Abstract
Plants contain light signaling systems and undergo metabolic perturbation and reprogramming under light stress in order to adapt to environmental changes. Flavonoids are one of the largest classes of natural phytochemical compounds having several biological functions conferring stress defense to plants and health benefits in animal diets. A recent study of phenylacylated-flavonoids (also called hydroxycinnamoylated-flavonoids) of natural accessions of Arabidopsis suggested that phenylacylation of flavonoids relates to selection under different natural light conditions. Phenylacylated-flavonoids which are decorated with hydroxycinnamoyl units, namely cinnamoyl, 4-coumaroyl, caffeoyl, feruloyl and sinapoyl moieties, are widely distributed in the plant kingdom. Currently, more than 400 phenylacylated flavonoids have been reported. Phenylacylation renders enhanced phytochemical functions such as ultraviolet-absorbance and antioxidant activity, although, the physiological role of phenylacylation of flavonoids in plants is largely unknown. In this review, we provide an overview of the occurrence and natural diversity of phenylacylated-flavonoids as well as postulating their biological functions both in planta and with respect to biological activity following their consumption.
Keywords: Flavonoid, Natural diversity, Phenelyacylation, Species conservation, UV-B
Introduction
Flavonoids are by far the major sub-class of phenylpropanoids, which are themselves the third largest class of plant secondary metabolites (specialized metabolites). Flavonoids comprise in excess of 8000 metabolites bearing a common diphenylpropane (C6–C3–C6) backbone (Tohge et al. 2013a, b). Several classes of flavonoids have been categorized on the basis of their core skeleton: C6–C3–C6 (flavonoids), (C6–C3–C6)2 (biflavonoids) and (C6–C3–C6)n (condensed tannins). The major flavonoids, in which two aromatic rings are linked via a three carbon chain (C6–C3–C6), have been reported to have manifold biological functions in the productive pollen fertility, signalling with microorganisms, auxin transport regulation and pigmentation, as well as in the protection of plants from various environment stresses, for example high light, UV-B irradiation and pathogens/pests (Taylor and Grotewold 2005; Tohge et al. 2013a, b; Nakabayashi et al. 2014). Additionally they exhibit a range of diverse functions including roles in developmental processes in plants (Taylor and Grotewold 2005; Silva-Navas et al. 2016; Tohge and Fernie 2016). On the other hand, flavonoids have several health beneficial effects including anti-retroviral, anti-hypertensive, anti-inflammatory, anti-aging and insulin-sensitizing activities, the reduction of the risk of a range of chronic diseases including cardiovascular disease, cancer and osteoporosis as well as inhibition of low-density lipoprotein (LDL) oxidation (Butelli et al. 2008; Crozier et al. 2009; Zhang et al. 2014; Tohge and Fernie 2016).
Some sub-classes of phenylpropanoids, such as syringyl-lignin and hydroxycinnamate biosynthesis are consequently widespread in plant lineages even in Angiosperms. On the other hand, some flavonoid biosynthetic genes are missing in several plant species such as algae and moss (Tohge et al. 2013a, b). Since orthologous genes of the early steps in flavonoid pathway are found even in liverworts and mosses (Winkel-Shirley 2001), the evolution of flavonoid biosynthesis appears to be as yet incompletely understood. However, the appearance of subclasses of flavonoids (Fig. 1), suggests that branches in the pathway have evolved in specific plant lineages over the course of evolution. The core flavonoid biosynthetic pathway is well documented. The main chemical backbone of flavonoids (flavonoid aglycones, Fig. 1) is synthesized from phenylalanine via a set of core biosynthetic genes including CHS, chalcone isomerase (CHI), flavanone 3-hydroxylase (F3H), dihydroflavonol 4-reductase (DFR), anthocyanidin synthase (ANS) that are relatively well-conserved among land plants. Specific steps make specialized flavonoids such as isoflavone synthase (IFS) and isoflavone reductase (IFR) which make isoflavones, flavonol synthase (FLS) required for flavonol biosynthesis and flavonoid 3′,5′-hydroxylase (F3′5′H) for production of 3′,5′-hydroxylated flavonoids. Six major types of flavonoid aglycones, flavonols (Fig. 1a e.g. kaempferol and quercetin), flavones (Fig. 1b e.g. apigenin and luteolin), anthocyanidins (Fig. 1c e.g. cyanidin, delphinidin and petunidin), flavanols (Fig. 1d e.g. catechin and epicatechin), isoflavones (Fig. 1e e.g. daidzein) and flavanones (Fig. 1f e.g. naringenin and sakuranetin) are the major sub-classes of the flavonoid family. In addition, additional steps (Zhang et al. 2014) are involved in further decoration such as glycosylation and acylation of flavonoids and these exhibit a large natural diversity and are commonly species-specific (Tohge et al. 2013a, b). Several gene families have been characterized functionally as being involved in flavonoid decoration. Flavonoid glycosyltransferase genes were found in the UDP-sugar-dependent glycosyltransferase 1 (UGT1) (Yonekura-Sakakibara and Hanada 2011) and glycoside hydrolase family 1-type (beta glucosidase, BGLU) (Matsuba et al. 2010; Ishihara et al. 2016) gene families. Similarly, flavone synthases (FNS-I and FNS-II) have been found from both P450 and 2-oxoglutarate/Fe(II)-dependent dioxygenase (2-ODD) gene families (Tohge et al. 2013a, b). Furthermore, flavonoid acyltransferases have been characterized from both the BEAT/AHCT/HCBT/DAT (BAHD) (D’Auria 2006; Tohge et al. 2015; Petersen 2016) and the serine carboxypeptidase-like (SCPL) (Tohge et al. 2016) gene families. Interestingly, the same or highly similar products can be formed by enzymes from different gene families using differently-activated substrates. Such evolutionary convergence of flavonoid decoration enzymes has been subject to natural selective pressures which generate mechanisms for tolerance to environmental stress such as that imposed by natural UV-B (280–315 nm) irradiation (Jenkins 2009; Tilbrook et al. 2013) and photo-oxidative damage induced by photo-inhibition (Steyn et al. 2002;). Such effects of irradiance in UV-B wavelength range on plants has been widely studied, since UV-B causes primary effects of non-specific damage to DNA, RNA, proteins and lipid membranes as well as secondary effects such as inhibition of photosynthesis and growth, induction of cell death, and deformities in flower and pollen morphologices. Plants have a UVR/COP/HY5 cascade light signalling system to protect them from harmful UV-B damage, which includes metabolic perturbation and reprogramming under light stress. Part of this metabolic reprograming induces production of phytoprotectants such as phenylpropanoids in land plants. Enhancing our understanding of gene conservation and the corresponding specificity of flavonoids in nature is important for future studies in which interesting natural variants are characterized at the molecular genetic, biochemical and physiological levels to lay the foundations for the design of stress resistant crop plants.
