Abstract
Microorganisms play a major role in biogeochemical cycles. As such they are attractive candidates for developing new or improving existing biotechnological applications, in order to deal with the accumulation and pollution of organic and inorganic compounds. Their ability to participate in bioremediation processes mainly depends on their capacity to metabolize toxic elements and catalyze reactions resulting in, for example, precipitation, biotransformation, dissolution, or sequestration. The contribution of genomics may be of prime importance to a thorough understanding of these metabolisms and the interactions of microorganisms with pollutants at the level of both single species and microbial communities. Such approaches should pave the way for the utilization of microorganisms to design new, efficient and environmentally sound remediation strategies, as exemplified by the case of arsenic contamination, which has been declared as a major risk for human health in various parts of the world.
Keywords: genomics, arsenic, bioremediation/phytoremediation, microorganism, ecosystem ecology
From Genes to Metagenomes
In over three billion years of evolution, microorganisms have colonized nearly all ecological niches, including the most extreme environments. Due to their multiple metabolic activities, they play a major part in biogeochemical cycles, affecting soil productivity or water quality (Madsen, 2011) and constitute an immense reservoir of genes with high potentials for biotechnology applications. For those reasons, microorganisms from the environment have aroused a strong interest since long before the microbial genomics era. A large number of enzymes and genes coding for biocatalyzers (cellulases, proteases, lipases/esterases, glycosidases, chitinases, xylanases, phosphatases) or for enzymes involved in vitamin and antibiotic biosynthesis have thus been isolated from environmental microorganisms (Colin et al., 2015; Jacques et al., 2017; Krüger et al., 2018). Many of these enzymes have been used for research, industrial or pharmaceutical applications (Madhavan et al., 2017) like, for instance, restriction enzymes and the Taq DNA polymerase that sparked a revolution in molecular biology techniques (Ishino and Ishino, 2014).
More than 20 years ago, thanks to the rise of molecular biology and the automation of DNA sequencing, microbiology embraced genomics, the ensemble of approaches which address the organization and activity of organisms within the scope of their full genome, acknowledging that no living system can be reduced to a single gene expressed at some time or another (Bertin et al., 2015). Since the very first genome sequence from a free-living organism, Haemophilus influenzae Rb (Fleischmann et al., 1995), the number of new microbial genome sequences published each year has grown exponentially to reach in 2014 a total of over 30,000 publicly available sequenced bacterial genomes (Land et al., 2015).
Yet, diversity data provided by molecular methods suggest that there remains in many ecosystems a vast majority of microorganisms belonging to taxa that have not been isolated in pure culture (Rashid and Stingl, 2015) and cultivation may be extremely difficult for a majority of them. Environmental genomic approaches could nonetheless provide access directly to the genome of uncultivated organisms like ‘Candidatus Desulforudis audaxviator,’ which practically represents the sole species present in a gold mine and can fix nitrogen using a cellular mechanism similar to that of Archaea (Chivian et al., 2008). Metagenomic analyses of nitrogen metabolism in anaerobic enriched cultures also led to the reconstruction of prokaryotic genomes such as Kuenenia stuttgartiensis (Strous et al., 2006), ‘Candidatus Nitrospira defluvii’ (Lücker et al., 2010) or the archaeon ‘Candidatus Methanoperedens nitroreducens’ (Haroon et al., 2013) involved in oceanic ammonium oxidation, nitrite oxidation in sewage treatment plant sludge and anaerobic oxidization of methane coupled to nitrate reduction, respectively. Similarly, the genome of an iron-oxidizer strain belonging to the Ferrovum genus was reconstructed from a mixed culture grown from samples collected in a mine water treatment plant (Ullrich et al., 2016).
Though molecular techniques associated with bioinformatic and genome-mining methods are invaluable tools to reveal the potential in genome data (Machado et al., 2017; Vallenet et al., 2017), cultivation remains an important challenge in microbiology, necessary for expanding our knowledge of microorganisms’ physiology and for bioremediation (Overmann et al., 2017). However, microorganisms from the environment may require essential nutrients or particular growth conditions, or may be extremely slow growers or obligate symbionts. Although tackling these issues generally demands strenuous efforts to design and test many isolation media, genome characterization may highlight metabolic characteristics of the targeted organism that could be leveraged to select and cultivate a given strain (Garza and Dutilh, 2015). This strategy allowed the isolation of the first nitrifying archaeon (Schleper et al., 2005) after an analysis of the Sargasso Sea metagenome (Venter et al., 2004) had detected on the same DNA fragment an Archaea-specific ribosomal gene and a gene coding for the ammonium monooxygenase, a key enzyme in nitrification. Subsequent physiological studies showed that the nitrification function was indeed expressed (Könneke et al., 2005). Another example is provided by the isolation of Leptospirillum ferrodiazotrophum (Tyson et al., 2005), which a previous metagenomic study had shown to be the only strain in an acid mine tailing to be able to fix nitrogen.
Beyond approaches centered on single organisms, the developments of genomics have rendered possible a global view of microbial communities that could help a better understanding of natural remediation processes and identifying candidate species for the design of bioremediation treatment plants. In this respect, high-throughput tools such as microarrays have allowed to address ecological questions related to the structure and function of microbial communities. Developed from the genomic data present in databases, such approaches may be helpful to study the diversity and dynamics of microbial populations using nucleic acids extraction and hybridization (Zhou et al., 2015). They were successfully used, for example, to examine the responses of microbial communities after the wreck of a drilling rig in the Gulf of Mexico had released about 5 million barrels of crude oil (Beazley et al., 2012). This study suggested that the microbial community of the rhizosphere in the affected coastal salt marsh could strongly contribute to hydrocarbon natural remediation. Recently, the combination of ribosomal 16S RNA gene high-throughput sequencing with DNA-based stable isotope probing in activated sludge samples incubated with Na213CO3 uncovered the dynamics of ammonium-oxidizing microorganism abundance and the relative importance of archaeal and bacterial ammonium oxidation activities in a waste water treatment plant (Pan et al., 2018).
In recent years, the development of high-throughput sequencing and assembly software has allowed to determine the complete genome sequence of uncultivated microorganisms from direct sequencing of metagenomic libraries or environmental DNA from complex microbial communities. Despite a number of critical issues regarding sampling, assembly or annotation (Teeling and Glöckner, 2012; Thomas et al., 2012), more than 10,000 metagenome projects are now referenced in the Genomes Online Database (Mukherjee et al., 2016). This number is expected to increase dramatically with such massive projects as the Earth Microbiome Project whose goal is to produce a global Gene Atlas of microbial communities encompassing an estimated 500,000 genomes (Gilbert et al., 2010; Thompson et al., 2017).
