings by severing the hypocotyl above its base, 3.5 cm from the cotyledons. Epicotyl cuttings were made from 50-d-old seedlings by severing the seedling above the cotyledons, 3.5 cm from the apex. All of the needles except for the apical tuft were removed. Cuttings were place in distilled water either with or without 10 μm IBA. IBA- plus NPA-treated samples were placed in distilled water with IBA (10 μm) and NPA (10 μm).
RNA Extraction
The bottom 0.5 cm was removed from the cuttings and quick frozen in liquid N2. The stem segments from cuttings from 30 seedlings were pooled for each time point and treatment. Total RNA was extracted as previously described (Hutchison et al., 1990).
Differential Display RT-PCR
Differential display RT-PCR was performed using a modification of the method of Liang and Pardee (1992), as described by Rhodes and Van Beneden (1996). All primers were obtained from Operon Technologies (Alameda, CA). Total RNA extracted from the stem segments was pretreated with DNase I (10 units/25 μg RNA) in 100 mm Tris-HCl (pH 8.3), 500 mm KCl, and 15 mm MgCl2 for 30 min at 37°C. After the segments were treated with DNase I the RNA was extracted once with DEPC-treated water-saturated phenol:chloroform (3:1) and once with chloroform. The RNA was then precipitated by adding 0.1 volume of 3 m sodium acetate (pH 5.2) and 3 volumes of 100% ethanol. Samples were centrifuged in a microfuge (Brinkmann) for 45 min at 4°C. After washing with 80% ethanol the pellet was redissolved in DEPC-treated water.
Total RNA (0.2 μg) plus the T11GC primer (1.5 μm) was heated to 80°C for 5 min in DEPC-treated water and cooled to 37°C for 5 min. The heated and reannealed RNA-primer mixtures were reverse transcribed in a 20-μL reaction containing 50 mm Tris-HCl (pH 8.3), 75 mm KCl, 3 mm MgCl2, 1 mm DTT, 500 μm each dNTP, 5 units of RNasin (Promega), and 300 units of Superscript reverse transcriptase (GIBCO-BRL) for 60 min at 37°C. Reactions were stopped by heating at 80°C for 10 min.
The cDNA was amplified in a 20-μL PCR containing 1× PCR buffer (Perkin-Elmer), 1.5 mm MgCl2, 5 μm each dNTP, 80 μm α-35S-dATP (5 μCi, Amersham), 0.5 μm each of primers OPD9 (5′-CTCTGGAGAC-3′) and OPD10 (5′-GGTCTACACC-3′), and 0.3 unit of AmpliTaq DNA polymerase (Perkin-Elmer). Cycling conditions were: initial denaturation at 94°C for 5 min, then 94°C for 90 s; 38°C for 40 s; and 72°C for 60 s for 40 cycles. After a final extension at 72°C for 5 min, the reactions were cooled to 4°C.
The PCR reactions were electrophoresed through a 6% denaturing acrylamide gel, and the bands were detected by autoradiography. PCR fragments were extracted from the gel by soaking the gel slice in sterile distilled water at 4°C overnight and by boiling for 15 min. The DNA was ethanol precipitated, redissolved in sterile distilled water, and reamplified using the above conditions. The reamplified PCR fragments were cloned into a TA-cloning vector prepared from SmaI digest pUC19 DNA, as described by Marchuk et al. (1991).
Full-Length cDNA Clones
A near-full-length cDNA clone was constructed in a two-step procedure. First, 5′-RACE clones were made essentially as described by Harvey and Darlison (1991). First-strand cDNA was reverse transcribed from total RNA using the anchor primer T11GC. A poly(A) tail was then added to the 3′ end of the first cDNA using terminal transferase (New England Biolabs). The DNA was amplified with the Promega primer adapter (5′-GTCGACTCTAGATTTTTTTTTTTTTTT-3′; Xba-PA) and primer based on the DD21.4-1 sequence (5′-GCATTTCATAGCAGG-3′, Exp3) and cloned as described above. 3′-RACE clones were made essentially as described by Schaefer (1995). First-strand cDNA was reverse transcribed from total RNA as described above, except the Xba-PA primer was used. PCR amplification was carried out with Xba-PA for the 3′ primer and a primer designed from the 5′-RACE clone sequence (5′-CCGGATCCTTCAGATCTCCCTGATTCTC-3′; Ptexp75RI) for the 5′ primer and Xba-PA for the 3′ primer. The amplified fragment was ligated into the EcoRI and XbaI sites of pBluescript (Stratagene).