Fig. 1.

Major flavonoid aglycones in land plants. a Flavonols, b flavones, c anthocyanidins, d flavanols, e isoflavones and f flavanones
Natural chemical diversity of functional phytochemicals often corresponds to the evolution of adaptation to specific environments, and therefore the understanding of natural diversity using a broad range of accessions offers a very powerful tool to investigate phytochemical functions in nature. Whilst studies of natural variation and diversity using accessions have been carried out in many plant species, including Arabidopsis, with regard to the accumulation of specific primary metabolites and glucosinolates (Schauer and Fernie 2006; Rowe et al. 2008; Carreno-Quintero et al. 2013; Wen et al. 2014; Schilmiller et al. 2015), few examples have focused on flavonoid biosynthesis (Routaboul et al. 2012; Ishihara et al. 2016; Tohge et al. 2016).
In a recent study, we isolated phenylacylated-flavonols amongst natural accessions of Arabidopsis and characterized the gene encoding flavonol phenylacyltransferase 2 (AtFPT2, At2g22960) responsible for the production of accession specific phenylacylated-flavonols. Additionally our UV-B stress experiments using transgenic plants expressing FPT2 and the gain-of-function NIL suggested that phenylacylation of flavonoids relates to the response to UV-B stress (Tohge et al. 2016). Phenylacylated-flavonoids are decorated with several hydroxycinnamoyl units (cinnamoyl, 4-coumaroyl, caffeoyl, feruloyl and sinapoyl). Furthermore, computational estimation of the most stable stereochemical structure of 6″-O-phenylacylflavonol resulted in a bridge piled structure between the aromatic rings A and sinapoyl ring like intramolecular copigmentation of phenylacylated anthocyanins (Bakowska-Barczak 2005; Tohge et al. 2016). Given that phenylacylation confers enhanced UV-absorbance, understanding the natural diversity in phenylacylated-flavonoids might provide insights into adaption to UV-B irradiance.
Phenylacylated-flavonoids are widely distributed in plant species including vegetables, trees and ornamental flowers. Genes encoding flavonoid-phenylacyltransferases were found in some of these species. These genes are known to have emerged via convergent evolution i.e. they have evolved independently in species of different lineages, but have highly similar functions. In this review, we detail recent advances in our understanding of (1) UV-B responses linked to the production of flavonoids, (2) studies of natural variance in the response to UV-B, (3) function of specific decorations of flavonoids under UV-B stress, and (4) broader natural diversity of phenylacylated-flavonoids in the plant kingdom.
Consequences of climate change on UV-B irradiance
Given a booming global population, a stable food supply under changing environmental conditions is becoming a priority, globally. The risk of UV-B damage is fundamental, although only 0.5% of UV-B (280–315 nm) irradiance reaches the earth’s surface (Blumthaler 1993). As we described above, UV-B causes several primary and secondary effects on organisms. When DNA absorbs UV-B energy, cyclobutane pyrimidine dimers (CPDs) form, leading to mutations (Garinis et al. 2005). In addition, proteins and lipids are damaged by UV-B irradiation itself (Kramer et al. 1991; Hideg et al. 2013). Oxidative stress, caused by an imbalance between reactive oxygen species (ROS) production and antioxidant scavenging capacity have been proposed as key factors in UV-B stress (Jenkins 2009). An overabundance of ROS production additionally damages cell components, resulting in apoptosis. The thylakoid membrane has been proposed as the source of free radicals generated from hydrogen peroxide (H2O2) cleavage in response to UV-B light (Hideg and Vass 1996), although the precise mechanism of ROS generation following exposure to UV-B irradiation remains unclear (Yannarelli et al. 2006; Jenkins 2009). ROS mediated signaling acts by adjusting gene expression, proteolysis and thioredoxin dynamics.
Given the profound effects of light that plants are subjected to, considerable selective pressure to generate tolerance mechanisms involved in both sensing and responding to natural UV-B irradiation (Sablowski et al. 1994; Jenkins 2009; Kusano et al. 2011; Tilbrook et al. 2013). Since the fractionation of UV-B irradiation after passing through the atmosphere is largely dependent on atmospheric conditions (for example cloud cover, snow, aerosol etc.), the final intensity of UV-B reaching the earth´s surface is highly unstable and variant day to day, although the total area of the ozone hole has decreased slightly since 2006 as a result of efforts to restrict the use of chlorofluorocarbons (Solomon et al. 2016). Such instability of UV-B irradiation could therefore impair crop yields greatly, since the detrimental effects described above combine to constrain food security considerably by impacting stabilities of food supplies, crop morphology and plant and seed yields (Mohammed and Tarpley 2010; Wargent and Jordan 2013).
The UV-B signalling response is mediated by the UVR8/COP/HY5 pathway
Our understanding of the signaling following UV-B irradiation in plants has been greatly enhanced by work on the UV-B photoreceptor of Arabidopsis (Arabidopsis thaliana) (AtUVR8) and the downstream signaling cascades it triggers (Rizzini et al. 2011). Current models of the signal transduction and transcriptional regulation network of the UV-B response in higher plants are summarised in Fig. 2a. UV-B photoreceptors have been studied in several plant species including desert poplar (Mao et al. 2015), grapeberry (Liu et al. 2015) and apple (Zhao et al. 2016). Interestingly a similar UVR8-mediated UV-B perception and acclimation response has been characterized functionally in Chlamydomonas (Tilbrook et al. 2016) suggesting that this response evolved early in the green plant lineage.
Fig. 2.