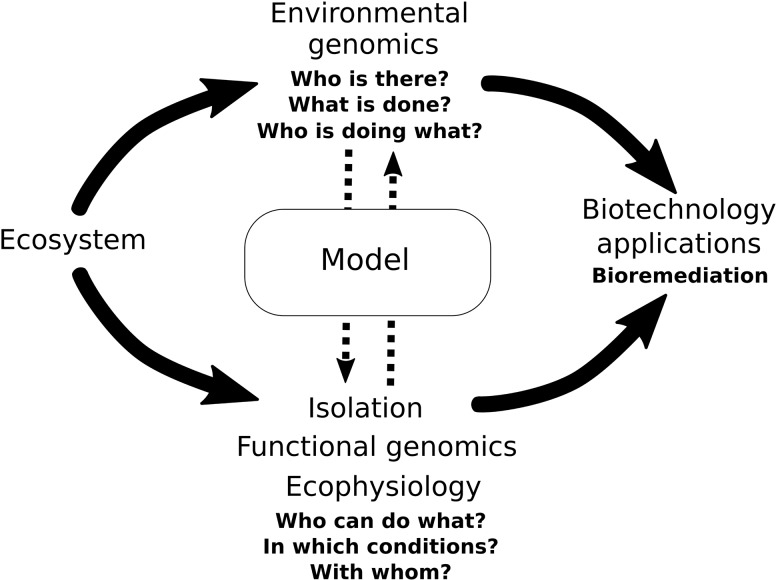
Environmental genomics now permits the study of the organisms in an ecosystem as a set of elements behaving within a complex network of interactions (Figure 1). For example, a genome-scale study of the complex symbiosis between the termite Macrotermes natalensis, its domesticated fungus and several gut bacterial communities demonstrated the cooperation between microorganisms in plant biomass conversion. The results showed that the insect provides the infrastructure allowing carbohydrate decomposition thanks to the functional complementarity between the fungus and the gut microbiota (Poulsen et al., 2014). More recently, the reconstruction of 2540 genomes using metagenomic data from 15 different sediment and groundwater environments allowed to highlight the key inter-organism interactions relevant to biogeochemical cycles in an aquifer in Colorado, United States (Anantharaman et al., 2016). Applying those approaches to various biotopes may thus provide valuable insights into the functioning of ecosystems, including polluted environments whose microbial communities could constitute prospective candidates for bioremediation.
FIGURE 1.
Multi-omic studies of microorganisms from the environment provide an integrated image or model of the microbial processes involved in the ecosystem’s functioning and that may be utilized for bioremediation or other biotechnology applications.
Arsenic Bioremediation and ‘Omics’ Approaches: a Case Study
Long-term exposure to arsenic represents a serious threat to human health worldwide (Nordstrom, 2002). Even though the occurrence of this element in drinking water constitutes the major source of exposure, recent studies on risks of arsenic accumulation in food revealed its presence in fish and crops cultivated with arsenic-contaminated waters (WHO, 2011; Jackson et al., 2012; Molin et al., 2015; Carlin et al., 2016). Numerous physico-chemical methods are commonly used for the treatment of arsenic-rich waters: coagulation/filtration, ion exchange, enhanced lime softening, adsorption and reverse osmosis (Ng et al., 2004; Nicomel et al., 2016). Over the last years, in a search for sustainable and cost-effective methods for water treatment, arsenic remediation turned to the potentialities of biological approaches. The use of rhizosphere microorganisms was recently investigated for their capacity to enhance phytoremediation of arsenic-contaminated environments (Ma et al., 2016). In particular, several arsenic-resistant microorganisms belonging to various genera, e.g., Bacillus, Achromobacter, Brevundimonas, Microbacterium, Ochrobactrum, Pseudomonas, Comamonas, Stenotrophomonas, Ensifer were reported to decrease toxic effects of arsenic and enhance plant growth by acting on arsenic mobilization and accumulation in plants (Cavalca et al., 2010; Ghosh et al., 2011; Wang et al., 2011; Yang et al., 2012; Pandey et al., 2013; Mallick et al., 2014, 2018; Mesa et al., 2017). The ability of fungi to resist, solubilize, transform or uptake metal species could also be used in mycoremediation of arsenic-contaminated soil (Singh et al., 2015; Srivastava et al., 2011). The production of volatile trimethylarsine by reductive methylation from inorganic and methylated arsenic compounds was reported in several fungal strains, e.g., Aspergillus glaucum, Candida humicola, Scopulariopsis brevicaulis, Gliocladium roseum, Penicillium gladioli, and Fusarium spp. (Cullen and Reimer, 1989; Lin, 2008). Bioaugmentation could thus represent a strategy to enhance the efficiency of As removal from waters and soils by the addition of specialized bacteria or fungi, either natural or genetically engineered, able to directly remove As by volatilization (Edvantoro et al., 2004; Chen P. et al., 2017) or indirectly through the formation of biogenic Fe-Mn oxides (Bai et al., 2016). However, despite an increasing interest for mycoremediation and rhizoremediation of arsenic contamination, still very little is known about their scalability.
To date, the bioremediation of arsenic-rich environments is mainly based on the use of microorganisms able to resist or metabolize arsenic through oxidoreduction reactions (Huang, 2014). Over the last decades the ecology of arsenic has been widely studied and several arsenic-metabolizing microorganisms isolated from various ecosystems have been characterized at the genomic level (Oremland and Stolz, 2003; Páez-Espino et al., 2009; Andres and Bertin, 2016). Herminiimonas arsenicoxydans was the first arsenic-metabolizing bacterium to be described. This β-proteobacterium isolated from an industrial wastewater treatment plant in Germany was shown to resist to high levels of arsenic and to oxidize arsenite, As(III), into arsenate, As(V) (Muller et al., 2007). Functional genomics demonstrated that this arsenic response is biphasic: H. arsenicoxydans activates the resistance response based in part on the induction of efflux mechanisms before inducing the detoxification processes leading to As(III) oxidation (Cleiss-Arnold et al., 2010; Koechler et al., 2010). Additionally, electron microscopy revealed that the strain is able to sequester arsenic within an exopolysaccharide (EPS) matrix (Muller et al., 2007). Thiomonas sp. 3As isolated from an abandoned mine in France was also shown to produce large amounts of EPS in the presence of arsenite, making it a good candidate for the development of bioremediation strategies relying on biofilm-based bioreactors (Arsène-Ploetze et al., 2010). A strain belonging to the Rhizobium genus isolated from an Australian gold mine was shown to carry arsenic resistance and detoxification genes on a large plasmid, which could provide an interesting genetic tool to transfer arsenic detoxification capacity into closely related plant-associated bacteria with the perspective of phytoremediation (Andres et al., 2013). More recently, the genome of two arsenite-oxidizing strains hyper-tolerant to arsenite was fully described: Halomonas A3H3 isolated from multicontaminated sediments in Mediterranean Sea (Koechler et al., 2013), and Pseudomonas xanthomarina S11 isolated from an arsenic-contaminated former gold mine in France (Koechler et al., 2015). Overall, the identification and the exploitation of microbial metabolic potentialities for arsenic-contaminated water treatment are considered an emerging challenge as mirrored by an increasing number of recent studies (Crognale et al., 2017). Among the available bacterial-driven processes, bioprecipitation, biosynthesis of adsorbent materials, biosorption and biovolatilization, involving several microorganisms (Table 1), are the most interestingly described for bioremediation of arsenic-contaminated waters (Fazi et al., 2016).
Table 1.