DNA Sequencing and Analysis
DNA was sequenced by the University of Maine DNA Sequencing Facility (Orono) using the ABI-Prism Dye Terminator Cycle Sequencing Ready Reaction kit with Amplitaq DNA Polymerase, FS (Perkin-Elmer). The reaction products were resolved and the sequence was determined using an ABI 373A DNA Stretch Sequencer. DNA sequences were analyzed using the Wisconsin Package software (version 8, Genetics Computer Group, Madison, WI). PCR primers were designed using Primer3 (Rozen and Skaletsky, 1997). The location of the signal peptide cleavage site was determined with SignalP (Nielsen et al., 1997).
Northern Blots
Northern blots were performed as previously described (Hutchison et al., 1990). Expansin mRNA was detected using the insert from clone DD21.4-1 as a probe. The actin probe was from clone pAc20.20, a loblolly pine partial cDNA clone obtained by 3′-RACE (accession no. AF085331; C. Diaz-Sala and K.W. Hutchison, unpublished data). 18S rRNA levels were detected using a larch 18S rRNA probe, as previously described (Hutchison et al., 1990). Radioactive DNA probes were made from PCR-amplified inserts of the respective clones according to the method of Feinberg and Vogelstein (1983). Autoradiograms were exposed for 5 to 9 d using a DuPont Lightning Plus intensifying screen. RNA levels were quantitated by scanning the autoradiograms with a densitometer (LKB, Bromma, Sweden) or by using ImagePC software (Scion, Frederick, MD).
RESULTS AND DISCUSSION
Expansin-Related Sequences Are Detected in Loblolly Pine Seedlings undergoing Adventitious Root Formation
To search for genes involved in adventitious rooting we used differential display RT-PCR to detect transcripts expressed after 24 h in auxin-treated hypocotyl stem segments but not found in untreated hypocotyl segments. Two clones (pDD21.3.3 and pDD21.4.1) were obtained from a gel-purified differential display-RT-PCR band that conformed to these criteria. These clones were sequenced and were 231 bp, exclusive of the RAPD primer-binding site. The sequence for clone pDD21.4.1 was deposited in the GenBank database (accession no. U64889). A BLAST search (Altschul et al., 1990) of the database showed a high level of similarity to the α-expansin gene family of monocots and dicots (Shcherban et al., 1995; and below).
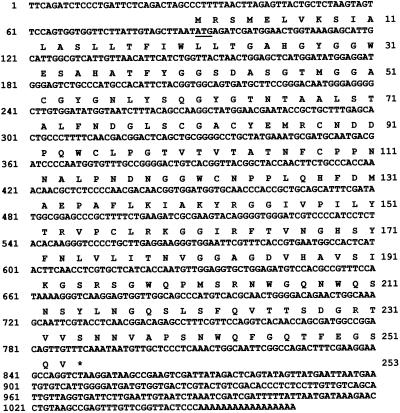
To obtain a full-length cDNA sequence a 5′-RACE was first constructed. The sequence of this clone can be found in the GenBank/EMBL/DDBJ databases (accession no. U64895). A unique primer was designed that annealed to the 5′ end of the loblolly pine expansin cDNA and used in a 3′-RACE reaction. This resulted in a PCR fragment of approximately 1 kb, which was subsequently cloned and sequenced. The complete sequence and translation of clone pCPEx7-8 is given in Figure 1 and has been deposited in the GenBank database (accession no. AF085330). The putative protein is translated from the 5′-most ATG. The total length of the sequence minus the poly(A) tail is 999 nucleotides.
Figure 1.
The DNA and deduced amino acid sequence for a loblolly pine expansin. The expansin cDNA sequence was submitted to the GenBank/EMBL DNA databases (accession no. AF085330). The putative translation initiation codon is underlined. The translation termination codon is indicated by an asterisk. Nucleotide positions are given by numbers to the left of the DNA sequence. Amino acid positions are indicated by numbers to the right of the amino acid sequence. The sequence includes the 5′ primer used for 3′-RACE cloning but does not include additional 5′ residues found in a 5′-RACE clone (accession no. U64895) from which the 5′ PCR primer was derived.