Model of UVR8/COP1 mediated UV-B signaling cascade. Current model of UVR8/COP1 mediated signaling cascade. PHY PHYTOCHROME A, CRY CRYPTOCHROME, UVR8 UV REPAIR DEFECTIVE 8, COP1 CONSTITUTIVE PHOTOMORPHOGENIC 1, DDB UV-damaged DNA binding protein, HY5 ELONGATED HYPOCOTYL 5, HYH HY5-HOMOLOG, CUL4 CULLIN 4, RUP REPRESSOR OF UV-B PHOTOMORPHOGENESIS
The genes involved in the UV-B signaling cascade have largely been characterized by forward genetic screening for UV-B irradiation-sensitive mutants. The principal genes identified as mediators of UV-B photomorphogenic responses are CONSTITUTIVE PHOTOMORPHOGENIC 1 (COP1, encoding a multi-functional E3-ubiquitin ligase) (Oravecz et al. 2006), ELONGATED HYPOCOTYL 5 (HY5, encoding a bZIP transcription factor) (Ulm et al. 2004), HY5-HOMOLOG (HYH) (Brown and Jenkins 2008) encoding another bZIP transcription factor and UV-B photoreceptor UVR8 (Rizzini et al. 2011). Interestingly, UV-B regulates the expression of many genes involved in diverse processes including metabolism, morphogenesis, photosynthesis, UV-protection and pest and pathogen defense (Kilian et al. 2007; Findlay and Jenkins 2016). Within this signalling cascade, the UV-B photoreceptor, UVR8, acts to up-regulate both HY5 and HYH, and down-regulate expression of COP1 which can balance photo-morphogenesis and high light responsive pathways. By contrast, the transcription factors, HY5 and HYH, control several MYB regulators such PRODUCTION OF ANTHOCYANIN PIGMENT 1 (PAP1/AtMYB75) and PRODUCTION OF FLAVONOL GLYCOSIDES (PFG1/MYB12) for anthocyanin and flavonol pathways respectively (Fig. 2a) (Tohge et al. 2005; Stracke et al. 2011). In addition, the UV-B responsive AtMYB4 was characterized as a negative regulator of phenylpropanoid pathway by its function in downregulating expression of the genes encoding cinnamate-4-hydroxylase (C4H) and 4-CoA ligase (4CL) which are key enzymatic steps in general phenylpropanoid metabolism in Arabidopsis. Expression of AtMYB4 represses the production of sinapoyl-malate which offers protection against UV irradiation. AtMYB4 belongs to quite an ancient family of genes encoding R2R3MYB transcriptional repressors. It may be that the earliest UV protecting agents were hydroxycinnamic acid-esters, such as chlorogenic acids. In specific cases, flavonoids may have taken over some or all of the roles of UV-protection but not in species belonging to the family Solanaceae, like tobacco which relies heavily on chlorogenic acids for UV-B protection. These regulators which are, in turn, under the regulation of HY5 suggested that the complex structure of light signalling cascade can influence metabolism in order to adapt to severe environments of higher UV-B intensity.
The metabolic hierarchy of the UV-B response
Despite the direct connections within the UV-B signaling cascade between UV-B irradiation and metabolic reprogramming, signaling has been reported to adopt a hierarchic multi-phasic response structure in several plant species (Casati et al. 2011; Kusano et al. 2011; Kim et al. 2012; Kaling et al. 2015). Indeed the reprogramming of metabolic pathways appears to contain many different factors including ROS effects, inhibition of photosynthesis, the imbalance of energy and central metabolism and large-scale changes in gene expression but also the production of phytochemical protectants. During the early stages of exposure to UV-B irradiation, plant cells are “primed” for higher primary metabolite levels via a mechanism involving metabolic reprogramming for efficient conservation of carbon to the aromatic amino acid (AAA) precursors for the production of phenylpropanoids such as hydroxycinnamates and flavonoids. Metabolite profiling over a time course following UV-irradiation allow us to suggest that ascorbate plays an important role in the short-term response to UV-B irradiation. Additionally, results suggested that both flavonoid and hydroxycinnamates play a more important role in the adaptive response of plants to UV-B exposure (Kusano et al. 2011). Observation of general reductions in sugar levels (Casati et al. 2011; Kusano et al. 2011; Kim et al. 2012) and the reduced expression of genes associated with photosynthesis, photorespiration and the Calvin–Benson cycle (Kilian et al. 2007) allowed us to suggest a consequent down-regulation of genes associated with cell wall synthesis, starch synthesis and lipid metabolism (Koch 1996; Kusano et al. 2011). On the other hand, the transcriptomic and metabolomic changes in the TCA cycle, nucleotide metabolism and amino acid biosynthesis suggested these changes are only partially in accordance with changes in the steady-state levels of intermediates which are closely associated with these processes.
Natural variance in the response to UV-B
Although many studies have reported the UV-B protective function of various phenylpropanoids focusing on the biological processes such as flower development, pollination and seed production (Rozema et al. 2009), few studies succeeded to establish any relationship between UV-B stress and different accessions. In a recent study of growth related traits in 345 Arabidopsis thaliana accessions in response to UV radiation stress, a mutation in phenylalanine ammonia lyase (PAL1) which encodes the first enzyme of phenylpropanoid biosynthesis was found to be associated with genetic polymorphism between UV-B sensitive and resistant accessions (Piofczyk et al. 2015). Interestingly the Arabidopsis pna-10 accession was characterized as a natural deletion mutant of the SNG1 (At2g22990) gene, which encodes an enzyme in the biosynthesis of sinapoyl-malate which is one of hydroxycinnamate derivatives (Li et al. 2010).
Recently we performed profiling of natural variation of floral secondary metabolite composition and abundance in 64 Arabidopsis accessions (Tohge et al. 2016). For this purpose a previously described liquid-chromatography/mass-spectrometry (LC/MS) protocol (Tohge and Fernie 2010), was used to profile 68 peaks which included 24 flavonoids and 18 unknown peaks which were putatively annotated as flavonol derivatives, by MS/MS analysis. Following purification of these compounds a battery of analytical chemical techniques including 1- and 2-D nuclear magnetic resonance (NMR) spectroscopy was employed, and one of the major peaks was identified as a novel phenylacylated-flavonol glycoside (flavonol-3-O-(2″-O-rhamnosyl-6″-O-sinapoyl)glucoside-7-O-rhamnoside) and given the trivial name saiginol A (Tohge et al. 2016). The other 17 peaks, subsequently named saiginiols B-R, were annotated as novel phenylacylated-flavonol glycosides (flavonol-3-O-(-6″-O-phenylacyl)glycoside-7-O-rhamnosides) according to the presence or absence of three possible aglycones (Fig. 1, kaempferol, quercetin and isorhamnetin) and as displaying one of three types of phenylacylation (Fig. 3, sinapoyl, caffeoyl and p-coumaroyl moieties) based on MS/MS fragmentation and elution time profiles. A total of 29 putative flavonol-phenylacylglycosides (annotated to date are 2″- or 4″-O-phenylacylated-flavonols) have been documented in Brassica vegetables (Cartea et al. 2011).
Fig. 3.