Microorganisms used in As-removal processes from waters.
| Microorganism | Process | Reference |
|---|---|---|
| Ralstonia eutropha | Bio-adsorption | Mondal et al., 2008 |
| Rhodopseuodomonas palustris | Bio-volatilization | Liu et al., 2011 |
| Sphingomonas desiccabilis | Bio-volatilization | Liu et al., 2011 |
| Bacillus idriensis | Bio-volatilization | Liu et al., 2011 |
| Cyanobacteria | Bio-volatilization | Yin et al., 2011 |
| Klebsiella oxytoca | Bio-synthesis of adsorbent materials | Casentini et al., 2015 |
| Mixed microbial community | Bio-precipitation | Omoregie et al., 2013 |
| Gallionella ferruginea and Leptothrix ochracea | Microbial Fe-oxidation coupled to As removal | Katsoyiannis and Zouboulis, 2004 |
| Desulfotomaculum auripigmentum | As and Fe–As sulfide precipitation driven by sulfate reducers | Newman et al., 1997 |
| Mixed Sulfate Reducing Bacteria | As removal driven by sulfate reduction processes | Teclu et al., 2008; Serrano and Leiva, 2017 |
| Mixed microbial community | Microbial Fe- and Mn-oxidation coupled to As removal | Thapa Chhetri et al., 2014 |
| Mixed microbial community | As removal via co-oxidation with Fe and sorption or co-precipitation with Fe(III) (oxyhydr)oxides | Nitzsche et al., 2015 |
| Mixed microbial community | Microbial Fe- and Mn-oxidation coupled to AsIII removal | Yang et al., 2014 |
| Mixed microbial community | As(III) microbial oxidation coupled to FeII oxidation | Kamei-Ishikawa et al., 2017 |
| Aliihoeflea sp. 2WW | As(III) microbial oxidation | Corsini et al., 2014 |
| Thiomonas arsenivorans | As(III) microbial oxidation | Wan et al., 2010; Dastidar and Wang, 2012 |
| Rhodococcus equi | As(III) microbial oxidation | Bag et al., 2010 |
| CAsO1 bacterial consortium | As(III) microbial oxidation | Battaglia-Brunet et al., 2002; Michel et al., 2007 |
| Ensifer adhaerens | As(III) microbial oxidation | Ito et al., 2012 |
| Mixed microbial community | As(III) microbial oxidation | Gude et al., 2018 |
| Mixed microbial community | As(III) microbial oxidation | Li et al., 2016 |
| Mixed microbial community | As(III) microbial oxidation | Sun et al., 2011 |
| Mixed microbial community | Anoxic As(III) microbial oxidation coupled with chemolithotrophic denitrification | Sun et al., 2010 |
In recent years, several environmental genomic studies of arsenic-contaminated ecosystems have been conducted (Huang et al., 2016) and the molecular mechanisms involved have been recently reviewed in detail (Andres and Bertin, 2016). A metagenomic study of an acid mine drainage in France yielded nearly complete reconstructions of seven microbial genomes, providing a better understanding of the arsenic metabolism and natural attenuation which significantly reduce arsenic concentration along the creek, thanks to arsenite oxidation followed by co-precipitation with iron and sulfur. This analysis led to the identification of the corresponding genes, in particular aio coding for arsenite oxidase in Thiomonas sp. and rus coding for rusticyanin in Acidithiobacillus sp. (Bertin et al., 2011). A comparative metagenomic study of sediments in two harbors on the Mediterranean French coast, focusing on sequence markers specific for sulfur-metabolizing bacteria uncovered a correspondence between biotic sulfate reduction and the abiotic production of highly soluble thioarsenical compounds. In combination with arsenate reduction these processes, which favor arsenic dispersion in the water column, could explain the higher mobility of arsenic observed on the most contaminated site (Plewniak et al., 2013). Recently, the assembly of 27 Micrarcheota and 12 Parvarchaeota new genomes from 12 acid mine drainage and hot spring metagenomes was reported in a study targeting Archaeal Richmond Mine Acidophilic Nanoorganisms. The analysis of these almost complete genomes suggests a possible contribution of these organisms to carbon and nitrogen cycling by organic matter degradation, as well as to iron oxidation (Chen L.-X. et al., 2017). Those studies suggest that arsenic bioremediation strategies could be based upon microbial communities with iron, sulfur, and arsenic metabolism capacities and highlight the importance of metabolisms other than those of metals in arsenic removal. In this respect, mixed microbial communities were tested for bio-precipitation capacity and arsenic removal coupled with iron and manganese oxidation in filtration systems (Table 1) and recently, the use of acid/metal-tolerant sulfate reducing bacteria was applied for arsenic removal from an acid mine drainage (Serrano and Leiva, 2017).
As(III) microbial oxidation can also be coupled to commonly used adsorption removal technology, without any chemicals addition nor toxic by-products (Bahar et al., 2013). The As(III)-oxidation potentialities of several As(III)-oxidizing microorganisms, such as Aliihoeflea sp. 2WW, Thiomonas arsenivorans strain b6, Ensifer adhaerens, Rhodococcus equi and other As(III)-oxidizing mixed bacterial populations as planktonic cells or associated with biofilms were successfully tested in lab-scale experiments for treating contaminated water (Table 1). Moreover, the anoxic As(III) microbial oxidation coupled with chemolithotrophic denitrification was successfully employed in the treatment of arsenic in bioreactors (Sun et al., 2010). To date, only one case study of full-scale treatment of arsenic contaminated groundwater using biological As(III) oxidation has been documented in the scientific literature (Katsoyiannis et al., 2008). This multi-stage treatment method was based on the biological oxidation of NH4+ and Mn(II) for the simultaneous As(III) oxidation and subsequent As(V) removal by coagulation. However, As removal is strongly dependent on Fe(II) and Mn(II) concentrations since the process relies on the sorption of As on iron and manganese oxides produced by autochthonous Fe(II)- and Mn(II)-oxidizing bacteria.
Although several studies demonstrated the efficacy of arsenic removal from water by microorganisms, these approaches are yet to be fully exploited for arsenic remediation, and knowledge about the diversity and distribution of functional genes controlling arsenic transformation in such processes is still quite fragmentary (Andres and Bertin, 2016; Crognale et al., 2017). The industrial application of arsenic removal from water still requires further evaluation in real situation of additional aspects such as the influence on microbial As(III) oxidation of geometric and hydraulic parameters in column systems or the requirement for carbon supply to support fast reactions. Although recent batch experiment works are addressing the question of the effects of nutrient sources and temperature in acid mine drainage (Tardy et al., 2018), there is still a want of further genomic and metagenomic studies of arsenic-contaminated ecosystems addressing not only the metabolisms of metals, arsenic and sulfurs but the full-range of microbial metabolic capacities. Such studies will be necessary for understanding the complex trophic interaction network of microorganisms in those ecosystems and for designing optimized artificial microbial communities that could be exploited in large-scale arsenic remediation systems.