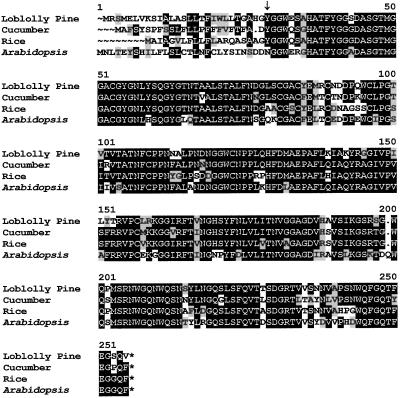
Based on a phylogenetic analysis of angiosperm expansin sequences, Shcherban et al. (1995) proposed that the expansins represented an ancient gene lineage and would be found in other land plants. Our results confirm this hypothesis. The predicted amino acid sequence of the loblolly pine α-expansin protein is highly conserved relative to those from the angiosperms Arabidopsis, cucumber, and rice (Fig. 2). There is little similarity in the first 27 residues of the peptide sequence. This segment constitutes the signal peptide identified using SignalP (Nielsen et al., 1997), and the predicted N terminus of the mature peptide at Y28 is one amino acid downstream of that found for a mature α-expansin protein (CuExS1) from cucumber (Shcherban et al., 1995). The mature peptide shows an average sequence identity of 80% to the rice, cucumber, and Arabidopsis α-expansins. The evolutionary relationship of the loblolly pine expansin to that of other plants was shown by Cho and Kende (1997), who used a partial cDNA sequence that we previously submitted to the GenBank database (accession no. U64892). Although we have not yet purified the protein, the strong sequence conservation with known α-expansins suggests that the loblolly pine expansins will be functionally equivalent. Preliminary genetic data suggest that there may be three family members that are expressed in hypocotyls in response to auxin treatment (S. McInnis, Z. Wang, and K.W. Hutchison, unpublished data).
Figure 2.
Alignment of loblolly pine amino acid sequence for expansin with angiosperm expansin sequences. Deduced amino acid sequences for expansins from loblolly pine (accession no. AF085330), Arabidopsis (accession no. U30481), cucumber (accession no. U30382), and rice (accession no. U85246) were aligned using the Pileup program from the Wisconsin Package DNA sequence. Output was produced using the program Boxshade (www.isrec.isb-sib.ch/software/BOX_form.html). Amino acids printed in inverse represent positions where at least 50% of the residues are identical. Amino acids that are similar to the consensus are shaded. The location of the putative N terminus of the mature peptide from loblolly pine as predicted by SignalP (Nielsen et al., 1997) is indicated by an arrow (↓).
Expression of Expansin Sequences during Adventitious Rooting
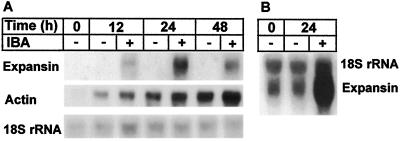
Expansin mRNA levels increased during the early stages of induction of adventitious root formation in response to the application of exogenous auxin (Fig. 3A). RNA was extracted at different times from hypocotyl and epicotyl stem segments in the presence and absence of IBA. The stem segments were fully elongated before they were exposed to IBA. Northern blots were probed using clone pDD21.4.1. Peak expression was achieved in 24 h, after which there was a slow decline in expansin mRNA levels. Nevertheless, after 5 d there was still a significant level of expansin mRNA in hypocotyl segments undergoing rooting (data not shown). In a second experiment, RNA was extracted from samples at 0 and 24 h in the presence and absence of IBA. In this experiment, with more RNA loaded onto the gel, a low level of expansin expression could be detected at 0 and 24 h in the absence of auxin (Fig. 3B). However, densitometric measurement of the autoradiogram indicated that exogenous auxin increases expansin mRNA levels by 50- to 100-fold. The low level of expansin expression may represent constitutive expression of the sequences or induction by the endogenous pool of auxin, which accumulates at the base of the cutting.
Figure 3.
Northern blot of hypocotyl RNA from loblolly pine seedling cuttings. A, RNA was extracted from the base of hypocotyl cuttings and placed in either distilled water (−) or distilled water plus 10 mm IBA (+) for the times indicated. One microgram of total RNA was electrophoresed in a 1% agarose gel, northern blotted, and hybridized with the clone pDD21.4.1. After exposure to x-ray film, the filter was erased and rehybridized with an 18S rRNA probe from eastern larch and with a loblolly pine partial cDNA clone for actin (accession no. AF085331), as described by Hutchison et al. (1990). B, RNA extracted from hypocotyl cuttings at the times indicated in an independent rooting experiment. Four micrograms of RNA was used per sample, and the filter was hybridized with the larch 18S rRNA probe and the expansin probe.