Phenylacyl moieties found in flavonoids
Following mQTL (metabolic quantitative trait locus) analysis using Near Isogenic Lines (NIL) harboring chromosome segmental substitution of Col-0 (saiginol non-producer) in C24 (saiginol producer) or the reciprocal substitutions of C24 in Col-0, and transcriptome analysis focusing solely on the mQTL region, only two genes were identified that displayed massively altered expression. These genes were subsequently renamed as putative flavonol phenylacyltransferases (AtFPT). The second candidate gene, FPT2 (At2g22960), was functionally characterized by metabolite profiling of 35S-driven overexpressing transgenic plants (Tohge et al. 2016). Sequence analysis revealed short or large gene deletions (2–600 bp) in non-producer accessions. Interestingly, the accessions deriving from high irradiance habitats (from latitudes between 16 and 43° North or altitudes above 1000 m) produced saiginols. Additionally phenylacylation renders flavonols much more efficient in UV-B absorption than their molecular precursors. Importantly, the levels of only the saiginols and not the overall levels of phenylpropanoids were altered in these accession demonstrating the speicificity of this modification. These results suggested that FPT2 and by implication the saiginols confer additional protection against UV-B. In further UV-B treatment experiments using transgenic plants expressing FPT2, and the gain-of-function NIL, saiginol producing lines showed higher silique production and seed production under UV-B irradiation (Tohge et al. 2016). This result suggested that phenylacylation renders flavonoids much more efficient in UV-B absorption and tolerance under UV-B irradiation.
Other plant phenylacyltransferase genes
Genes encoding flavonoid phenylacyltransferases which transfer hydroxycinnamoyl moieties to flavonoid glycosides were first identified within the BAHD gene family. The BAHD gene super family includes a large number of acyltransferases using CoA activated acids. Phenylacyl-CoA: anthocyanin-3-O-phenylacyltransferase activities were reported, at an early stage, in some plant species such as Silene dioica (Kamsteeg et al. 1980) and Matthiola incana (Teusch et al. 1987). Subsequently, anthocyanin-5-O-Glc-6‴-O-phenylacyltransferase in Gentiana triflora (Fujiwara et al. 1998) and Perilla frutescens (Yonekura-Sakakibara et al. 2000) and anthocyanin-3-O-rhamnosylglucoside p-coumaroyltransferase in Iris hollandica (Yoshihara et al. 2006) were identified. In Arabidopsis, two genes encoding anthocyanin-3-O-glucoside p-coumaroyltransferases were characterized (Luo et al. 2007). Recently flavonoid-3-O-rutinoside-4‴-phenylacyltransferase (SlFdAT1, Solyc12g088170) was identified using a combined metabolomics and transcriptomics integrative approach using Del/Ros1 expressing transgenic purple tomato (Tohge et al. 2015).
In contrast, the AtFPT2 gene corresponding to saiginol production found in Arabidopsis accessions encodes a serine carboxypeptidase-like (SCPL) gene (Tohge et al. 2016). Amongst SPCL genes only the phenylacyltransferase AtFPT2 and AtSAT (anthocyanin-3-O-(2″-O-xylosyl)glucoside p-coumaroyltransferase (Fraser et al. 2007) have been characterized to date. Both AtFPT2 and AtSAT genes are found within genomic regions recently evolved via tandem duplication of SCPL genes. Flavonoid phenylacyltransferase genes are conserved only within closely related species but not between plant species having similar functions of phenylacyltransferase. Interestingly, both BAHD and SCPL genes which could be encoding phenylacyltransferase are found not only in flavonoid biosynthesis but also in biosynthesis of other UV-B protectants such as sinapoyl-derivatives in Brassica plants and chlorogenic acids in Solanaceous plants. A hydroxycinnamoylCoA: quinate hydroxycinnamoyl transferase (HQT) which is responsible for the synthesis of chlorogenates in Solanaceous plants is an anciently evolved BAHD enzyme. HQT and a hydroxycinnamoyl-CoA transferase (HCT) stand out from other BAHD acyl-transferases (Sonnante et al. 2010) as having evolved early and retained their structural differences over long periods of evolutionary time unlike the other more versatile BAHDs. Despite of this fact, Brassica phenylacyltransferase genes are assumed to have relatively recently evolved under natural selective pressure such as light selection following their speciation.
Natural diversity of phenylacylated-flavonoids in plants
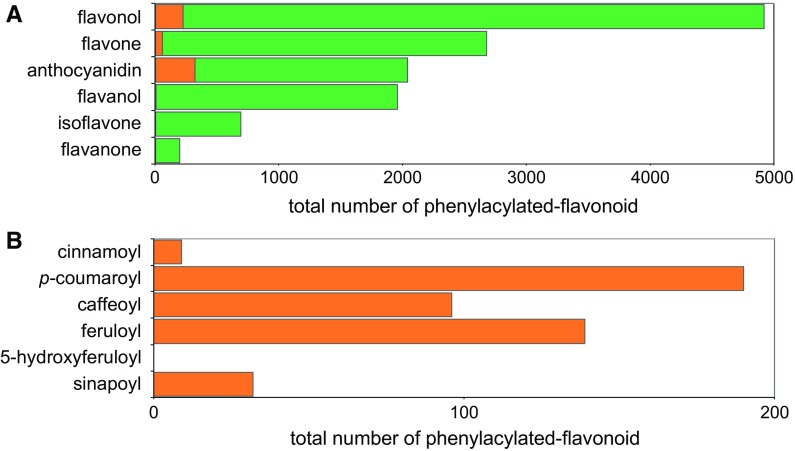
Given the species specificity of phenylacyltransferase genes, the understanding of natural diversity of phenylacylated-flavonoids has become an important means by which to study the relationship between gene functional- and chemical-diversities in nature. In order to check natural diversity of phenylacylated-flavonoids, the KNApSAcK database (http://kanaya.naist.jp/KNApSAcK/) (Afendi et al. 2012) which is one of the largest databases of known literature based phytochemicals was used. Major flavonoids were searched for aglycones and phenylacyl moieties described in Figs. 1 and 3. A total of 2846 flavonoids in 293 plant species were found (total 12,469 pairs of flavonoids per species). Interestingly, flavonols (4.7% of 4914 compounds) and anthocyanins (18.5% of 2038 compounds) exhibited a much higher proportion of phenylacylation than other classes of flavonoids, flavone (2.0% of 2673 compounds), flavanol (0.3% of 1959 compounds), isoflavone (0.1% of 690 compounds) and flavanone (0.0% of 195 compounds) (Fig. 4a). Phenylacylated-flavonoids are, therefore, mainly found in conserved classes of flavonoids such as anthocyanins and flavonols. Additionally p-coumaroyl, caffeoyl and feruloyl moieties are the major decoration form for phenylacylation whilst no 5-feruloyl-flavonoids [only one fond in purple lisianthusa by our literature survey (Markham et al. 2000)] were found amongst currently reported compounds (Fig. 4b). The comparisons of flavonoids and phenylacylated-flavonoids by compound class or decoration type showed clear differences between compound classes. However, in comparisons of total flavonoids and phenylacylated-flavonoids per species, no clear correlation between the taxonomic tree or speciation was observed (Fig. 5). Solanales, Gentianales, Campanulids and Brassicaceae plants exhibit a relatively high ratio of phenylacylated-flavonoids, but their closely related species did not share this trend. Our analysis of the natural diversity of phenylacylated-flavonols suggests that flavonoid-phenylacylation has likely evolved for certain flavonoid classes independently in different plant species. We contend that further understanding of natural diversity of phenylacylated-flavonoids and functional characterization of phenylacyltransferases will provide an overview of phytochemical evolution in relation to plant speciation and natural selections such as light selection.