Conclusions and Perspectives
At the interface between molecular biology and ecology, environmental genomic DNA sequencing techniques allow to reach, beyond the mere description of a simple organism, the characterization of complex microbial communities including organisms recalcitrant to isolation and culture. In association with global functional approaches – metatranscriptomics, metaproteomics, metabolomics including stable-isotope probing (Fischer et al., 2016; Musat et al., 2016; Vogt et al., 2016; Zuñiga et al., 2017) – these techniques help increasing our knowledge of the functioning of ecosystems. Additionally, the sequencing depth attained by these new technologies can give access to the less represented species of an ecosystem (the rare biosphere). Allowing fast and inexpensive massive characterization of microbial communities, they could also be an asset for the continuous monitoring of microbial communities involved in bioremediation processes to avoid changes that could compromise the efficiency of the treatment (Lovley, 2003; Stenuit et al., 2008; Techtmann and Hazen, 2016). In combination with the indispensable experimentations in the laboratory and in the field, these approaches require the development of efficient reproducible sampling and extraction methods as well as of robust and new computing solutions for storing, exchanging, and analyzing the huge amounts of data they produce. Indeed, power analysis and sample size requirements estimation for high-throughput sequencing data demand computations of much higher complexity than classical statistical analyses and must be fine-tuned to the type of problem that is being addressed (Pasolli et al., 2016; Li et al., 2017). It is moreover necessary that all published studies include complementary data (meta-data) which should be collected for every genome/metagenome to permit the proper exploitation of data (Satinsky et al., 2013) as defined by the Genomic Standard Consortium (Yilmaz et al., 2011).
The public access on sites like the EBI Metagenomics (Mitchell et al., 2017) to thousands of metagenomic samples combined with big data analysis, data mining algorithms and metabolic modeling constitutes an unprecedented opportunity to study and understand how the different components of an ecosystem may function together in relation with environmental biotic and abiotic factors, largely surpassing mere inventories of biological objects. A better understanding of the concerned organisms, of their spatial and temporal distribution, of the adaptive and evolutive processes at stake and of the metabolic interactions they develop should thus provide an integrated image of the microbial communities and metabolic functions involved in the microbiological processes underlying arsenic removal from water. Using ad hoc predictive models, such knowledge may be expected to permit the optimal utilization of microorganisms’ properties in biotechnological applications and bioremediation processes.
Author Contributions
PB and FP organized the content of the entire manuscript and wrote the genomics section. SC and SR wrote the section on bioremediation.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Anantharaman K., Brown C. T., Hug L. A., Sharon I., Castelle C. J., Probst A. J., et al. (2016). Thousands of microbial genomes shed light on interconnected biogeochemical processes in an aquifer system. Nat. Commun. 7:13219. 10.1038/ncomms13219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andres J., Arsène-Ploetze F., Barbe V., Brochier-Armanet C., Cleiss-Arnold J., Coppée J. Y., et al. (2013). Life in an arsenic-containing gold mine: genome and physiology of the autotrophic arsenite-oxidizing bacterium Rhizobium sp, NT-26. Genome Biol. Evol. 5 934–953. 10.1093/gbe/evt061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andres J., Bertin P. (2016). The microbial genomics of arsenic. FEMS Microbiol. Rev. 40 299–322. 10.1093/femsre/fuv050 [DOI] [PubMed] [Google Scholar]
- Arsène-Ploetze F., Koechler S., Marchal M., Coppe J., Chandler M., Bonnefoy V., et al. (2010). Structure, function, and evolution of the Thiomonas spp. genome. PLoS Genet. 6:e1000859. 10.1371/journal.pgen.1000859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bag P., Bhattacharya P., Chowdhury R. (2010). Bio-detoxification of arsenic laden groundwater through a packed bed column of a continuous flow reactor using immobilized cells. Soil Sediment Contam. 19 455–466. 10.1080/15320383.2010.486050 [DOI] [Google Scholar]
- Bahar M. M., Megharaj M., Naidu R. (2013). Bioremediation of arsenic-contaminated water: recent advances and future prospects. Water Air Soil Pollut. 224:1722 10.1007/s11270-013-1722-y [DOI] [Google Scholar]
- Bai Y., Chang Y., Liang J., Chen C., Qu J. (2016). Treatment of groundwater containing Mn(II). Fe(II), As(III) and Sb(III) by bioaugmented quartz-sand filters. Water Res. 106 126–134. 10.1016/j.watres.2016.09.040 [DOI] [PubMed] [Google Scholar]
- Battaglia-Brunet F., Dictor M., Garrido F., Crouzet C., Morin D. (2002). An arsenic(III)-oxidizing bacterial population: selection, characterization, and performance in reactors. J. Appl. Microbiol. 93 656–667. 10.1046/j.1365-2672.2002.01726.x [DOI] [PubMed] [Google Scholar]
- Beazley M. J., Martinez R. J., Rajan S., Powell J., Piceno Y. M., Tom L. M., et al. (2012). Microbial community analysis of a coastal salt marsh affected by the Deepwater Horizon Oil spill. PLoS One 7:e41305. 10.1371/journal.pone.0041305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertin P., Michotey V., Normand P. (2015). “Contributions of descriptive and functional genomics to microbial ecology,” in Environmental Microbiology: Fundamentals and Applications: Microbial Ecology, eds Bertrand J. C., et al. (Berlin: Springer; ), 831–846. [Google Scholar]
- Bertin P. N., Heinrich-Salmeron A., Pelletier E., Goulhen-Chollet F., Arsène-Ploetze F., Gallien S., et al. (2011). Metabolic diversity among main microorganisms inside an arsenic-rich ecosystem revealed by meta- and proteo-genomics. ISME J. 5 1735–1747. 10.1038/ismej.2011.51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlin D. J., Naujokas M. F., Bradham K. D., Cowden J., Heacock M., Henry H. F., et al. (2016). Arsenic and environmental health: state of the science and future research opportunities. Environ. Health Perspect. 124 890–899. 10.1289/ehp.1510209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casentini B., Rossetti S., Gallo M., Baldi F. (2015). “Potentialities of biogenerated iron hydroxides nanoparticles in arsenic water treatment,” in Proceeding of the 6th European Bioremediation Conference, Chania. [Google Scholar]