The large increase in expansin mRNA levels in response to exogenous auxin is not a result of a generalized increase in transcription. Probing the same blot with a loblolly pine actin probe showed an overall increase in actin mRNA levels during the course of the experiment and a slight increase in response to auxin addition (Fig. 3A). We also showed that expression of neither Phe ammonia lyase nor a cyclin-dependent kinase is auxin induced in the bases of hypocotyl and epicotyl cuttings and that actin expression may be inhibited by auxin in epicotyls (Greenwood et al., 1997). The importance of the small increase in actin mRNA levels in response to auxin is therefore difficult to assess at this time.
Regulation of expansin expression appears to be complex, although some of the complexity may be due to different regulatory schemes for different members of the expansin gene family. Rose et al. (1987) reported the up-regulation of one tomato expansin by ethylene during fruit ripening. In rice expansin expression in the internode region is increased by GA (Cho and Kende, 1997). In this case, three different family members showed a response to GA addition, but the kinetics of the response was unique for each sequence. Downes and Crowell (1998) reported that a β-expansin from cotton responds to the addition of cytokinins. Our observation that in pine expansins increase in response to exogenous auxin during adventitious root formation adds yet another hormone and developmental circumstance to the list of controls of the expansin gene family. An increase of expansin expression in response to auxin in cucumber hypocotyls has also been reported (Shieh et al., 1997).
Fleming et al. (1997) showed that topical application of expansin to tomato apical meristems could induce localized tissue expansion and morphogenesis of leaf primordia. These data suggest a role for expansin in organogenesis. It is possible that expansin is playing a similar role in adventitious root formation. This is most likely to occur during the earliest events in adventitious rooting in the rooting-competent cells within the stem. Expansin expression increases during and is maximal within the first 2 d of rooting induction and coincides with the observed dedifferentiation of cambial cells. An additional correlation is that the auxin-dependent 100-fold increase in expansin levels occurs during the first 2 d of the rooting process, a time when the rooting process is also sensitive to NPA inhibition (Diaz-Sala et al., 1996).
Cosgrove (1996) proposed that there are a variety of cell- and organ-specific expansins that can respond to a variety of unique environmental and hormonal signals. We do not know whether the patterns of expansin expression that we observe are the result of a coordinated expression of all members of a gene family or only some members. Experiments are in progress to test these alternatives. Work is also in progress to look at the cellular distribution of expansin expression in the cuttings and at the level of expansin protein. These data demonstrate that expansin is found in gymnosperms, as well as the angiosperms, is highly conserved, and mRNA levels respond to auxin.
Abbreviations:
- DEPC
diethyl pyrocarbonate
- IBA
indole-3-butyric acid
- NPA
N-(1-naphthyl)phthalamic acid
- RACE
rapid amplification of cDNA ends
- RT-PCR
reverse transcription-PCR
Footnotes
This work was supported by a grant from the Maine Agriculture and Forestry Experiment Station (K.W.H.) and the North Carolina State Loblolly and Slash Pine Rooted Cutting Project (M.S.G.). This is Maine Agriculture and Forestry Experiment Station publication no. 2321.
LITERATURE CITED
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Cho HT, Kende H (1997) Expression of expansin genes is correlated with growth in deepwater rice. Plant Cell 1661–1671 [DOI] [PMC free article] [PubMed]
- Cosgrove DJ. Plant cell enlargement and the action of expansins. Bioessays. 1996;8:533–540. doi: 10.1002/bies.950180704. [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. Assembly and enlargement of the primary cell wall in plants. Annu Rev Cell Dev Biol. 1997;3:171–201. doi: 10.1146/annurev.cellbio.13.1.171. [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. Cell wall loosening by expansins. Plant Physiol. 1998;18:333–339. doi: 10.1104/pp.118.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove DJ, Li ZC. Role of expansin in cell enlargement of oat coleoptiles. Analysis of developmental gradients and photocontrol. Plant Physiol. 1993;03:1321–1328. doi: 10.1104/pp.103.4.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Sala C, Hutchison K, Goldfarb B, Greenwood MS. Maturation-related loss of rooting competence by loblolly pine stem cuttings: the role of auxin transport, metabolism and tissue sensitivity. Physiol Plant. 1996;97:481–490. [Google Scholar]