Fig. 4.
Natural diversity and ratio of flavonoid with/without phenylacyl moieties in land plants. a Total number of flavonoid and phenylacylated-flavonoid in each sub-class of flavonoids, b total number of phenylacylated-flavonoid in each decoration type. Flavonoid entries found in KNApSAcK (http://kanaya.naist.jp/knapsack_jsp/top.html) were used. Green and orange indicate non-phenylacylated- and phenylacylated-flavonoid, respectively. (Color figure online)
Fig. 5.
Natural diversity and ratio of total flavonoid and phenylacylated flavonoid per family in land plants. Flavonoid entries found in KNApSAcK (http://kanaya.naist.jp/knapsack_jsp/top.html) were used
Conclusions
To adapt under light stress conditions, land plants have a light signaling system including induction of flavonoid production which can mitigate light stress. In a recent study of natural accessions of Arabidopsis, phenylacylation of flavonoids was found to confer enhanced phytochemical functions as UV-B protectants in plants. In our analysis of natural diversity of phenylacylated-flavonoids, this type of flavonoid decoration was found in some specific flavonoid class such as anthocyanin and flavonol. Additionally they were observed in several specific plant species. Given the fact that we assume this is due to the functional convergence of phenylacyltransferase then the identification of genes encoding flavonoid-phenylacyltransferases and studies of natural diversity of phenylacylated-flavonoids will be highly valuable in order to secure food yields. However functional characterization of phenylacylflavonoids are still not well documented, and the investigation of associations of phenylacylation of flavonoids with other stress conditions such as where there are limitations on dark fixation of photosynthesis but incident light is high, as in cold, high light conditions, will thus be an important topic for future research. Additionally since flavonoids offer a legion of other protective functions, both in planta and following dietary intake by animals, it will be important to assess the role of phenylacylated flavonoids in these contexts.
Acknowledgements
Open access funding provided by Max Planck Society. Funding from the Max-Planck-Society (to TT, LS and ARF) is gratefully acknowledged. We thank to National Council for Scientific and Technological Development CNPq-Brazil for financially supported to LPS.
Abbreviations
- ANS
Anthocyanidin synthase
- BAHD
BEAT/AHCT/HCBT/DAT
- BGLU
Beta glucosidase
- CHI
Chalcone isomerase
- CHS
Naringenin-chalcone synthase
- COP1
CONSTITUTIVE PHOTOMORPHOGENIC 1
- CPD
Cyclobutane pyrimidine dimers
- CRY
CRYPTOCHROME
- CUL4
CULLIN 4
- DDB
UV-damaged DNA binding protein
- DFR
Dihydroflavonol 4-reductase
- EBG
Early biosynthetic genes
- F3′5′H
Flavonoid 3′,5′-hydroxylase
- F3H
Flavanone 3-hydroxylase
- FLS
Flavonol synthase
- FPT
Flavonol phenylacyltransferase
- HY5
ELONGATED HYPOCOTYL 5
- HYH
HY5-HOMOLOG
- IFR
Isoflavone reductase
- IFS
Isoflavone synthase
- LBG
Late biosynthetic genes
- LDL
Low-density lipoprotein
- NIL
Near isogenic lines
- NMR
Nuclear magnetic resonance
- PAL
Phenylalanine ammonia lyase
- PHY
PHYTOCHROME A
- QTL
Quantitative trait locus
- RUP
REPRESSOR OF UV-B PHOTOMORPHOGENESIS
- SCPL
Serine carboxypeptidase-like
- UGT1
UDP-sugar-dependent glycosyltransferase 1
- UV
Ultra-violet
- UVR8
UV REPAIR DEFECTIVE 8
References
- Afendi FM, Okada T, Yamazaki M, Hirai-Morita A, Nakamura Y, Nakamura K, Ikeda S, Takahashi H, Altaf-Ul-Amin M, Darusman LK, Saito K, Kanaya S. KNApSAcK family databases: integrated metabolite-plant species databases for multifaceted plant research. Plant Cell Physiol. 2012;53:e1. doi: 10.1093/pcp/pcr165. [DOI] [PubMed] [Google Scholar]
- Bakowska-Barczak A. Acylated anthocyanins as stable, natural food colorants—a review. Pol J Food Nutr Sci. 2005;14:107–115. [Google Scholar]
- Blumthaler M (1993) Solar UV measurements. In: Tevini M (ed) UV-B radiation and ozone depletion: effects on humans, animals, plants, microorganisms, and materials. Lewis Publishers, Raton, USA. pp 71–94
- Brown BA, Jenkins GI. UV-B signaling pathways with different fluence-rate response profiles are distinguished in mature Arabidopsis leaf tissue by requirement for UVR8, HY5, and HYH. Plant Physiol. 2008;146:576–588. doi: 10.1104/pp.107.108456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butelli E, Titta L, Giorgio M, Mock HP, Matros A, Peterek S, Schijlen EGWM, Hall RD, Bovy AG, Luo J, Martin C. Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors. Nat Biotechnol. 2008;26:1301–1308. doi: 10.1038/nbt.1506. [DOI] [PubMed] [Google Scholar]
- Carreno-Quintero N, Bouwmeester HJ, Keurentjes JJB. Genetic analysis of metabolome-phenotype interactions: from model to crop species. Trends Genet. 2013;29:41–50. doi: 10.1016/j.tig.2012.09.006. [DOI] [PubMed] [Google Scholar]
- Cartea ME, Francisco M, Soengas P, Velasco P. Phenolic compounds in Brassica vegetables. Molecules. 2011;16:251–280. doi: 10.3390/molecules16010251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casati P, Campi M, Morrow DJ, Fernandes JF, Walbot V. Transcriptomic, proteomic and metabolomic analysis of UV-B signaling in maize. BMC Genom. 2011;12:321. doi: 10.1186/1471-2164-12-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crozier A, Jaganath IB, Clifford MN. Dietary phenolics: chemistry, bioavailability and effects on health. Nat Prod Rep. 2009;26:1001–1043. doi: 10.1039/b802662a. [DOI] [PubMed] [Google Scholar]