- Cavalca L., Zanchi R., Corsini A., Colombo M., Romagnoli C., Canzi E., et al. (2010). Arsenic-resistant bacteria associated with roots of the wild Cirsium arvense (L.) plant from an arsenic polluted soil, and screening of potential plant growth-promoting characteristics. Syst. Appl. Microbiol. 33 154–164. 10.1016/j.syapm.2010.02.004 [DOI] [PubMed] [Google Scholar]
- Chen L.-X., Méndez-García C., Dombrowski N., Servín-Garcidueñas L. E., Eloe-Fadrosh E. A., Fang B.-Z., et al. (2017). Metabolic versatility of small archaea Micrarchaeota and Parvarchaeota. ISME J. 12 756–775. 10.1038/s41396-017-0002-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P., Li J., Wang H.-Y., Zheng R.-L., Sun G.-X. (2017). Evaluation of bioaugmentation and biostimulation on arsenic remediation in soil through biovolatilization. Environ. Sci. Pollut. Res. 24 21739–21749. 10.1007/s11356-017-9816-5 [DOI] [PubMed] [Google Scholar]
- Chivian D., Brodie E. L., Alm E. J., Culley D. E., Dehal P. S., DeSantis T. Z., et al. (2008). Environmental genomics reveals a single-species ecosystem deep within earth. Science 322 275–278. 10.1126/science.1155495 [DOI] [PubMed] [Google Scholar]
- Cleiss-Arnold J., Koechler S., Proux C., Fardeau M. L., Dillies M. A., Coppee J. Y., et al. (2010). Temporal transcriptomic response during arsenic stress in Herminiimonas arsenicoxydans. BMC Genomics 11:709. 10.1186/1471-2164-11-709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colin P. Y., Kintses B., Gielen F., Miton C. M., Fischer G., Mohamed M. F., et al. (2015). Ultrahigh-throughput discovery of promiscuous enzymes by picodroplet functional metagenomics. Nat. Commun. 6:10008. 10.1038/ncomms10008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corsini A., Zaccheo P., Muyzer G., Andreoni V., Cavalca L. (2014). Arsenic transforming abilities of groundwater bacteria and the combined use of Aliihoeflea sp. strain 2WW and goethite in metalloid removal. J. Hazard. Mater. 269 89–97. 10.1016/j.jhazmat.2013.12.037 [DOI] [PubMed] [Google Scholar]
- Crognale S., Amalfitano S., Casentini B., Fazi S., Petruccioli M., Rossetti S. (2017). Arsenic-related microorganisms in groundwater?: a review on distribution, metabolic activities and potential use in arsenic removal processes. Rev. Environ. Sci. Biotechnol. 16 647–665. 10.1007/s11157-017-9448-8 [DOI] [Google Scholar]
- Cullen W. R., Reimer K. J. (1989). Arsenic-speciation in the environment. Chem. Rev. 89 713–763. 10.1021/cr00094a002 [DOI] [Google Scholar]
- Dastidar A., Wang Y.-T. (2012). Modeling arsenite oxidation by chemoautotrophic Thiomonas arsenivorans strain b6 in a packed-bed bioreactor. Sci. Total Environ. 432 113–121. 10.1016/j.scitotenv.2012.05.051 [DOI] [PubMed] [Google Scholar]
- Edvantoro B. B., Naidu R., Megharaj M., Merrington G., Singleton I. (2004). Microbial formation of volatile arsenic in cattle dip site soils contaminated with arsenic and DDT. Appl. Soil Ecol. 25 207–217. 10.1016/j.apsoil.2003.09.006 [DOI] [Google Scholar]
- Fazi S., Amalfitano S., Casentini B., Davolos D., Pietrangeli B., Crognale S., et al. (2016). Arsenic removal from naturally contaminated waters: a review of methods combining chemical and biological treatments. Rend. Lincei 27 51–58. 10.1007/s12210-015-0461-y [DOI] [Google Scholar]
- Fischer A., Manefield M., Bombach P. (2016). Application of stable isotope tools for evaluating natural and stimulated biodegradation of organic pollutants in field studies. Curr. Opin. Biotechnol. 41 99–107. 10.1016/j.copbio.2016.04.026 [DOI] [PubMed] [Google Scholar]
- Fleischmann R. D., Adams M. D., White O., Clayton R. A., Kirkness E. F., Kerlavage A. R., et al. (1995). Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269 496–512. 10.1126/science.7542800 [DOI] [PubMed] [Google Scholar]
- Garza D. R., Dutilh B. E. (2015). From cultured to uncultured genome sequences: metagenomics and modeling microbial ecosystems. Cell. Mol. Life Sci. 72 4287–4308. 10.1007/s00018-015-2004-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh P., Rathinasabapathi B., Ma L. Q. (2011). Arsenic-resistant bacteria solubilized arsenic in the growth media and increased growth of arsenic hyperaccumulator Pteris vittata L. Bioresour. Technol. 102 8756–8761. 10.1016/j.biortech.2011.07.064 [DOI] [PubMed] [Google Scholar]
- Gilbert J. A., Meyer F., Antonopoulos D., Balaji P., Brown C. T., Brown C. T., et al. (2010). Meeting report: the terabase metagenomics workshop and the vision of an earth microbiome project. Stand. Genomic Sci. 3 243–248. 10.4056/sigs.1433550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gude J. C. J., Rietveld L. C., van Halem D. (2018). Biological As(III) oxidation in rapid sand filters. J. Water Process Eng. 21 107–115. 10.1016/j.jwpe.2017.12.003 [DOI] [Google Scholar]
- Haroon M. F., Hu S., Shi Y., Imerlfort M., Keller J., Hugenholtz P., et al. (2013). Anaerobic oxidation of methane coupled to nitrate redcution in a novel archaeal lineage. Nature 500 567–570. 10.1038/nature12375 [DOI] [PubMed] [Google Scholar]
- Huang J. (2014). Impact of microorganisms on arsenic biogeochemistry?: a review. Water Air Soil Pollut. 225:1848 10.1007/s11270-013-1848-y [DOI] [Google Scholar]
- Huang L.-N., Kuang J.-L., Shu W.-S. (2016). Microbial ecology and evolution in the acid mine drainage model system. Trends Microbiol. 24 581–593. 10.1016/j.tim.2016.03.004 [DOI] [PubMed] [Google Scholar]
- Ishino S., Ishino Y. (2014). DNA polymerases as useful reagents for biotechnology – the history of developmental research in the field. Front. Microbiol. 5:465 10.3389/fmicb.2014.00465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito A., Miura J.-I., Ishikawa N., Umita T. (2012). Biological oxidation of arsenite in synthetic groundwater using immobilised bacteria. Water Res. 46 4825–4831. 10.1016/j.watres.2012.06.013 [DOI] [PubMed] [Google Scholar]
- Jackson B. P., Taylor V. F., Karagas M. R., Punshon T., Cottingham K. L. (2012). Arsenic, organic foods, and brown rice syrup. Environ. Health Perspect. 120 623–626. 10.1289/ehp.1104619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacques P., Béchet M., Bigan M., Caly D., Chataigné G., Coutte F., et al. (2017). High-throughput strategies for the discovery and engineering of enzymes for biocatalysis. Bioprocess Biosyst. Eng. 40 161–180. 10.1007/s00449-016-1690-x [DOI] [PubMed] [Google Scholar]