- Downes BP, Crowell DN. Cytokinin regulates the expression of a soybean beta-expansin gene by a post-transcriptional mechanism. Plant Mol Biol. 1998;7:437–444. doi: 10.1023/a:1005920732211. [DOI] [PubMed] [Google Scholar]
- Feinberg AP, Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Fleming AJ, McQueen-Mason S, Mandel T, Kuhlemeier C. Induction of leaf primordia by the cell wall protein expansin. Science. 1997;76:1415–1418. [Google Scholar]
- Greenwood MS, Diaz-Sala C, Singer PB, Decker A, Hutchison KW (1997) Differential gene expression during maturation-caused decline in adventitious rooting ability in loblolly pine (Pinus taeda L.). In A Altman, Y Waisel, eds, Biology of Root Formation and Development. Plenum Press, New York, pp 203–208
- Greenwood MS, Hopper CA, Hutchison KW. Maturation in larch. I. Effect of age on shoot growth, foliar characteristics, and DNA methylation. Plant Physiol. 1989;90:406–412. doi: 10.1104/pp.90.2.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwood MS, Hutchison KW (1993) Maturation as a developmental process. In MR Ahuja, WJ Libby, eds, Clonal Forestry: Genetics, Biotechnology and Application. Springer Verlag, New York, pp 14–33
- Greenwood MS, Weir RJ. Genetic variation in rooting ability of loblolly pine cuttings: effects of auxin and family on rooting by hypocotyl cuttings. Tree Physiol. 1995;15:41–45. doi: 10.1093/treephys/15.1.41. [DOI] [PubMed] [Google Scholar]
- Harvey RJ, Darlison MB. Random-primed cDNA synthesis facilitates the isolation of multiple 5′-cDNA ends by RACE. Nucleic Acids Res. 1991;19:4002. doi: 10.1093/nar/19.14.4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison KW, Sherman CB, Greenwood MS, Weber J, Smith SS, Singer PB, Greenwood MS. Maturation in larch. II. Effects of age on photosynthesis and gene expression in developing foliage. Plant Physiol. 1990;94:1308–1315. doi: 10.1104/pp.94.3.1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang P, Pardee AB. Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science. 1992;257:967–971. doi: 10.1126/science.1354393. [DOI] [PubMed] [Google Scholar]
- Marchuk D, Drumm M, Saulino A, Collins FS. Construction of T-vectors, a rapid and general system for direct cloning of unmodified PCR products. Nucleic Acids Res. 1991;19:1154. doi: 10.1093/nar/19.5.1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQueen-Mason S. Expansins and cell wall expansion. J Exp Bot. 1995;46:1639–1650. [Google Scholar]
- McQueen-Mason SJ, Durachko DM, Cosgrove DJ. Two endogenous proteins that induce cell wall extension in plants. Plant Cell. 1992;4:1425–1433. doi: 10.1105/tpc.4.11.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Engin. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- Poethig RS. Phase change and the regulation of shoot morphogenesis in plants. Science. 1990;250:923–930. doi: 10.1126/science.250.4983.923. [DOI] [PubMed] [Google Scholar]
- Rayle DL, Cleland RE. The acid growth theory of auxin-induced cell elongation is alive and well. Plant Physiol. 1992;99:1271–1274. doi: 10.1104/pp.99.4.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes LD, Van Beneden RJ. Gene expression analysis in aquatic animals using differential display polymerase chain reaction. In: Ostander GK, editor. Techniques in Aquatic Toxicology. Boca Raton, FL: CRC Press; 1996. pp. 161–183. [Google Scholar]
- Rose JKC, Lee HH, Bennett AB. Expression of a divergent expansin gene is fruit-specific and ripening-regulated. Proc Natl Acad Sci USA. 1997;4:5955–5960. doi: 10.1073/pnas.94.11.5955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S, Skaletsky HJ (1997) Primer3. Primer3 Input (primer3.cgi v 0.2c). http://www.genome.wi.mit.edu/genome_software/other/primer3.html(September 23, 1997)
- Schaefer BC. Revolutions in rapid amplification of cDNA ends: new strategies for polymerase chain reaction cloning of full-length cDNA ends. Anal Biochem. 1995;227:255–273. doi: 10.1006/abio.1995.1279. [DOI] [PubMed] [Google Scholar]
- Shcherban TY, Shi J, Durachko DM, Guiltinan MJ, McQueen-Mason SJ, Shieh M, Cosgrove DJ. Molecular cloning and sequence analysis of expansins: a highly conserved, multigene family of proteins that mediates cell wall extension in plants. Proc Natl Acad Sci USA. 1995;92:9245–9249. doi: 10.1073/pnas.92.20.9245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shieh M, Shi J, Cosgrove DJ. Developmental, hormonal and light regulation of the transcript for the cell wall-loosening protein expansin (abstract no. 341) Plant Physiol. 1997;114:S-85. [Google Scholar]