- D’Auria JC. Acyltransferases in plants: a good time to be BAHD. Curr Opin Plant Biol. 2006;9:331–340. doi: 10.1016/j.pbi.2006.03.016. [DOI] [PubMed] [Google Scholar]
- Findlay KM, Jenkins GI. Regulation of UVR8 photoreceptor dimer/monomer photo-equilibrium in Arabidopsis plants grown under photoperiodic conditions. Plant Cell Environ. 2016;39:1706–1714. doi: 10.1111/pce.12724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser CM, Thompson MG, Shirley AM, Ralph J, Schoenherr JA, Sinlapadech T, Hall MC, Chapple C. Related Arabidopsis serine carboxypeptidase-like sinapoylglucose acyltransferases display distinct but overlapping substrate specificities. Plant Physiol. 2007;144:1986–1999. doi: 10.1104/pp.107.098970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara H, Tanaka Y, Yonekura-Sakakibara K, Fukuchi-Mizutani M, Nakao M, Fukui Y, Yamaguchi M, Ashikari T, Kusumi T. cDNA cloning, gene expression and subcellular localization of anthocyanin 5-aromatic acyltransferase from Gentiana triflora. Plant J. 1998;16:421–431. doi: 10.1046/j.1365-313x.1998.00312.x. [DOI] [PubMed] [Google Scholar]
- Garinis GA, Mitchell JR, Moorhouse MJ, Hanada K, de Waard H, Vandeputte D, Jans J, Brand K, Smid M, van der Spek PJ, Hoeijmakers JHA, Kanaar R, van der Horst GTJ. Transcriptome analysis reveals cyclobutane pyrimidine dimers as a major source of UV-induced DNA breaks. EMBO J. 2005;24:3952–3962. doi: 10.1038/sj.emboj.7600849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hideg E, Vass I. UV-B induced free radical production in plant leaves and isolated thylakoid membranes. Plant Sci. 1996;115:251–260. doi: 10.1016/0168-9452(96)04364-6. [DOI] [Google Scholar]
- Hideg E, Jansen MAK, Strid A. UV-B exposure, ROS, and stress: inseparable companions or loosely linked associates? Trends Plant Sci. 2013;18:107–115. doi: 10.1016/j.tplants.2012.09.003. [DOI] [PubMed] [Google Scholar]
- Ishihara H, Tohge T, Viehöver P, Fernie A, Weisshaar B, Stracke R. Natural variation in flavonol accumulation in Arabidopsis is determined by the flavonol glucosyltransferase BGLU6. J Exp Bot. 2016;67:1505–1517. doi: 10.1093/jxb/erv546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins GI. Signal transduction in responses to UV-B radiation. Annu Rev Plant Biol. 2009;60:407–431. doi: 10.1146/annurev.arplant.59.032607.092953. [DOI] [PubMed] [Google Scholar]
- Kaling M, Kanawati B, Ghirardo A, Albert A, Winkler JB, Heller W, Barta C, Loreto F, Schmitt-Kopplin P, Schnitzler JP. UV-B mediated metabolic rearrangements in poplar revealed by non-targeted metabolomics. Plant Cell Environ. 2015;38:892–904. doi: 10.1111/pce.12348. [DOI] [PubMed] [Google Scholar]
- Kamsteeg J, VanBrederode J, Kuipers-Tierei E, Van Nigtevecht G. Identification, properties and genetic control of UDP-Lrhamnose: anthocyanidin 3-O-glucoside, 6-O-rhamnosyltransferase isolated from the petals of red campion (Silene dioica) Z Naturforsch Sect C Biosci. 1980;33:249–257. [Google Scholar]
- Kilian J, Whitehead D, Horak J, Wanke D, Weinl S, Batistic O, D’Angelo C, Bornberg-Bauer E, Kudla J, Harter K. The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J. 2007;50:347–363. doi: 10.1111/j.1365-313X.2007.03052.x. [DOI] [PubMed] [Google Scholar]
- Kim S, Yun EJ, Hossain MA, Lee H, Kim KH. Global profiling of ultraviolet-induced metabolic disruption in Melissa officinalis by using gas chromatography-mass spectrometry. Anal Bioanal Chem. 2012;404:553–562. doi: 10.1007/s00216-012-6142-0. [DOI] [PubMed] [Google Scholar]
- Koch KE. Carbohydrate-modulated gene expression in plants. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:509–540. doi: 10.1146/annurev.arplant.47.1.509. [DOI] [PubMed] [Google Scholar]
- Kramer GF, Norman HA, Krizek DT, Mirecki RM. Influence of UV-B radiation on polyamines, lipid-peroxidation and membrane-lipids in cucumber. Phytochemistry. 1991;30:2101–2108. doi: 10.1016/0031-9422(91)83595-C. [DOI] [Google Scholar]
- Kusano M, Tohge T, Fukushima A, Kobayashi M, Hayashi N, Otsuki H, Kondou Y, Goto H, Kawashima M, Matsuda F, Niida R, Matsui M, Saito K, Fernie AR. Metabolomics reveals comprehensive reprogramming involving two independent metabolic responses of Arabidopsis to UV-B light. Plant J. 2011;67:354–369. doi: 10.1111/j.1365-313X.2011.04599.x. [DOI] [PubMed] [Google Scholar]
- Li X, Bergelson J, Chapple C. The ARABIDOPSIS accession Pna-10 is a naturally occurring sng1 deletion mutant. Mol Plant. 2010;3:91–100. doi: 10.1093/mp/ssp090. [DOI] [PubMed] [Google Scholar]
- Liu LL, Gregan S, Winefield C, Jordan B. From UVR8 to flavonol synthase: UV-B-induced gene expression in Sauvignon blanc grape berry. Plant Cell Environ. 2015;38:905–919. doi: 10.1111/pce.12349. [DOI] [PubMed] [Google Scholar]
- Luo J, Nishiyama Y, Fuell C, Taguchi G, Elliott K, Hill L, Tanaka Y, Kitayama M, Yamazaki M, Bailey P, Parr A, Michael AJ, Saito K, Martin C. Convergent evolution in the BAHD family of acyl transferases: identification and characterization of anthocyanin acyl transferases from Arabidopsis thaliana. Plant J. 2007;50:678–695. doi: 10.1111/j.1365-313X.2007.03079.x. [DOI] [PubMed] [Google Scholar]