- Kamei-Ishikawa N., Segawa N., Yamazaki D., Ito A., Umita T. (2017). Arsenic removal from arsenic-contaminated water by biological arsenite oxidation and chemical ferrous iron oxidation using a down-flow hanging sponge reactor. Water Supply 17 1249–1259. 10.2166/ws.2017.025 [DOI] [Google Scholar]
- Katsoyiannis I. A., Zikoudi A., Hug S. J. (2008). Arsenic removal from groundwaters containing iron, ammonium, manganese and phosphate: a case study from a treatment unit in northern Greece. Desalination 224 330–339. 10.1016/j.desal.2007.06.014 [DOI] [Google Scholar]
- Katsoyiannis I. A., Zouboulis A. I. (2004). Application of biological processes for the removal of arsenic from groundwaters. Water Res. 38 17–26. 10.1016/j.watres.2003.09.011 [DOI] [PubMed] [Google Scholar]
- Koechler S., Arsène-Ploetze F., Brochier C., Cholet F., Jost B., Lièvremont D., et al. (2015). Constitutive arsenite oxidase expression detected in the arsenic hypertolerant Pseudomonas xanthomarina S11. Res. Microbiol. 166 205–214. 10.1016/j.resmic.2015.02.010 [DOI] [PubMed] [Google Scholar]
- Koechler S., Plewniak F., Barbe V., Battaglia-Brunet F., Jost B., Joulian C., et al. (2013). Genome sequence of Halomonas sp. strain A3H3, isolated from arsenic-rich marine sediments. Genome Announc. 1:e00819-13. 10.1128/genomeA.00819-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koechler S., Cleiss-Arnold J., Proux C., Sismeiro O., Dillies M.-A., Goulhen-Chollet F., et al. (2010). Multiple controls affect arsenite oxidase gene expression in Herminiimonas arsenicoxydans. BMC Microbiol. 10:53. 10.1186/1471-2180-10-53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Könneke M., Bernhard A. E., de la Torre J. R., Walker C. B., Waterbury J. B., Stahl D. A. (2005). Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 7058 543–546. 10.1038/nature03911 [DOI] [PubMed] [Google Scholar]
- Krüger A., Schäfers C., Schröder C., Antranikian G. (2018). Towards a sustainable biobased industry – Highlighting the impact of extremophiles. New Biotechnol. 40 144–153. 10.1016/j.nbt.2017.05.002 [DOI] [PubMed] [Google Scholar]
- Land M., Hauser L., Jun S.-R., Nookaew I., Leuze M. R., Ahn T.-H., et al. (2015). Insights from 20 years of bacterial genome sequencing. Funct. Integr. Genomics 15 141–161. 10.1007/s10142-015-0433-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C.-I., Samuels D. C., Zhao Y.-Y., Shyr Y., Guo Y. (2017). Power and sample size calculations for high-throughput sequencing-based experiments. Brief. Bioinform. 5 1752–1779. 10.1093/bib/bbx061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Zeng X. C., He Z., Chen X., Guoji E., Han Y., et al. (2016). Long-term performance of rapid oxidation of arsenite in simulated groundwater using a population of arsenite-oxidizing microorganisms in a bioreactor. Water Res. 101 393–401. 10.1016/j.watres.2016.05.058 [DOI] [PubMed] [Google Scholar]
- Lin Z.-Q. (2008). “Volatilization,” in Ecological Processes. Encyclopedia of Ecology Vol. 5 eds Jorgensen S. E., Fath B. D. (Oxford: Elsevier; ), 3700–3705. 10.1016/B978-008045405-4.00302-5 [DOI] [Google Scholar]
- Liu S., Zhang F., Chen J., Sun G. (2011). Arsenic removal from contaminated soil via biovolatilization by genetically engineered bacteria under laboratory conditions. J. Environ. Sci. 23 1544–1550. 10.1016/S1001-0742(10)60570-0 [DOI] [PubMed] [Google Scholar]
- Lovley D. R. (2003). Cleaning up with genomics: applying molecular biology to bioremediation. Nat. Rev. Microbiol. 1 35–44. 10.1038/nrmicro731 [DOI] [PubMed] [Google Scholar]
- Lücker S., Wagner M., Maixner F., Pelletier E., Koch H., Vacherie B., et al. (2010). A Nitrospira metagenome illuminates the physiology and evolution of globally important nitrite-oxidizing bacteria. Proc. Natl. Acad. Sci. U.S.A. 107 13479–13484. 10.1073/pnas.1003860107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Rajkumar M., Zhang C., Freitas H. (2016). Beneficial role of bacterial endophytes in heavy metal phytoremediation. J. Environ. Manage. 174 14–25. 10.1016/j.jenvman.2016.02.047 [DOI] [PubMed] [Google Scholar]
- Machado H., Tuttle R. N., Jensen P. R. (2017). Omics-based natural product discovery and the lexicon of genone mining. Curr. Opin. Microbiol. 39 136–142. 10.1016/j.mib.2017.10.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhavan A., Sindhu R., Binod P., Sukumaran R., Pandey A. (2017). Strategies for design of improved biocatalysts for industrial applications. Bioresour. Technol. 245 1304–1313. 10.1016/j.biortech.2017.05.031 [DOI] [PubMed] [Google Scholar]
- Madsen E. L. (2011). Microorganisms and their roles in fundamental biogeochemical cycles. Curr. Opin. Biotechnol. 22 456–464. 10.1016/j.copbio.2011.01.008 [DOI] [PubMed] [Google Scholar]
- Mallick I., Bhattacharyya C., Mukherji S., Dey D., Sarkar S. C., Mukhopadhyay U. K., et al. (2018). Effective rhizoinoculation and biofilmformation by arsenic immobilizing halophilic plant growth promoting bacteria (PGPB) isolated from mangrove rhizosphere: a step towards arsenic rhizoremediation. Sci. Total Environ. 61 1239–1250. 10.1016/j.scitotenv.2017.07.234 [DOI] [PubMed] [Google Scholar]
- Mallick I., Tofajjen Hossain S. K., Sinha S., Mukherjee S. K. (2014). Brevibacillus sp. KUMAs2, a bacterial isolate for possible bioremediation of arsenic in rhizosphere. Ecotoxicol. Environ. Saf. 107 236–244. 10.1016/j.ecoenv.2014.06.007 [DOI] [PubMed] [Google Scholar]
- Mesa V., Navazas A., Gonzalez-Gill R., Gonzalez A., Weyens N., Lauga B., et al. (2017). Use of endophytic and rhizosphere bacteria to improve phytoremediation of arsenic-contaminated industrial soils by Autochthonous Betula celtiberica. Appl. Environ. Microbiol. 83:e03411-16. 10.1128/AEM.03411-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel C., Jean M., Coulon S., Dictor M.-C., Delorme F., Morin D., et al. (2007). Biofilms of As(III)-oxidising bacteria: formation and activity studies for bioremediation process development. Appl. Microbiol. Biotechnol. 77 457–467. 10.1007/s00253-007-1169-4 [DOI] [PubMed] [Google Scholar]
- Mitchell A. L., Scheremetjew M., Denise H., Potter S., Tarkowska A., Qureshi M., et al. (2017). EBI Metagenomics in 2017: enriching the analysis of microbial communities, from sequence reads to assemblies. Nucleic Acids Res. 46 D726–D735. 10.1093/nar/gkx967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molin M., Ulven S. M., Meltzer H. M., Alexander J. (2015). Arsenic in the human food chain, biotransformation and toxicology—review focusing on seafood arsenic. J. Trace Elem. Med. Biol. 31 249–259. 10.1016/j.jtemb.2015.01.010 [DOI] [PubMed] [Google Scholar]
- Mondal P., Majumder C. B., Mohanty B. (2008). Treatment of arsenic contaminated water in a batch reactor by using Ralstonia eutropha MTCC 2487 and granular activated carbon. J. Hazard. Mater. 153 588–599. 10.1016/j.jhazmat.2007.09.028 [DOI] [PubMed] [Google Scholar]
- Mukherjee S., Stamatis D., Bertsch J., Ovchinnikova G., Verezemska O., Isbandi M., et al. (2016). Genomes OnLine Database (GOLD) v.6: data updates and feature enhancements. Nucleic Acids Res. 45 D446–D456. 10.1093/nar/gkw992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller D., Médigue C., Koechler S., Barbe V., Barakat M., Talla E., et al. (2007). A tale of two oxidation states?: bacterial colonization of arsenic-rich environments. PLoS Genet. 3:e53. 10.1371/journal.pgen.0030053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musat N., Musat F., Weber P. K., Pett-Ridge J. (2016). Tracking microbial interactions with NanoSIMS. Curr. Opin. Biotechnol. 41 114–121. 10.1016/j.copbio.2016.06.007 [DOI] [PubMed] [Google Scholar]
- Newman D. K., Kennedy E. K., Coates J. D., Ahmann D., Ellis D. J., Lovley D. R., et al. (1997). Dissimilatory arsenate and sulfate reduction in Desulfotomaculum auripigmentum sp. nov. Arch. Microbiol. 168 380–388. 10.1007/s002030050512 [DOI] [PubMed] [Google Scholar]
- Ng K. S., Ujang Z., Le-Clech P. (2004). Arsenic removal technologies for drinking water treatment. Rev. Environ. Sci. Biotechnol. 3 43–53. 10.1023/B:RESB.0000040054.28151.84 [DOI] [Google Scholar]
- Nicomel N. R., Leus K., Folens K., Van Der Voort P., Du Laing G. (2016). Technologies for arsenic removal from water: current status and future perspectives. Int. J. Environ. Res. Public Health 13 1–24. 10.3390/ijerph13010062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitzsche K. S., Weigold P., Lösekann-Behrens T., Kappler A., Behrens S. (2015). Microbial community composition of a household sand filter used for arsenic, iron, and manganese removal from groundwater in Vietnam. Chemosphere 138 47–59. 10.1016/j.chemosphere.2015.05.032 [DOI] [PubMed] [Google Scholar]
- Nordstrom D. K. (2002). Public health—worldwide occurrences of arsenic in groundwater. Science 296 2143–2145. 10.1126/science.1072375 [DOI] [PubMed] [Google Scholar]
- Omoregie E. O., Couture R., Van Cappellen P., Corkhill C. L., Charnock J. M., Polya D. A., et al. (2013). Arsenic bioremediation by biogenic iron oxides and sulfides. Appl. Environ. Microbiol. 79 4325–4335. 10.1128/AEM.00683-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oremland R. S., Stolz J. F. (2003). The ecology of arsenic. Science 300 939–944. 10.1126/science.1081903 [DOI] [PubMed] [Google Scholar]
- Overmann J., Abt B., Sikorski J. (2017). Present and future of culturing bacteria. Annu. Rev. Microbiol. 71 711–730. 10.1146/annurev-micro-090816-093449 [DOI] [PubMed] [Google Scholar]
- Páez-Espino D., Tamames J., de Lorenzo V., Canovas D. (2009). Microbial responses to environmental arsenic. Biometals 22 117–130. 10.1007/s10534-008-9195-y [DOI] [PubMed] [Google Scholar]
- Pan K.-L., Gao J.-F., Li H.-Y., Fan X.-Y., Li D.-C., Jiang H. (2018). Ammonia-oxidizing bacteria dominate ammonia oxidation in a full-scale wastewater treatment plant revealed by DNA-based stable isotope probing. Bioresour. Technol. 256 152–159. 10.1016/j.biortech.2018.02.012 [DOI] [PubMed] [Google Scholar]
- Pandey S., Ghosh P. K., Ghosh S., De T. K., Maiti T. K. (2013). Role of heavy metal resistant Ochrobactrum sp. and Bacillus spp. strains in bioremediation of a rice cultivar and their PGPR like activities. J. Microbiol. 51 11–17. 10.1007/s12275-013-2330-7 [DOI] [PubMed] [Google Scholar]
- Pasolli E., Truong D. T., Malik F., Waldron L., Segata N. (2016). Machine learning meta-analysis of large metagenomic datasets: tools and biological insights. PLoS Comput. Biol. 12:e1004977. 10.1371/journal.pcbi.1004977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plewniak F., Koechler S., Navet B., Dugat-Bony E., Bouchez O., Peyret P., et al. (2013). Metagenomic insights into microbial metabolism affecting arsenic dispersion in Mediterranean marine sediments. Mol. Ecol. 22 4870–4883. 10.1111/mec.12432 [DOI] [PubMed] [Google Scholar]
- Poulsen M., Hub H., Li C., Chen Z., Xu L., Otani S., et al. (2014). Complementary symbiont contributions to plant decomposition in a fungus-farming termite. Proc. Natl. Acad. Sci. U.S.A. 111 14500–14505. 10.1073/pnas.1319718111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashid M., Stingl U. (2015). Contemporary molecular tools in microbial ecology and their application to advancing biotechnology. Biotechnol. Adv. 33 1755–1773. 10.1016/j.biotechadv.2015.09.005 [DOI] [PubMed] [Google Scholar]
- Satinsky B. M., Gifford S. M., Crump B. C., Moran M. A. (2013). Use of internal standards for quantitative metatranscriptome and metagenome analysis. Methods Enzymol. 531 237–250. 10.1016/B978-0-12-407863-5.00012-5 [DOI] [PubMed] [Google Scholar]
- Schleper C., Jurgens G., Jonuscheit M. (2005). Genomic studies of uncultivated archaea. Nat. Rev. Microbiol. 3 479–488. 10.1038/nrmicro1159 [DOI] [PubMed] [Google Scholar]
- Serrano J., Leiva E. (2017). Removal of arsenic using acid/metal-tolerant sulfate reducing bacteria: a new approach for bioremediation of high-arsenic acid minewaters. Water 9:994 10.3390/w9120994 [DOI] [Google Scholar]
- Singh M., Srivastava P. K., Verma P. C., Kharwar R. N., Singh N., Tripathi R. D. (2015). Soil fungi for mycoremediation of arsenic pollution in agriculture soils. J. Appl. Microbiol. 119 1278–1290. 10.1111/jam.12948 [DOI] [PubMed] [Google Scholar]
- Srivastava P. K., Vaish A., Dwivedi S., Chakrabarty D., Singh N., Tripathi R. D. (2011). Biological removal of arsenic pollution by soil fungi. Sci. Total Environ. 409 2430–2442. 10.1016/j.scitotenv.2011.03.002 [DOI] [PubMed] [Google Scholar]
- Stenuit B., Eyers L., Schuler L., Agathos S. N., George I. (2008). Emerging high-throughput approaches to analyze bioremediation of sites contaminated with hazardous and / or recalcitrant wastes. Biotechnol. Adv. 26 561–575. 10.1016/j.biotechadv.2008.07.004 [DOI] [PubMed] [Google Scholar]