- Mao K, Wang L, Li YY, Wu RL. Molecular cloning and functional analysis of UV RESISTANCE LOCUS 8 (PeUVR8) from Populus euphratica. PLoS ONE. 2015;10:e0132390. doi: 10.1371/journal.pone.0132390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markham KR, Gould KS, Winefield CS, Mitchell KA, Bloor SJ, Boase MR. Anthocyanic vacuolar inclusions—their nature and significance in flower colouration. Phytochemistry. 2000;55:327–336. doi: 10.1016/S0031-9422(00)00246-6. [DOI] [PubMed] [Google Scholar]
- Matsuba Y, Sasaki N, Tera M, Okamura M, Abe Y, Okamoto E, Nakamura H, Funabashi H, Takatsu M, Saito M, Matsuoka H, Nagasawa K, Ozeki Y. A novel glucosylation reaction on anthocyanins catalyzed by acyl-glucose-dependent glucosyltransferase in the petals of carnation and delphinium. Plant Cell. 2010;22:3374–3389. doi: 10.1105/tpc.110.077487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammed AR, Tarpley L. Differential response of Southern US rice (Oryza sativa L.) cultivars to ultraviolet-B radiation. J Agron Crop Sci. 2010;196:286–295. [Google Scholar]
- Nakabayashi R, Yonekura-Sakakibara K, Urano K, Suzuki M, Yamada Y, Nishizawa T, Matsuda F, Kojima M, Sakakibara H, Shinozaki K, Michael AJ, Tohge T, Yamazaki M, Saito K. Enhancement of oxidative and drought tolerance in Arabidopsis by overaccumulation of antioxidant flavonoids. Plant J. 2014;77:367–379. doi: 10.1111/tpj.12388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oravecz A, Baumann A, Mate Z, Brzezinska A, Molinier J, Oakeley EJ, Adam E, Schafer E, Nagy F, Ulm R. CONSTITUTIVELY PHOTOMORPHOGENIC1 is required for the UV-B response in Arabidopsis. Plant Cell. 2006;18:1975–1990. doi: 10.1105/tpc.105.040097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen M. Hydroxycinnamoyltransferases in plant metabolism. Phytochem Rev. 2016;15:699–727. doi: 10.1007/s11101-015-9417-1. [DOI] [Google Scholar]
- Piofczyk T, Jeena G, Pecinka A. Arabidopsis thaliana natural variation reveals connections between UV radiation stress and plant pathogen-like defense responses. Plant Physiol Biochem. 2015;93:34–43. doi: 10.1016/j.plaphy.2015.01.011. [DOI] [PubMed] [Google Scholar]
- Rizzini L, Favory J-J, Cloix C, Faggionato D, O’Hara A, Kaiserli E, Baumeister R, Schaefer E, Nagy F, Jenkins GI, Ulm R. Perception of UV-B by the Arabidopsis UVR8 protein. Science. 2011;332:103–106. doi: 10.1126/science.1200660. [DOI] [PubMed] [Google Scholar]
- Routaboul JM, Dubos C, Beck G, Marquis C, Bidzinski P, Loudet O, Lepiniec L. Metabolite profiling and quantitative genetics of natural variation for flavonoids in Arabidopsis. J Exp Bot. 2012;63:3749–3764. doi: 10.1093/jxb/ers067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe HC, Hansen BG, Halkier BA, Kliebenstein DJ. Biochemical networks and epistasis shape the Arabidopsis thaliana metabolome. Plant Cell. 2008;20:1199–1216. doi: 10.1105/tpc.108.058131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozema J, Blokker P, Fuertes MAM, Broekman R. UV-B absorbing compounds in present-day and fossil pollen, spores, cuticles, seed coats and wood: evaluation of a proxy for solar UV radiation. Photochem Photobiol Sci. 2009;8:1233–1243. doi: 10.1039/b904515e. [DOI] [PubMed] [Google Scholar]
- Sablowski RWM, Moyano E, Culianezmacia FA, Schuch W, Martin C, Bevan M. A flower-specific Myb protein activates transcription of phenylpropanoid biosynthetic genes. EMBO J. 1994;13:128–137. doi: 10.1002/j.1460-2075.1994.tb06242.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer N, Fernie AR. Plant metabolomics: towards biological function and mechanism. Trends Plant Sci. 2006;11:508–516. doi: 10.1016/j.tplants.2006.08.007. [DOI] [PubMed] [Google Scholar]
- Schilmiller AL, Moghe GD, Fan P, Ghosh B, Ning J, Jones AD, Last RL. Functionally divergent alleles and duplicated Loci encoding an acyltransferase contribute to acylsugar metabolite diversity in solanum trichomes. Plant Cell. 2015;27:1002–1017. doi: 10.1105/tpc.15.00087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva-Navas J, Moreno-Risueño MA, Manzano C, Téllez-Robledo B, Navarro-Neila S, Carrasco V, Pollmann S, Gallego FJ, del Pozo JC. Flavonols mediate root phototropism and growth through regulation of proliferation-to-differentiation transition. Plant Cell. 2016;28:1372–1387. doi: 10.1105/tpc.15.00857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon S, Ivy DJ, Kinnison D, Mills MJ, Neely RR, III, Schmidt A. Emergence of healing in the Antarctic ozone layer. Science. 2016;353:269–274. doi: 10.1126/science.aae0061. [DOI] [PubMed] [Google Scholar]
- Sonnante G, D’Amore R, Blanco E, Pierri CL, De Palma M, Luo J, Tucci M, Martin C. Novel hydroxycinnamoyl-coenzyme A quinate transferase genes from artichoke are involved in the synthesis of chlorogenic acid. Plant Physiol. 2010;153:1224–1238. doi: 10.1104/pp.109.150144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steyn WJ, Wand SJE, Holcroft DM, Jacobs G. Anthocyanins in vegetative tissues: a proposed unified function in photoprotection. N Phytol. 2002;155:349–361. doi: 10.1046/j.1469-8137.2002.00482.x. [DOI] [PubMed] [Google Scholar]
- Stracke R, Favory J-J, Gruber H, Bartelniewoehner L, Bartels S, Binkert M, Funk M, Weisshaar B, Ulm R. The Arabidopsis bZIP transcription factor HY5 regulates expression of the PFG1/MYB12 gene in response to light and ultraviolet-B radiation. Plant Cell Environ. 2011;33:88–103. doi: 10.1111/j.1365-3040.2009.02061.x. [DOI] [PubMed] [Google Scholar]