- Strous M., Pelletier E., Mangenot S., Rattei T., Lehner A., Taylor M. W., et al. (2006). Deciphering the evolution and metabolism of an anammox bacterium from a community genome. Nature 440 790–794. 10.1038/nature04647 [DOI] [PubMed] [Google Scholar]
- Sun W., Sierra-alvarez R., Field J. A. (2011). Long term performance of an arsenite-oxidizing-chlorate-reducing microbial consortium in an upflow anaerobic sludge bed (UASB) bioreactor. Bioresour. Technol. 102 5010–5016. 10.1016/j.biortech.2011.01.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W., Sierra-Alvarez R., Hsu I., Rowlette P., Field J. A. (2010). Anoxic oxidation of arsenite linked to chemolithotrophic denitrification in continuous bioreactors. Biotechnol. Bioeng. 105 909–917. 10.1002/bit.22611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardy V., Casiot C., Fernandez-Rojo L., Resongles E., Desoeuvre A., Joulian C., et al. (2018). Temperature and nutrients as drivers of microbially mediated arsenic oxidation and removal from acid mine drainage. Appl. Microbiol. Biotechnol. 102 2413–2424. 10.1007/s00253-017-8716-4 [DOI] [PubMed] [Google Scholar]
- Techtmann S. M., Hazen T. C. (2016). Metagenomic applications in environmental monitoring and bioremediation. J. Ind. Microbiol. Biotechnol. 43 1345–1354. 10.1007/s10295-016-1809-8 [DOI] [PubMed] [Google Scholar]
- Teclu D., Tivchev G., Liang M., Wallis M. (2008). Bioremoval of arsenic species from contaminated waters by sulphate-reducing bacteria. Water Res. 42 4885–4893. 10.1016/j.watres.2008.09.010 [DOI] [PubMed] [Google Scholar]
- Teeling H., Glöckner F. O. (2012). Current opportunities and challenges in microbialmetagenome analysis - a bioinformatic perspective. Brief. Bioinform. 13 728–742. 10.1093/bib/bbs039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thapa Chhetri R., Suzuki I., Fujita T., Takeda M., Koizumi J., Fujikawa Y., et al. (2014). Bacterial diversity in biological filtration system for the simultaneous removal of arsenic, iron and manganese from groundwater. J. Water Environ. Technol. 12 135–149. 10.2965/jwet.2014.135 [DOI] [Google Scholar]
- Thomas T., Gilbert J., Meyer F. (2012). Metagenomics - a guide from sampling to data analysis. Microb. Inform. Exp. 2:3. 10.1186/2042-5783-2-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson L. R., Sanders J. G., McDonald D., Amir A., Ladau J., Locey K. J., et al. (2017). A communal catalogue reveals Earth’s multiscale microbial diversity. Nature 551 457–463. 10.1038/nature24621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyson G. W., Lo I., Baker B. J., Allen E. E., Hugenholtz P., Banfield J. F. (2005). Genome-directed isolation of the key nitrogen fixer Leptospirillum ferrodiazotrophum sp. nov. from an acidophilic microbial community. Appl. Environ. Microbiol. 71 6319–6324. 10.1128/AEM.71.10.6319-6324.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullrich S. R., Poehlein A., Tischler J. S., González C., Ossandon F. J., Daniel R., et al. (2016). Genome analysis of the biotechnologically relevant acidophilic iron oxidising strain JA12 indicates phylogenetic and metabolic diversity within the novel genus “Ferrovum”. PLoS One 1:e0146832. 10.1371/journal.pone.0146832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallenet D., Calteau A., Cruveiller S., Gachet M., Lajus A., Josso A., et al. (2017). MicroScope in 2017: an expanding and evolving integrated resource for community expertise of microbial genomes. Nucleic Acids Res. 45 D517–D528. 10.1093/nar/gkw1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter J. C., Remington K., Heidelberg J. F., Halpern A. L., Rusch D., Eisen J. A., et al. (2004). Environmental genome shotgun sequencing of the Sargasso Sea. Science 304 66–74. 10.1126/science.1093857 [DOI] [PubMed] [Google Scholar]
- Vogt C., Dorer C., Musat F., Richnow H. H. (2016). Multi-element isotope fractionation concepts to characterize the biodegradation of hydrocarbons — from enzymes to the environment. Curr. Opin. Biotechnol. 41 90–98. 10.1016/j.copbio.2016.04.027 [DOI] [PubMed] [Google Scholar]
- Wan J., Klein J., Simon S., Joulian C., Dictor M.-C., Deluchat V., et al. (2010). AsIII oxidation by Thiomonas arsenivorans in up-flow fixed-bed reactors coupled to As sequestration onto zero-valent iron-coated sand. Water Res. 44 5098–5108. 10.1016/j.watres.2010.08.044 [DOI] [PubMed] [Google Scholar]
- Wang Q., Xiong D., Zhao P., Yu X., Tu B., Wang G. (2011). Effect of applying an arsenic resistant and plant growth-promoting rhizobacterium to enhance soil arsenic phytoremediation by Populus deltoides LH05-17. J. Appl. Microbiol. 111 1065–1074. 10.1111/j.1365-2672.2011.05142.x [DOI] [PubMed] [Google Scholar]
- WHO (2011). Evaluation of Certain Contaminants in Food. Seventy-Second Report of the Joint FAO/ WHO Expert Committee on Food Additives. WHO Technical Report, No. 959. Rome: World Health Organization. [Google Scholar]
- Yang L., Li X., Chu Z., Ren Y., Zhang J. (2014). Distribution and genetic diversity of the microorganisms in the biofilter for the simultaneous removal of arsenic, iron and manganese from simulated groundwater. Bioresour. Technol. 156 384–388. 10.1016/j.biortech.2014.01.067 [DOI] [PubMed] [Google Scholar]
- Yang Q., Tu S., Wang G., Liao X., Yan X. (2012). Effectiveness of applying of arsenate reducing bacteria to enhance arsenic removal from polluted soils by Pteris vittata L. Int. J. Phytorem. 14 89–99. 10.1080/15226510903567471 [DOI] [PubMed] [Google Scholar]
- Yilmaz P., Kottmann R., Field D., Knight R., Cole J. R., Amaral-Zettler L., et al. (2011). Minimum information about a marker gene sequence (MIMARKS) and minimum information about any (x) sequence (MIxS) specifications. Nat. Biotechnol. 29 415–420. 10.1038/nbt.1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X.-X., Chen J., Qin J., Sun G.-X., Rosen B. P., Zhu Y.-G. (2011). Biotransformation and volatilization of arsenic by three photosynthetic cyanobacteria. Plant Physiol. 156 1631–1638. 10.1104/pp.111.178947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., He Z., Yang Y., Deng Y., Tringe S. G., Alvarez-Cohen L. (2015). High-throughput metagenomic technologies for complex microbial community analysis: open and closed formats. mBio 6:e02288-14. 10.1128/mBio.02288-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuñiga C., Zaramela L., Zengler K. (2017). Elucidation of complexity and prediction of interactions in microbial communities. Microb. Biotechnol. 10 1500–1522. 10.1111/1751-7915.12855 [DOI] [PMC free article] [PubMed] [Google Scholar]