- Taylor LP, Grotewold E. Flavonoids as developmental regulators. Curr Opin Plant Biol. 2005;8:317–323. doi: 10.1016/j.pbi.2005.03.005. [DOI] [PubMed] [Google Scholar]
- Teusch M, Forkmann G, Seyffert W. Genetic control of hydroxycinnamoyl-coenzyme a: Anthocyanidin 3-glycoside-hydroxycinnamoyltransferase from petals of Matthiola incana. Phytochemistry. 1987;26:991–994. doi: 10.1016/S0031-9422(00)82333-X. [DOI] [Google Scholar]
- Tilbrook K, Arongaus AB, Binkert M, Heijde M, Yin R, Ulm R. The UVR8 UV-B photoreceptor: perception, signaling and response. Arabidopsis Book. 2013;11:e0164. doi: 10.1199/tab.0164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilbrook K, Dubois M, Crocco CD, Yin R, Chappuis R, Allorent G, Schmid-Siegert E, Goldschmidt-Clermont M, Ulm R. UV-B perception and acclimation in Chlamydomonas reinhardtii. Plant Cell. 2016;28:966–983. doi: 10.1105/tpc.15.00287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tohge T, Fernie AR. Combining genetic diversity, informatics and metabolomics to facilitate annotation of plant gene function. Nat Protoc. 2010;5:1210–1227. doi: 10.1038/nprot.2010.82. [DOI] [PubMed] [Google Scholar]
- Tohge T, Fernie A. Specialized metabolites of the flavonol class mediate root phototropism and growth. Mol Plant. 2016;9:1554–1555. doi: 10.1016/j.molp.2016.10.019. [DOI] [PubMed] [Google Scholar]
- Tohge T, Nishiyama Y, Hirai MY, Yano M, Nakajima J, Awazuhara M, Inoue E, Takahashi H, Goodenowe DB, Kitayama M, Noji M, Yamazaki M, Saito K. Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor. Plant J. 2005;42:218–235. doi: 10.1111/j.1365-313X.2005.02371.x. [DOI] [PubMed] [Google Scholar]
- Tohge T, Watanabe M, Hoefgen R, Fernie AR. The evolution of phenylpropanoid metabolism in the green lineage. Crit Rev Biochem Mol Biol. 2013;48:123–152. doi: 10.3109/10409238.2012.758083. [DOI] [PubMed] [Google Scholar]
- Tohge T, Watanabe M, Hoefgen R, Fernie AR. Shikimate and phenylalanine biosynthesis in the green lineage. Front Plant Sci. 2013;4:62. doi: 10.3389/fpls.2013.00062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tohge T, Zhang Y, Peterek S, Matros A, Rallapalli G, Tandrón Y, Butelli E, Kallam K, Hertkorn N, Mock H, Martin C, Fernie A. Ectopic expression of snapdragon transcription factors facilitates the identification of genes encoding enzymes of anthocyanin decoration in tomato. Plant J. 2015;83:686–704. doi: 10.1111/tpj.12920. [DOI] [PubMed] [Google Scholar]
- Tohge T, Wendenburg R, Ishihara H, Nakabayashi R, Watanabe M, Sulpice R, Hoefgen R, Takayama H, Saito K, Stitt M, Fernie A. Characterization of a recently evolved flavonol-phenylacyltransferase gene provides signatures of natural light selection in Brassicaceae. Nat Commun. 2016;7:12399. doi: 10.1038/ncomms12399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulm R, Baumann A, Oravecz A, Mate Z, Adam E, Oakeley EJ, Schafer E, Nagy F. Genome-wide analysis of gene expression reveals function of the bZIP transcription factor HY5 in the UV-B response of Arabidopsis. Proc Natl Acad Sci USA. 2004;101:1397–1402. doi: 10.1073/pnas.0308044100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wargent JJ, Jordan BR. From ozone depletion to agriculture: understanding the role of UV radiation in sustainable crop production. N Phytol. 2013;197:1058–1076. doi: 10.1111/nph.12132. [DOI] [PubMed] [Google Scholar]
- Wen W, Li D, Li X, Gao Y, Li W, Li H, Liu J, Liu H, Chen W, Luo J, Yan J. Metabolome-based genome-wide association study of maize kernel leads to novel biochemical insights. Nat Commun. 2014;5:3438. doi: 10.1038/ncomms4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkel-Shirley B. Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 2001;126:485–493. doi: 10.1104/pp.126.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yannarelli GG, Noriega GO, Batlle A, Tomaro ML. Heme oxygenase up-regulation in ultraviolet-B irradiated soybean plants involves reactive oxygen species. Planta. 2006;224:1154–1162. doi: 10.1007/s00425-006-0297-x. [DOI] [PubMed] [Google Scholar]
- Yonekura-Sakakibara K, Hanada K. An evolutionary view of functional diversity in family 1 glycosyltransferases. Plant J. 2011;66:182–193. doi: 10.1111/j.1365-313X.2011.04493.x. [DOI] [PubMed] [Google Scholar]
- Yonekura-Sakakibara K, Tanaka Y, Fukuchi-Mizutani M, Fujiwara H, Fukui Y, Ashikari T, Murakami Y, Yamaguchi M, Kusumi T. Molecular and biochemical characterization of a novel hydroxycinnamoyl-CoA: anthocyanin 3-O-glucoside-6″-O-acyltransferase from Perilla frutescens. Plant Cell Physiol. 2000;41:495–502. doi: 10.1093/pcp/41.4.495. [DOI] [PubMed] [Google Scholar]
- Yoshihara N, Imayama T, Matsuo Y, Fukuchi-Mizutani M, Tanaka Y, Ino I, Yabuya T. Characterization of cDNA clones encoding anthocyanin 3-p-coumaroyltransferase from Iris hollandica. Plant Science. 2006;171:632–639. doi: 10.1016/j.plantsci.2006.06.005. [DOI] [Google Scholar]
- Zhang Y, Butelli E, Martin C. Engineering anthocyanin biosynthesis in plants. Curr Opin Plant Biol. 2014;19:81–90. doi: 10.1016/j.pbi.2014.05.011. [DOI] [PubMed] [Google Scholar]
- Zhao C, Mao K, You CX, Zhao XY, Wang SH, Li YY, Hao YJ. Molecular cloning and functional analysis of a UV-B photoreceptor gene, MdUVR8 (UV Resistance Locus 8), from apple. Plant Sci. 2016;247:115–126. doi: 10.1016/j.plantsci.2016.03.006. [DOI] [PubMed] [Google Scholar]